Abstract
The in vitro activity of the novel antimicrobial peptide dendrimer G3KL was evaluated against 32 Acinetobacter baumannii (including 10 OXA-23, 7 OXA-24, and 11 OXA-58 carbapenemase producers) and 35 Pseudomonas aeruginosa (including 18 VIM and 3 IMP carbapenemase producers) strains and compared to the activities of standard antibiotics. Overall, both species collections showed MIC50/90 values of 8/8 μg/ml and minimum bactericidal concentrations at which 50% or 90% of strains tested are killed (MBC50/90) of 8/8 μg/ml. G3KL is a promising molecule with antibacterial activity against multidrug-resistant and extensively drug-resistant A. baumannii and P. aeruginosa isolates.
TEXT
The spread of Acinetobacter baumannii and Pseudomonas aeruginosa isolates that are resistant to carbapenem antibiotics due to the production of carbapenemases represents a serious threat (1). These strains are usually multidrug-resistant (MDR) due to the coexpression of mechanisms involving other classes of antibiotics, thus drastically limiting our therapeutic armamentarium (2–4). In particular, extensively drug-resistant (XDR) isolates are commonly detected worldwide (5), whereas the prevalence of pandrug-resistant (PDR) isolates is increasing worryingly in several countries (6–8). Therefore, novel antimicrobial strategies need to be rapidly developed.
Recently, there has been a rising interest in evaluating naturally occurring or synthetic antimicrobial peptides (AMPs) with activity against prokaryotic membranes. This attention is due to their wide spectrum of activity against both Gram-positive and Gram-negative species, potent bactericidal activity, and ability to bypass common mechanisms of resistance that affect standard antibiotics (9, 10). However, several reasons have so far limited the clinical implementation of AMPs: (i) high susceptibility to degradation by endogenous and microbial proteases; (ii) toxicity due to the high concentration necessary to inhibit bacteria; and (iii) short half-life because of high protein binding (11). Several authors have modified AMPs to obtain proteolytically resistant versions, mostly by sequence variations and the use of d-amino acids (12–15). However, redesigning the peptide chain topology, in particular by introducing multiple branching points to obtain synthetic AMP dendrimers (AMPDs), seems a promising solution to overcome all of the the aforementioned problems (16–18).
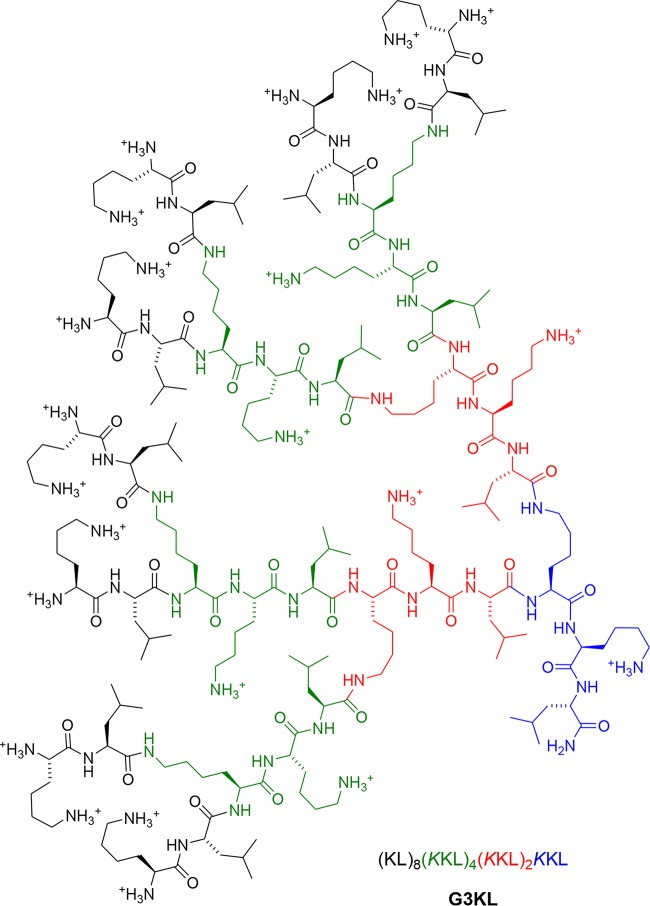
G3KL is a novel AMP dendrimer (AMPD) developed at the Department of Chemistry and Biochemistry of the University of Bern (Switzerland) by sequence optimization of an initial hit compound identified by screening a combinatorial library of dendrimers using a tailored high-throughput screening assay and presumed to act as a membrane-disrupting agent (19–22). Its activity requires a dendritic topology and only natural lysine and leucine residues alternating in the branches (Fig. 1). This novel AMPD has demonstrated in vitro activity against several Gram-negative strains, low toxicity to human red blood cells (minimal hemolytic concentration of 840 μg/ml versus >2,000 μg/ml for polymyxin B), stability in human serum (half-life [t1/2] of ∼18 h and MICs of 2 and 4 μg/ml for P. aeruginosa PAO1 in Mueller-Hinton medium with or without 30% human serum, respectively), and easy preparation by standard solid-phase peptide synthesis; attempts to identify G3KL-resistant strains were also unsuccessful (21).
FIG 1.
Molecular structure of the novel antimicrobial peptide dendrimer (AMPD) G3KL [amino acid sequence (KL)8(KKL)4(KKL)2KKL, where K is lysine, K is branched lysine, and L is leucine].
In the present work, we analyzed the in vitro activity of G3KL against 32 A. baumannii and 35 P. aeruginosa isolates collected in different countries during diverse periods. Species identification was confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Bruker), whereas MICs for different classes of antibiotics were obtained in cation-adjusted Mueller-Hinton II (CAMHII) broth (BBL) using the microdilution GNX2F panels (Trek Diagnostics Systems). MICs were interpreted according to the CLSI criteria (23). P. aeruginosa ATCC 27853 was used as the control. For G3KL, MICs were also achieved in microdilution in CAMHII broth using both polystyrene and polypropylene 96-well plates (24). The minimum bactericidal concentration (MBC) for G3KL was then obtained by culturing 30 μl of the broth from the endpoint well and from the log2 dilution above the MIC onto CAMHII agar plates (BBL) (25). For several strains, carbapenemase genes were already characterized (26–29); the presence of β-lactamase genes (including class A, B, and D carbapenemases) in the remaining isolates was analyzed by implementing the CT103XL microarray (CheckPoints). Phenotypic analysis indicated that among the 32 A. baumannii isolates, there were 5 MDR, 24 XDR, and 1 PDR isolate, whereas the 35 P. aeruginosa isolates included 8 MDR, 19 XDR, and 2 PDR strains (5). Moreover, several of the the strains tested produced the most frequently detected class D (10 OXA-23, 7 OXA-24, and 11 OXA-58) and class B (18 VIM and 3 IMP) carbapenemases described in A. baumannii and P. aeruginosa, respectively (see Tables S1 and S2 in the supplemental material) (1). As shown in Table 1, the isolates of both species possessed very high MICs for β-lactams (e.g., MIC50 values for meropenem of ≥16 μg/ml), aminoglycosides (e.g., MIC50 values for gentamicin of ≥16 μg/ml), and ciprofloxacin (MIC50 values of ≥4 μg/ml); the only standard antibiotics maintaining in vitro activity (≥80%) were colistin and polymyxin B.
TABLE 1.
Overall phenotypic results for standard antibiotics and G3KL antimicrobial peptide dendrimer tested against the collection of 67 isolates
| Antimicrobial | MIC (or MBC where indicated) (μg/ml) and susceptibility results for isolates of: |
|||||||
|---|---|---|---|---|---|---|---|---|
|
A. baumannii (n = 32)a |
P. aeruginosa (n = 35)b |
|||||||
| MIC50 | MIC90 | MIC range | S (%)c | MIC50 | MIC90 | MIC range | S (%)c | |
| Piperacillin-tazobactam | ≥128 | ≥128 | ≤8–≥128 | 9.4 | ≥128 | ≥128 | ≤8–≥128 | 25.7 |
| Ticarcillin-clavulanate | ≥256 | ≥256 | ≤16–≥256 | 6.3 | ≥256 | ≥256 | ≤16–≥256 | 8.6 |
| Ceftazidime | ≥32 | ≥32 | 1–≥32 | 6.3 | ≥32 | ≥32 | ≤1–≥32 | 20.0 |
| Cefepime | 16 | ≥32 | ≤2–≥32 | 25.0 | ≥32 | ≥32 | ≤2–≥32 | 25.7 |
| Aztreonam | ≥32 | ≥32 | 16–≥32 | NA | ≥32 | ≥32 | ≤2–≥32 | 17.1 |
| Imipenem | ≥16 | ≥16 | ≤1–≥16 | 15.3 | ≥16 | ≥16 | ≤1–≥16 | 25.7 |
| Meropenem | ≥16 | ≥16 | ≤1–≥16 | 11.5 | ≥16 | ≥16 | ≤1–≥16 | 25.7 |
| Doripenem | ≥4 | ≥4 | 0.25–≥4 | 18.8 | ≥4 | ≥4 | ≤0.125–≥4 | 25.7 |
| Gentamicin | ≥16 | ≥16 | ≤1–≥16 | 6.3 | ≥16 | ≥16 | ≤1–≥16 | 42.9 |
| Amikacin | ≥64 | ≥64 | ≤4–≥64 | 18.8 | 16 | ≥64 | ≤4–≥64 | 51.4 |
| Ciprofloxacin | ≥4 | ≥4 | ≤0.25–≥4 | 6.3 | ≥4 | ≥4 | ≤0.25–≥4 | 31.4 |
| Tigecycline | 1 | 4 | ≤0.25–4 | NA | 8 | ≥16 | 2–≥16 | NA |
| Colistin | 0.5 | 1 | ≤0.25–≥8 | 96.9 | 2 | ≥8 | 0.5–≥8 | 80.0 |
| Polymyxin B | 0.5 | 2 | ≤0.25–≥8 | 90.6 | 1 | ≥8 | 1–≥8 | 82.9 |
| G3KL | ||||||||
| MICs | 8 (8)d | 8 (8) | 4–16 (4–8) | NA | 8 (8) | 8 (8) | 4–64 (2–32) | NA |
| MBCs | 8 (8) | 8 (8) | 4–16 (4–8) | NA | 8 (8) | 8 (8) | 4–128 (4–32) | NA |
Includes 5 MDR isolates (4 OXA-58 producers and 1 carbapenemase negative), 24 XDR isolates (10 OXA-23, 6 OXA-24, and 7 OXA-58 producers and 1 carbapenemase negative), and 1 PDR isolate (OXA-24 producer) (see Table S1 in the supplemental material).
Includes 8 MDR isolates (2 VIM and 1 IMP producer and 5 carbapenemase negative), 19 XDR isolates (15 VIM and 2 IMP producers and 2 carbapenemase negative), and 2 PDR isolates (1 VIM producer and 1 carbapenemase negative) (see Table S2 in the supplemental material).
S, susceptible according to CLSI criteria of 2014 (23); NA, not applicable.
Results in parentheses are those obtained with polypropylene 96-well plates.
For G3KL, polypropylene plates were used to avoid any hypothetical attachment (and therefore effect on the MICs) of the positively charged AMPD to the surface of the polystyrene wells (21). Overall, G3KL showed MIC50/90 values of 8/8 μg/ml and MBCs at which 50% or 90% of strains tested are killed (MBC50/90) of 8/8 μg/ml for both A. baumannii and P. aeruginosa strains regardless of the plate used, though slightly lower MIC/MBC ranges were observed for polypropylene plates (Table 1). Given the fact that the recorded MBCs were mostly equal to the corresponding MICs (overall, 64 out of 67 strains tested with polypropylene plates and 51 out of 67 strains tested with polystyrene plates; see Tables S1 and S2 in the supplemental material), G3KL has a putative bactericidal effect (30).
Previous studies have assessed the MICs and MBCs of modified AMPs against A. baumannii and P. aeruginosa, these being in general higher than those obtained for G3KL (e.g., for P. aeruginosa, MICs of 150 and MBCs of 20 to >300 μg/ml were reported in references 12 and 13). However, in one study, five polycationic AMPs were tested against A. baumannii, and MIC50 values similar to those found here for G3KL were obtained, although the MIC90/MBC90 values (both ranged from 16 to 32 μg/ml) were overall higher than for G3KL (31). In another study, 15 AMPs were tested against colistin-resistant and -susceptible A. baumannii strains, and only a few yielded MIC50/90 values similar to those for G3KL; moreover, many had significantly different MICs for the two groups of isolates, a phenomenon that we observed for G3KL (Table 1) (14). Sanchez-Gomez et al. assessed 11 AMPs against P. aeruginosa, with only one lactoferrin analogue having MIC50/90 and MBC50/90 values equal to those of G3KL (15). However, since this is a derivative of a naturally occurring linear peptide, in vivo proteolysis is likely to happen. Very recently, several dendrimers have also been evaluated. Lind et al. reported MICs of 41 to 149 μM (i.e., ∼88 to 300 μg/ml) for three dendrimers against P. aeruginosa ATCC 27853 (32), whereas Bahar et al. observed no viable cells of P. aeruginosa PAO1 and PDO300 strains after treating them with a dendrimer at a concentration of 50 μM (i.e., ∼77 μg/ml) (33).
Overall, we note that there is a scarcity of data regarding the in vitro performance of recently designed AMPDs against P. aeruginosa and A. baumannii. More importantly, for most of the aforementioned studies (with either AMPs or AMPDs), small numbers of strains or only laboratory controls were tested. In contrast, we evaluated the activity of G3KL against a large collection of difficult-to-treat isolates (including those producing carbapenemases) that are frequently faced in the contemporary international clinical scenario.
In conclusion, the AMPD G3KL possesses promising in vitro activity against A. baumannii and P. aeruginosa, yielding results that are not apparently different for wild-type, MDR, XDR, and PDR isolates (either producing or not producing carbapenemases). Since G3KL does not resemble other molecules in nature, has low MIC/MBC values compared to those of other AMPs and AMPDs, and has little toxicity for red blood cells in vitro, it seems plausible that this molecule should be further investigated with tissue culture assays and animal studies to support future pharmacological formulations and potential clinical applications.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Swiss National Science Foundation (SNSF; project numbers 153377 to A.E. and 159941 to J.-L.R. and partially by project 138094 to S.L.L.). João Pires is a Ph.D. student (2014 to 2017) supported by SNSF (project number 153377 to A.E.).
We thank Sara Droz (Institute for Infectious Diseases, University of Bern) for providing some of the isolates used in this study.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01853-15.
REFERENCES
- 1.Diene SM, Rolain JM. 2014. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin Microbiol Infect 20:831–838. doi: 10.1111/1469-0691.12655. [DOI] [PubMed] [Google Scholar]
- 2.Spellberg B, Bonomo RA. 2015. Combination therapy for extreme drug-resistant Acinetobacter baumannii: ready for prime time? Crit Care Med 43:1332–1334. doi: 10.1097/CCM.0000000000001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doi Y, Murray GL, Peleg AY. 2015. Acinetobacter baumannii: evolution of antimicrobial resistance-treatment options. Semin Respir Crit Care Med 36:85-98. doi: 10.1055/s-0034-1398388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viale P, Giannella M, Tedeschi S, Lewis R. 2015. Treatment of MDR-Gram negative infections in the 21st century: a never ending threat for clinicians. Curr Opin Pharmacol 24:30–37. doi: 10.1016/j.coph.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 6.Agodi A, Voulgari E, Barchitta M, Quattrocchi A, Bellocchi P, Poulou A, Santangelo C, Castiglione G, Giaquinta L, Romeo MA, Vrioni G, Tsakris A. 2014. Spread of a carbapenem- and colistin-resistant Acinetobacter baumannii ST2 clonal strain causing outbreaks in two Sicilian hospitals. J Hosp Infect 86:260–266. doi: 10.1016/j.jhin.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Durante-Mangoni E, Del Franco M, Andini R, Bernardo M, Giannouli M, Zarrilli R. 2015. Emergence of colistin resistance without loss of fitness and virulence after prolonged colistin administration in a patient with extensively drug-resistant Acinetobacter baumannii. Diagn Microbiol Infect Dis 82:222–226. doi: 10.1016/j.diagmicrobio.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 8.Lopez-Rojas R, McConnell MJ, Jimenez-Mejias ME, Dominguez-Herrera J, Fernandez-Cuenca F, Pachon J. 2013. Colistin resistance in a clinical Acinetobacter baumannii strain appearing after colistin treatment: effect on virulence and bacterial fitness. Antimicrob Agents Chemother 57:4587–4589. doi: 10.1128/AAC.00543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung RC, Ng TB, Wong JH. 2015. Marine peptides: bioactivities and applications. Mar Drugs 13:4006–4043. doi: 10.3390/md13074006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mojsoska B, Jenssen H. 2015. Peptides and peptidomimetics for antimicrobial drug design. Pharmaceuticals 8:366–415. doi: 10.3390/ph8030366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters BM, Shirtliff ME, Jabra-Rizk MA. 2010. Antimicrobial peptides: primeval molecules or future drugs? PLoS Pathog 6:e1001067. doi: 10.1371/journal.ppat.1001067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGrath DM, Barbu EM, Driessen WHP, Lasco TM, Tarrand JJ, Okhuysen PC, Kontoyiannis DP, Sidman RL, Pasqualini R, Arap W. 2013. Mechanism of action and initial evaluation of a membrane active all-D-enantiomer antimicrobial peptidomimetic. Proc Natl Acad Sci U S A 110:3477–3482. doi: 10.1073/pnas.1221924110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueno S, Minaba M, Nishiuchi Y, Taichi M, Tamada Y, Yamazaki T, Kato Y. 2011. Generation of novel cationic antimicrobial peptides from natural non-antimicrobial sequences by acid-amide substitution. Ann Clin Microbiol Antimicrob 10:11. doi: 10.1186/1476-0711-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vila-Farres X, Garcia de la Maria C, López-Rojas R, Pachón J, Giralt E, Vila J. 2012. In vitro activity of several antimicrobial peptides against colistin-susceptible and colistin-resistant Acinetobacter baumannii. Clin Microbiol Infect 18:383–387. doi: 10.1111/j.1469-0691.2011.03581.x. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez-Gomez S, Ferrer-Espada R, Stewart PS, Pitts B, Lohner K, Martinez de Tejada G. 2015. Antimicrobial activity of synthetic cationic peptides and lipopeptides derived from human lactoferricin against Pseudomonas aeruginosa planktonic cultures and biofilms. BMC Microbiol 15:137. doi: 10.1186/s12866-015-0473-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mintzer MA, Dane EL, O'Toole GA, Grinstaff MW. 2012. Exploiting dendrimer multivalency to combat emerging and re-emerging infectious diseases. Mol Pharm 9:342–354. doi: 10.1021/mp2005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reymond JL, Darbre T. 2012. Peptide and glycopeptide dendrimer apple trees as enzyme models and for biomedical applications. Org Biomol Chem 10:1483–1492. doi: 10.1039/c2ob06938e. [DOI] [PubMed] [Google Scholar]
- 18.Reymond JL, Bergmann M, Darbre T. 2013. Glycopeptide dendrimers as Pseudomonas aeruginosa biofilm inhibitors. Chem Soc Rev 42:4814–4822. doi: 10.1039/c3cs35504g. [DOI] [PubMed] [Google Scholar]
- 19.Fluxa VS, Maillard N, Page MG, Reymond JL. 2011. Bead diffusion assay for discovering antimicrobial cyclic peptides. Chem Commun (Camb) 47:1434–1436. doi: 10.1039/C0CC04670A. [DOI] [PubMed] [Google Scholar]
- 20.Stach M, Maillard N, Kadam RU, Kalbermatter D, Meury M, Page MGP, Fotiadis D, Darbre T, Reymond J-L. 2012. Membrane disrupting antimicrobial peptide dendrimers with multiple amino termini. Medchemcomm 3:86–89. doi: 10.1039/C1MD00272D. [DOI] [Google Scholar]
- 21.Stach M, Siriwardena TN, Kohler T, van Delden C, Darbre T, Reymond JL. 2014. Combining topology and sequence design for the discovery of potent antimicrobial peptide dendrimers against multidrug-resistant Pseudomonas aeruginosa. Angew Chem Int Ed Engl 53:12827–12831. doi: 10.1002/anie.201409270. [DOI] [PubMed] [Google Scholar]
- 22.Ravi HK, Stach M, Soares TA, Darbre T, Reymond JL, Cascella M. 2013. Electrostatics and flexibility drive membrane recognition and early penetration by the antimicrobial peptide dendrimer bH1. Chem Commun (Camb) 49:8821–8823. doi: 10.1039/c3cc44912b. [DOI] [PubMed] [Google Scholar]
- 23.CLSI. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. M100-S24. CLSI, Wayne, PA. [Google Scholar]
- 24.CLSI. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed Document M7-A9. CLSI, Wayne, PA. [Google Scholar]
- 25.Mahon CR, Manuselis G. 2000. Textbook of diagnostic microbiology, 2nd ed W.B. Saunders Company, St. Louis, MO. [Google Scholar]
- 26.Principe L, Piazza A, Giani T, Bracco S, Caltagirone MS, Arena F, Nucleo E, Tammaro F, Rossolini GM, Pagani L, Luzzaro F. 2014. Epidemic diffusion of OXA-23-producing Acinetobacter baumannii isolates in Italy: results of the first cross-sectional countrywide survey. J Clin Microbiol 52:3004–3010. doi: 10.1128/JCM.00291-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Docquier JD, Luzzaro F, Amicosante G, Toniolo A, Rossolini GM. 2001. Multidrug-resistant Pseudomonas aeruginosa producing PER-1 extended-spectrum serine-β-lactamase and VIM-2 metallo-β-lactamase. Emerg Infect Dis 7:910–911. doi: 10.3201/eid0705.010528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D'Andrea MM, Giani T, D'Arezzo S, Capone A, Petrosillo N, Visca P, Luzzaro F, Rossolini GM. 2009. Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 53:3528–3533. doi: 10.1128/AAC.00178-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luzzaro F, Ortisi G, Larosa M, Drago M, Brigante G, Gesu G. 2011. Prevalence and epidemiology of microbial pathogens causing bloodstream infections: results of the OASIS multicenter study. Diagn Microbiol Infect Dis 69:363–369. doi: 10.1016/j.diagmicrobio.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 30.French GL. 2006. Bactericidal agents in the treatment of MRSA infections—the potential role of daptomycin. J Antimicrob Chemother 58:1107–1117. doi: 10.1093/jac/dkl393. [DOI] [PubMed] [Google Scholar]
- 31.Giacometti A, Cirioni O, Del Prete MS, Barchiesi F, Paggi AM, Petrelli E, Scalise G. 2000. Comparative activities of polycationic peptides and clinically used antimicrobial agents against multidrug-resistant nosocomial isolates of Acinetobacter baumannii. J Antimicrob Chemother 46:807–810. doi: 10.1093/jac/46.5.807. [DOI] [PubMed] [Google Scholar]
- 32.Lind TK, Polcyn P, Zielinska P, Cardenas M, Urbanczyk-Lipkowska Z. 2015. On the antimicrobial activity of various peptide-based dendrimers of similar architecture. Molecules 20:738–753. doi: 10.3390/molecules20010738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bahar AA, Liu Z, Totsingan F, Buitrago C, Kallenbach N, Ren D. 2015. Synthetic dendrimeric peptide active against biofilm and persister cells of Pseudomonas aeruginosa. Appl Microbiol Biotechnol 99:8125–8135. doi: 10.1007/s00253-015-6645-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.