Abstract
Recently, bioMérieux, France, introduced the Rapidec Carba NP test kit for rapid detection of carbapenemase-producing Gram-negative bacteria. This kit was evaluated in this study, and we report sensitivity, specificity, and positive and negative predictive values of 92.6%, 96.2%, 95.83%, and 92.6%, respectively. The test was easy to perform and interpret and relatively inexpensive ($5/Rs 300 per test) and provides a practical solution for early detection of carbapenemase-producing, multidrug-resistant Gram-negative bacteria.
TEXT
Carbapenems are the most effective antimicrobial agents against Gram-negative bacteria and have a broad spectrum of antibacterial activity against members of the Enterobacteriaceae, Pseudomonas aeruginosa, Acinetobacter baumannii complex, etc. They are generally used as last-resort drugs for the treatment of infections caused by multidrug-resistant bacteria (1). Mechanisms of resistance to carbapenems include production of β-lactamases, efflux pumps, and mutations that alter the expression and/or function of porins and penicillin-binding proteins. Carbapenemases are specific β-lactamases with the ability to hydrolyze carbapenems, and their production appears to be the most important cause of carbapenem resistance. It is well documented that there is widespread distribution of carbapenemases in Gram-negative bacteria (1). An increasing number of Ambler class A carbapenemases (e.g., KPC and GES enzymes), class B metallo-β-lactamases (e.g., VIM, IMP, and NDM β-lactamases), and class D carbapenemases (e.g., OXA-23, -48, -51, and -143) have recently emerged (2). Carbapenem-resistant Gram-negative bacilli remain a formidable threat, as few antimicrobial agents are reliably active and very little is expected to be available in the near future to combat this situation.
The detection of carbapenemases in clinical microbiology labs is challenging because phenotypic tests like the modified Hodge test and triple-disc test (Rosco Diagnostics, Denmark) are time-consuming and difficult to interpret. PCR-based molecular methods, UV spectrophotometry, and MALDI-TOF MS (matrix-assisted laser desorption ionization–time of flight mass spectrometry) assays have overall good sensitivity and specificity but require trained personnel and good infrastructure and are expensive (3).
Recently bioMérieux (La Balme-les-Grottes, France) introduced the Rapidec Carba NP (carbapenemase Nordmann-Poirel) kit to detect carbapenemases from bacterial colonies grown on recommended selective or nonselective agar plates. The manufacturer claims detection of carbapenemase producing bacteria in 30 min to 2 h, with sensitivity and specificity of 97.8% compared with molecular techniques (www.biomerieux-diagnostics.com/sites/clinic/files/9308689-002-gb-a-rapidec-carba-np.pdf). This kit was evaluated by Poirel and Nordmann (4) and Dortet et al. (5) in 2015 using precharacterized bacterial isolates at the Emerging Antibiotic Resistance Unit, University of Fribourg, Switzerland, and the National Reference Center for Antibiotic Resistance, France, respectively. However, to the best of our knowledge, no independent field trial of this kit has been reported by any laboratory worldwide to date, and this study was planned with the aim of evaluating the field efficacy of the Rapidec Carba NP kit in a resource-limited bacteriology laboratory of a developing country like India.
The strains used in this study were collected from various clinical samples of patients admitted to L.L.R & Associated Hospitals (10 hospitals with different superspecialties), GSVM Medical College, Kanpur, and District Hospital, Kanpur, India. All isolated nonduplicate strains were initially screened for carbapenem sensitivity by the disc diffusion method; nonsusceptibility to carbapenems was confirmed by MIC estimation using E strips, and the results were interpreted using the Clinical and Laboratory Standards Institute guidelines (6). Environmental and surveillance samples were not included in our study. A panel of 100 bacterial isolates were selected for testing: 50 containing different classes of carbapenemases (NDM, KPC, VIM, IMP, and OXA-48, all confirmed by PCR [7] and sequence analysis) and 50 non-carbapenemase producers, of which 25 were sensitive to carbapenem and 25 were resistant to at least one of the carbapenems tested. In the latter group, phenotypic tests for carbapenemase production (modified Hodge test and KPC/MBL confirmation kit; Rosco Diagnostics, Denmark) were also negative; furthermore, to rule out the presence of any carbapenemase activity, carbapenem hydrolysis was performed spectrophotometrically as described earlier (8), and it was concluded that the carbapenem resistance observed in this group was due to some other mechanism (Table 1). The clonality of the strains of the family Enterobacteriaceae was determined by enterobacterial repetitive intergenic consensus (ERIC) PCR, whereas for nonfermenters, repetitive extragenic palindromic (REP) PCR was performed.
TABLE 1.
Results of the Rapidec Carba NP test with carbapenemase-producing and non-carbapenemase-producing Gram-negative bacteria
| Species (no. of isolates tested) | Carbapenemase detected (no. of isolates tested) | MIC (μg/ml) of imipenem | Rapidec Carba NP test result |
|---|---|---|---|
| Carbapenemase-producing bacteria, Rapidec Carba NP positive (46)a | |||
| Escherichia coli (23) | NDM (12) | 4–128 | + |
| OXA-48 (6) | 1–8 | + | |
| KPC (2) | 16–32 | + | |
| VIM (2) | 32 | + | |
| IMP (1) | 16 | + | |
| Pseudomonas aeruginosa (10) | NDM (5) | 4–64 | + |
| IMP (3) | 8–32 | + | |
| KPC (2) | 8–16 | + | |
| Acinetobacter baumannii (5) | NDM (2) | 16–32 | + |
| KPC (2) | 4–8 | + | |
| IMP (1) | 8 | + | |
| Klebsiella pneumoniae (3) | NDM (1) | 32 | + |
| KPC (2) | 8–16 | + | |
| Proteus mirabilis (2) | NDM (1) | 32 | + |
| KPC (1) | 32 | + | |
| Citrobacter freundii (1) | NDM (1) | 16 | + |
| Enterobacter cloacae (1) | OXA- 48 (1) | 8 | + |
| Stenotrophomonas maltophilia (1) | NDM (1) | 8 | + |
| Carbapenemase-producing bacteria, Rapidec Carba NP negative (4)a | |||
| Escherichia coli (3) | OXA- 48 (2) | 4–8 | − |
| IMP (1) | 4 | − | |
| Klebsiella pneumoniae (1) | OXA- 48 (1) | 4 | − |
| Non-carbapenemase-producing, carbapenem-resistant bacteria, Rapidec Carba NP positive (2)b | |||
| Escherichia coli (1) | 4 | + | |
| Pseudomonas aeruginosa (1) | 8 | + | |
| Non-carbapenemase-producing bacteria, Rapidec Carba NP negative (48)b | |||
| Escherichia coli (19) | 0.06–8 | − | |
| Pseudomonas aeruginosa (9) | 0.06–32 | − | |
| Acinetobacter baumannii (6) | 0.12–16 | − | |
| Klebsiella pneumoniae (8) | 0.06–32 | − | |
| Proteus mirabilis (2) | 0.12–4 | − | |
| Citrobacter freundii (2) | 0.06–4 | − | |
| Enterobacter cloacae (2) | 0.12–4 | − |
PCR- and imipenem hydrolysis-positive strains.
PCR- and imipenem hydrolysis-negative strains.
The test was performed as recommended by the manufacturer. A loopful (10-μl loop) of a bacterial colony was picked up from overnight-incubated Mueller-Hinton agar plates (bioMérieux) and mixed into API suspension medium (provided with kit); the bacterial suspension was then transferred to wells in the test strip and incubated at 37°C. Visual reading of the test strip was done after 30 min and after 2 h, if necessary. A positive test corresponded to a color change from red to yellow-orange, whereas a red color indicated a negative result (Fig. 1). The test strips were read by two independent blinded readers to determine interrater reliability. Any discrepancy between the two readings was resolved by an independent blinded third reader, whose reading was taken as final.
FIG 1.
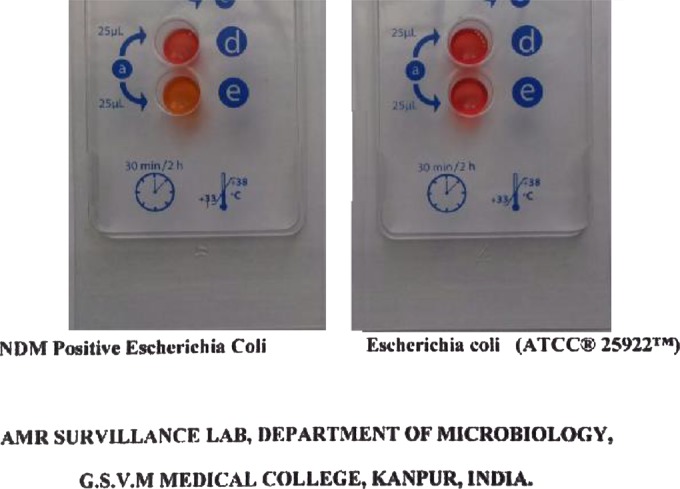
Rapidec Carba NP test results. The yellow-orange color in well E with NDM-positive Escherichia coli indicates a positive result, whereas the red color with Escherichia coli ATCC 25922 indicates a negative result.
The kit detected carbapenemase activity in 46 out of 50 PCR-confirmed strains. However, it could not detect carbapenemase activity in 3 OXA-48-positive strains (two Escherichia coli isolates [nonclonal] and one Klebsiella pneumoniae isolate) and one IMP-positive Escherichia coli strain. False-positive results were also obtained with two carbapenemase-negative carbapenem-resistant strains (one Escherichia coli and one Pseudomonas aeruginosa isolate). None of the carbapenem-sensitive bacteria were positive. Overall, the kit showed good efficacy (Table 2).
TABLE 2.
Diagnostic accuracy of Rapidec Carba NP
| Statistical index | Value (%) | 95% confidence interval (%) |
|---|---|---|
| Sensitivity | 92.00 | 80.77–97.78 |
| Specificity | 96.00 | 86.29–99.51 |
| Positive predictive value | 95.83 | 85.75–99.49 |
| Negative predictive value | 92.31 | 81.46–97.86 |
To date, the Rapidec Carba NP kit had been evaluated in only two studies. Poirel and Nordmann (4) tested this kit with precharacterized strains and documented a sensitivity and specificity of 96%; they also reported false-negative results with group D carbapenemases, which is in accordance with the fact that group D carbapenemases are known to lack significant carbapenemase activity. In a similar study from the National Reference Center for Antibiotic Resistance, France, Dortet et al. (5) documented a sensitivity of 99% and specificity of 100%.
To conclude, the Rapidec Carba NP test was simple, easy to perform and interpret, and relatively inexpensive ($5/Rs 300 per test) and in most cases gave results in <30 min. The kit was found to be well suited for rapid detection of NDM, the most common carbapenemase isolated in the Indian subcontinent (9). The kit can also be used to screen the strains before expensive molecular tests, like PCR and gene sequencing, are performed for identification of the carbapenemase genes. This kit has a high degree of sensitivity and specificity and provides a practical solution for early detection of carbapenemase-producing, multidrug-resistant Gram-negative bacteria in peripheral microbiological laboratories. Further, it will help in implementation of infection prevention and control measures, e.g., patient isolation and quarantine, to limit the spread of carbapenemase producers and avoid outbreaks in health care settings.
ACKNOWLEDGMENTS
The strains used in this study were isolated and characterized as part of an extramural research project entitled “A study on molecular epidemiology, diagnosis and therapeutic option of carbapenemase producing Gram-negative bacteria,” IRIS no. 2011-13870, funded by the Indian Council of Medical Research, New Delhi, India.
We have no conflicts of interest to declare.
REFERENCES
- 1.Garg A, Garg J, Swaroop A, Kala S, Rao YK, Kumar A, Singh RK, Tripathi P. 2011. Emerging carbapenemases: bringing a step closer to extremely drug resistant bacteria. Int J Curr Res 3:35–38. [Google Scholar]
- 2.Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. 2011. Carbapenems: past, present, and future. Antimicrob Agents Chemother 55:4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dortet L, Bréchard L, Cuzon G, Poirel L, Nordmann P. 2014. Strategy for rapid detection of carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 58:2441–2445. doi: 10.1128/AAC.01239-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poirel L, Nordmann P. 2015. Rapidec Carba NP test for rapid detection of carbapenemase producers. J Clin Microbiol 53:3003–3008. doi: 10.1128/JCM.00977-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dortet L, Agathine A, Naas T, Cuzon G, Poirel L, Nordmann P. 2015. Evaluation of the RAPIDEC® CARBA NP, the Rapid CARB Screen® and the Carba NP test for biochemical detection of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 70:3014–3022. doi: 10.1093/jac/dkv213. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing: 23rd informational supplement. Document M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 7.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Bernabeu S, Poirel L, Nordmann P. 2012. Spectrophotometry-based detection of carbapenemase producers among Enterobacteriaceae. Diagn Microbiol Infect Dis 74:88–90. doi: 10.1016/j.diagmicrobio.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 9.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


