Abstract
Malaria control is hindered by the evolution and spread of resistance to antimalarials, necessitating multiple changes to drug policies over time. A comprehensive antimalarial drug resistance surveillance program is vital for detecting the potential emergence of resistance to antimalarials, including current artemisinin-based combination therapies. An antimalarial drug resistance surveillance study involving 203 Plasmodium falciparum malaria-positive children was conducted in western Kenya between 2010 and 2013. Specimens from enrolled children were analyzed in vitro for sensitivity to chloroquine (CQ), amodiaquine (AQ), mefloquine (MQ), lumefantrine, and artemisinin derivatives (artesunate and dihydroartemisinin) and for drug resistance allele polymorphisms in P. falciparum crt (Pfcrt), Pfmdr-1, and the K13 propeller domain (K13). We observed a significant increase in the proportion of samples with the Pfcrt wild-type (CVMNK) genotype, from 61.2% in 2010 to 93.0% in 2013 (P < 0.0001), and higher proportions of parasites with elevated sensitivity to CQ in vitro. The majority of isolates harbored the wild-type N allele in Pfmdr-1 codon 86 (93.5%), with only 7 (3.50%) samples with the N86Y mutant allele (the mutant nucleotide is underlined). Likewise, most isolates harbored the wild-type Pfmdr-1 D1246 allele (79.8%), with only 12 (6.38%) specimens with the D1246Y mutant allele and 26 (13.8%) with mixed alleles. All the samples had a single copy of the Pfmdr-1 gene (mean of 0.907 ± 0.141 copies). None of the sequenced parasites had mutations in K13. Our results suggest that artemisinin is likely to remain highly efficacious and that CQ sensitivity appears to be on the rise in western Kenya.
INTRODUCTION
Antimalarial drug resistance has hampered progress in malaria control and has led to several changes in drug policies over time. To minimize the potential emergence of drug resistance, the World Health Organization (WHO) recommends the use of artemisinin-based combination therapy (ACT), which consists of an artemisinin derivative coformulated with another class of antimalarial (1). In Kenya, the use of chloroquine (CQ) as the first line of malaria treatment was discontinued in the late 1990s due to a high percentage of treatment failures. CQ was replaced by sulfadoxine-pyrimethamine (SP) as the first line of treatment, with amodiaquine (AQ) as a second choice. However, the use of SP was short-lived, as the majority of circulating parasites rapidly developed resistance to SP (reviewed in reference 2). Therefore, in April 2004, the ACT artemether-lumefantrine (AL; Coartem) was recommended as the first-line therapy for the treatment of uncomplicated Plasmodium falciparum malaria in Kenya, although this was not fully implemented until late 2006, when the country was finally able to undertake delivery of large quantities of the drug. Thus, between 2004 and 2006, AQ was briefly used as the first-line treatment for malaria in Kenya. Dihydroartemisinin-piperaquine (DHAP) is the current second-line antimalarial drug treatment, while artesunate-amodiaquine (ASAQ) is registered for use in the private sector along with AL.
Genetic markers of resistance to many antimalarial drugs have been elucidated (reviewed in reference 3). These constitute a complex pattern of single nucleotide polymorphisms (SNPs) in the pertinent genes. For example, SNPs at codons 86, 184, 1034, 1042, and 1246 in the P. falciparum multidrug resistance 1 gene (Pfmdr-1) have been shown to modulate sensitivities to different antimalarial drugs, including CQ and mefloquine (MQ) (4, 5). Preliminary findings from studies involving AQ monotherapy suggest that this drug selects for the Pfmdr1 N86Y SNP (the mutant nucleotide is underlined) (6–10). Experimental studies have shown that an increase in the copy number of the Pfmdr-1 gene is associated with increased resistance to MQ (11), lumefantrine (LU), and artemisinin (12). Mutations in codons 72 to 76 (CVMNK) of the P. falciparum chloroquine resistance transporter gene (Pfcrt) have been associated with resistance to CQ and AQ (13; reviewed in reference 14), with the K76T point mutation being the most predictive of CQ resistance. Resistance to SP is conferred by the accumulation of SNPs in two genes that encode enzymes involved in P. falciparum folate metabolism, dihydrofolate reductase and dihydropteroate synthase, which are targeted by pyrimethamine and sulfadoxine, respectively.
The current first-line treatments for uncomplicated malaria in nearly all countries where malaria is endemic are ACTs. However, recent studies in Southeast Asia have demonstrated delayed parasite clearance during therapeutic efficacy studies with ACTs and artemisinin monotherapy (15–18), implying that parasite isolates in this region are developing resistance to artemisinin. A genetic marker of artemisinin resistance was recently identified to consist of polymorphisms in the propeller domain of the P. falciparum kelch protein, found on chromosome 13 (K13) (19). Several other studies have confirmed this observation, linking a number of mutations in the propeller domain to delayed parasite clearance in patients treated with artemisinin (20, 21).
Interestingly, molecular epidemiological studies have revealed that the withdrawal of an antimalarial from a population due to high treatment failure rates can lead to the reemergence of drug-sensitive parasites in that population over time. For example, 10 years after the discontinuation of CQ for malaria treatment in Malawi, several studies demonstrated the return of CQ-sensitive parasites and an increase in the prevalence of parasites harboring the wild-type (WT) Pfcrt K76 codon (22, 23). Similar trends were later reported from studies conducted in other parts of Malawi (24, 25), Tanzania (26, 27), and Kenya (28–31), raising the possibility of the selected reintroduction of CQ for malaria treatment. In contrast, countries that delayed the withdrawal of CQ for malaria treatment demonstrated no significant decreases in the prevalence of CQ resistance parasites (reviewed in reference 32), implying that drug pressure plays a major role in the maintenance of drug-resistant parasites in a population.
The emerging concern of artemisinin resistance in Southeast Asia poses a major threat to the global effort toward the effective control and case management of malaria caused by P. falciparum. The possibility of the spread of this phenotype to other parts of the world makes it imperative that countries establish comprehensive surveillance programs to detect the potential emergence of resistance to artemisinin and its partner drugs. In addition, it is important to monitor the dynamics of previously utilized drugs such as CQ and SP, given the demonstrated possibility of reemergence of drug-sensitive parasites after the withdrawal of failing drugs. The WHO recommends that countries set up surveillance mechanisms to monitor the emergence of resistance to antimalarial drugs by using data from therapeutic efficacy studies complemented with molecular marker data (33). Although therapeutic efficacy studies are the gold-standard method for confirmation of drug resistance, they are expensive and labor-intensive. On the other hand, in vitro drug sensitivity assays and molecular surveillance for genetic markers are relatively inexpensive and can provide data on the emergence and temporal dynamics of genetic markers of drug resistance, sometimes even before clinical resistance becomes established. These assays have been used to detect reduced drug sensitivity to several antimalarial drugs, including CQ, MQ, quinine, lumefantrine, AQ, and SP.
We report findings from an in vitro drug sensitivity surveillance study conducted in western Kenya between 2010 and 2013 in which parasite sensitivity to CQ, AQ, lumefantrine, artesunate (ARS), and dihydroartemisinin (DHA) was determined and samples were genotyped for drug resistance polymorphisms in Pfcrt, Pfmdr-1, and the K13 propeller domain.
MATERIALS AND METHODS
Study sites, patient recruitment, and sample collection.
This study was conducted in Siaya and Bondo District Hospitals in western Kenya between 2010 and 2013. These hospitals are located in moderate- to high-malaria-transmission areas with widespread coverage of long-lasting insecticide-treated bed nets (LLINs). The two hospitals are ∼60 km from the Kenya Medical Research Institute (KEMRI) in Kisian, where the laboratory studies were conducted. The Bondo District Hospital outpatient department attends to ∼50 patients on a single day during peak malaria season. Siaya District Hospital is the only major hospital serving Siaya County, and the outpatient department examines >40 patients per day during peak malaria transmission seasons.
Children between the ages of 2 and 7 years with an axillary temperature of ≥37.5°C or a history of fever in the previous 24 h with no recent treatment (at least in the last 7 days) were eligible for enrollment in the study. All eligible children were referred to study personnel for further screening. The children were weighed, and the height and axillary temperature were measured by study personnel. A few drops of finger prick blood were collected and used for the microscopic determination of malaria infection, parasitemia, and hemoglobin levels. Hemoglobin levels were measured by using the HaemoCue photometer (HaemoCue AB, Angelholm, Sweden). Additional inclusion criteria included a slide confirmation of P. falciparum infection and a parasitemia level of ≥2,000 asexual parasites/μl of blood. Written informed consent to participate in the study was obtained from the parents/guardians of children who participated in this study. Blood samples were collected into EDTA Microtainers for in vitro drug sensitivity assays and on Whatman grade 3 filter paper for genotyping assays. This study was approved by the institutional review boards of KEMRI, Nairobi, Kenya, and the Centers for Disease Control and Prevention (CDC), Atlanta, GA.
In vitro drug sensitivity assays.
In vitro drug susceptibility testing was performed for CQ, AQ, MQ, lumefantrine, ARS, and DHA (obtained from the World Wide Antimalarial Resistance Network [WWARN]), using the SYBR green I fluorometric method as described previously (34). Briefly, blood samples were diluted appropriately to 1% parasitemia and 2% hematocrit using culture medium (RPMI 1640 supplemented with 25 mM HEPES buffer). The samples were added to 96-well microtiter plates with different concentrations of test drugs and cultured at 37°C under standard culture conditions. After 72 h of incubation, SYBR green dye was added to culture plates, and the plates were incubated for 1 h. Plates were analyzed on a fluorescence reader (M200; Tecan, Mänedorf, Switzerland) to determine growth inhibition (34). The amount of SYBR green dye incorporated decreases as parasite growth declines. A sigmoid, 4-parameter concentration inhibition model was applied to generate 50% inhibitory concentration (IC50) estimations using the In Vitro Analysis and Reporting Tool (IVART) developed by the WWARN (35). The geometric mean IC50 was calculated for each drug. The CQ IC50, MQ IC50, and DHA IC50 for laboratory-cultured sensitive P. falciparum isolate 3D7 were tested in every experiment as a control.
Genotyping for resistance markers.
Genomic DNA was isolated by using the QIAamp blood minikit (Qiagen Inc., CA, USA). The Pfcrt gene was amplified and sequenced for drug resistance markers associated with CQ (codons 72 to 76) and AQ (codon 76) resistance. Similarly, the Pfmdr-1 gene was genotyped at codons 86, 184, 1042, and 1246. Direct Sanger sequencing was utilized for SNPs as previously described (36). Sequencing of the nested purified PCR products was performed by using a BigDye Terminator v3.1 cycle sequencing kit on an iCycler thermal cycler (Bio-Rad, CA, USA). The reaction mixtures were precipitated in 70% ethanol to clean up dye terminators, rehydrated in 10 μl HiDi formamide, and then sequenced on a 3130xl ABI genetic analyzer (ABI Prism, CA, USA). In addition, the K13 propeller domain associated with artemisinin resistance was sequenced as recently described (37). Sequence analysis was performed by using Geneious R7 (Biomatters, Auckland, New Zealand).
Pfmdr-1 gene copy number determination.
The Pfmdr-1 copy number was assessed by using a previously described protocol (11). Only samples that were singly infected (monoclonal infections) were used for this determination. The P. falciparum β-tubulin gene was used as the reference gene for relative quantification of Pfmdr-1 gene copy numbers. For relative quantification, the 3D7 parasite strain was used as a single-copy control in addition to the W2Mef (2 copies) and Dd2 (3 to 4 copies) strains, which were included in every run. All samples were tested in triplicate. The reactions were set up with a 12.5-μl total reaction mixture volume containing 6.25 μl of Platinum 2× buffer (Life Technologies, New York, NY, USA), 300 nM each forward and reverse Pfmdr-1 primer, 100 nM each β-tubulin primer, 100 nM β-tubulin probe, and 150 μM Pfmdr-1 probe. Two microliters of template DNA was used. The assay was performed by using the Agilent Mx3005 real-time PCR instrument (Agilent Technologies, CA, USA), using the following PCR parameters: 50 cycles of 95°C for 15 s and 58°C for 1 min. The copy number was determined by using the relative quantification module in MxPro3005 software (Agilent Technologies, CA, USA), using the comparative ΔΔCT method, and rounded to the nearest integer (11).
Statistics.
Data and statistical analyses were performed by using PASW Statistics for Windows version 18.0 (SPSS Inc., Chicago, IL, USA) and R version 3.1.2 (R Foundation for Statistical Computing, Vienna, Austria). Differences in subject characteristics were analyzed by using analysis of variance (ANOVA) for continuous, normally distributed variables and a Pearson chi-square test for categorical variables. IC50s and parasite densities were log transformed, and annual IC50s are summarized as geometric means and standard deviations. Differences in IC50 geometric means over time were tested with linear regression models controlling for age. Fisher's exact test was used to analyze changes in mutation prevalence over time. Finally, linear regressions were run on log-transformed IC50s to test for differences in haplotypes using singly infected samples. These results are reported as geometric mean ratios (GMRs).
RESULTS
Characteristics of study subjects.
A total of 203 children were enrolled in the study during 4 years of continuous surveillance (Table 1). Due to low patient enrollments in the first 2 years of the study, the study was amended to include children below the age of 2 years. Only 8 samples were obtained from Bondo District Hospital due to staffing problems in that hospital. We observed significant differences over the 4 years of the study in the ages, heights, weights, auxiliary temperatures, and parasite densities of the enrolled patients. No differences in the proportions of males and females or in the hemoglobin levels were observed.
TABLE 1.
Sample population summary
| Patient parameter | Value for yr |
P value | |||
|---|---|---|---|---|---|
| 2010 | 2011 | 2012 | 2013 | ||
| No. (%) of individuals of gender | |||||
| Female | 16 (42.11) | 17 (40.48) | 17 (43.59) | 35 (41.67) | 0.9938 |
| Male | 22 (57.89) | 25 (59.52) | 22 (56.41) | 49 (58.33) | |
| Mean age (yr) (SD; total no. of patients)a | 3.70 (1.19; 42) | 3.36 (1.41; 39) | 2.72 (1.60; 84) | 3.64 (1.23; 38) | 0.0007 |
| Mean height (cm) (SD; total no. of patients) | 99.29 (10.87; 42) | 87.21 (13.52; 38) | 86.11 (14.75; 84) | 96.62 (13.90; 37) | <0.0001 |
| Mean wt (kg) (SD; total no. of patients) | 21.40 (9.22; 42) | 13.02 (4.46; 39) | 12.60 (7.28; 84) | 15.06 (3.14; 38) | <0.0001 |
| Mean axillary temp (°C) (SD; total no. of patients) | 38.51 (0.82; 42) | 37.92 (0.99; 39) | 38.26 (1.07; 84) | 38.30 (0.72; 38) | 0.0425 |
| Mean hemoglobin level (g/dl) (SD; total no. of patients) | 9.53 (2.07; 42) | 9.62 (2.04; 39) | 10.04 (2.11; 84) | 9.27 (2.14; 38) | 0.2778 |
| Mean parasite density [ln(parasites/μl)] (SD; total no. of patients) | 10.95 (0.96; 42) | 10.37 (1.63; 39) | 11.03 (1.37; 84) | 10.37 (1.12; 38) | 0.0110 |
Enrollment criteria were amended in 2012 to include children below the age of 2 years.
Pfcrt genotyping.
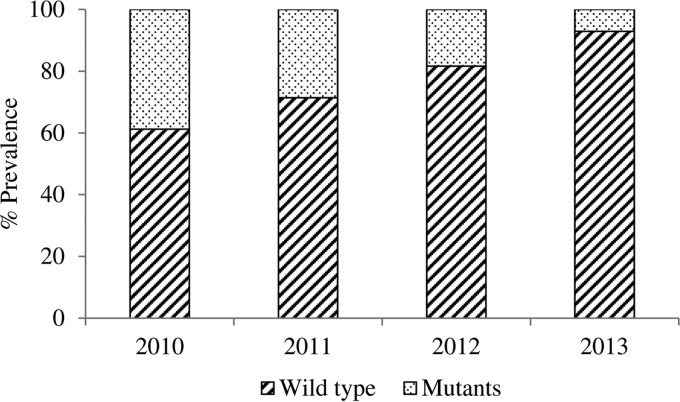
The SNP analysis of the Pfcrt gene was successfully performed for codons 72 to 76 on all 203 samples. Mutations were observed in the M74I, N75E, and K76T codons, giving the CVIET mutant genotype. Parasite isolates were divided into three groups: those that harbored the CVIET mutant haplotype (9.80%), those that harbored the wild-type CVMNK haplotype (76.5%), and those that harbored a mixture of both mutant and wild-type genotypes (mixed) (13.7%). The prevalence of the observed mutant genotype was calculated by combining isolates with only mutant codons with isolates with mixed genotypes. We observed a significant increase in the prevalence of the wild-type CVMNK genotype, from 61.2% in 2010 to 71.4% in 2011, 81.3% in 2012, and 93.0% in 2013 (P = 0.0001) (Fig. 1).
FIG 1.
Temporal increase in the prevalence of the Pfcrt wild-type genotype between 2010 and 2013. The prevalence of the observed mutant genotype was calculated by combining isolates with pure mutant codons with isolates with mixed genotypes (mutant and wild type). A temporal increase in the prevalence of the wild-type CVMNK genotype, from 61.22% in 2010 to 92.94% in 2013, was observed.
Pfmdr-1 genotyping and gene copy number.
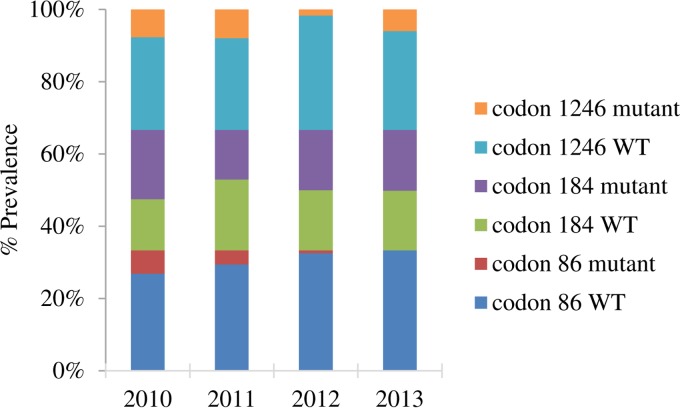
Mutations in the Pfmdr-1 gene were observed only at codons 86 (n = 200), 184 (n = 193), and 1246 (n = 194), and isolates were classified as having a mutant, wild-type, or mixed (harboring both mutant and wild-type codons) genotype for each locus. The majority of samples harbored the wild-type N86 allele (93.5%), with only 7 (3.50%) samples harboring the mutant Y allele (N86Y; 4 samples in 2010, 3 in 2011, 0 in 2012, and 0 in 2013). Likewise, the majority of samples harbored the wild-type D1246 allele (79.8%), with only 12 (6.38%) samples having the D1246Y mutant allele and 26 (13.8%) having mixed alleles. Codon 184 was evenly distributed among the mutant (37.3%), wild-type (37.3%), and mixed (25.4%) alleles. The prevalence of the observed alleles was calculated by combining isolates with pure mutant codons together with isolates with mixed genotypes. We observed a significant decrease in the prevalence of the N86Y mutant allele (P = 0.0001), but no significant differences were observed in the prevalence of mutations at codon 184 (P = 0.5120) or 1246 (P = 0.0680) (Fig. 2).
FIG 2.
Distribution of SNPs in the Pfmdr-1 gene over time. Mutations in the Pfmdr-1 gene were observed only at codons 86 (n = 200), 184 (n = 193), and 1246 (n = 194). The isolates were classified as having mutant, wild-type (WT), or mixed (harboring both mutant and wild-type alleles) genotypes for each locus. The prevalence of the observed alleles was calculated by combining isolates with pure mutant codons with isolates with mixed genotypes. The majority of the samples harbored the wild-type N86 and the wild-type D1246 alleles. Codon 184 was evenly distributed among the samples. We observed a significant decrease in the prevalence of the N86Y mutant allele (P = 0.0001) over the study period. No significant differences were observed in the prevalence of mutations at codon 184 (P = 0.5120) or 1246 (P = 0.0680).
Out of the 203 samples, 41 had monoclonal infections, i.e., infected with only one detectable genotype, and were successfully amplified for Pfmdr-1 copy number determination. These 41 samples harbored a single copy of the Pfmdr-1 gene (mean of 0.907 ± 0.141 copies).
K13 propeller domain genotyping.
One hundred forty-seven samples (72%) were successfully sequenced for K13 mutations. Our sequencing results did not reveal any SNPs in the K13 propeller domain in the parasite isolates tested.
In vitro drug sensitivity assays.
The yearly IC50s for CQ, AQ, MQ, ARS, and DHA are shown in Table 2. The geometric mean CQ IC50, MQ IC50, and DHA IC50 for laboratory-cultured P. falciparum isolate 3D7 were 18.22 nM, 41.22 nM, and 10.20 nM, respectively. Results from the LU in vitro sensitivity assays were disregarded for further analysis due to large inconsistencies in the majority of the experiments. We observed a significant decline in the yearly IC50s for CQ, from 31.77 nM in 2010 to 19.85 nM in 2013. We also observed differences in the IC50s for ARS and DHA, but no obvious trend was observed. No statistical differences were seen for MQ and AQ.
TABLE 2.
Yearly IC50s from 2010 to 2013
| Drug | Isolate type | Geometric mean IC50 (nM) (SD; no. of samples) for yr |
P valuea | |||
|---|---|---|---|---|---|---|
| 2010 | 2011 | 2012 | 2013 | |||
| CQ | Clinical | 31.77 (4.5; 30) | 23.42 (4.5; 39) | 21.09 (4.9; 35) | 19.85 (7.1; 70) | 0.0061 |
| 3D7 | 21.32 (6.78; 25) | 16.28 (12.01; 42) | 19.77 (15.11; 23) | 17.79 (17.93; 53) | 0.111 | |
| MQ | Clinical | 36.19 (5.5; 29) | 27.20 (6.4; 39) | 26.57 (10.2; 33) | 32.23 (8.4; 69) | 0.8896 |
| 3D7 | 77.39 (108.73; 25) | 34.96 (19.05; 42) | 36.89 (52.56; 23) | 36.64 (27.24; 55) | 0.428 | |
| AQ | Clinical | 19.94 (4.0; 31) | 16.14 (4.0; 39) | 9.92 (5.9; 33) | 12.44 (8.6; 59) | 0.0576 |
| ARS | Clinical | 2.76 (4.7; 30) | 4.08 (10.5; 38) | 1.74 (15.5; 34) | 0.78 (10.1; 71) | <0.0001 |
| DHA | Clinical | 5.87 (10.1; 29) | 16.3 (14.9; 39) | 14.38 (21.3; 34) | 7.2 (10.7; 67) | 0.0028 |
| 3D7 | 9.14 (8.79; 18) | 19.69 (63.65; 41) | 13.36 (42.55; 19) | 7.03 (11.25; 55) | 0.159 | |
The statistical test controlled for age and log-transformed parasite density, except for laboratory isolate 3D7.
Association of observed polymorphisms and in vitro drug sensitivity.
These analyses were performed by using samples that harbored either pure mutant or pure wild-type strains with no mixed infections. The CQ IC50s were shown to be significantly different depending on the isolates' Pfcrt genotype (Table 3). Parasites that harbored the mutant Pfcrt CVIET haplotype had significantly higher CQ IC50s than did those with the wild-type CVMNK haplotype (P < 0.0001). Similarly, the geometric mean CQ IC50 was significantly higher for the parasites that harbored the Pfmdr-1 N86Y mutation than for those that harbored the wild-type N86 allele (P = 0.0111). Five of these seven samples (71.4%) also harbored the CVIET genotype. A significantly higher MQ IC50 was also observed for parasites that harbored the Pfmdr-1 Y184F mutation (P = 0.0008) than for isolates that harbored the wild-type Y184 allele. No significant differences were observed for the other drugs (Table 3).
TABLE 3.
Association of genotype and drug sensitivity values
| Drug | IC50 (nM) |
|||||||
|---|---|---|---|---|---|---|---|---|
|
Pfmdr-1 |
Pfcrt |
|||||||
| N86 | 86Y | Y184 | 184F | D1246 | 1246Y | CVNMK | CVIET | |
| CQ | 19.55 | 91.21a | 18.75 | 24.03 | 19.72 | 44.66 | 16.18 | 76.43a |
| MQ | 31.29 | 14.04 | 20.45 | 43.17a | 32.43 | 17.53 | 32.27 | 19.18 |
| AQ | 11.96 | 22.34 | 11.44 | 14.11 | 12.11 | 13.65 | 11.99 | 15.08 |
| ARS | 1.59 | 1.61 | 1.58 | 2.00 | 1.59 | 1.22 | 1.51 | 1.24 |
| DHA | 7.67 | 9.72 | 7.55 | 7.49 | 8.14 | 5.63 | 8.27 | 5.53 |
A significant difference in the IC50 was observed for comparisons between parasites harboring the wild-type genotype and those harboring mutant genotypes.
DISCUSSION
In this report, an antimalarial surveillance study was conducted between 2010 and 2013 to monitor the in vitro drug sensitivity patterns and genetic polymorphisms associated with CQ, AQ, MQ, LU, and artemisinin derivatives (ARS and DHA) in western Kenya. Three main observations were made: (i) none of the parasites sequenced had any mutations in the K13 propeller domain known to be associated with artemisinin resistance, (ii) the proportion of parasites with increased in vitro sensitivity to CQ appears to be increasing in this region of Kenya, and (iii) gene mutations associated with CQ resistance (Pfcrt CVIET and Pfmdr-1 N86Y) are on the decline.
A total of at least 12 polymorphisms in the K13 propeller domain were recently implicated in artemisinin resistance in Southeast Asia (19–21, 37). Similar to previous studies conducted in Africa (38–40), we did not observe any mutations in the propeller domain of the K13 gene that we sequenced. Some studies in Africa have observed a low frequency of other mutations, some of which are found in the propeller domain, in parasites from Africa (38–40). To date, many of the therapeutic efficacy studies conducted in Africa have indicated that artemisinin-based combination therapies are still efficacious (21, 41, 42). Therefore, the role of the novel African K13 mutations in artemisinin resistance, if any, is still to be elucidated. Other secondary loci, in addition to the K13 propeller mutations, are likely to play a role in resistance to artemisinin in Asia (reviewed in reference 43). Our results suggest that artemisinin is still efficacious in this region of Kenya, although therapeutic efficacy studies will be required to conclusively determine this.
We observed a decline in the prevalence of genetic polymorphisms associated with CQ resistance in both the Pfcrt (CVIET; from 39.0% in 2010 to 7.00% in 2013) and Pfmdr-1 (codon N86Y; from 18.0% in 2010 to 0.00% in 2013) genes. An increase in the in vitro drug sensitivity to CQ was also observed, as supported by the decline in the CQ IC50s over the study period. These data suggest that there may be a reversion to CQ sensitivity in parasites from this region of Kenya. The first report of a decline in the prevalence of CQ-resistant parasites in Africa was documented in Malawi, 10 years after CQ withdrawal (22, 24). Similar trends were later reported in studies conducted in Malawi (24, 25), in Tanzania (26), and along the coast of Kenya (28–30). In Kenya, this decline was first reported in Kilifi in 2006, 13 years after the discontinuation of CQ use (28), in which the prevalence of the Pfcrt K76T mutation was shown to decrease from ∼95% in 1993 to ∼60% in 2006. Studies from western Kenya demonstrated that in 2008, the prevalence of parasites with the Pfcrt CVIET genotype was 72.4%, which was shown to decline to 32.1% by 2011 (44). During our study period, we observed a reduction of the prevalence of this genotype to 7% by 2013, demonstrating that the prevalence of this genotype is on the decline. In 2007, the prevalence of parasites with the Pfmdr-1 N86Y mutation in western Kenya was shown to be ∼69% (45). However, a steady decline in the prevalence of this mutation was subsequently observed, from 42% in 2008 to 30% in 2009, 28% in 2010, and 15% in 2011 (44). Our study showed a decline in prevalence from 18% in 2010 to 0% by 2013. This apparent decline in the prevalence of drug-resistant genotypes may be explained by the fact that the driving force for the selection of antimalarial resistance SNPs is drug usage, and therefore, the decrease in drug pressure (CQ) after a policy change to artemether-lumefantrine therapy in 2006 could have helped reduce the CQ drug pressure, resulting in the reversal of CQ resistance-associated genotypes to the more sensitive genotypes. It has been proposed that parasites with Pfcrt mutations incur a fitness cost in the absence of CQ (25; reviewed in reference 43), hence the return of CQ-sensitive parasites in many African countries that have discontinued the use of CQ. Another potential explanation for the return of parasites with the wild-type Pfcrt and Pfmdr-1 N86 alleles may be attributed to LU, the partner drug in AL (Coartem). Previous studies demonstrated that LU may select for CQ-sensitive alleles such as the Pfmdr-1 N86 and Pfcrt K76 alleles, and these alleles have been associated with AL treatment failure (46, 47). Our study and recent studies in western Kenya show a significant increase in the prevalence of the WT allele at codon K76 of Pfcrt and an upward trend in the prevalence of WT alleles in Pfmdr-1 codons N86 and N1246 in parasites collected after the introduction of ACT in Kenya (31). Therefore, the use of AL as the first line of malaria treatment in Kenya may be contributing to the selection of these wild-type alleles.
Characterization of the sensitivity profiles of other artemisinin partner drugs, such as LU, AQ, MQ, and SP, is important for any drug resistance surveillance study. We did not observe any changes in the MQ IC50s. This was not surprising given that MQ is not in use in Kenya. On the other hand, we observed a steady decline in the AQ IC50 from 2010 to 2012, which slightly increased in 2013. Both the Pfcrt K76T and Pfmdr-1 N86Y mutations are also associated with AQ resistance (3, 44). The observed decline in the prevalence of parasites with these mutations suggests that most of the circulating parasites in this region are AQ sensitive. The use of AQ monotherapy for treatment was widespread in western Kenya, both unofficially from 2000 and officially between 2004 and 2006, and interestingly, AQ usage in this region was documented in a community-based study in 2007 (45). This could explain the still high levels of prevalence of parasites harboring the Pfcrt K76T mutant in circulation in 2007 (94%) (45), as AQ usage could have continued the selection pressure for this polymorphism even after CQ discontinuation. By 2008, the use of ACT was fully implemented, leading to reductions in both CQ and AQ drug pressure, hence the decline in the polymorphisms associated with resistance to these drugs, as we observed in 2013. However, because ASAQ is currently registered for use in the private sector in Kenya, it is important to continue to monitor the response of circulating parasites to AQ in this region.
Increase Pfmdr-1 copy numbers (gene amplification) are more common in Southeast Asia and a rare occurrence in Kenya and have been associated with decreased sensitivity to drugs such as mefloquine, artesunate, and AL. We did not observe any increase in the Pfmdr-1 copy number during the time period of our study. However, a recent study from Kenya showed that a few samples, collected after ACT introduction, had increased Pfmdr-1 copy numbers and that this increase was associated with decreased in vitro susceptibility to AQ and piperaquine (48), in contrast to a previous study which did not observe any association between Pfmdr-1 copy number increases and AQ resistance (49). This can be explained by the fact that only a few samples in Kenya show multiple Pfmdr-1 genes.
It is expected that parasites with drug resistance mutations also demonstrate increased IC50s in in vitro drug sensitivity assays, used as surrogate assays for resistance. In our study, parasites that harbored the Pfcrt CVIET mutant haplotype had significantly increased CQ IC50s compared to those of the wild-type parasites. Although the majority of the samples over the study period harbored the wild-type genotype, the minority that possessed the CVIET mutations demonstrated elevated CQ IC50s. The CQ IC50 was also found to be higher for the parasites that harbored the Pfmdr-1 N86Y mutant than for those with the wild-type N86 codon. Previous studies demonstrated strong associations between the combined Pfcrt CVIET and Pfmdr-1 N86Y mutations and increased CQ IC50 (44). Indeed, the highest mean CQ IC50 was observed with the seven samples that harbored both the Pfcrt CVIET and Pfmdr-1 N86Y mutations. These results underscore the association of drug-resistant genotypic markers with in vitro drug sensitivity assays, which serve as good alternatives/surrogates for therapeutic efficacy studies, depending on the drug mechanism.
However, recent studies have indicated that traditional in vitro drug sensitivity assays, as utilized in our study, are inappropriate for the determination of artemisinin resistance. Those studies demonstrated that the IC50s of the artemisinin derivatives tested did not correlate with the in vivo parasite clearance rate (50). Therefore, alternative in vitro assays have been proposed, such as the ring-stage survival assay for the in vitro determination of the artemisinin resistance phenotype (51). While we observed differences in the yearly IC50s of the two artemisinin derivatives tested (ARS and DHA), no clear pattern (increase or decrease) could be determined, making these results inconclusive.
In conclusion, the results from our study suggest that artemisinin is still efficacious and that CQ-sensitive parasite strains are increasing in proportion compared to CQ-resistant strains in this part of Kenya. Since lumefantrine, the partner drug in AL, has been shown to select for CQ-sensitive parasites, continued monitoring for drug resistance, including artemisinin and partner drugs, is important to inform malaria control programs.
ACKNOWLEDGMENTS
This work was supported by the World Wide Antimalarial Resistance Network (WWARN), by the Division of Malaria Control, Ministry of Health, Kenya, through a grant from the United Kingdom Department for International Development (DfID), by the Centers for Disease Control and Prevention Antimicrobial Resistance Working Group, and by the Atlanta Research and Education Foundation. S.A.O. was partially supported by a CDC-based American Society for Microbiology postdoctoral fellowship.
This study would not have been possible without the willingness of all the children and their parents/guardians to participate in this study; for this we are grateful. We appreciate the hard work of all the CDC and KEMRI field workers and laboratory staff who worked on this study.
The use of trade names and names of commercial sources is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention or the U.S. Department of Health and Human Services. The findings and conclusions in this presentation are those of the authors and do not necessarily represent those of the Centers for Disease Control and Prevention. This paper is published with the approval of the KEMRI director.
REFERENCES
- 1.Anonymous. 2006. WHO calls for an immediate halt to provision of single-drug artemisinin malaria pills. New malaria treatment guidelines issued by WHO. Saudi Med J 27:574–575. [PubMed] [Google Scholar]
- 2.Sridaran S, McClintock SK, Syphard LM, Herman KM, Barnwell JW, Udhayakumar V. 2010. Anti-folate drug resistance in Africa: meta-analysis of reported dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) mutant genotype frequencies in African Plasmodium falciparum parasite populations. Malar J 9:247. doi: 10.1186/1475-2875-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Picot S, Olliaro P, de Monbrison F, Bienvenu AL, Price RN, Ringwald P. 2009. A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. Malar J 8:89. doi: 10.1186/1475-2875-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foote SJ, Kyle DE, Martin RK, Oduola AM, Forsyth K, Kemp DJ, Cowman AF. 1990. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature 345:255–258. doi: 10.1038/345255a0. [DOI] [PubMed] [Google Scholar]
- 5.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. 2000. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 6.Duraisingh MT, Jones P, Sambou I, von Seidlein L, Pinder M, Warhurst DC. 2000. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol Biochem Parasitol 108:13–23. doi: 10.1016/S0166-6851(00)00201-2. [DOI] [PubMed] [Google Scholar]
- 7.Humphreys GS, Merinopoulos I, Ahmed J, Whitty CJ, Mutabingwa TK, Sutherland CJ, Hallett RL. 2007. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother 51:991–997. doi: 10.1128/AAC.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martensson A, Stromberg J, Sisowath C, Msellem MI, Gil JP, Montgomery SM, Olliaro P, Ali AS, Bjorkman A. 2005. Efficacy of artesunate plus amodiaquine versus that of artemether-lumefantrine for the treatment of uncomplicated childhood Plasmodium falciparum malaria in Zanzibar, Tanzania. Clin Infect Dis 41:1079–1086. doi: 10.1086/444460. [DOI] [PubMed] [Google Scholar]
- 9.Sisowath C, Ferreira PE, Bustamante LY, Dahlstrom S, Martensson A, Bjorkman A, Krishna S, Gil JP. 2007. The role of pfmdr1 in Plasmodium falciparum tolerance to artemether-lumefantrine in Africa. Trop Med Int Health 12:736–742. doi: 10.1111/j.1365-3156.2007.01843.x. [DOI] [PubMed] [Google Scholar]
- 10.Sisowath C, Stromberg J, Martensson A, Msellem M, Obondo C, Bjorkman A, Gil JP. 2005. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem). J Infect Dis 191:1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 11.Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. 2006. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis 194:528–535. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, Coulibaly D, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med 344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 14.Wellems TE, Plowe CV. 2001. Chloroquine-resistant malaria. J Infect Dis 184:770–776. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- 15.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, ler Moo C, Al-Saai S, Dondorp AM, Lwin KM, Singhasivanon P, Day NP, White NJ, Anderson TJ, Nosten F. 2012. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet 379:1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amaratunga C, Mao S, Sreng S, Suon S, Fairhurst RM. 2013. Slow parasite clearance rates in response to artemether in patients with severe malaria. Lancet Infect Dis 13:113–114. doi: 10.1016/S1473-3099(12)70347-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amaratunga C, Sreng S, Suon S, Phelps ES, Stepniewska K, Lim P, Zhou C, Mao S, Anderson JM, Lindegardh N, Jiang H, Song J, Su XZ, White NJ, Dondorp AM, Anderson TJ, Fay MP, Mu J, Duong S, Fairhurst RM. 2012. Artemisinin-resistant Plasmodium falciparum in Pursat province, western Cambodia: a parasite clearance rate study. Lancet Infect Dis 12:851–858. doi: 10.1016/S1473-3099(12)70181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Menard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale JC, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Menard D. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takala-Harrison S, Jacob CG, Arze C, Cummings MP, Silva JC, Dondorp AM, Fukuda MM, Hien TT, Mayxay M, Noedl H, Nosten F, Kyaw MP, Nhien NT, Imwong M, Bethell D, Se Y, Lon C, Tyner SD, Saunders DL, Ariey F, Mercereau-Puijalon O, Menard D, Newton PN, Khanthavong M, Hongvanthong B, Starzengruber P, Fuehrer HP, Swoboda P, Khan WA, Phyo AP, Nyunt MM, Nyunt MH, Brown TS, Adams M, Pepin CS, Bailey J, Tan JC, Ferdig MT, Clark TG, Miotto O, MacInnis B, Kwiatkowski DP, White NJ, Ringwald P, Plowe CV. 2015. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J Infect Dis 211:670–679. doi: 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han KT, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, et al. . 2014. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, Kazembe PN, Djimde AA, Kouriba B, Taylor TE, Plowe CV. 2003. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis 187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- 23.Mita T, Kaneko A, Lum JK, Bwijo B, Takechi M, Zungu IL, Tsukahara T, Tanabe K, Kobayakawa T, Bjorkman A. 2003. Recovery of chloroquine sensitivity and low prevalence of the Plasmodium falciparum chloroquine resistance transporter gene mutation K76T following the discontinuance of chloroquine use in Malawi. Am J Trop Med Hyg 68:413–415. [PubMed] [Google Scholar]
- 24.Frosch AE, Laufer MK, Mathanga DP, Takala-Harrison S, Skarbinski J, Claassen CW, Dzinjalamala FK, Plowe CV. 2014. Return of widespread chloroquine-sensitive Plasmodium falciparum to Malawi. J Infect Dis 210:1110–1114. doi: 10.1093/infdis/jiu216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laufer MK, Takala-Harrison S, Dzinjalamala FK, Stine OC, Taylor TE, Plowe CV. 2010. Return of chloroquine-susceptible falciparum malaria in Malawi was a reexpansion of diverse susceptible parasites. J Infect Dis 202:801–808. doi: 10.1086/655659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohammed A, Ndaro A, Kalinga A, Manjurano A, Mosha JF, Mosha DF, van Zwetselaar M, Koenderink JB, Mosha FW, Alifrangis M, Reyburn H, Roper C, Kavishe RA. 2013. Trends in chloroquine resistance marker, Pfcrt-K76T mutation ten years after chloroquine withdrawal in Tanzania. Malar J 12:415. doi: 10.1186/1475-2875-12-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malmberg M, Ngasala B, Ferreira PE, Larsson E, Jovel I, Hjalmarsson A, Petzold M, Premji Z, Gil JP, Bjorkman A, Martensson A. 2013. Temporal trends of molecular markers associated with artemether-lumefantrine tolerance/resistance in Bagamoyo district, Tanzania. Malar J 12:103. doi: 10.1186/1475-2875-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mwai L, Ochong E, Abdirahman A, Kiara SM, Ward S, Kokwaro G, Sasi P, Marsh K, Borrmann S, Mackinnon M, Nzila A. 2009. Chloroquine resistance before and after its withdrawal in Kenya. Malar J 8:106. doi: 10.1186/1475-2875-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mang'era CM, Mbai FN, Omedo IA, Mireji PO, Omar SA. 2012. Changes in genotypes of Plasmodium falciparum human malaria parasite following withdrawal of chloroquine in Tiwi, Kenya. Acta Trop 123:202–207. doi: 10.1016/j.actatropica.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Okombo J, Kamau AW, Marsh K, Sutherland CJ, Ochola-Oyier LI. 2014. Temporal trends in prevalence of Plasmodium falciparum drug resistance alleles over two decades of changing antimalarial policy in coastal Kenya. Int J Parasitol Drugs Drug Resist 4:152–163. doi: 10.1016/j.ijpddr.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Achieng AO, Muiruri P, Ingasia LA, Opot BH, Juma DW, Yeda R, Ngalah BS, Ogutu BR, Andagalu B, Akala HM, Kamau E. 2015. Temporal trends in prevalence of Plasmodium falciparum molecular markers selected for by artemether-lumefantrine treatment in pre-ACT and post-ACT parasites in western Kenya. Int J Parasitol Drugs Drug Resist 5:92–99. doi: 10.1016/j.ijpddr.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frosch AE, Venkatesan M, Laufer MK. 2011. Patterns of chloroquine use and resistance in sub-Saharan Africa: a systematic review of household survey and molecular data. Malar J 10:116. doi: 10.1186/1475-2875-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.World Health Organization. 2010. Global report on antimalarial drug efficacy and drug resistance: 2000-2010. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 34.Bacon DJ, Latour C, Lucas C, Colina O, Ringwald P, Picot S. 2007. Comparison of a SYBR green I-based assay with a histidine-rich protein II enzyme-linked immunosorbent assay for in vitro antimalarial drug efficacy testing and application to clinical isolates. Antimicrob Agents Chemother 51:1172–1178. doi: 10.1128/AAC.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woodrow CJ, Dahlstrom S, Cooksey R, Flegg JA, Le Nagard H, Mentre F, Murillo C, Menard D, Nosten F, Sriprawat K, Musset L, Quashie NB, Lim P, Fairhurst RM, Nsobya SL, Sinou V, Noedl H, Pradines B, Johnson JD, Guerin PJ, Sibley CH, Le Bras J. 2013. High-throughput analysis of antimalarial susceptibility data by the WorldWide Antimalarial Resistance Network (WWARN) in vitro analysis and reporting tool. Antimicrob Agents Chemother 57:3121–3130. doi: 10.1128/AAC.02350-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alam MT, de Souza DK, Vinayak S, Griffing SM, Poe AC, Duah NO, Ghansah A, Asamoa K, Slutsker L, Wilson MD, Barnwell JW, Udhayakumar V, Koram KA. 2011. Selective sweeps and genetic lineages of Plasmodium falciparum drug-resistant alleles in Ghana. J Infect Dis 203:220–227. doi: 10.1093/infdis/jiq038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talundzic E, Okoth SA, Congpuong K, Plucinski MM, Morton L, Goldman IF, Kachur PS, Wongsrichanalai C, Satimai W, Barnwell JW, Udhayakumar V. 2015. Selection and spread of artemisinin-resistant alleles in Thailand prior to the global artemisinin resistance containment campaign. PLoS Pathog 11:e1004789. doi: 10.1371/journal.ppat.1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isozumi R, Uemura H, Kimata I, Ichinose Y, Logedi J, Omar AH, Kaneko A. 2015. Novel mutations in K13 propeller gene of artemisinin-resistant Plasmodium falciparum. Emerg Infect Dis 21:490–492. doi: 10.3201/eid2103.140898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamau E, Campino S, Amenga-Etego L, Drury E, Ishengoma D, Johnson K, Mumba D, Kekre M, Yavo W, Mead D, Bouyou-Akotet M, Apinjoh T, Golassa L, Randrianarivelojosia M, Andagalu B, Maiga-Ascofare O, Amambua-Ngwa A, Tindana P, Ghansah A, MacInnis B, Kwiatkowski D, Djimde AA. 2015. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J Infect Dis 211:1352–1355. doi: 10.1093/infdis/jiu608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor SM, Parobek CM, DeConti DK, Kayentao K, Coulibaly SO, Greenwood BM, Tagbor H, Williams J, Bojang K, Njie F, Desai M, Kariuki S, Gutman J, Mathanga DP, Martensson A, Ngasala B, Conrad MD, Rosenthal PJ, Tshefu AK, Moormann AM, Vulule JM, Doumbo OK, Ter Kuile FO, Meshnick SR, Bailey JA, Juliano JJ. 2015. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis 211:680–688. doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maiga AW, Fofana B, Sagara I, Dembele D, Dara A, Traore OB, Toure S, Sanogo K, Dama S, Sidibe B, Kone A, Thera MA, Plowe CV, Doumbo OK, Djimde AA. 2012. No evidence of delayed parasite clearance after oral artesunate treatment of uncomplicated falciparum malaria in Mali. Am J Trop Med Hyg 87:23–28. doi: 10.4269/ajtmh.2012.12-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plucinski MM, Talundzic E, Morton L, Dimbu PR, Macaia AP, Fortes F, Goldman I, Lucchi N, Stennies G, MacArthur JR, Udhayakumar V. 2015. Efficacy of artemether-lumefantrine and dihydroartemisinin-piperaquine for treatment of uncomplicated malaria in children in Zaire and Uige provinces, Angola. Antimicrob Agents Chemother 59:437–443. doi: 10.1128/AAC.04181-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takala-Harrison S, Laufer MK. 2015. Antimalarial drug resistance in Africa: key lessons for the future. Ann N Y Acad Sci 1342:62–67. doi: 10.1111/nyas.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eyase FL, Akala HM, Ingasia L, Cheruiyot A, Omondi A, Okudo C, Juma D, Yeda R, Andagalu B, Wanja E, Kamau E, Schnabel D, Bulimo W, Waters NC, Walsh DS, Johnson JD. 2013. The role of Pfmdr1 and Pfcrt in changing chloroquine, amodiaquine, mefloquine and lumefantrine susceptibility in western-Kenya P. falciparum samples during 2008-2011. PLoS One 8:e64299. doi: 10.1371/journal.pone.0064299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah M, Omosun Y, Lal A, Odero C, Gatei W, Otieno K, Gimnig JE, Kuile FT, Hawley WA, Nahlen B, Kariuki S, Walker E, Slutsker L, Hamel M, Shi YP. 2015. Assessment of molecular markers for anti-malarial drug resistance after the introduction and scale-up of malaria control interventions in western Kenya. Malar J 14:75. doi: 10.1186/s12936-015-0588-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malmberg M, Ferreira PE, Tarning J, Ursing J, Ngasala B, Bjorkman A, Martensson A, Gil JP. 2013. Plasmodium falciparum drug resistance phenotype as assessed by patient antimalarial drug levels and its association with pfmdr1 polymorphisms. J Infect Dis 207:842–847. doi: 10.1093/infdis/jis747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venkatesan M, Gadalla NB, Stepniewska K, Dahal P, Nsanzabana C, Moriera C, Price RN, Martensson A, Rosenthal PJ, Dorsey G, Sutherland CJ, Guerin P, Davis TM, Menard D, Adam I, Ademowo G, Arze C, Baliraine FN, Berens-Riha N, Bjorkman A, Borrmann S, Checchi F, Desai M, Dhorda M, Djimde AA, El-Sayed BB, Eshetu T, Eyase F, Falade C, Faucher JF, Froberg G, Grivoyannis A, Hamour S, Houze S, Johnson J, Kamugisha E, Kariuki S, Kiechel JR, Kironde F, Kofoed PE, LeBras J, Malmberg M, Mwai L, Ngasala B, Nosten F, Nsobya SL, Nzila A, Oguike M, Otienoburu SD, Ogutu B, et al. . 2014. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P. falciparum malaria after artemether-lumefantrine and artesunate-amodiaquine. Am J Trop Med Hyg 91:833–843. doi: 10.4269/ajtmh.14-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ngalah BS, Ingasia LA, Cheruiyot AC, Chebon LJ, Juma DW, Muiruri P, Onyango I, Ogony J, Yeda RA, Cheruiyot J, Mbuba E, Mwangoka G, Achieng AO, Ng'ang'a Z, Andagalu B, Akala HM, Kamau E. 2015. Analysis of major genome loci underlying artemisinin resistance and pfmdr1 copy number in pre- and post-ACTs in western Kenya. Sci Rep 5:8308. doi: 10.1038/srep08308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmgren G, Gil JP, Ferreira PM, Veiga MI, Obonyo CO, Bjorkman A. 2006. Amodiaquine resistant Plasmodium falciparum malaria in vivo is associated with selection of pfcrt 76T and pfmdr1 86Y. Infect Genet Evol 6:309–314. doi: 10.1016/j.meegid.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 50.Klonis N, Xie SC, McCaw JM, Crespo-Ortiz MP, Zaloumis SG, Simpson JA, Tilley L. 2013. Altered temporal response of malaria parasites determines differential sensitivity to artemisinin. Proc Natl Acad Sci U S A 110:5157–5162. doi: 10.1073/pnas.1217452110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, Lim P, Mao S, Sopha C, Sam B, Anderson JM, Duong S, Chuor CM, Taylor WR, Suon S, Mercereau-Puijalon O, Fairhurst RM, Menard D. 2013. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis 13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]