Abstract
In Enterobacteriaceae, the blaNDM genes have been found in many different genetic contexts, and a wide diversity of plasmid scaffolds bearing those genes has been found. In August 2013, we identified NDM-1-producing Escherichia coli and Enterobacter hormaechei strains from a single rectal swab sample from a patient hospitalized in Rio de Janeiro, Brazil, who had no history of travel abroad. Complete DNA sequencing using the Illumina platform and annotation of the two plasmids harboring the blaNDM-1 gene, one from each strain, showed that they belonged to incompatibility groups IncFIIK and IncX3 and harbored a novel transposon named Tn3000. Similar genetic structures have been identified among other isolates in Brazil but also on plasmids from other continents. Our findings suggest that the blaNDM-1 gene may be transmitted by Tn3000 in different parts of the world.
INTRODUCTION
Since the original description of NDM-1 carbapenemase in Escherichia coli and Klebsiella pneumoniae (1), 11 variants of this enzyme have been reported, with NDM-1 being the most prevalent (2). These enzymes have now been detected worldwide in Enterobacteriaceae (3), in Pseudomonas aeruginosa (4), and in many different Acinetobacter species (5). It has been proposed that the dissemination of the blaNDM-1 gene among Acinetobacter strains is mediated by a composite transposon designated Tn125, with two ISAba125 copies bracketing the resistance gene module (6). Although in Acinetobacter the blaNDM-1 has most frequently been found chromosomally located, some reports have described this gene located on plasmids (7, 8).
In Enterobacteriaceae, the blaNDM genes have been found mainly on plasmids (9). In contrast to the more conserved genetic environment observed in Acinetobacter spp., many different genetic contexts have been described in Enterobacteriaceae, with a wide diversity of plasmids harboring blaNDM genes (10–13). Among NDM variants described to date, all but NDM-2 and NDM-14 were detected in Enterobacteriaceae (14). Most of the sequences available in GenBank have a complete or truncated ISAba125 upstream and the bleMBL gene downstream from the blaNDM gene. Many different mobile elements have been found bracketing these genes and can potentially mobilize them (15). Three examples of genetic elements bearing the blaNDM-1 gene are (i) the Tn125 transposon (6), originally described in Acinetobacter but now detected in Enterobacteriaceae (16), (ii) the one detailed under GenBank accession no. KP900016 (17), in which an IS5 family transposase is located upstream from a truncated ISAba125 and the blaNDM-1 gene and is also found 6.064 kb downstream from the blaNDM-1 gene, bracketing a 9.476 kb genetic element, and (iii) the one detailed under GenBank accession no. KR059865 (18), in which IS3000 (IS3 family) is found 2.479 kb upstream from the blaNDM-1 gene and a TnAsn3-like tnpA, also from IS3 family, is found 4.757 kb downstream from the blaNDM-1 gene, bracketing a 12.802-kb genetic element.
There are few reports on genes other than blaNDM-1 which include complete mobile elements both upstream and downstream from the blaNDM gene. In GenBank deposit AB898038 (19), an IS6 family transposase truncates the ISAba125 and an unknown transposase is present 2.367 kb downstream from the blaNDM-3 gene. In K. pneumoniae plasmid pJEG027 (20), an IS5 family transposase truncates the ISAba125 and IS26 is found 2.189 kb downstream from the blaNDM-4 gene. A similar genetic structure is present in GenBank deposits KP826705 (unpublished) and KP178355 (21), containing, respectively, the blaNDM-7 and blaNDM-5 genes.
In Brazil, the first NDM-positive strain was reported in 2013, bearing a chromosomally located blaNDM-1 gene in Providencia rettgeri (22). Subsequently, plasmid-borne blaNDM-1 genes were identified in Enterobacter hormaechei (23), Enterobacter cloacae, P. rettgeri, K. pneumoniae (24), and Acinetobacter baumannii (25), but the sequences of these plasmids remain unknown. In E. hormaechei, the plasmid was reported be ∼420 to 490 kb (23), while in E. cloacae, P. rettgeri, and K. pneumoniae, the plasmid was reported to be ∼230 kb (24) and in A. baumannii the estimated plasmid size was 100 kb (25).
In this study, we aimed to characterize the genetic environment surrounding the blaNDM-1 gene in two Enterobacteriaceae species, E. coli and E. hormaechei, which were simultaneously recovered from a rectal swab from a hospitalized patient who had never traveled outside Brazil. Our investigation revealed that in both isolates, the blaNDM-1 gene was carried in an original transposon structure.
MATERIALS AND METHODS
Bacterial strains.
Two NDM-producing strains, E. hormaechei E0083033-1 and E. coli E0083033-2, recovered from the same rectal swab sample from a pediatric patient on August 2013 in Rio de Janeiro, Brazil, were used in this study. The patient was under treatment for acute lymphoblastic leukemia and was admitted at Children's Hospital for 2 days for skin-tunneled central venous catheter placement. She had no history of previous infections or colonization by carbapenem-resistant Enterobacteriaceae (CRE), but since she had been previously hospitalized in another institution, according to institutional infection control recommendations, a rectal swab sample was collected for CRE surveillance.
Species identification.
Identification of species was done by mass spectrometry (MS) using the Vitek MS system (bioMérieux), as recommended by the manufacturer.
Molecular identification was performed by partial sequencing of the gyrB gene, as previously described (26). The identification of the Enterobacter strains at the species level was confirmed by partial sequencing of the hsp60 gene, as previously described (27, 28), except that Platinum Taq DNA polymerase was used in PCRs and DNA sequences were obtained using BigDye Terminator version 3.1 and a 3130xl genetic analyzer (Applied Biosystems), according to the manufacturer's instructions. Contigs were assembled using DNABaser program version 3.4.5 (Heracle Biosoft) and subsequently compared to the sequences from the type strains available at GenBank, using the BLAST program.
Detection of carbapenemase-encoding genes by PCR and sequencing.
Multiplex PCRs for the blaNDM, blaOXA-48, blaKPC, blaIMP, blaVIM, and blaSPM genes were performed as previously described (29), except that primers 27F (AGAGTTTGATYMTGGCTCAG) and 1492R (GGTTACCTTGTTACGACTT) were included in order to amplify the 16S rRNA gene as an internal control (30). For full-length amplification of the blaNDM-1 gene, primers NDM-L-bleo-FW (5′-TGGGTCGAGGTCAGGATAGG) and NDM-R-Aba-125-RV (5′-GCTTTTGAAACTGTCGCACCT) were designed using Primer-BLAST. Amplicons were sequenced and assembled as described above.
Plasmid extraction, transformation, and conjugation assays.
Plasmid DNA was obtained from the wild-type (WT) strains by alkaline extraction (31) and subsequently used to transform E. coli TOP10 (Invitrogen) by electroporation. Transformants were selected on LB agar containing ceftazidime (4 mg/liter). Conjugation experiments were performed using WT strains as donors and E. coli J53 as the recipient, as described previously (32). Transconjugants were selected on LB agar containing ceftazidime (4 mg/liter) plus sodium azide (125 mg/liter). The presence of the blaNDM-1 gene in transformants and transconjugants was confirmed by PCR (29).
Estimation of plasmid size was performed after 0.7% agarose gel electrophoresis, using a curve obtained by plotting the distance (millimeters) from the origin against the decimal logarithm of the plasmid size (154 kb, 66.2 kb, 37.6 kb, and 7.4 kb) from the reference strain E. coli 39R861 (33).
Antimicrobial susceptibility profile of WT strains and their transformants.
Antimicrobial susceptibility profiles were determined by broth microdilution (34) using cation-adjusted Mueller-Hinton broth (Becton-Dickinson) and Etest strips for fosfomycin and aztreonam. E. coli ATCC 25922 was used as a control. Results were interpreted according to the M100-S25 document from CLSI (35), except for tigecycline and fosfomycin, for which results were interpreted according to the EUCAST breakpoints (36). For polymyxin B, the colistin criteria from EUCAST were applied. The disk diffusion method (35, 37) was used to test for ampicillin susceptibility, with and without the addition of 10 μl of a 0.1 M EDTA solution to the disks in order to inhibit the NDM-1 activity. A blank disk containing only 0.1 M EDTA was also included as control.
Complete plasmid sequencing, assembly, annotation, and analysis.
Plasmid DNA was extracted (31) from transformants grown overnight at 37°C in an orbital shaker in LB broth containing imipenem (1 mg/liter). DNA samples were tagmented using the Nextera DNA sample preparation kit before fragments of ∼2,000 bp were captured, purified, and sequenced using a MiSeq Reagent Nano kit, v2 (500 cycles), in MiSeq equipment from Illumina. Sequences were assembled de novo in contigs using the SeqMan NGen program version 4.0 (DNAStar) and subsequently aligned using SeqMan Pro version 10.1.1 (DNAStar). Open reading frames (ORFs) were predicted and annotated using RAST (http://rast.nmpdr.org/) (38). Manual curation and sequence similarity searches directed against the GenBank database were carried out using the ARTEMIS genome browser and annotation tool (39). Insertion sequences were manually reviewed, directing searches against the IS Finder database (https://www-is.biotoul.fr/) (40). The full plasmid sequences were compared to those available at GenBank using BLAST.
Nucleotide sequence accession numbers.
The complete nucleotide sequences of the pEh1A and pEc2A plasmids were deposited in GenBank under accession numbers KR822246 and KR822247, respectively.
RESULTS
Species identification and screening for carbapenemase-encoding genes.
Identification using the Vitek MS system identified the E0083033-1 strain as E. cloacae complex with 99% confidence. When the gyrB partial sequence (1,138 bp) was compared to those pertaining to the reference strains published by Brady et al. (41), the highest similarity (96%) was obtained with E. hormaechei strain CCUG 27126. The partial sequence of the hsp60 gene (341 bp) was identical to that from the type strain of “E. hormaechei subsp. steigerwaltii” DSMZ16691. The Vitek MS system (bioMérieux) identified strain E0083033-2 as E. coli with 99% confidence, which was further confirmed by sequencing of the partial gyrB sequence (1,138 bp). When the WT strains were tested by multiplex PCR for detection of carbapenemase-encoding genes, both were positive for blaNDM and negative for the other genes evaluated. Full sequencing of amplicons identified the blaNDM-1 gene in both strains.
Plasmid profile, transformation, and conjugation assays.
E. hormaechei strain E00383033-1 possessed five plasmid bands (ca. 130 kb, ca. 90 kb, ca. 70 kb, ca. 7 kb, and ca. 6 kb), while the E. coli transconjugant and transformant strains showed only a single plasmid band of approximately 90 kb (data not shown). E. coli strain E0083033-2 exhibited two plasmid bands (160 kb and ca. 70 kb), while the transformant and the transconjugant possessed a single plasmid band of approximately 70 kb (data not shown). The plasmids carrying the blaNDM-1 gene were successfully transferred by conjugation, at a frequency of 5.3 × 10−1 with E. hormaechei strain E0083033-1 as the donor and at a frequency of 6.0 × 10−1 with E. coli strain E0083033-2 as the donor.
Antimicrobial susceptibility profiles.
The transformant obtained with plasmid DNA extracted from E. hormaechei E0083033-1 as the donor showed resistance to all β-lactams tested except aztreonam. It remained susceptible to aminoglycosides, fluoroquinolones, rifampin, and chloramphenicol (Table 1).
TABLE 1.
MICs for wild-type strains and their transformants
| Antimicrobial | MIC (μg/ml) for straina: |
||||
|---|---|---|---|---|---|
| E0083033-1 | TF1A | E0083033-2 | TF2A | TOP10 | |
| Ampicillin | ≥2,056 | ≥2,056 | ≥2,056 | ≥2,056 | 8 |
| Aztreonam | 64 | 0.094 | 0.064 | 0.125 | 0.094 |
| Cefepime | ≥64 | 32 | ≥64 | ≥64 | 0.06 |
| Cefoxitin | ≥1,024 | 512 | ≥1,024 | ≥1,024 | 8 |
| Ceftazidime | ≥64 | ≥64 | ≥64 | ≥64 | 0.25 |
| Ceftriaxone | ≥64 | ≥64 | ≥64 | ≥64 | 0.06 |
| Ertapenem | 64 | 16 | 64 | 16 | 0.015 |
| Imipenem | 64 | 32 | 32 | 32 | 0.25 |
| Meropenem | 32 | 16 | 32 | 16 | 0.015 |
| Amikacin | 8 | 4 | 16 | 4 | 2 |
| Gentamicin | 2 | 0.5 | 0.5 | 0.25 | 0.5 |
| Kanamycin | 32 | 4 | 32 | 32 | 2 |
| Tobramycin | 16 | 0.5 | 16 | 8 | 0.25 |
| Ciprofloxacin | 1 | 0.004 | 0.5 | 0.016 | 0.004 |
| Levofloxacin | 0.5 | ≤0.008 | 0.25 | ≤0.008 | 0.015 |
| Chloramphenicol | 4 | 2 | 4 | 2 | 2 |
| Fosfomycin | 0.75 | 0.38 | 0.5 | 0.38 | 0.38 |
| Tigecycline | 0.25 | 0.03 | 0.25 | 0.125 | 0.5 |
| Polymyxin B | 1 | 0.5 | 1 | 0.25 | 0.5 |
| Rifampin | 512 | 8 | 512 | 512 | 8 |
E0083033-1, WT E. hormaechei strain; TF1A, transformant derived from E. hormaechei E0083033-1; E0083033-2, WT E. coli strain; TF2A, transformant derived from E. coli E0083033-2.
E. coli strain E0083033-2 and its transformant were resistant to all β-lactams tested except aztreonam. MICs of tobramycin, amikacin, kanamycin, ciprofloxacin, and rifampin for the corresponding transformants were 2- to 64-fold increased, while those of chloramphenicol and gentamicin were unchanged (Table 1).
No inhibition zones were observed with blank disks containing 0.1 M EDTA or ampicillin disks when testing the transformant harboring pEc2A. Of note, an inhibition zone of 19 mm in diameter was observed with the ampicillin disk with addition of 0.1 M EDTA when testing the transformant containing pEh1A, in which the only antimicrobial resistance gene is blaNDM-1.
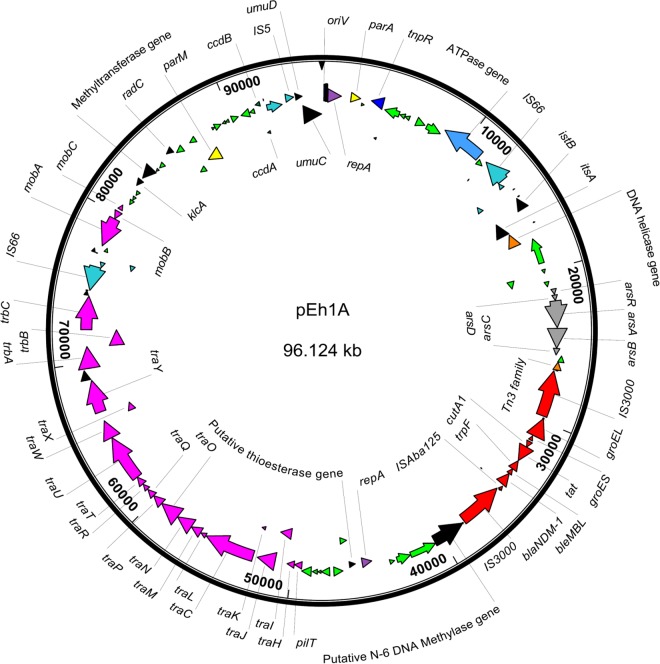
Plasmid pEh1A sequence analysis.
The complete DNA sequence of plasmid pEh1A from E. hormaechei E0083033-1 was obtained, with an average depth of coverage of 470. It is a circular 96,124-bp plasmid with a G+C content of 53.1% and carries a total of 100 open reading frames (Fig. 1). DNA sequence comparison with sequences available in GenBank revealed a similarity index of 99% with two IncF plasmids, one from “E. hormaechei subsp. oharae” recovered in Brazil (GenBank accession no. NG_041719.1) (23) and plasmid pKPX-1 from K. pneumoniae recovered in Taiwan from a patient with a history of hospitalization in India (GenBank accession no. AP012055.1) (42). The pEh1A DNA sequence differed from that of E. hormaechei by the presence of a 40-bp repeat region at position 70886 (GenBank accession no. NG_041719.1) (23) downstream of the parA gene and the lack of a 1,370-bp fragment (partial sequence of the second copy of IS3000).
FIG 1.
Circular map of IncFIIK plasmid pEh1A from E. hormaechei. Genes encoding hypothetical proteins are in green, genes encoding the conjugation apparatus are in pink, and genes from Tn3000 are in red.
Comparison of the 250,444-bp plasmid pKPX-1 (42) showed that it contains all gene clusters and operons found in plasmid pEh1A (96,124 bp). These two plasmids differed in the ordering of operons, as the arsenic resistance operon is inverted with respect to the blaNDM-1 gene in pEh1A. They also differed by the presence of a gene coding for a hypothetical protein and a truncated tnpA gene, both occurring downstream of the arsenic operon in pEh1A, and by the presence of a tnpR gene truncating IS3000 downstream of the groEL gene. The nucleotide sequences from the two plasmids share 93.8% similarity (90,184 bp identical over the 96,124 bp of pEh1A).
The sequences of the oriV and repA genes (nucleotide positions 1 to 1276) from plasmid pEh1A were compared to those previously studied by Villa et al. (43). The highest similarity index (99%; 1,273/1,276) was observed with plasmid pKF3-94 (GenBank accession no. FJ876826.1) (44), belonging to the IncFIIK group. The oriV region from pEh1A possessed two DnaA boxes upstream from the repA gene, with an AT-rich region of 63.3% (nucleotide positions 146 to 224 bp) and five iterons characterized by GGTG(T/G)(G/T) nucleotide sequences distant from each other by 15 or 16 bases (nucleotide positions 245 to 335).
Looking at the features related to plasmid transfer and stability, plasmid pEh1A carries tra and trb operons, which enable conjugal transfer. A ccdAB operon encoding a toxin/antitoxin system involved in postsegregation killing of plasmid-free cells was also identified. A complete arsenic resistance operon was identified at nucleotide positions 21296 to 25604.
The plasmid has a single copy of the blaNDM-1 gene flanked upstream by a truncated ISAba125 and downstream by the bleMBL gene, encoding resistance to bleomycin. That overall structure containing the blaNDM-1 gene was designated transposon Tn3000.
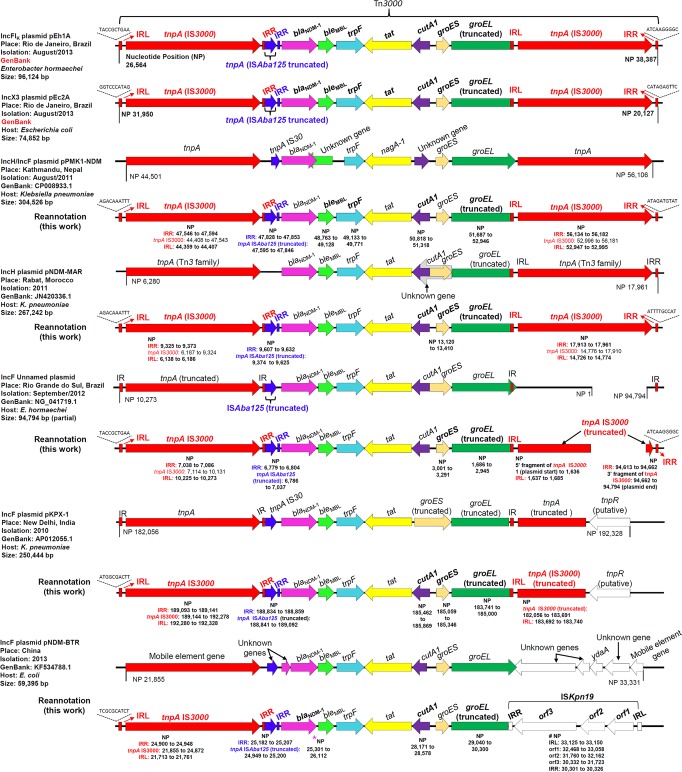
The Tn3000 transposon is conserved among plasmids from different continents.
Transposon Tn3000 is 11,823 bp long and is bracketed by two copies of IS3000. The first copy truncates the 5′ portion of the ISAba125 upstream of the blaNDM-1 gene. Downstream of the blaNDM-1 gene, the bleMBL gene was present, followed by genes encoding a phosphoribosylanthranilate isomerase (trpF), a twin-arginine translocation pathway signal protein (tat), and a divalent ion tolerance protein (cutA1). The groEL and groES genes were also part of Tn3000, but the groEL gene was truncated at its 3′ extremity by insertion of a second copy of IS3000. The Tn3000 nucleotide sequences identified on plasmids pEh1A and pEc2A were 100% identical. In silico analysis revealed sequences showing high similarities with Tn3000 in five plasmid sequences (Fig. 2) originating from isolates distributed over different continents. In silico analysis revealed that transposon Tn3000 was 99.9% identical to sequences identified on plasmids from incompatibility groups IncF and IncH originating from K. pneumoniae from Nepal (GenBank accession no. CP008933.1) (45) and Morocco (GenBank accession no. JN420336.1) (13). Therefore, those two plasmid sequences also harbored transposon Tn3000 (Fig. 2).
FIG 2.
Comparison of Tn3000 transposons of plasmids detected in different continents. *, a single-base-pair deletion in the blaNDM-1 gene at position 25509 created a stop codon at positions 25532 to 25534. #, a single-base-pair deletion in the orf2 gene from ISKpn19 at position 32162 altered the reading frame originally described. Gene names in bold indicate revision of the original annotation.
Two other plasmids, one from Porto Alegre, Brazil (GenBank accession no. NG_041719.1) (23), and one from New Delhi, India (pKPX-1; GenBank accession no. AP012055.1), also harbored transposon Tn3000, but the right-hand copy of IS3000 was truncated in those two cases. In the plasmid from Brazil, two inverted repeats (IRs) from the second copy of IS3000 were identified, but the tnpA gene lacked a fragment of 1,370 bp (Fig. 2).
In another plasmid (pNDM-BTR) from China (unpublished; GenBank accession no. KF534788.1), the left-hand extremity of transposon Tn3000 was conserved but the second copy of IS3000 located at the right extremity was aborted, truncated by ISKpn19 (Fig. 2).
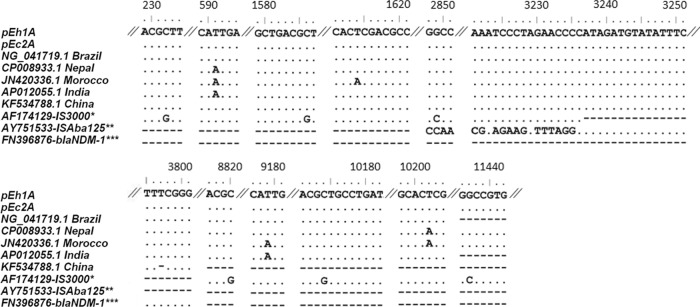
Transposon Tn3000 identified on plasmids from Brazil recovered in 2013 and described in this study was closely related to that identified from isolates from Nepal and Morocco, differing by 3 and 5 bp, respectively (Fig. 3).
FIG 3.
Polymorphisms in the Tn3000 transposon in unique plasmids detected in different continents. A dot indicates a nucleotide identical to that from the Tn3000 of the pEh1A plasmid in a given position. A dash indicates the absence of a nucleotide in a given position compared to Tn3000 of the pEh1A plasmid in a given position. Nucleotide numbering refers to the Tn3000 sequence. *, original IS3000 GenBank deposit; **, original ISAba125 GenBank deposit; ***, original blaNDM-1 gene GenBank deposit.
In none of the plasmid sequences analyzed were direct repeats flanking the Tn3000 transposon observed, suggesting that this structure may have been acquired by homologous recombination rather than by transposition.
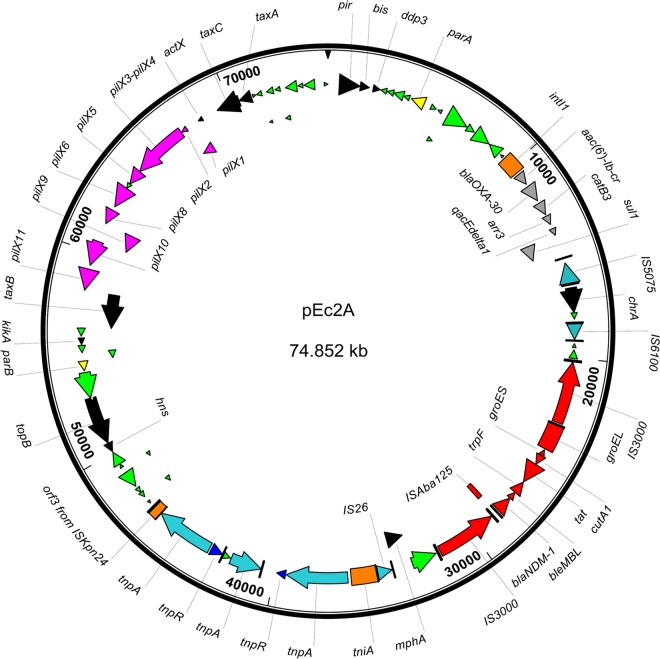
Plasmid pEc2A sequence analysis.
The complete DNA sequence from pEc2A from E. coli E0083033-2 was obtained, with an average depth of coverage of 2,771. It is a circular 74,852-bp plasmid with a 50.2% G+C content and 85 ORFs (Fig. 4).
FIG 4.
Circular map of IncX3 plasmid pEc2A plasmid from E. coli. Genes encoding hypothetical proteins are in green, genes encoding the conjugation apparatus are in pink, and genes from Tn3000 are in red.
The 29.5-kb backbone structure of plasmid pEc2A is typical of IncX plasmids, with genes encoding replication-associated proteins (pir, bis, parA, hns, and topB). It has a complete pilX operon, encoding a conjugation apparatus, and also taxA, taxB, and taxC genes, implicated in plasmid transfer. The taxC gene sequence was compared to IncX plasmids recently reviewed (46), and the highest similarity was observed with IncX3 plasmids pEC14_35 (GenBank accession no. JN935899) (95.4%) (47) and pIncX-SHV (GenBank accession no. JN247852) (95.3%) (48).
Plasmid pEc2A has a single copy of the blaNDM-1 gene flanked upstream by a truncated ISAba125 and downstream by the bleMBL gene. As observed for the IncF plasmid pEh1A, the blaNDM-1 gene occurred within Tn3000.
Plasmid pEc2A has a class 1 integron that is 99% similar to In37 (GenBank accession no. AY259086) (49). It possesses a variable region encompassing four gene cassettes, namely, aac(6′)-lb-cr, blaOXA-30, catB3, and arr3. MICs of tobramycin, amikacin, kanamycin, ciprofloxacin, and rifampin in the transformants harboring plasmid pEc2A were 2- to 64-fold increased compared to those for E. coli TOP10, while no elevation in chloramphenicol and gentamicin MICs was observed (Table 1).
DISCUSSION
The present study describes a new genetic element harboring blaNDM-1, Tn3000, which was found on plasmids of distinct incompatibility groups detected in different continents. Upon isolation of NDM-1-producing E. coli and E. hormaechei from a single rectal swab, our first hypothesis was that plasmid transfer occurred between these enterobacterial species, but plasmid analysis showed sizes that were significantly different. We subsequently introduced both plasmids into a single E. coli TOP10 strain and observed that they replicated and coexisted stably, which suggested different incompatibility groups. DNA sequence analysis confirmed that they belonged to different incompatibility groups: IncFIIK and IncX3. These are the first complete sequences of blaNDM-1-carrying plasmids from Brazil. blaNDM-1 has so far been found on plasmids of incompatibility groups IncF, IncH, IncL, IncM, and IncX (7, 50), as well as untypeable ones. Plasmid pEh1A, belongs to the IncFIIK incompatibility group and was found from “E. hormaechei subsp. steigerwaltii” in 2013; it is highly similar to the partial sequence of a plasmid isolated from “E. hormaechei subsp. oharae” in 2012 (23) in Porto Alegre, 1,571 km away from Rio de Janeiro.
The pEh1A IncFIIK plasmid has genes commonly found in IncF plasmid backbones, such as repA, parA, resD, and ccdAB, but is unusual in having an arsenic resistance operon (arsR, arsD, arsA, arsB, and arsC) instead of a mercury resistance operon (51).
The genetic structure observed in the plasmid extracted from E. coli (pEc2A) is as described by Norman and colleagues (52): pir-bis-par-hns-topB-pilX-actX-taxCA. The antimicrobial resistance genetic determinants located on the plasmid were embedded into two distinct genetic structures, namely, In37 and Tn3000. Concerning the In37 integron, the increased MICs of tobramycin, amikacin, kanamycin, ciprofloxacin, ampicillin, and rifampin observed for the transformant harboring plasmid pEc2A were consistent with the expression of gene cassettes driven by the Pc promoter. Of note, there was likely a lack of expression of the third gene cassette in In37 (catB3), as indicated by the low MICs observed for chloramphenicol in both the wild type and the transformant. If we consider that the genes upstream (blaOXA-30) and downstream (arr3) of the catB3 gene are expressed, the lack of chloramphenicol MIC elevation is most probably due to a posttranscriptional attenuation, as previously reported by Stokes and Hall (53).
The pEc2A plasmid isolated from E. coli belongs to the IncX3 incompatibility group. This suggests considerable potential for dissemination of blaNDM-1 in Brazil, as recently reported from China (54) and the United Arab Emirates (55).
We have found that the same genetic structure Tn3000 is present in plasmids of different sizes and incompatibility groups detected during the period from 2010 to 2013 in different countries and continents. IS3000 was originally described by Sabaté et al. (56). It was found in the In60 integron but oriented in the opposite direction of gene cassettes. These authors detected the presence of In60 containing IS3000 in a total of 30 E. coli and Salmonella species strains isolated from unrelated sources, but they were not able to demonstrate the occurrence of transposition events using a positive-selection vector strategy (57). One possibility to explain the presence of this element in different plasmids would be homologous recombination, but in this case the regions flanking IS3000 would be identical in different plasmids. This is not the case in the plasmids we have described or cited. If IS3000 and Tn3000 are not mobile elements, it would be hard to explain how they could be found flanked by different structures.
The presence of a truncated IS3000 in the 3′ portion of Tn3000 in plasmid pKPX-1 from India indicates that Tn3000 is the ancestral structure. Its insertion into this plasmid preceded a second transposition event that resulted in truncation of the IS3000. The full Tn3000 transposon sequence was found in two others plasmids, pPMK1-NDM (GenBank accession no. CP008933.1) (45) and pNDM-MAR (GenBank accession no. JN420336.1) (13), from Nepal and Morocco, respectively. If we consider that the Tn3000 sequence from the plasmids we described in this work, which were isolated in Rio de Janeiro, Brazil, in August 2013, is identical to that from the plasmid isolated in Porto Alegre, Brazil, in September 2012, the frequency of mutations in Tn3000 is less than one per 11 months. Zhao et al. (44) analyzed 110 strains harboring three plasmids with lengths of 70,057 to 147,416 bp by Illumina sequence analysis. When they compared the full plasmid sequences obtained in different years from different strains, they found 331 to 1,256 SNPs, depending on the plasmid studied (44). If we extrapolate this number to a pair of strains and a 11.8-kb structure as in the case of Tn3000, this range would be from 1 to 2.8 SNPs in 4 years in a 11.8-kb fragment. Consequently our finding of no SNPs when comparing the DNA sequences from Tn3000 in the plasmids isolated in Brazil 11 months apart is consistent with the findings of Zhao et al. (44). If we use these mutation rates to calculate the evolutionary distance in years between Tn3000 detected in Brazil and those detected in different continents, the smallest distance would be with the element from Nepal, with the time required to accumulate the three observed SNPs being 4.3 to 12 years. The plasmid isolated in Nepal was detected in August 2011. If we compare the Tn3000 DNA sequence from Brazil to that from Morocco, also detected in 2011, there are five SNPs, and their evolutive distance would be 7.1 to 20 years.
Conclusions.
In summary, we have described the first two complete plasmid sequences harboring blaNDM-1 from Brazil and have described a new transposon, designated Tn3000, which appears to mediate the transfer of blaNDM-1 among plasmids from different incompatibility groups in Brazil, Nepal, Morocco, and India.
ACKNOWLEDGMENTS
This work was supported by CAPES and the Fleury Institute.
We thank Michael S. Gilmore, Department of Microbiology and Immunobiology, Harvard Medical School, for reviewing the manuscript.
We declare no conflicts of interest.
REFERENCES
- 1.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zou D, Huang Y, Zhao X, Liu W, Dong D, Li H, Wang X, Huang S, Wei X, Yan X, Yang Z, Tong Y, Huang L, Yuan J. 2015. A novel New Delhi metallo-beta-lactamase variant, NDM-14, isolated in a Chinese hospital possesses increased enzymatic activity against carbapenems. Antimicrob Agents Chemother 59:2450–2453. doi: 10.1128/AAC.05168-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nordmann P. 2014. Carbapenemase-producing Enterobacteriaceae: overview of a major public health challenge. Med Mal Infect 44:51–56. doi: 10.1016/j.medmal.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Zafer MM, Amin M, El Mahallawy H, Ashour MS, Al Agamy M. 2014. First report of NDM-1-producing Pseudomonas aeruginosa in Egypt. Int J Infect Dis 29:80–81. doi: 10.1016/j.ijid.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Zhang R, Hu YY, Yang XF, Gu DX, Zhou HW, Hu QF, Zhao K, Yu SF, Chen GX. 2014. Emergence of NDM-producing non-baumannii Acinetobacter spp. isolated from China. Eur J Clin Microbiol Infect Dis 33:853–860. doi: 10.1007/s10096-013-2024-4. [DOI] [PubMed] [Google Scholar]
- 6.Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, Nordmann P. 2012. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob Agents Chemother 56:1087–1089. doi: 10.1128/AAC.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, Hao Q, Yang X, Xiao X, Luan C, Yang Y, Cui Y, Yang R, Gao GF, Song Y, Zhu B. 2012. Novel plasmid and its variant harboring both a blaNDM-1 gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob Agents Chemother 56:1698–1702. doi: 10.1128/AAC.06199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang WJ, Lu Z, Schwarz S, Zhang RM, Wang XM, Si W, Yu S, Chen L, Liu S. 2013. Complete sequence of the blaNDM-1-carrying plasmid pNDM-AB from Acinetobacter baumannii of food animal origin. J Antimicrob Chemother 68:1681–1682. doi: 10.1093/jac/dkt066. [DOI] [PubMed] [Google Scholar]
- 9.Carattoli A. 2013. Plasmids and the spread of resistance. Int J Med Microbiol 303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Carattoli A, Villa L, Poirel L, Bonnin RA, Nordmann P. 2012. Evolution of IncA/C blaCMY-(2)-carrying plasmids by acquisition of the blaNDM-(1) carbapenemase gene. Antimicrob Agents Chemother 56:783–786. doi: 10.1128/AAC.05116-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfeifer Y, Wilharm G, Zander E, Wichelhaus TA, Gottig S, Hunfeld KP, Seifert H, Witte W, Higgins PG. 2011. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J Antimicrob Chemother 66:1998–2001. doi: 10.1093/jac/dkr256. [DOI] [PubMed] [Google Scholar]
- 12.Sekizuka T, Matsui M, Yamane K, Takeuchi F, Ohnishi M, Hishinuma A, Arakawa Y, Kuroda M. 2011. Complete sequencing of the blaNDM-1-positive IncA/C plasmid from Escherichia coli ST38 isolate suggests a possible origin from plant pathogens. PLoS One 6:e25334. doi: 10.1371/journal.pone.0025334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villa L, Poirel L, Nordmann P, Carta C, Carattoli A. 2012. Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J Antimicrob Chemother 67:1645–1650. doi: 10.1093/jac/dks114. [DOI] [PubMed] [Google Scholar]
- 14.Shrestha B, Tada T, Miyoshi-Akiyama T, Shimada K, Ohara H, Kirikae T, Pokhrel BM. 2015. Identification of a novel NDM variant, NDM-13, from a multidrug-resistant Escherichia coli clinical isolate in Nepal. Antimicrob Agents Chemother 59:5847–5850. doi: 10.1128/AAC.00332-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Partridge SR, Iredell JR. 2012. Genetic contexts of blaNDM-1. Antimicrob Agents Chemother 56:6065–6067. (Author reply, 56:6071.) doi: 10.1128/AAC.00117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, Li H, Feng J, Li Y, Chen X, Guo X, Chen W, Wang L, Lin L, Yang H, Yang W, Wang J, Zhou D, Liu C, Yin Z. 2015. NDM-1 encoded by a pNDM-BJ01-like plasmid p3SP-NDM in clinical Enterobacter aerogenes. Front Microbiol 6:294. doi: 10.3389/fmicb.2015.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun F, Yin Z, Feng J, Qiu Y, Zhang D, Luo W, Yang H, Yang W, Wang J, Chen W, Xia P, Zhou D. 2015. Production of plasmid-encoding NDM-1 in clinical Raoultella ornithinolytica and Leclercia adecarboxylata from China. Front Microbiol 6:458. doi: 10.3389/fmicb.2015.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Q, Fang L, Fu Y, Du X, Shen Y, Yu Y. 2015. Dissemination of NDM-1-producing Enterobacteriaceae mediated by the IncX3-type plasmid. PLoS One 10:e0129454. doi: 10.1371/journal.pone.0129454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tada T, Miyoshi-Akiyama T, Shimada K, Kirikae T. 2014. Biochemical analysis of metallo-beta-lactamase NDM-3 from a multidrug-resistant Escherichia coli strain isolated in Japan. Antimicrob Agents Chemother 58:3538–3540. doi: 10.1128/AAC.02793-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Espedido BA, Dimitrijovski B, van Hal SJ, Jensen SO. 8 June 2015. The use of whole-genome sequencing for molecular epidemiology and antimicrobial surveillance: identifying the role of IncX3 plasmids and the spread of blaNDM-4-like genes in the Enterobacteriaceae. J Clin Pathol doi: 10.1136/jclinpath-2015-203044. [DOI] [PubMed] [Google Scholar]
- 21.Wailan AM, Paterson DL, Caffery M, Sowden D, Sidjabat HE. 2015. Draft genome sequence of NDM-5-producing Escherichia coli sequence type 648 and genetic context of blaNDM-5 in Australia. Genome Announc 3:00194–15. doi: 10.1128/genomeA.00194-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carvalho-Assef AP, Pereira PS, Albano RM, Beriao GC, Chagas TP, Timm LN, Da Silva RC, Falci DR, Asensi MD. 2013. Isolation of NDM-producing Providencia rettgeri in Brazil. J Antimicrob Chemother 68:2956–2957. doi: 10.1093/jac/dkt298. [DOI] [PubMed] [Google Scholar]
- 23.Carvalho-Assef AP, Pereira PS, Albano RM, Beriao GC, Tavares CP, Chagas TP, Marques EA, Timm LN, Da Silva RC, Falci DR, Asensi MD. 2014. Detection of NDM-1-, CTX-M-15, and qnrB4-producing Enterobacter hormaechei isolates in Brazil. Antimicrob Agents Chemother 58:2475–2476. doi: 10.1128/AAC.02804-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quiles MG, Rocchetti TT, Fehlberg LC, Kusano EJ, Chebabo A, Pereira RM, Gales AC, Pignatari AC. 2015. Unusual association of NDM-1 with KPC-2 and armA among Brazilian Enterobacteriaceae isolates. Braz J Med Biol Res 48:174–177. doi: 10.1590/1414-431X20144154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pillonetto M, Arend L, Vespero EC, Pelisson M, Chagas TP, Carvalho-Assef AP, Asensi MD. 2014. First report of NDM-1-producing Acinetobacter baumannii sequence type 25 in Brazil. Antimicrob Agents Chemother 58:7592–7594. doi: 10.1128/AAC.03444-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukushima M, Kakinuma K, Kawaguchi R. 2002. Phylogenetic analysis of Salmonella, Shigella, and Escherichia coli strains on the basis of the gyrB gene sequence. J Clin Microbiol 40:2779–2785. doi: 10.1128/JCM.40.8.2779-2785.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann H, Roggenkamp A. 2003. Population genetics of the nomenspecies Enterobacter cloacae. Appl Environ Microbiol 69:5306–5318. doi: 10.1128/AEM.69.9.5306-5318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann H, Stindl S, Ludwig W, Stumpf A, Mehlen A, Monget D, Pierard D, Ziesing S, Heesemann J, Roggenkamp A, Schleifer KH. 2005. Enterobacter hormaechei subsp. oharae subsp. nov., E. hormaechei subsp. hormaechei comb. nov., and E. hormaechei subsp. steigerwaltii subsp. nov., three new subspecies of clinical importance. J Clin Microbiol 43:3297–3303. doi: 10.1128/JCM.43.7.3297-3303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Maiwald M. 2004. Broad-range PCR for detection and identification of bacteria, p 379–390. In Persing DH, Tenover FC (ed), Molecular microbiology: diagnostic principles and practice. ASM Press, Washington, DC. [Google Scholar]
- 31.Birnboim HC, Doly J. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casali N, Preston A (ed). 2003. E. coli plasmid vectors: methods and applications. Humana Press, Totowa, NJ. [Google Scholar]
- 33.Macrina FL, Kopecko DJ, Jones KR, Ayers DJ, McCowen SM. 1978. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid 1:417–420. doi: 10.1016/0147-619X(78)90056-2. [DOI] [PubMed] [Google Scholar]
- 34.CLSI. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 10th ed CLSI document M07-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 35.CLSI. 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI document M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 36.EUCAST. 2015. Breakpoint tables for interpretation of MICs and zone diameters, version 5.0. European Committee on Antimicrobial Susceptibility Testing, Basel, Switzerland. [Google Scholar]
- 37.CLSI. 2015. Performance standards for antimicrobial disk susceptibility tests; approved standard, 12th ed CLSI document M02-A12. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 38.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 40.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brady C, Cleenwerck I, Venter S, Coutinho T, De Vos P. 2013. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst Appl Microbiol 36:309–319. doi: 10.1016/j.syapm.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 42.Huang TW, Chen TL, Chen YT, Lauderdale TL, Liao TL, Lee YT, Chen CP, Liu YM, Lin AC, Chang YH, Wu KM, Kirby R, Lai JF, Tan MC, Siu LK, Chang CM, Fung CP, Tsai SF. 2013. Copy number change of the NDM-1 sequence in a multidrug-resistant Klebsiella pneumoniae clinical isolate. PLoS One 8:e62774. doi: 10.1371/journal.pone.0062774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villa L, Garcia-Fernandez A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 44.Zhao F, Bai J, Wu J, Liu J, Zhou M, Xia S, Wang S, Yao X, Yi H, Lin M, Gao S, Zhou T, Xu Z, Niu Y, Bao Q. 2010. Sequencing and genetic variation of multidrug resistance plasmids in Klebsiella pneumoniae. PLoS One 5:e10141. doi: 10.1371/journal.pone.0010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoesser N, Giess A, Batty EM, Sheppard AE, Walker AS, Wilson DJ, Didelot X, Bashir A, Sebra R, Kasarskis A, Sthapit B, Shakya M, Kelly D, Pollard AJ, Peto TE, Crook DW, Donnelly P, Thorson S, Amatya P, Joshi S. 2014. Genome sequencing of an extended series of NDM-producing Klebsiella pneumoniae isolates from neonatal infections in a Nepali hospital characterizes the extent of community- versus hospital-associated transmission in an endemic setting. Antimicrob Agents Chemother 58:7347–7357. doi: 10.1128/AAC.03900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L, Chavda KD, Fraimow HS, Mediavilla JR, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob Agents Chemother 57:269–276. doi: 10.1128/AAC.01648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. 2012. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Garcia-Fernandez A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, Mancini C, Carattoli A. 2012. Klebsiella pneumoniae ST258 producing KPC-3 identified in italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob Agents Chemother 56:2143–2145. doi: 10.1128/AAC.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang M, Tran JH, Jacoby GA, Zhang Y, Wang F, Hooper DC. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob Agents Chemother 47:2242–2248. doi: 10.1128/AAC.47.7.2242-2248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother 55:5403–5407. doi: 10.1128/AAC.00585-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szczepanowski R, Braun S, Riedel V, Schneiker S, Krahn I, Puhler A, Schluter A. 2005. The 120 592 bp IncF plasmid pRSB107 isolated from a sewage-treatment plant encodes nine different antibiotic-resistance determinants, two iron-acquisition systems and other putative virulence-associated functions. Microbiology 151:1095–1111. doi: 10.1099/mic.0.27773-0. [DOI] [PubMed] [Google Scholar]
- 52.Norman A, Hansen LH, She Q, Sorensen SJ. 2008. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60:59–74. doi: 10.1016/j.plasmid.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 53.Stokes HW, Hall RM. 1991. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation. Plasmid 26:10–19. doi: 10.1016/0147-619X(91)90032-R. [DOI] [PubMed] [Google Scholar]
- 54.Ho P-L, Li Z, Lo W-U, Cheung Y-Y, Lin C-H, Sham P-C, Cheng VC-C, Ng T-K, Que T-L, Chow K-H. 2012. Identification and characterization of a novel incompatibility group X3 plasmid carrying blaNDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg Microbes Infect 1:6. doi: 10.1038/emi.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sonnevend A, Al Baloushi A, Ghazawi A, Hashmey R, Girgis S, Hamadeh MB, Al Haj M, Pal T. 2013. Emergence and spread of NDM-1 producer Enterobacteriaceae with contribution of IncX3 plasmids in the United Arab Emirates. J Med Microbiol 62:1044–1050. doi: 10.1099/jmm.0.059014-0. [DOI] [PubMed] [Google Scholar]
- 56.Sabate M, Navarro F, Miro E, Campoy S, Mirelis B, Barbe J, Prats G. 2002. Novel complex sul1-type integron in Escherichia coli carrying blaCTX-M-9. Antimicrob Agents Chemother 46:2656–2661. doi: 10.1128/AAC.46.8.2656-2661.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon R, Hotte B, Klauke B, Kosier B. 1991. Isolation and characterization of insertion sequence elements from gram-negative bacteria by using new broad-host-range, positive selection vectors. J Bacteriol 173:1502–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]