Abstract
In many Gram-negative pathogens, mutations in the key cell wall-recycling enzyme AmpD (N-acetyl-anhydromuramyl-l-alanine amidase) affect the activity of the regulator AmpR, which leads to the expression of AmpC β-lactamase, conferring resistance to expanded-spectrum cephalosporin antibiotics. Burkholderia cepacia complex (Bcc) species also have these Amp homologs; however, the regulatory circuitry and the nature of causal ampD mutations remain to be explored. A total of 92 ampD mutants were obtained, representing four types of mutations: single nucleotide substitution (causing an amino acid substitution or antitermination of the enzyme), duplication, deletion, and IS element insertion. Duplication, which can go through reversion, was the most frequent type. Intriguingly, mutations in ampD led to the induction of two β-lactamases, AmpC and PenB. Coregulation of AmpC and PenB in B. cenocepacia, and likely also in many Bcc species with the same gene organization, poses a serious threat to human health. This resistance mechanism is of evolutionary optimization in that ampD is highly prone to mutations allowing rapid response to antibiotic challenge, and many of the mutations are reversible in order to resume cell wall recycling when the antibiotic challenge is relieved.
INTRODUCTION
Burkholderia cepacia complex (Bcc) is a group of bacteria comprised of at least 17 closely related species, including Burkholderia cepacia, B. cenocepacia, B. multivorans, and B. vietnamiensis (1–4). Members of the Bcc are known as significant cystic fibrosis (CF) pathogens that can cause rapid clinical deterioration with necrotizing pneumonia and sepsis resulting in early death, which is called “cepacia syndrome” (5, 6). Although all Bcc species have been isolated from CF infection, B. cenocepacia and B. multivorans have been shown to be most responsible for the severity of the infection (7–9). Bcc infections occur beyond CF, as demonstrated through reports of infections in immunocompromised patients such as those with cancer or HIV and also among immunocompetent individuals (10, 11). Infections involving Bcc species are generally difficult to cure because of their intrinsic multidrug resistance (12, 13). Current therapies often include expanded-spectrum cephalosporins, including ceftazidime, which is one of a few antimicrobial agents effective against the infection (12, 14). However, the genomes of Bcc species contain various β-lactamase genes, including that coding for AmpC, which potentially has hydrolytic activity for expanded-spectrum cephalosporins, and AmpD, a key cell wall-recycling enzyme (N-acetyl-anhydromuramyl-l-alanine amidase) (15). Mutations in ampD gene indirectly result in overexpression of ampC in many Gram-negative pathogens, including Pseudomonas aeruginosa, Enterobacter cloacae, and Citrobacter freundii (16–19); therefore, similar regulation is expected in Bcc species.
Bacterial cell walls are efficiently recycled during growth. For example, Escherichia coli recycles 90% of the peptidoglycan degradation products produced during growth in each generation (20). Exposure to β-lactam antibiotics can disrupt the cell wall-recycling system (21); in many bacteria, changes in peptidoglycan metabolite levels serve as a mechanism for detecting the antibiotics that lead to the regulation of ampC through the LysR-type transcriptional regulator AmpR (18, 19, 22). AmpR is a tetramer that operates as both a repressor and an activator depending on the ligand, which is determined by the balance between cell wall synthesis and degradation (23). When AmpR binds UDP-MurNAc-pentapeptide, the precursor of peptidoglycan, at the D-Ala-D-Ala motif, expression of ampC is repressed. In contrast, AmpR becomes an activator when it binds peptidoglycan degradation products 1,6-anhydroMurNAc-peptides. The levels of 1,6-anhydroMurNAc-peptides increase when β-lactam antibiotics are present or when AmpD, which degrades them as part of the recycling process, is not functional due to mutation in the gene (16). Some β-lactams, including cefoxitin and imipenem, are strong inducers of AmpR-mediated AmpC β-lactamase expression, while others, including ceftazidime, cefotaxime, and aztreonam, are known to be poor inducers. Different levels of inhibition of low-molecular-weight penicillin-binding proteins (PBPs) by different antibiotics may be partly responsible for this difference in gene induction (24, 25). In response to poor inducers that do not trigger the regular regulatory mechanism, mutations in ampD may play a pivotal role in inducing the expression of ampC (25).
In this study, we demonstrate cell wall-recycling-linked β-lactamase regulation in B. cenocepacia, which simultaneously induces two enzymes, AmpC and PenB (26), due to ampD mutations. We profiled the repertoire of mutations in ampD, showing that the gene particularly favors reversible duplication mutations, suggesting that this regulatory system is the result of optimized evolution for increased survival against dynamic antibiotic challenges.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli strains used for molecular cloning and conjugation were grown in Luria Bertani (LB) medium, and B. cenocepacia strains were grown in Iso-sensitest medium (Oxoid, Basingstoke, United Kingdom) at 37°C (27). Tetracycline was used at 10 μg/ml for E. coli, and the antibiotics used for B. cenocepacia strains were as follows: kanamycin, 50 μg/ml; tetracycline, 150 μg/ml; and ceftazidime, 8 μg/ml.
Isolation of ceftazidime-resistant mutants.
A single colony of B. cenocepacia strain J2315 grown on Iso-sensitest agar at 37°C for 2 days was used to inoculate 3 ml of Iso-sensitest broth, and the inoculum was incubated with shaking (250 rpm) for 18 h at 37°C. Bacterial cells were washed with fresh broth and diluted in the same broth to approximately 107 CFU/ml. Diluted bacterial suspensions of 100 μl were spread on Iso-sensitest agar containing 8 μg/ml ceftazidime (four times the MIC of B. cenocepacia strain J2315) and incubated for 48 h at 37°C. Visible colonies were streaked on the same selective agar plate for confirmation of the acquired antibiotic resistance.
Measurement of MIC.
The MIC values were measured by using the agar dilution method (28), using Iso-sensitest agar instead of Mueller-Hinton (MH) agar. For the agar dilution method, a single colony of each B. cenocepacia strain grown on Iso-sensitest agar at 37°C for 2 days was inoculated in 3 ml of Iso-sensitest broth, and inoculums were incubated in a shaking incubator at 37°C for 18 h. Overnight cultures were diluted with Iso-sensitest broth and adjusted to 1 × 107 CFU/ml. One microliter of diluted bacterial suspension (approximately 104 bacterial cells) was dropped onto each Iso-sensitest agar plate containing an antibiotic using a multichannel pipette. After incubation at 37°C for 20 h, the lowest concentration at which there was no visible colonies was determined as the MIC value for the antibiotic. Adjusted concentrations of 1 × 107 CFU/ml of cell suspensions were confirmed by spreading 100 μl of serial dilutions on Iso-sensitest agar, incubating, and counting visible colonies.
Mapping mutations conferring ceftazidime resistance using whole-genome sequencing.
To identify genes involved in ceftazidime resistance, the genomes of two ceftazidime-resistant isolates that had different MIC values were sequenced using Illumina HiSeq2000. Data trimming, mapping reads to reference genome, and single-nucleotide polymorphism (SNP) analysis were performed using CLC Genomics Workbench 6 software (CLC-bio, Aarhus, Denmark).
Nucleotide sequence analysis of the ampD gene.
Genomic DNA of each ceftazidime-resistant mutant was extracted using a Wizard Genomic DNA purification kit (Promega, Madison, WI, USA) and was used as the PCR template. The coding sequence region of ampD (591 bp) and short flanking regions (314 bp upstream of the start codon and 447 bp downstream of the stop codon) were PCR amplified using primers ampD-F (5′-CCGATGCGACAGATTCTTCT-3′) and ampD-R (5′-AAAGCTCCTGGTGTTGGATG-3′). PCRs were performed in a 50-μl reaction mixture containing 1.0 unit of KOD FX Neo polymerase (Toyobo, Osaka, Japan), 25 μl of 2× PCR buffer for KOD FX Neo, 0.4 mM deoxynucleoside triphosphates (dNTPs), 100 ng template genomic DNA, and 0.3 μM each primer. A three-step PCR was conducted as follows: predenaturation step (94°C for 2 min), amplification step (35 cycles of 98°C for 10 s, 60°C for 30 s, and 68°C for 60 s), and final extension step (68°C for 7 min) using the C1000 Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA). Next, 1,352-bp PCR amplicons were purified using a PCR purification kit (Qiagen, Hilden, Germany) and sequenced by a 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA) in both directions using the primer pair ampD-F and ampD-R.
Direct repeat pairs in the coding sequence of ampD were searched using UGENE software (www.ugene.unipro.ru) (29). The parameters used were as follows: minimum repeat length, 6 bp; repeats identity, 100%; minimum distance between repeats, 0; and maximum distance between repeats, 30.
Complementation of ampD mutants with the wild-type gene.
The whole ampD operon (2,409 bp) was PCR amplified using KOD FX Neo polymerase and primers ampD-CF (5′-ATATATGGTACCGCCTTGCCTTCGTAGTCG-3′) and ampD-CR (5′-ATATATAAGCTTGCCCTGAGAACCCTGTCC-3′), containing a KpnI and HindIII recognition site (underlined) at the end, respectively. The PCR mixture was prepared as described above, and the PCR was conducted as follows: predenaturation step (94°C for 2 min), amplification step (35 cycles of 98°C for 10 s, 60°C for 30 s, and 68°C for 90 s), and final extension step (68°C for 7 min). Purified PCR products treated with KpnI and HindIII were ligated with the broad-host-range vector pRK415 (30) in E. coli strain DH5a. Sequence-confirmed plasmids were transformed into E. coli strain S17-1 (31) through the conventional method (32). The S17-1 strain harboring the plasmid was conjugated with ceftazidime-resistant B. cenocepacia mutants on Iso-sensitest agar plates supplemented with 50 μg/ml kanamycin and 150 μg/ml tetracycline. After incubation at 37°C for 2 days, successful conjugants were obtained and confirmed by restriction pattern analysis of the plasmid.
Reversion test.
A ceftazidime-resistant mutant harboring the 21-bp duplication in ampD was grown on Iso-sensitest agar supplemented with 8 μg/ml ceftazidime at 37°C for 2 days. A single colony was inoculated in 3 ml of Iso-sensitest broth without ceftazidime and incubated with shaking (250 rpm) at 37°C overnight. Then, 30 μl of this inoculum was diluted 1:100 in fresh broth and again incubated overnight; this incubation step was repeated for up to 30 days. After every 5 days, genomic DNA was extracted from 1 ml of each inoculum, and approximately 1 × 102 cells were spread on an Iso-sensitest agar plate with or without 8 μg/ml ceftazidime to measure the ratio between bacterial cells that had a reversion and duplication mutants. Genomic DNA from each time point was used as a template for PCR using KOD FX Neo polymerase and the primer pair ampD-IF (5′-GTTCGACGAGGCGCAATAC-3′) and ampD-IR (5′-AAGCGTTGCCAATCGAAAT-3′) to determine whether a reversion occurred in the ampD sequence. PCR was conducted as follows: predenaturation step (94°C for 2 min), amplification step (35 cycles of 98°C for 10 s, 60°C for 30 s, and 68°C for 30 s), and a final extension step (68°C for 7 min).
qRT-PCR analysis.
Quantitative real-time PCR (qRT-PCR) was conducted with strain J2315 to measure the expression levels of ampC (BCAS0156), penB (BCAM2165), and penR (BCAM2166), using gyrB (BCAL0421) as a reference gene (33). For the levels induced by ampD mutations, the wild-type strain and selected mutants were grown on Iso-sensitest medium to the mid-log phase (optical density at 600 nm [OD600] of 1.0) without antibiotic pressure before isolating total RNA. To determine the expression levels induced by antibiotics, the wild-type strain was grown to an OD600 of 0.5 without antibiotics; a subinhibitory concentration of each antibiotic (ampicillin, 100 μg/ml; ceftazidime, 0.5 μg/ml; cefotaxime, 10 μg/ml; meropenem, 2 μg/ml) was then added, and the cells were grown further to an OD600 of 1.0. All harvested bacterial cultures were treated with RNAprotect bacteria reagent (Qiagen, Hilden, Germany) to prevent alterations in the transcriptome. RNA samples were prepared using an Easy-spin total RNA extraction kit (Intron, South Korea) and treated with RQ1 RNase-Free DNase (Promega, Madison, WI, USA). cDNA was synthesized with 2 μg of DNase-treated RNA using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen Life Technologies, Carlsbad, CA, USA). Diluted cDNA was used as the template to perform quantitative real-time PCR using iQ SYBR green Supermix (Bio-Rad, Hercules, CA, USA) and the Bio-Rad CFX96 Real-Time System C1000 Touch Thermal Cycler according to the manufacturer's instructions. Primer pairs used for the PCR of each gene were as follows: ampC, 5′-ATTCAATGCGACACGCTTC-3′ and 5′-GGAATCGCGTACTGCTTCAT-3′; penB, 5′-AGTACGTTCAAGGCGATGCT-3′ and 5′-GGCGAATAGTTGACGAGGTC-3′; penR, 5′-CGGCTGTACGCTGTTTACG-3′ and 5′-GAACTGCTTGAGCACCGTTT-3′; and gyrB, 5′-CTGCTGCTCACGTTCCTGTA-3′ and 5′-TTCAGATACCGCTCGTCCTT-3′. Fold changes were calculated using the comparative threshold cycle (CT) method (34).
RESULTS AND DISCUSSION
Mutations in ampD conferring ceftazidime resistance in B. cenocepacia.
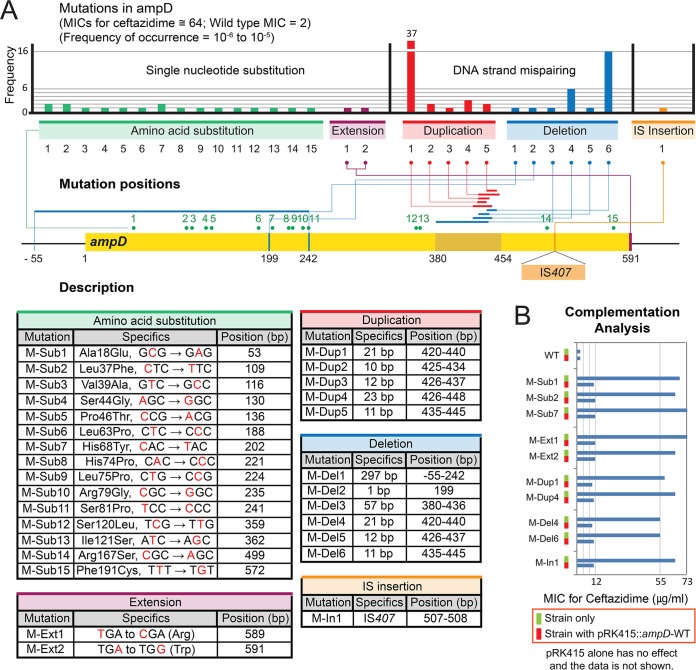
B. cenocepacia strain J2315, the strain originating from a CF patient in the United Kingdom (35), was subjected to a large selection scheme using ceftazidime at a concentration of 8 μg/ml, which is four times the MIC (2 μg/ml) of the strain. From 106 bacterial cells that were spread on a selection agar plate, 1 to 10 colonies emerged after 48 h of incubation, showing a frequency range of 10−6 to 10−5 (Fig. 1A). Two of the mutants were randomly selected, and their whole genomes were sequenced to locate the mutations (see Materials and Methods). In both genomes, mutations were mapped to a single gene, ampD (BCAL3430). We collected up to 92 ampD mutants, and the MICs of ceftazidime for these mutants were measured to be around 64 μg/ml (Fig. 1A).
FIG 1.
Mutations in ampD conferring resistance to ceftazidime. (A) Map of the mutations on ampD. Positions of single-nucleotide substitution mutations resulting in amino acid substitutions or protein extension, duplications, deletions, and IS insertion are mapped. The numbers of occurrences of each mutation are shown in a bar graph. All mutations exhibited an MIC value around 64 μg/ml. Details of each mutation are provided in tables below the map. (B) Complementation analysis with the wild-type ampD. Restoration of ceftazidime susceptibility by intact ampD is observed with all mutations, but data for a few representative mutations are shown.
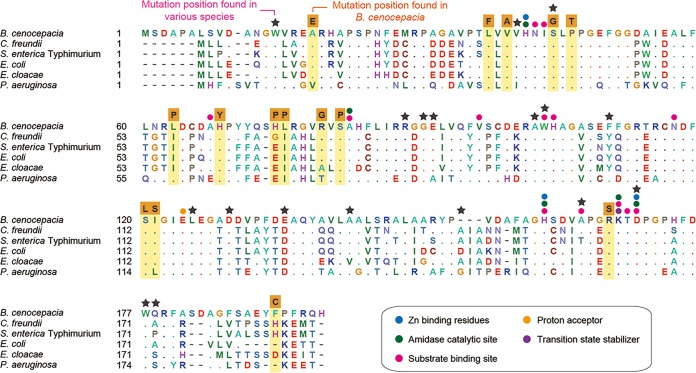
We identified four types of mutations in ampD: substitution, duplication, deletion, and IS (insertion sequence) insertion (Fig. 1A). When each of the mutants was complemented with a plasmid harboring the wild-type ampD, ceftazidime sensitivity was restored (Fig. 1B). This result suggests that those coselected ampD mutations (see Materials and Methods) are solely responsible for the development of the ceftazidime resistance. It is noteworthy that substitutions were not the most frequent mutation type in ampD, as they were in Burkholderia thailandensis penL (penA in the study by Yi et al. [36] is renamed penL here, following the nomenclature guidelines by Poirel et al. [26]), coding for a class A β-lactamase, in response to ceftazidime (36). Fifteen substitution mutations were spread along the coding region of ampD (Fig. 1A). Protein sequence alignment of AmpD from strain J2315 with that from other species revealed that most substitutions occurred in highly conserved positions or are closely associated with functional domains, suggesting functional interference caused by these mutations (Fig. 2). Amino acid substitutions have been reported in many positions in AmpD from various species (37–39) (Fig. 2). Among the 15 amino acid substitution positions identified in B. cenocepacia AmpD in the present study, one (Ser44Gly) matched with that previously identified in E. coli (Ser37Arg in this case) (38) (Fig. 2). In addition to these amino acid substitutions, two nucleotide substitution mutations occurred at the stop codon, converting it into a codon for Arg or Trp, extending the enzyme by 135 amino acid residues (Fig. 1A).
FIG 2.
Alignment of AmpD enzymes. Amino acid sequences of AmpD from various bacteria are aligned. The positions of substitutions on AmpD from B. cenocepacia strain J2315 are highlighted. The resultant amino acid from each substitution event is shown above the column in an orange box. The functional residues identified by the Conserved Domain Database (CDD) from NCBI (45) are denoted by colored circles. Previously identified ampD mutation positions from various species (39) are denoted by stars. Abbreviations: C. freundii, Citrobacter freundii; S. enterica Typhimurium, Salmonella enterica serovar Typhimurium LT2; E. coli, Escherichia coli K-12; E. cloacae, Enterobacter cloacae; P. aeruginosa, Pseudomonas aeruginosa PAO1.
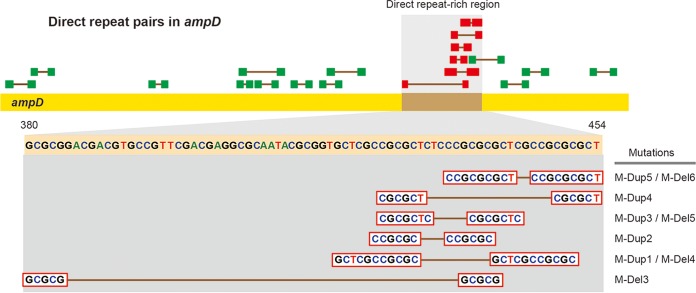
Among the mutations, duplications were the most abundant type, occurring in 45 of 92 mutants (Fig. 1A). Duplications of 10, 11, 12, 21, and 23 bp were identified in a small region of the gene (bp 420 to 448) (Fig. 1A). Duplications of nucleotides that are a multiple of 3 (12 and 21 bp) cause in-frame insertions, while others (10, 11, and 23 bp) cause frameshifts, both resulting in null mutations. A pair of direct repeats was found associated with each of the duplications (Fig. 3). Duplication or deletion of a region between the sequences of a direct repeat pair is known to be caused by DNA strand slippage during the DNA replication process (40). Genetic modifications mediated by this process have been well established in bacteria, including Burkholderia (41). The frequency of DNA slippage is correlated with the length of the repeats but inversely correlated with the distance between the repeat pair (40). Accordingly, the 21-bp duplication was the most frequent among the five duplications due to the longest direct repeats associated with the template DNA (Fig. 3).
FIG 3.
Distribution of direct repeat pairs in ampD. Direct repeat pairs present in the coding sequence of ampD are shown in green or red boxes. The pairs in red boxes are those associated with duplications or deletions, and they are in the region with the highest concentration of repeats in the gene.
In addition to duplications, deletion mutations can occur around repeat sequences. We found three deletions of 11, 12, and 21 bp with the same direct repeat pairs that mediated duplications (Fig. 3). The 11-bp deletion occurred most frequently due to long direct repeats and a short distance between them. In addition, there was a 1-bp deletion and a large deletion of 297 bp involving the upstream region (Fig. 1A). In both deletions, no repeat sequences were found, suggesting the involvement of a different mechanism.
In addition to these mutations, an insertion mutation with an IS element, IS407, was obtained (Fig. 1A). IS407 is known to be involved in genome modification in Burkholderia (42), and 13 copies were previously identified in the B. cenocepacia J2315 genome (10 in Chr 1, 1 in Chr 2, 1 in Chr 3, and 1 in the plasmid). A similar case of IS element-mediated disruption of ampD has been reported in P. aeruginosa with IS1669 (43), suggesting that this IS-mediated mechanism is an effective common option for the disruption of ampD.
Reversibility of duplications in ampD.
Duplications have been demonstrated to go through reversion when selective pressure for the wild-type gene is applied (41). A strain with a duplication mutation, M-Dup1, was used to determine if the mutation goes through reversion when antibiotic pressure is removed. After 15 days of incubation without ceftazidime, no reversion was observed; however, after 20 days of incubation, the number of ceftazidime-resistant colonies started to decrease and the wild-type strain began to be detected and continued to increase steadily during incubation (Fig. 4). In contrast, a control strain with M-Sub1 did not show any signs of reversion after 30 days of incubation (data not shown). These results demonstrated that ampD disrupted by a duplication mutation can be restored to resume cell wall recycling for efficient use of peptidoglycan metabolites when antibiotic challenge is relieved.
FIG 4.
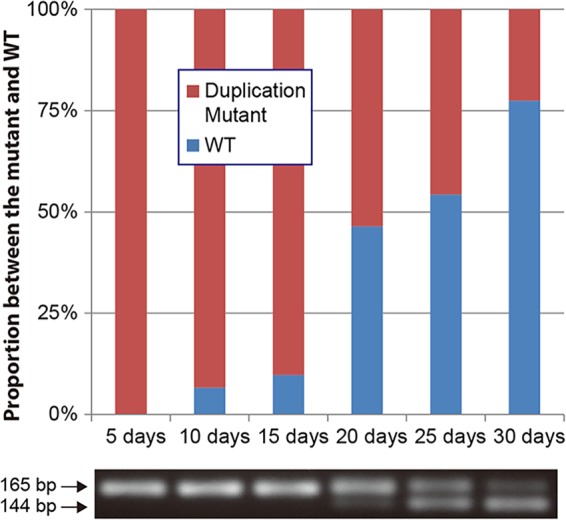
Reversion of a duplication mutation. The proportional variation between the wild type and a strain with a duplication mutation (M-Dup1) during the course of a 30-day incubation without antibiotic pressure is shown in a bar graph. Changes in PCR products containing the duplication region are shown below the graph.
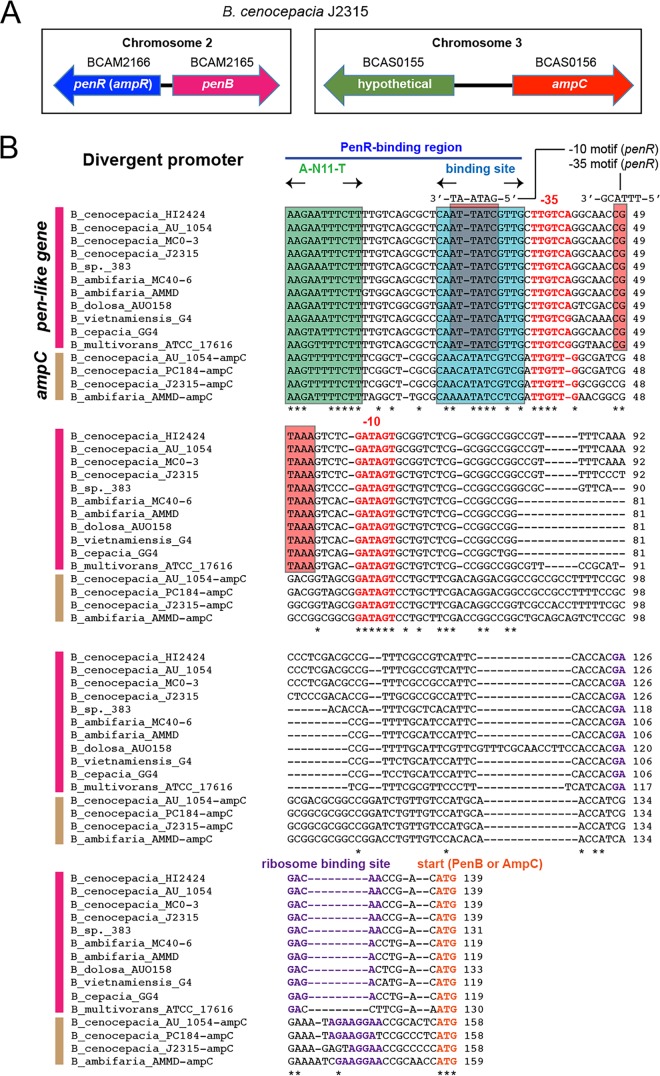
Coregulation of penB and ampC.
Because mutations in ampD lead to ceftazidime resistance in many Gram-negative pathogens, resulting in overexpression of ampC, a similar regulatory system was expected to be present in B. cenocepacia. However, in B. cenocepacia and many Bcc species, ampC and its regulator, ampR, named penR in Burkholderia genomes (55% amino acid identity with AmpR in P. aeruginosa), are not associated, sharing a divergent promoter, as in other previously studied species (Fig. 5A). In Bcc species, penB is instead associated with the ampR homolog. Intriguingly, qRT-PCR analyses showed that expression of penB and the orphaned ampC was increased in ampD mutants (Table 1). A slight reduction in expression level was observed in penR in most strains except the strain with M-Dup1, which had the highest overexpression of penB and ampC (Table 1). There were different levels of gene expression depending on the mutation type in ampD, with M-Dup1, M-Del6, and M-In1 conferring distinctively higher expression than other mutations (Table 1). This pattern may be due to the remnant activity of the mutant AmpD enzymes; while duplication, deletion, and IS insertion mutations are likely null mutations, substitutions or extension mutations may allow some levels of residual function. Consistent with previous studies in other bacterial species, ceftazidime and cefotaxime were poorer inducers than amoxicillin and meropenem (Table 1), suggesting that ampD mutations are required for bacterial survival during selection with ceftazidime pressure.
FIG 5.
Promoters regulated by the AmpR homolog, PenR. (A) Gene organization involving penR, penB, and ampC in B. cenocepacia. In B. cenocepacia, including many Bcc species, pen-like genes rather than ampC are linked to the regulator, and ampC is orphaned in another chromosome. (B) Alignment of the promoters of pen-like genes and ampC from various bacteria. Start codons, ribosome-binding sequences, and −10 and −35 sequences are in bold. The conserved regions of the putative PenR DNA-binding sites are denoted in boxes.
TABLE 1.
RT-PCR gene expression analysis with strains of B. cenocepacia J2315
| Strain | Expression relative to that of the WTa |
||
|---|---|---|---|
| ampCb | penB | penR (ampRc) | |
| WT and mutants | |||
| WT | 1 | 1 | 1 |
| M-Dup1d | 8,724.7 ± 2,861.7 | 750.3 ± 413.2 | 1.1 ± 0.2 |
| M-Del6 | 8,110.0 ± 2,846.1 | 493.8 ± 82.8 | 0.8 ± 0.2 |
| M-In1 | 5,494.6 ± 1,780.3 | 407.2 ± 24.8 | 0.7 ± 0.2 |
| M-Sub5 | 2,489.1 ± 640.4 | 243.5 ± 4.7 | 0.5 ± 0.1 |
| M-Sub8 | 1,999.0 ± 666.6 | 241.4 ± 38.7 | 0.4 ± 0.2 |
| M-Sub14 | 1,508.0 ± 441.9 | 234.1 ± 18.7 | 0.4 ± 0.1 |
| M-Ext2 | 600.4 ± 131.7 | 135.5 ± 20.5 | 0.4 ± 0.0 |
| WT with inductione using: | |||
| AMX | 62.2 ± 8.9 | 40.4 ± 2.1 | 0.6 ± 0.1 |
| CAZ | 12.4 ± 1.0 | 14.3 ± 1.6 | 0.8 ± 0.1 |
| CTXM | 23.4 ± 6.1 | 38.6 ± 3.6 | 0.4 ± 0.1 |
| MER | 409.1 ± 76.4 | 139.5 ± 6.9 | 1.1 ± 0.1 |
Wild-type strain.
Target gene for the expression analysis.
penR in B. cenocepacia J2315 is an ampR homolog.
ampD mutation in the strain.
Abbreviations for antibiotics used in the induction: AMX, amoxicillin; CAZ, ceftazidime; CTXM, cefotaxime; MER, meropenem.
Consistent with the gene expression patterns, both ampC and penB genes have conserved cis elements for regulation in the promoters: an LysR-type regulator binding site (A-N11-T) and an inverted repeat sequence in the region right upstream of the −35 sequence, which is bound by AmpR in its activator conformation due to a ligand of 1,6-anhydroMurNAc-peptides (Fig. 5B) (23).
MICs of randomly selected mutant strains for a set of β-lactam antibiotics demonstrated combined activities of AmpC and PenB (Table 2). Our results showed the induced enzyme activities toward penicillin (ampicillin), expanded-spectrum cephalosporins (ceftazidime and cefotaxime), and carbapenem (meropenem) (Table 2). An inhibitor of β-lactamases, clavulanate, had no inhibitory effect on these combined enzyme activities (Table 2). To distinguish the observed activities between AmpC and PenB, the genes for the enzymes were separately cloned into plasmid pRK415K and were tested for their activities in B. thailandensis with and without its own PenL β-lactamase (Table 3). The results showed that the hydrolytic activity toward ampicillin is mostly from PenB, while that toward expanded-spectrum cephalosporins is from both enzymes. PenB also exhibited a weak activity toward meropenem (Table 3). The data suggest that PenB from B. cenocepacia strain J2315 is similar to those in strains 09-54 and 212 among variable PenB enzymes produced by the species (26).
TABLE 2.
MICs of B. cenocepacia strains for β-lactam antibiotics with and without the inhibitor clavulanate
| Strain | MIC (μg/ml)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| AMPb | AMP-CLAc | CAZ | CAZ-CLA | CTXM | CTXM-CLA | MER | MER-CLA | |
| WTd | 4,000 | 4,000 | 2 | 1 | 20 | 15 | 4 | 4 |
| M-Dup1e | 6,000 | 6,000 | 60 | 60 | 800 | 600 | 18 | 15 |
| M-Del1 | 5,500 | 5,000 | 55 | 50 | 700 | 600 | 17 | 15 |
| M-Sub2 | 6,000 | 6,000 | 60 | 60 | 800 | 800 | 15 | 15 |
| M-Sub4 | 6,000 | 6,000 | 60 | 60 | 800 | 700 | 15 | 15 |
| M-Sub5 | 6,000 | 6,000 | 60 | 60 | 750 | 700 | 18 | 18 |
| M-Sub8 | 7,000 | 6,000 | 70 | 60 | 800 | 800 | 18 | 18 |
| M-Sub14 | 7,000 | 6,000 | 65 | 60 | 800 | 700 | 18 | 18 |
MICs were determined by the agar dilution method.
Abbreviations for the antibiotics and the inhibitor (CLA) used: AMP, ampicillin; CLA, clavulanate; CAZ, ceftazidime; CTXM, cefotaxime; MER, meropenem.
CLA was used at a concentration of 1 μg/ml.
Wild-type strain.
ampD mutations in the strain.
TABLE 3.
MICs of B. thailandensis strains for β-lactam antibiotics with and without penB or ampC genes from B. cenocepacia strain J2315
| B. thailandensis strain | MIC (μg/ml)a |
|||
|---|---|---|---|---|
| AMP | CAZ | CTXM | MER | |
| E264 strainsb | ||||
| E264 | 28 | 1.5 | 7 | 1 |
| E264/pRK415K | 28 | 1.5 | 7 | 1 |
| E264/pRK415K::penB | 256 | 2 | 14.7 | 1.5 |
| E264/pRK415K::ampC | 32 | 2 | 13.3 | 1 |
| E264 (ΔpenA) strainsc | ||||
| E264 (ΔpenA) | 4 | 0.75 | 0.9 | 1 |
| E264 (ΔpenA)/pRK415K | 4 | 0.75 | 0.9 | 1 |
| E264 (ΔpenA)/pRK415K::penB | 256 | 1.5 | 9.3 | 1.2 |
| E264 (ΔpenA)/pRK415K::ampC | 7.3 | 1.5 | 5.3 | 1 |
MICs were determined by the agar dilution method. Abbreviations: AMP, ampicillin; CAZ, ceftazidime; CTXM, cefotaxime; MER, meropenem.
Wild-type strain E264 is the host for the plasmid pRK415K. Wild-type penA and ampC genes are from B. cenocepacia strain J2315.
A mutant strain of E264 that lacks penL is the host for the plasmid pRK415K. Wild-type penA and ampC genes are from B. cenocepacia strain J2315.
Conclusions.
The frequency of resistance mutations in ampD was estimated to be 10−6 to 10−5 (Fig. 1A), 2 orders of magnitude higher than that for substitution mutations in B. thailandensis PenL conferring resistance to ceftazidime, which was previously estimated to be 10−8 to 10−7 (36). This result suggests that ampD mutations are facilitated under antibiotic pressure (44). The preference for duplications in ampD is due to the presence of a small region with a large number of direct repeats, causing high susceptibility to DNA slippage in both forward and reverse reactions (Fig. 3). Reversion of ampD mutations is beneficial to bacteria in order to resume resource-saving cell wall recycling when antibiotics are no longer present. Furthermore, the induction system regulates two enzymes with slightly different substrate spectra, AmpC and PenB, which poses a serious threat to human health. In conclusion, B. cenocepacia has a highly advanced ampD mutation-mediated antibiotic induction system. The development of therapeutic means to target cell wall recycling-linked antibiotic resistance will be an important future research direction against Bcc infections.
ACKNOWLEDGMENT
This work was supported by a grant (NRF-2015R1A2A2A01004007) from the Core Research Program of the National Research Foundation (NRF) of the Republic of Korea to H.S.K.
REFERENCES
- 1.Coenye T, Vandamme P, Govan JR, LiPuma JJ. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J Clin Microbiol 39:3427–3436. doi: 10.1128/JCM.39.10.3427-3436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipuma JJ. 2005. Update on the Burkholderia cepacia complex. Curr Opin Pulm Med 11:528–533. doi: 10.1097/01.mcp.0000181475.85187.ed. [DOI] [PubMed] [Google Scholar]
- 3.Vanlaere E, Lipuma JJ, Baldwin A, Henry D, De Brandt E, Mahenthiralingam E, Speert D, Dowson C, Vandamme P. 2008. Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int J Syst Evol Microbiol 58:1580–1590. doi: 10.1099/ijs.0.65634-0. [DOI] [PubMed] [Google Scholar]
- 4.Vanlaere E, Baldwin A, Gevers D, Henry D, De Brandt E, LiPuma JJ, Mahenthiralingam E, Speert DP, Dowson C, Vandamme P. 2009. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp nov and Burkholderia lata sp nov. Int J Syst Evol Microbiol 59:102–111. doi: 10.1099/ijs.0.001123-0. [DOI] [PubMed] [Google Scholar]
- 5.Mahenthiralingam E, Baldwin A, Dowson CG. 2008. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J Appl Microbiol 104:1539–1551. doi: 10.1111/j.1365-2672.2007.03706.x. [DOI] [PubMed] [Google Scholar]
- 6.Gibson RL, Burns JL, Ramsey BW. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am J Respir Crit Care Med 168:918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- 7.Loutet SA, Valvano MA. 2010. A decade of Burkholderia cenocepacia virulence determinant research. Infect Immun 78:4088–4100. doi: 10.1128/IAI.00212-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LiPuma JJ, Spilker T, Gill LH, Campbell PW III, Liu L, Mahenthiralingam E. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am J Respir Crit Care Med 164:92–96. doi: 10.1164/ajrccm.164.1.2011153. [DOI] [PubMed] [Google Scholar]
- 9.Drevinek P, Mahenthiralingam E. 2010. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect 16:821–830. doi: 10.1111/j.1469-0691.2010.03237.x. [DOI] [PubMed] [Google Scholar]
- 10.Mann T, Ben-David D, Zlotkin A, Shachar D, Keller N, Toren A, Nagler A, Smollan G, Barzilai A, Rahav G. 2010. An outbreak of Burkholderia cenocepacia bacteremia in immunocompromised oncology patients. Infection 38:187–194. doi: 10.1007/s15010-010-0017-0. [DOI] [PubMed] [Google Scholar]
- 11.Marioni G, Rinaldi R, Ottaviano G, Marchese-Ragona R, Savastano M, Staffieri A. 2006. Cervical necrotizing fasciitis: a novel clinical presentation of Burkholderia cepacia infection. J Infect 53:e219–e222. doi: 10.1016/j.jinf.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 12.Leitao JH, Sousa SA, Cunha MV, Salgado MJ, Melo-Cristino J, Barreto MC, Sa-Correia I. 2008. Variation of the antimicrobial susceptibility profiles of Burkholderia cepacia complex clonal isolates obtained from chronically infected cystic fibrosis patients: a five-year survey in the major Portuguese treatment center. Eur J Clin Microbiol Infect Dis 27:1101–1111. doi: 10.1007/s10096-008-0552-0. [DOI] [PubMed] [Google Scholar]
- 13.Speert DP. 2002. Advances in Burkholderia cepacia complex. Paediatr Respir Rev 3:230–235. doi: 10.1016/S1526-0542(02)00185-9. [DOI] [PubMed] [Google Scholar]
- 14.Aaron SD, Ferris W, Henry DA, Speert DP, Macdonald NE. 2000. Multiple combination bactericidal antibiotic testing for patients with cystic fibrosis infected with Burkholderia cepacia. Am J Respir Crit Care Med 161:1206–1212. doi: 10.1164/ajrccm.161.4.9907147. [DOI] [PubMed] [Google Scholar]
- 15.Holden MT, Seth-Smith HM, Crossman LC, Sebaihia M, Bentley SD, Cerdeno-Tarraga AM, Thomson NR, Bason N, Quail MA, Sharp S, Cherevach I, Churcher C, Goodhead I, Hauser H, Holroyd N, Mungall K, Scott P, Walker D, White B, Rose H, Iversen P, Mil-Homens D, Rocha EP, Fialho AM, Baldwin A, Dowson C, Barrell BG, Govan JR, Vandamme P, Hart CA, Mahenthiralingam E, Parkhill J. 2009. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol 191:261–277. doi: 10.1128/JB.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson JW, Fisher JF, Mobashery S. 2013. Bacterial cell-wall recycling. Ann N Y Acad Sci 1277:54–75. doi: 10.1111/j.1749-6632.2012.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidtke AJ, Hanson ND. 2006. Model system to evaluate the effect of ampD mutations on AmpC-mediated beta-lactam resistance. Antimicrob Agents Chemother 50:2030–2037. doi: 10.1128/AAC.01458-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopp U, Wiedemann B, Lindquist S, Normark S. 1993. Sequences of wild-type and mutant ampD genes of Citrobacter freundii and Enterobacter cloacae. Antimicrob Agents Chemother 37:224–228. doi: 10.1128/AAC.37.2.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juan C, Macia MD, Gutierrez O, Vidal C, Perez JL, Oliver A. 2005. Molecular mechanisms of beta-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother 49:4733–4738. doi: 10.1128/AAC.49.11.4733-4738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park JT. 1995. Why does Escherichia coli recycle its cell wall peptides? Mol Microbiol 17:421–426. doi: 10.1111/j.1365-2958.1995.mmi_17030421.x. [DOI] [PubMed] [Google Scholar]
- 21.Cho H, Uehara T, Bernhardt Thomas G. 2014. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 159:1300–1311. doi: 10.1016/j.cell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindberg F, Lindquist S, Normark S. 1987. Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii beta-lactamase. J Bacteriol 169:1923–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vadlamani G, Thomas MD, Patel TR, Donald LJ, Reeve TM, Stetefeld J, Standing KG, Vocadlo DJ, Mark BL. 2015. The β-lactamase gene regulator AmpR is a tetramer that recognizes and binds the d-Ala-d-Ala motif of its repressor UDP-N-acetylmuramic acid (MurNAc)-pentapeptide. J Biol Chem 290:2630–2643. doi: 10.1074/jbc.M114.618199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders CC, Bradford PA, Ehrhardt AF, Bush K, Young KD, Henderson TA, Sanders WE. 1997. Penicillin-binding proteins and induction of AmpC beta-lactamase. Antimicrob Agents Chemother 41:2013–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee M, Hesek D, Blázquez B, Lastochkin E, Boggess B, Fisher JF, Mobashery S. 2015. Catalytic spectrum of the penicillin-binding protein 4 of Pseudomonas aeruginosa, a nexus for the induction of β-lactam antibiotic resistance. J Am Chem Soc 137:190–200. doi: 10.1021/ja5111706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poirel L, Rodriguez-Martinez JM, Plesiat P, Nordmann P. 2009. Naturally occurring class A beta-lactamases from the Burkholderia cepacia complex. Antimicrob Agents Chemother 53:876–882. doi: 10.1128/AAC.00946-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sass A, Marchbank A, Tullis E, Lipuma JJ, Mahenthiralingam E. 2011. Spontaneous and evolutionary changes in the antibiotic resistance of Burkholderia cenocepacia observed by global gene expression analysis. BMC Genomics 12:373. doi: 10.1186/1471-2164-12-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 29.Okonechnikov K, Golosova O, Fursov M, UGENE team . 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 30.Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 31.Simon R, Priefer U, Puhler A. 1983. Vector plasmids for in vivo and in vitro manipulations of gram-negative bacteria. Springer-Verlag KG, Berlin, Germany. [Google Scholar]
- 32.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 33.Peeters E, Sass A, Mahenthiralingam E, Nelis H, Coenye T. 2010. Transcriptional response of Burkholderia cenocepacia J2315 sessile cells to treatments with high doses of hydrogen peroxide and sodium hypochlorite. BMC Genomics 11:90. doi: 10.1186/1471-2164-11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 35.Mahenthiralingam E, Coenye T, Chung JW, Speert DP, Govan JR, Taylor P, Vandamme P. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J Clin Microbiol 38:910–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yi H, Cho KH, Cho YS, Kim K, Nierman WC, Kim HS. 2012. Twelve positions in a beta-lactamase that can expand its substrate spectrum with a single amino acid substitution. PLoS One 7:e37585. doi: 10.1371/journal.pone.0037585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stapleton P, Shannon K, Phillips I. 1995. DNA sequence differences of ampD mutants of Citrobacter freundii. Antimicrob Agents Chemother 39:2494–2498. doi: 10.1128/AAC.39.11.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrosino JF, Pendleton AR, Weiner JH, Rosenberg SM. 2002. Chromosomal system for studying AmpC-mediated beta-lactam resistance mutation in Escherichia coli. Antimicrob Agents Chemother 46:1535–1539. doi: 10.1128/AAC.46.5.1535-1539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carrasco-Lopez C, Rojas-Altuve A, Zhang W, Hesek D, Lee M, Barbe S, Andre I, Ferrer P, Silva-Martin N, Castro GR, Martinez-Ripoll M, Mobashery S, Hermoso JA. 2011. Crystal structures of bacterial peptidoglycan amidase AmpD and an unprecedented activation mechanism. J Biol Chem 286:31714–31722. doi: 10.1074/jbc.M111.264366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gore JM, Ran FA, Ornston LN. 2006. Deletion mutations caused by DNA strand slippage in Acinetobacter baylyi. Appl Environ Microbiol 72:5239–5245. doi: 10.1128/AEM.00283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi H, Song H, Hwang J, Kim K, Nierman WC, Kim HS. 2014. The tandem repeats enabling reversible switching between the two phases of beta-lactamase substrate spectrum. PLoS Genet 10:e1004640. doi: 10.1371/journal.pgen.1004640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song H, Hwang J, Yi H, Ulrich RL, Yu Y, Nierman WC, Kim HS. 2010. The early stage of bacterial genome-reductive evolution in the host. PLoS Pathog 6:e1000922. doi: 10.1371/journal.ppat.1000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bagge N, Ciofu O, Hentzer M, Campbell JIA, Givskov M, Hoiby N. 2002. Constitutive high expression of chromosomal beta-lactamase in Pseudomonas aeruginosa caused by a new insertion sequence (IS1669) located in ampD. Antimicrob Agents Chemother 46:3406–3411. doi: 10.1128/AAC.46.11.3406-3411.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Denamur E, Matic I. 2006. Evolution of mutation rates in bacteria. Mol Microbiol 60:820–827. doi: 10.1111/j.1365-2958.2006.05150.x. [DOI] [PubMed] [Google Scholar]
- 45.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]