Abstract
Preexposure prophylaxis (PrEP) using antiretroviral drugs is effective in reducing the risk of human immunodeficiency virus type 1 (HIV-1) infection, but adherence to the PrEP regimen is needed. To improve adherence, a long-acting injectable formulation of the nonnucleoside reverse transcriptase (RT) inhibitor rilpivirine (RPV LA) has been developed. However, there are concerns that PrEP may select for drug-resistant mutations during preexisting or breakthrough infections, which could promote the spread of drug resistance and limit options for antiretroviral therapy. To address this concern, we administered RPV LA to macaques infected with simian immunodeficiency virus containing HIV-1 RT (RT-SHIV). Peak plasma RPV levels were equivalent to those reported in human trials and waned over time after dosing. RPV LA resulted in a 2-log decrease in plasma viremia, and the therapeutic effect was maintained for 15 weeks, until plasma drug concentrations dropped below 25 ng/ml. RT mutations E138G and E138Q were detected in single clones from plasma virus in separate animals only at one time point, and no resistance mutations were detected in viral RNA isolated from tissues. Wild-type and E138Q RT-SHIV displayed similar RPV susceptibilities in vitro, whereas E138G conferred 2-fold resistance to RPV. Overall, selection of RPV-resistant variants was rare in an RT-SHIV macaque model despite prolonged exposure to slowly decreasing RPV concentrations following injection of RPV LA.
INTRODUCTION
Despite increasing use of antiretroviral therapy (ART), which suppresses viral replication and reduces the risk of human immunodeficiency virus type 1 (HIV-1) transmission, an estimated two million new HIV-1 infections still occur annually worldwide (1). While research continues to define effective, vaccine-elicited protective immune responses, preexposure prophylaxis (PrEP) has proven effective in reducing HIV-1 transmission. Thus far, clinical trials have exhibited a 44 to 75% reduction in HIV-1 infections in individuals treated with tenofovir (TDF) with or without emtricitabine (FTC) (2–5). These successes led to FDA approval of daily oral TDF-FTC PrEP for high-risk populations.
One prominent issue with PrEP is dependence of efficacy on adherence. Data from clinical trials has shown that patients with detectable plasma drug levels indicative of adherence have reduced risk of HIV-1 infection, while poor PrEP adherence confers little protection (2, 4, 6–8). Although recent work suggests that sexual event-driven TDF-FTC administration can be effective in men (9–11), sporadic PrEP adherence can lower PrEP efficacy and promote selection of drug-resistant variants. To reduce pill burden, long-acting injectable nanoparticle formulations of the nonnucleoside reverse transcriptase inhibitor (NNRTI) rilpivirine (RPV) and an experimental integrase inhibitor, cabotegravir, have been developed (12, 13). Injectable PrEP formulations create a depot at the injection site and provide drug release for weeks to months, thus eliminating the need for daily drug administration. Long-acting injectable medications have been used successfully to administer antipsychotics, contraceptives, hormone replacements, and cancer treatments (14–17).
Rilpivirine is the most recently approved NNRTI for ART in combination with other antiretrovirals in patients with plasma viremia of less than 100,000 copies of HIV-1 RNA/ml (18, 19). Clinical trials have shown RPV-TDF-FTC to be noninferior to and more tolerable than efavirenz-based ART (18). During these trials, patients failing RPV-based ART also tended to select unique NNRTI-associated resistance mutations in reverse transcriptase (RT) compared with those on an efavirenz-based regimen (20). Long-acting rilpivirine (RPV LA) is an injectable nanoparticle formulation and has been shown to be safe and tolerable, with detectable drug concentrations maintained in plasma and tissues weeks after a single injection (21–24). Multiple clinical trials are under way to test safety and acceptability of RPV LA as PrEP in HIV-1-uninfected men and women (13).
Of concern with RPV LA PrEP is the development of drug resistance if an individual with an undetected HIV-1 infection receives PrEP or someone on PrEP becomes infected, resulting in treatment with RPV monotherapy. Additionally, while patients on a pill-based regimen can cease PrEP and rapidly clear the drug, injectable medications will require weeks to reach undetectable concentrations. These situations may increase the selection of drug-resistant mutations (DRMs), promoting spread of drug resistance and limiting future ART options through cross-resistance (25). Resistance analyses of breakthrough infections in TDF-FTC clinical trials revealed that selection of DRMs was rare in patients who became infected after receiving PrEP, but DRMs did develop in the few individuals that were HIV-1+ before initiating PrEP (26–28).
Although the development of DRMs during TDF-FTC PrEP clinical trials was rare (26–28), there are currently no published data on the resistance outcome of long-acting PrEP as monotherapy. We report here on a pilot study to explore the selection of drug resistance by RPV LA in RT-SHIV-infected macaques. RT-SHIVmne is a chimeric simian immunodeficiency virus (SIV) that contains the HIV-1 reverse transcriptase coding region (29, 30). SIV is not susceptible to NNRTIs due to sequence differences within the RT coding region, but NNRTI sensitivity is established by swapping the SIV and HIV-1 RT coding regions (31, 32). RT-SHIV macaque models have been used to study HIV-1 ART, drug resistance, PrEP, and persistence (30, 32–48). In this study, we treated RT-SHIV-infected macaques with RPV LA and measured plasma viremia, drug concentrations, and drug-resistant isolates over 35 weeks. Our data show that viremia was suppressed by RPV LA monotherapy, which rebounded to pretherapy levels as plasma drug concentrations waned. However, resistance to RPV in RT-SHIV was difficult to select both in vitro and in vivo.
MATERIALS AND METHODS
Cell culture and antiretroviral inhibitors.
293T and TZM-bl (49) cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals) and 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 0.292 mg/ml of l-glutamine (P-S-G; Life Technologies). GHOST-R3/X4/R5 cells (50) were maintained in the same medium as described above with the addition of 100 μg/ml of Geneticin (Life Technologies), 100 μg/ml of hygromycin (Life Technologies), and 0.5 μg/ml of puromycin (EMD Millipore). CEMx174 cells were cultured in RPMI 1640 medium supplemented with 10% FBS and P-S-G. All cell lines were incubated at 37°C with 5% CO2.
RPV was acquired from the NIH AIDS Reagent Program. RPV LA was donated by Janssen Sciences UC Ireland.
Virus production and titer determination.
Plasmids encoding HIV-1LAI with silent restriction sites within RT (51) and RT-SHIVmne (30) were used for virus production by transfection into 293T cells with Lipofectamine 2000 (Life Technologies) and stored at −80°C. Mutations were introduced into the plasmids by site-directed mutagenesis using either the QuikChange II XL kit (Agilent Technologies) or the Q5 site-directed mutagenesis kit (New England BioLabs). HIV-1 titers were determined on GHOST cells with modifications from previous work (50), using an LSRII flow cytometer (BD Biosciences). Titers of RT-SHIV stocks were determined using TZM-bl indicator cells and β-galactosidase staining as previously described (29, 52).
Animals.
Two juvenile pigtailed macaques (Macaca nemestrina) were housed at the University of Pittsburgh in accordance with American Association of Accreditation of Laboratory Animal Care standards. All procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee. The animals were negative for serum antibodies to HIV type 2, SIV, type D retrovirus, and simian T-lymphotropic virus type 1 at study initiation. Animals were infected intravenously with 1 × 105 infectious units (IU) of RT-SHIVmne. At 6 and 8 weeks postinfection, animals were treated with 200 mg/kg of RPV LA by intramuscular (i.m.) injection near the scapula. This dosing regimen was determined by prior pharmacokinetic analysis that showed that a single 50-mg/kg dose of RPV LA did not result in plasma drug concentrations above 16 ng/ml within 24 h of administration, consistent with our previous observations of rapid metabolism of NNRTIs in pigtailed macaques (data not shown). Blood was drawn under anesthesia weekly or bimonthly. Animals were euthanized at 35 weeks postinfection, and multiple tissues, including axillary lymph nodes (LN) and ileum, were immediately flash frozen and stored in liquid nitrogen.
T cell counts and viral RNA isolation.
Plasma was separated from EDTA-treated whole blood by centrifugation at 400 × g for 10 min and stored at −80°C until processed. Viral RNA (vRNA) was isolated from plasma as previously described (53). Briefly, plasma was mixed with Tris-buffered saline (Sigma) and pelleted by centrifugation at 21,000 × g for 1 h. Virus was then suspended in 3 M guanidinium chloride, 50 mM Tris-HCl (pH 7.6), 1 mM CaCl2, and 1 mg/ml of proteinase K and incubated at 42°C for 1 h, followed by addition of 5.7 M guanidine thiocyanate, 50 mM Tris-HCl (pH 7.6), 1 mM EDTA, and 600 μg/ml of glycogen and a 5-min incubation at room temperature. RNA was washed with isopropanol and ethanol and samples were suspended in 10 mM Tris-HCl (pH 8.0). Tissue vRNA isolation was performed as previously described (54).
Peripheral blood mononuclear cells (PBMCs) were isolated from samples using lymphocyte separation medium (Corning) and treated with ACK lysis buffer (Life Technologies) to remove red blood cells. PBMCs were stained with NHP T lymphocyte cocktail (BD Biosciences) with antibodies against CD3, CD4, and CD8. CD3+ CD4+ and CD3+ CD8+ cell populations were measured by flow cytometry. Isotype cocktail C (BD Biosciences) was used for isotype controls.
RPV plasma and tissue levels.
Rilpivirine was extracted from monkey plasma using protein precipitation followed by liquid chromatograph-tandem mass spectrometry (LC-MS/MS) analysis. Calibration standards and quality control (QC) samples were prepared in monkey plasma (EDTA) on the day of analysis. Fifty microliters of each plasma sample was mixed with 50 μl of a 50:50 methanol-water mixture containing amprenavir-d4 as the internal standard. Then, 300 μl of methanol was added to each sample. Following vortex and centrifugation steps, the resulting supernatant was transferred to a 96-well plate for LC-MS/MS analysis. Tissues were homogenized in 1 ml of 70:30 acetonitrile–1 mM ammonium phosphate (pH 7.4) with a Precellys hard tissue grinding kit tube (Cayman Chemical), followed by a similar plasma preparation procedure. Tissue weights ranged from 84.2 to 95.4 mg. Analyte concentrations from tissue homogenates were normalized to tissue weight. A tissue density of 1 g/ml was used to convert concentrations into nanograms per milliliter. A Shimadzu high-performance liquid chromatography system was used for separation, and an AB SCIEX API 5000 mass spectrometer (AB SCIEX) equipped with a turbo spray interface was used as the detector. The samples were analyzed with a set of calibration standards and QC samples, with a dynamic range of 0.5 to 2,000 ng/ml. The precision and accuracy of the calibration standards and QC samples were within the acceptable range of 15%.
Viral RNA quantitation.
Viral RNA was measured by generation of cDNA from vRNA isolated from plasma or tissues by the SuperScript III first-strand synthesis kit (Life Technologies) with the SIVgag-R primer (5′-CACTAGGTGTCTCTGCACTATCTGTTTTG-3′). Quantitative PCR was performed on cDNA using SsoFast probes SuperMix (Bio-Rad), SIVgag-F (5′-GTCTGCGTCATCTGGTGCATTC-3′) and SIVgag-R primers, and the SIVgag-probe (5′-FAM-CTTCCTCAGTGTGTTTCACTTTCTCTTCTGCG-3′ 6-carboxytetramethylrhodamine [TAMRA]). CCR5 primers and probe were previously described (54). Reaction conditions were 1 cycle of 95°C for 2 min followed by 45 cycles of 95°C for 15 s and 60°C for 1 min.
Viral RNA sequencing.
The RT coding region was amplified from viral cDNA from plasma or tissues using nested PCR. The first round of PCR was performed with the Platinum Taq DNA polymerase high-fidelity kit (Life Technologies), using primers ZA01 (5′-CTAGATCTGAATTTGCCTGCCC-3′) and ZA02 (5′-TGTAACAGGAATAGAGTTAGGTCC-3′) with the following reaction conditions: 94°C for 2 min; 40 cycles of 94°C for 15 s, 49°C for 30 s, and 68°C for 2 min; and 1 cycle of 68°C for 5 min. DNA was purified using the ExoSAP-IT PCR cleanup kit (Affymetrix) per the manufacturer's instructions. The second round of PCR amplification was performed with the Platinum PCR Supermix kit (Life Technologies), using primers RT19 (5′-GCAAAAGGATTAAAGGGACAA-3) and RT22 (5′-GGGTAATCCAAATTTGAATACCAATCCT-3′) with the following reaction conditions: 94°C for 2 min; 26 cycles of 94°C for 15 s, 63°C for 30 s with −0.5°C increments per cycle, and 72°C for 2 min; 15 cycles of 94°C for 15 s, 50°C for 30 s, and 72°C for 2 min; and 1 cycle of 72°C for 5 min. The PCR product was then cleaned using the Wizard SV gel and PCR cleanup system (Promega) and cloned into TOPO vectors using the pCR 2.1-TOPO TA cloning kit (Life Technologies) per the manufacturer's instructions. Bacterial colonies were screened for full-length RT sequence by PCR. RT-containing TOPO vectors were isolated from overnight bacterial cultures using the QiaPrep Spin Miniprep kit (Qiagen). Sequences were analyzed using DNASTAR (Lasergene).
RT-SHIV in vitro resistance selection.
CEMx174 cells were infected with wild-type (WT) RT-SHIV at a multiplicity of infection (MOI) of 0.05 in medium and incubated at 37°C for 2 h. Cells and virus were then resuspended in medium containing 0.1 nM RPV. Cultures were passaged every 2 to 3 days in new medium. If cytopathic effects were apparent, vRNA was isolated from culture supernatant using the RNeasy minikit (Qiagen) and sequenced as described above. If the RT sequence from vRNA isolated from the supernatant was WT, then 50 μl of culture supernatant was used to infect fresh CEMx174 cells and the RPV concentration was doubled to begin a new round of selection.
Drug susceptibility assay.
Drug susceptibility assays were performed as previously described, with minor modifications (29). TZM-bl cells were seeded at 5 × 103 cells in 96-well cell culture-treated white plates (PerkinElmer). The following day, virus and serial drug dilutions were prepared in phenol red-free DMEM (Life Technologies) supplemented with P-S-G and 10% FBS or different amounts of human or macaque serum. Medium containing virus (MOI of 0.05) and drug dilutions in a total volume of 0.2 ml was added to each well in triplicate. Wells with virus and no drug were used as 100% infection controls. Plates were incubated at 37°C for 48 h. Luciferase activity was measured using Britelite Plus reagent (PerkinElmer) on a Luminoskan Ascent microplate luminometer (Thermo Scientific). Relative luciferase units (RLU) were converted to percent infection by dividing the RLU of each drug dilution by the RLU of the 100% infection control. Wells containing cells with no virus and no drug were used to normalize for background luciferase output. The effective concentration to inhibit 50% of virus replication (EC50) was calculated using PRISM 6 (GraphPad). Specifically, EC50 was calculated by log transforming drug concentrations and using a four-parameter variable slope nonlinear regression for curve fitting analysis.
RESULTS
RPV LA treatment of RT-SHIV-infected macaques.
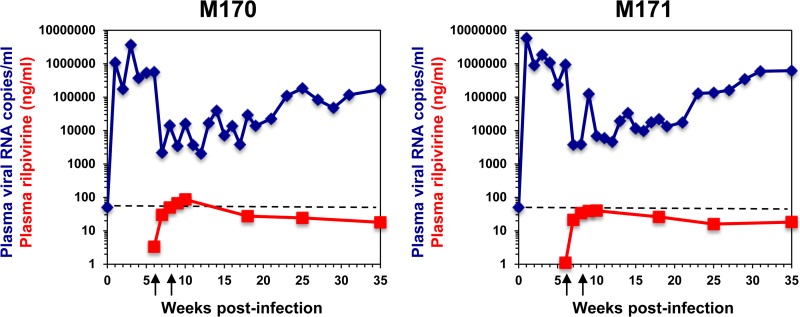
To assess the potential for selection of drug-resistant virus in vivo during RPV LA treatment, two pigtailed macaques were infected with RT-SHIV at week 0. Both animals showed peak viremia between 1 × 106 and 1 × 107 copies of vRNA/ml, which declined to a set point of 5 × 105 to 9 × 105 vRNA copies/ml (Fig. 1). Based on pharmacokinetic results (data not shown), animals were treated with two 200-mg/kg i.m. doses of RPV LA at weeks 6 and 8 postinfection. Posttreatment, there was an immediate ∼2-log decline in plasma viremia, which was maintained at approximately 1.35 × 104 vRNA copies/ml until 21 weeks postinfection. Viral loads slowly rebounded thereafter to pretherapy set point values (Fig. 1). There was a transient decrease in the percentage of CD4+ T cells in both animals after infection, followed by a steady posttreatment increase for the remainder of the study (data not shown). The only adverse events attributed to RPV LA treatment were redness and swelling at the injection sites.
FIG 1.
Plasma RT-SHIV levels in two macaques treated with RPV LA monotherapy. Animals were infected at week 0 and received two i.m. doses of RPV LA at 6 and 8 weeks postinfection (designated by arrows under the x axis). Plasma viremia (blue lines) and RPV concentrations (red lines) were measured. The dashed lines indicate the limit of detection of the qRT-PCR assay (50 RNA copies/ml).
Although a standard protective level of RPV has not been defined, the in vitro EC50 of RPV is 0.1 to 0.7 nM (55, 56), and our dosing strategy was designed to achieve plasma RPV concentrations equivalent to that reported for humans: 50 ng/ml (136 nM) (22, 56). Maximum plasma RPV concentrations of 86.5 ng/ml and 40 ng/ml (214.7 nM and 99.3 nM) were detected at week 10 postinfection in animals M170 and M171, respectively (Fig. 1). Plasma RPV concentrations were greater than 15 ng/ml (40.9 nM) in both animals up to 29 weeks after the first RPV LA injection. Virus rebound appeared to be associated with declining drug concentrations, although there was not a significant correlation between plasma RPV concentrations and viral loads by linear regression (R2 = 0.15). RPV concentrations in the axillary LN and ileum were 4- to 421-fold higher than in plasma at the time of necropsy (week 35 postinfection) (Table 1). As tissue homogenate concentrations are averaged across a heterogeneous matrix, these data do not differentiate between extracellular or intracellular drug exposure or intracellular drug exposure between cell types. Therefore, the averaged concentration could significantly overrepresent the concentration of drug in mononuclear cells. Since it is difficult to isolate mononuclear cells from tissue without extracellular leaching of drug during the isolation process, intracellular drug concentrations in tissues, and the resulting pharmacodynamic effect, are an area of continued study.
TABLE 1.
RPV concentrations in plasma and tissues of animals at 35 weeks postinfection
| Site | RPV concn (ng/ml) |
|
|---|---|---|
| Animal M170 | Animal M171 | |
| Plasma | 18 | 18 |
| Axillary LN | 155 | 7,590 |
| Ileum | 136 | 80 |
Plasma RT-SHIV drug resistance detection.
To identify selection of RPV-resistant variants, we sequenced the RT coding region of plasma virus at week 6 prior to therapy and at weeks 9, 13, 21, and 25 postinfection. At all time points, the full-length consensus RT sequence matched the starting RT sequence, indicating a lack of detectable mutations that constituted more than 20% of the virus population (data not shown). However, partial and full clonal RT regions were sequenced and revealed many minor viral variants in the plasma (Table 2). Based on Poisson distribution, the probability of detecting variants present at 5% frequency in a virus population with 90% certainty requires analysis of at least 45 clones. Hence, 45 clones were sequenced at 6, 9, 13, and 25 weeks postinfection. The majority of clonal sequences was either WT or had mutations in RT amino acids 1 to 250 that are not associated with known drug resistance. E138G was identified in a single clone at both weeks 9 and 13 in animal M170 but was not detectable later. E138Q was identified in a single clone at week 25 in animal M171. T69A and K65R are NRTI-associated resistance mutations that were also identified, but they were also detected at frequencies as low as E138G/Q.
TABLE 2.
Mutations identified in RT of plasma RT-SHIV RNA
| Animal | Mutations in RT-SHIV RTa (no. of clones) |
||||
|---|---|---|---|---|---|
| Wk 6b | Wk 9 | Wk 13 | Wk 21 | Wk 25 | |
| M170 | WT (14) | WT (25) | WT (19) | WT (4) | WT (24) |
| Non-DRM (31) | Non-DRM (19)c | Non-DRM (25) | Non-DRM (6) | Non-DRM (19) | |
| R125G, E138G (1) | S68G, E138G (1) | K65R (1) | |||
| M171 | WT (22) | WT (26) | WT (24) | WT (5) | WT (21) |
| Non-DRM (23) | Non-DRM (19) | Non-DRM (20) | Non-DRM (5) | Non-DRM (19) | |
| T69A (1) | E138Q, P157-, L213F, V244C (1) | ||||
Results represent a mixture of full-length and partial (amino acids 1 to 250) RT sequences. WT, wild type; non-DRM, mutations not associated with drug resistance; -, codon deletion; underline, known RPV-associated resistance mutations.
Prior to RPV-LA administration (6 and 8 weeks postinfection).
One sequence at this time point for this animal contained A33V.
To determine if E138G or E138Q conferred RPV resistance in RT-SHIV, the mutations were made in both HIV-1 and RT-SHIV and were tested for susceptibility to RPV compared to that of WT viruses in vitro. WT HIV-1 and RT-SHIV displayed similar RPV susceptibilities, with EC50s of 0.67 ± 0.08 and 0.35 ± 0.06 nM (Table 3). E138G conferred a slight increase in EC50 compared to WT virus in both HIV-1 and RT-SHIV backbones: EC50s of 0.94 ± 0.03 and 0.76 ± 0.12 nM (1.4- and 2.2-fold changes), respectively. Conversely, E138Q conferred a 4.3-fold EC50 increase above WT HIV-1 (2.85 ± 0.08 nM) and no difference from WT RT-SHIV (0.30 ± 0.05 nM). These EC50s for WT, E138G, and E138Q HIV-1 were similar to values previously reported (55, 57).
TABLE 3.
RPV susceptibilities of HIV-1 and RT-SHIV with select mutations identified in plasma and tissue vRNA
| Virus | Mutation(s) in RT sequence | EC50 (nM)a | Fold change from WT virusb |
|---|---|---|---|
| HIV-1 | WT | 0.67 ± 0.08 | 1.0 |
| E138G | 0.94 ± 0.03 | 1.4 | |
| E138Q | 2.85 ± 0.08 | 4.3 | |
| A33V | 0.42 ± 0.07 | 0.6 | |
| Y181H | 0.17 ± 0.02 | 0.3 | |
| A33V, Y181H | 0.13 ± 0.01 | 0.2 | |
| RT-SHIV | WT | 0.35 ± 0.06 | 1.0 |
| E138G | 0.76 ± 0.12 | 2.2 | |
| E138Q | 0.30 ± 0.05 | 0.8 |
Values are means and standard deviations from 3 independent experiments, each performed in triplicate.
Fold change is calculated as the ratio of the mean EC50 of mutant to WT virus.
Tissue RT-SHIV drug resistance detection.
Because no consistent RT mutations were identified in plasma virus from either animal, vRNA isolated from axillary LN and ileum obtained at 35 weeks postinfection was clonally sequenced. Axillary LN were chosen because they were the lymphoid tissues closest to the injection site. Tissues of the gastrointestinal tract were also sampled because they are known to harbor large numbers of infected cells, particularly in the terminal ileum (58). High levels of RT-SHIV RNA copies were measured by quantitative reverse transcription-PCR (qRT-PCR) in both the axillary LN and ileum taken at necropsy from both animals (Table 4). While measurements were not taken prior to drug exposure or during peak plasma drug concentrations, the results suggest that significant viral transcription occurs in the LN and ileum despite high RPV levels at those sites (Table 1).
TABLE 4.
RNA copies of RT-SHIV gag and macaque CD4 measured from tissue RNA
| Animal | Tissuea | No. of copiesb |
gag/CD4 ratio | |
|---|---|---|---|---|
| gag | CD4 | |||
| M170 | Axillary LN | 28 ± 0.5 | 327 ± 10 | 0.09 |
| Ileum | 334 ± 36 | 126,237 ± 58,548 | 0.003 | |
| M171 | Axillary LN | 10 ± 0.8 | 29 ± 2 | 0.3 |
| Ileum | 7,863 ± 367 | 54,343 ± 493 | 0.1 | |
Tissues taken at 35 weeks postinfection.
Means ± standard deviations (×106), measured by qRT-PCR in duplicate.
No known DRMs were detected in 20 clones from either tissue from both animals (Table 5). However, the A33V mutation was identified in 45% of ileum clones from both animals and 80% of axillary LN clones from animal M170. This mutation also was identified in a single clone in the plasma for M170 at week 9 (Table 2). Y181H was present and linked to A33V in two clones from the ileum of animal M171. While Y181H has not previously been reported in association with NNRTI resistance, Y181C, Y181I, and Y181V mutations display RPV resistance (55). Interestingly, we detected G112D in a single clone obtained from one axillary LN from animal M171. This mutation confers 2-fold resistance to RPV and was selected by an RPV analog in vitro (K. Melody and Z. Ambrose, unpublished data).
TABLE 5.
Mutations identified in RT of RT-SHIV RNA isolated from tissues
| Animal | Tissuea | Mutation(s) in RT-SHIV RTb (no. of clones) |
|---|---|---|
| M170 | Axillary LN | WT (1) |
| A33V (10) | ||
| M16I, A33V (2) | ||
| A33V, I135T (2) | ||
| A33V, V179A (1) | ||
| A33V, I195V, R206G (1) | ||
| Q85R, H96R (1) | ||
| P217S (1) | ||
| K220E (1) | ||
| Ileum | WT (8) | |
| A33V (10) | ||
| A33V, D218G (1) | ||
| A33V, V245A (1) | ||
| M171 | Axillary LN | WT (12) |
| N54S (1) | ||
| D76G (1) | ||
| E79G (1) | ||
| G112D (1) | ||
| G155R (1) | ||
| S163G, G196E (1) | ||
| H208R, D250G (1) | ||
| G242. (1) | ||
| Ileum | WT (14) | |
| A33V, Q145R (2) | ||
| A33V, K173R (1) | ||
| A33V, Y181H (2) | ||
| A33V, T200A (1) |
Tissues taken at 35 weeks postinfection.
Sequences represent amino acids 1 to 250 of RT. WT, wild type; ., stop codon.
Due to the high frequency of the A33V mutation in the viruses isolated from tissues and the association of RPV resistance with mutations at position 181, the amino acid substitutions A33V and Y181H were made separately and together in HIV-1 and tested for RPV sensitivity. A33V HIV-1 did not confer RPV resistance, while Y181H alone or with A33V conferred hypersusceptibility to RPV compared to WT HIV-1: 0.3- and 0.2-fold, respectively (Table 3).
Effects of human and macaque plasma on RPV inhibition of HIV-1 and RT-SHIV.
As RPV inhibition of HIV-1 was greatly decreased by the presence of high human serum proteins in vitro (55), the effect of macaque serum on RT-SHIV inhibition by RPV was investigated in cell culture. Similar to previously published results in which 50% human serum increased the EC50 of RPV against HIV-1 18.5-fold (55), 25% human serum reduced RPV inhibition of HIV-1 in vitro in our study 12-fold (Table 6). Similarly, addition of macaque serum increased the EC50 of RPV against RT-SHIV compared to the values with medium with FBS (Table 6). The results suggest that macaque serum proteins do not impact RPV activity against RT-SHIV more than human serum proteins.
TABLE 6.
Effects of human and macaque serum on RPV EC50
| Virus | Serum | EC50 (nM)a | Fold changeb |
|---|---|---|---|
| HIV-1 | 10% FBS | 0.54 ± 0.05 | 1 |
| 5% human | 1.34 ± 0.10 | 3 | |
| 10% human | 2.45 ± 0.44 | 5 | |
| 25% human | 6.72 ± 0.39 | 12 | |
| RT-SHIV | 10% FBS | 0.42 ± 0.08 | 1 |
| 5% macaque | 0.45 ± 0.21 | 1 | |
| 10% macaque | 1.16 ± 0.15 | 3 | |
| 25% macaque | 3.17 ± 1.37 | 8 |
Means ± standard deviations from triplicates from one experiment.
Compared to assay performed with 10% FBS.
Selection of RPV resistance in RT-SHIV was also difficult to achieve in CEMx174 cells using the same virus stock as used to infect the macaques. RT-SHIV-infected cells were exposed to increasing concentrations of RPV for 350 days (0.1 to 409.6 nM), and vRNA isolated from supernatants from all time points had WT sequence in the RT coding region (data not shown). This is in contrast to selection of Y181C or K103N in RT-SHIV in CEMx174 cells by nevirapine (NVP) or efavirenz (EFV), respectively (29).
DISCUSSION
Although global HIV-1 incidence is decreasing in large part due to rollout of ART, an effective vaccine or cure is not yet available. In lieu of a vaccine, FDA-approved PrEP comprised of two NRTIs can reduce infections in high-risk populations (2–5, 7). However, the most significant barrier to PrEP efficacy is patient adherence, as >90% protection is observed in patients with consistently detectable plasma drug concentrations, whereas no protection is seen in participants showing undetectable drug concentrations (2, 4, 6, 8, 59, 60). To improve adherence, long-acting injectable nanoparticle formulations of RPV have been developed and are currently being evaluated in clinical trials (13). However, inappropriate use of PrEP by suboptimal dosing or in individuals who are HIV-1+ may lead to development of drug resistance. The development of DRMs during PrEP could limit future therapy options, as was the case in single-dose NVP trials to prevent mother-to-child transmission of HIV-1 (61).
As there are no currently published data on the effect of RPV LA on HIV-1 resistance selection, we designed a pilot study to explore whether RPV LA monotherapy could select for drug resistance in a preexisting RT-SHIV infection in macaques. Although untreated macaques were not included as a comparison control, the peaks and set points of plasma viremia in the two animals in this study were similar to those in our previous study (30). While RPV LA dosing of and metabolism in macaques were different than those in humans, its administration in this study led to plasma RPV concentrations detected in the animals that were comparable to concentrations reported for humans who received 600-mg RPV LA doses (22, 24). RPV LA treatment displayed a therapeutic effect with approximate 2-log decreases in plasma viremia 1 week after treatment that was sustained for roughly 15 weeks after the first injection, and viremia increased as plasma RPV concentrations dropped below 25 ng/ml (68 nM). Axillary LN and ileum RPV concentrations were at least 4.5-fold greater than plasma concentrations 29 weeks after the first injection, which is consistent with a previous report showing that RPV LA achieves higher concentrations in lymph nodes than in plasma (21). While tissues were not assessed at earlier time points for vRNA and drug concentrations, it is likely that tissue RPV concentrations were higher at earlier time points, and it is clear that drug concentrations were not sufficient to completely suppress virus replication in tissues or blood, as evidenced by high RT-SHIV RNA levels.
Despite waning RPV plasma concentrations and lack of complete virus suppression, which may be seen in noncompliant individuals and suggesting suboptimal in vivo drug inhibition, persistent DRMs were not selected in the plasma or tissues of either animal after RPV LA administration. This is in contrast to our previous studies, in which EFV monotherapy was administered over 4 days in RT-SHIV-infected macaques and rapidly selected the NNRTI resistance mutation K103N in the plasma virus that affected the efficacy of subsequent combination therapy containing EFV, particularly in two animals with high plasma viremia levels similar to the animals in this study (30, 40, 62). Another study using a different strain of RT-SHIV in rhesus macaques also showed that K103N and other DRMs arise during EFV monotherapy (34). The RT mutation K103N confers approximately 20- to 35-fold resistance to HIV-1 in vitro and arises in HIV-infected individuals on EFV-based ART (55, 63).
In contrast, K103N and other single DRMs selected by NNRTIs approved earlier than RPV, such as Y181C, confer no or low-level (i.e., 1- to 3-fold) resistance to RPV (55). K103N was not detected in patients failing RPV-containing ART in two clinical trials (20). In fact, with the exception of K101P, Y181I, andY181V, which confer significant resistance to RPV in vitro (52-, 15-, and 12-fold changes above WT HIV-1, respectively), HIV-1 with any other single NNRTI-associated resistance mutation has no or low-level resistance to RPV (55). Unlike K103N and Y181C, the mutations K101P and Y181I/V require at least two base changes to be made and therefore are likely more difficult to develop. In addition, the accumulation of multiple NNRTI resistance mutations, as occurs in treatment-experienced HIV-1+ individuals, can enhance RPV resistance (20, 55, 64, 65), which is less likely to occur prior to therapy or to be transmitted to newly infected individuals (66). Thus, it appears that isolates that are highly resistant to RPV are difficult to develop compared to mutants that arise during use of other NNRTIs, such as EFV and NVP.
Similarly, in a macaque study investigating sustained release of the novel NNRTI MIV-150 from intravaginal rings (IVR) in RT-SHIV-infected rhesus macaques, DRMs were not detected by clonal sequencing of plasma virus after 42 days in 5/6 animals using the IVR; however, DRM selection did occur when animals were treated systemically with MIV-150 (39). One IVR-treated animal had a single plasma clone containing Y181I, while all other clones were WT. Similar results were found in LN at day 57, and no DRMs were detected in vRNA isolated from the cervix or vagina, the site of drug release.
Recently, one individual unexpectedly became infected with HIV-1 in the lowest RPV LA dose arm (300 mg) of the SSAT 040 trial (1/66 in the overall study and 1/20 in the 300-mg dose arm), which evaluated the pharmacokinetics of the drug in HIV-negative individuals (67). K101E was selected in the RT of plasma HIV-1 after seroconversion (i.e., K101E HIV-1 was not transmitted to this individual), and K101E HIV-1 clones had 4-fold resistance to RPV in vitro compared to that of WT virus. K101E or E138K/Q was the most common RT mutation detected in patients failing RPV-containing therapy in the ECHO, THRIVE, and STaR studies, with E138K constituting the majority of detected NNRTI-associated resistance mutations (20, 68). Molecular modeling studies of RPV bound to the crystal structure of HIV-1 RT show the formation of a salt bridge between E138 and K101 in the WT p66 subunit (69). This interaction was lost with the substitution of the low-level RPV-resistant mutations 138A/G/K/R/Q or 101E, suggesting that disruption of the K101-E138 salt bridge causes low-level RPV resistance (70). A higher prevalence of E138 mutations, particularly E138K/G, in HIV-1 from individuals failing RPV-containing therapy is unknown but may be due to mutational bias (71) or G-to-A hypermutation (72). The E138Q/G mutations that were selected by RPV LA in the macaques in our study and the K101E mutation that was selected in the individual who failed RPV LA were likely stochastic events.
While this is a small pilot study, the data are encouraging that drug resistance may be difficult to develop in HIV-1 RT during RPV LA monotherapy compared to NNRTIs approved prior to RPV. The single DRMs that we did detect remained a minor species despite persistent drug levels. Future studies looking at a larger group of animals with different viremia levels and comparing daily oral RPV dosing to dosing with RPV LA with different lengths of sustained released are warranted to understand how they influence the development of drug resistance. In addition, the effect of development of minority, low-level RPV-resistant viruses on subsequent combination therapy should be addressed.
ACKNOWLEDGMENTS
We thank the veterinary staff, especially Chris Janssen, for animal care and support. In addition, we appreciate the generous contribution of RPV LA by Janssen for these studies and helpful discussions with Peter Williams, Johan Vingerhoets, and Nic Sluis-Cremer. We also thank Kerri Penrose for technical assistance.
Funding for this work was provided by the Bill and Melinda Gates Foundation (grant 1019228 to Z.A. and J.W.M.), the UNC Center for AIDS Research P30 (grant AI50410 to A.D.M.K.), and the National Institutes of Health (T32 grant AI065380 to K.M.).
REFERENCES
- 1.WHO. 2014. Global update on the health sector response to HIV, 2014. WHO, Geneva, Switzerland. [Google Scholar]
- 2.Choopanya K, Martin M, Suntharasamai P, Sangkum U, Mock PA, Leethochawalit M, Chiamwongpaet S, Kitisin P, Natrujirote P, Kittimunkong S, Chuachoowong R, Gvetadze RJ, McNicholl JM, Paxton LA, Curlin ME, Hendrix CW, Vanichseni S, Bangkok Tenofovir Study Group . 2013. Antiretroviral prophylaxis for HIV infection in injecting drug users in Bangkok, Thailand (the Bangkok Tenofovir Study): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet 381:2083–2090. doi: 10.1016/S0140-6736(13)61127-7. [DOI] [PubMed] [Google Scholar]
- 3.Baeten JM, Donnell D, Ndase P, Mugo NR, Campbell JD, Wangisi J, Tappero JW, Bukusi EA, Cohen CR, Katabira E, Ronald A, Tumwesigye E, Were E, Fife KH, Kiarie J, Farquhar C, John-Stewart G, Kakia A, Odoyo J, Mucunguzi A, Nakku-Joloba E, Twesigye R, Ngure K, Apaka C, Tamooh H, Gabona F, Mujugira A, Panteleeff D, Thomas KK, Kidoguchi L, Krows M, Revall J, Morrison S, Haugen H, Emmanuel-Ogier M, Ondrejcek L, Coombs RW, Frenkel L, Hendrix C, Bumpus NN, Bangsberg D, Haberer JE, Stevens WS, Lingappa JR, Celum C, Partners PrEP Study Team. 2012. Antiretroviral prophylaxis for HIV prevention in heterosexual men and women. N Engl J Med 367:399–410. doi: 10.1056/NEJMoa1108524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grant RM, Lama JR, Anderson PL, McMahan V, Liu AY, Vargas L, Goicochea P, Casapia M, Guanira-Carranza JV, Ramirez-Cardich ME, Montoya-Herrera O, Fernandez T, Veloso VG, Buchbinder SP, Chariyalertsak S, Schechter M, Bekker LG, Mayer KH, Kallas EG, Amico KR, Mulligan K, Bushman LR, Hance RJ, Ganoza C, Defechereux P, Postle B, Wang F, McConnell JJ, Zheng JH, Lee J, Rooney JF, Jaffe HS, Martinez AI, Burns DN, Glidden DV, iPrEx Study Team. 2010. Preexposure chemoprophylaxis for HIV prevention in men who have sex with men. N Engl J Med 363:2587–2599. doi: 10.1056/NEJMoa1011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thigpen MC, Kebaabetswe PM, Paxton LA, Smith DK, Rose CE, Segolodi TM, Henderson FL, Pathak SR, Soud FA, Chillag KL, Mutanhaurwa R, Chirwa LI, Kasonde M, Abebe D, Buliva E, Gvetadze RJ, Johnson S, Sukalac T, Thomas VT, Hart C, Johnson JA, Malotte CK, Hendrix CW, Brooks JT, TDF2 Study Group. 2012. Antiretroviral preexposure prophylaxis for heterosexual HIV transmission in Botswana. N Engl J Med 367:423–434. doi: 10.1056/NEJMoa1110711. [DOI] [PubMed] [Google Scholar]
- 6.Van Damme L, Corneli A, Ahmed K, Agot K, Lombaard J, Kapiga S, Malahleha M, Owino F, Manongi R, Onyango J, Temu L, Monedi MC, Mak'Oketch P, Makanda M, Reblin I, Makatu SE, Saylor L, Kiernan H, Kirkendale S, Wong C, Grant R, Kashuba A, Nanda K, Mandala J, Fransen K, Deese J, Crucitti T, Mastro TD, Taylor D, FEM-PrEP Study Group. 2012. Preexposure prophylaxis for HIV infection among African women. N Engl J Med 367:411–422. doi: 10.1056/NEJMoa1202614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdool Karim Q, Abdool Karim SS, Frohlich JA, Grobler AC, Baxter C, Mansoor LE, Kharsany AB, Sibeko S, Mlisana KP, Omar Z, Gengiah TN, Maarschalk S, Arulappan N, Mlotshwa M, Morris L, Taylor D, CAPRISA 004 Trial Group. 2010. Effectiveness and safety of tenofovir gel, an antiretroviral microbicide, for the prevention of HIV infection in women. Science 329:1168–1174. doi: 10.1126/science.1193748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnell D, Baeten JM, Bumpus NN, Brantley J, Bangsberg DR, Haberer JE, Mujugira A, Mugo N, Ndase P, Hendrix C, Celum C. 2014. HIV protective efficacy and correlates of tenofovir blood concentrations in a clinical trial of PrEP for HIV prevention. J Acquir Immune Defic Syndr 66:340–348. doi: 10.1097/QAI.0000000000000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.García-Lerma JG, Otten RA, Qari SH, Jackson E, Cong ME, Masciotra S, Luo W, Kim C, Adams DR, Monsour M, Lipscomb J, Johnson JA, Delinsky D, Schinazi RF, Janssen R, Folks TM, Heneine W. 2008. Prevention of rectal SHIV transmission in macaques by daily or intermittent prophylaxis with emtricitabine and tenofovir. PLoS Med 5:e28. doi: 10.1371/journal.pmed.0050028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radzio J, Aung W, Holder A, Martin A, Sweeney E, Mitchell J, Bachman S, Pau CP, Heneine W, Garcia-Lerma JG. 2012. Prevention of vaginal SHIV transmission in macaques by a coitally-dependent Truvada regimen. PLoS One 7:e50632. doi: 10.1371/journal.pone.0050632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Lerma JG, Cong ME, Mitchell J, Youngpairoj AS, Zheng Q, Masciotra S, Martin A, Kuklenyik Z, Holder A, Lipscomb J, Pau CP, Barr JR, Hanson DL, Otten R, Paxton L, Folks TM, Heneine W. 2010. Intermittent prophylaxis with oral Truvada protects macaques from rectal SHIV infection. Sci Transl Med 2:14ra14. [DOI] [PubMed] [Google Scholar]
- 12.Margolis DA, Boffito M. 2015. Long-acting antiviral agents for HIV treatment. Curr Opin HIV AIDS 10:246–252. doi: 10.1097/COH.0000000000000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson A, McGowan I. 2015. Long-acting rilpivirine for HIV prevention. Curr Opin HIV AIDS 10:253–257. doi: 10.1097/COH.0000000000000160. [DOI] [PubMed] [Google Scholar]
- 14.Bera RB. 2014. Patient outcomes within schizophrenia treatment: a look at the role of long-acting injectable antipsychotics. J Clin Psychiatry 75(Suppl 2):S30–S33. [DOI] [PubMed] [Google Scholar]
- 15.Fennell C, Sartorius G, Ly LP, Turner L, Liu PY, Conway AJ, Handelsman DJ. 2010. Randomized cross-over clinical trial of injectable vs. implantable depot testosterone for maintenance of testosterone replacement therapy in androgen deficient men. Clin Endocrinol (Oxf) 73:102–109. [DOI] [PubMed] [Google Scholar]
- 16.Marberger M, Kaisary AV, Shore ND, Karlin GS, Savulsky C, Mis R, Leuratti C, Germa JR. 2010. Effectiveness, pharmacokinetics, and safety of a new sustained-release leuprolide acetate 3.75-mg depot formulation for testosterone suppression in patients with prostate cancer: a phase III, open-label, international multicenter study. Clin Ther 32:744–757. doi: 10.1016/j.clinthera.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Winner B, Peipert JF, Zhao Q, Buckel C, Madden T, Allsworth JE, Secura GM. 2012. Effectiveness of long-acting reversible contraception. N Engl J Med 366:1998–2007. doi: 10.1056/NEJMoa1110855. [DOI] [PubMed] [Google Scholar]
- 18.Cohen CJ, Molina JM, Cassetti I, Chetchotisakd P, Lazzarin A, Orkin C, Rhame F, Stellbrink HJ, Li T, Crauwels H, Rimsky L, Vanveggel S, Williams P, Boven K, ECHO, THRIVE study groups. 2013. Week 96 efficacy and safety of rilpivirine in treatment-naive, HIV-1 patients in two phase III randomized trials. AIDS 27:939–950. doi: 10.1097/QAD.0b013e32835cee6e. [DOI] [PubMed] [Google Scholar]
- 19.Janssen Therapeutics 2011. Edurant package insert. Janssen Therapeutics, Titusville, NJ. [Google Scholar]
- 20.Rimsky L, Vingerhoets J, Van Eygen V, Eron J, Clotet B, Hoogstoel A, Boven K, Picchio G. 2012. Genotypic and phenotypic characterization of HIV-1 isolates obtained from patients on rilpivirine therapy experiencing virologic failure in the phase 3 ECHO and THRIVE studies: 48-week analysis. J Acquir Immune Defic Syndr 59:39–46. doi: 10.1097/QAI.0b013e31823df4da. [DOI] [PubMed] [Google Scholar]
- 21.van't Klooster G, Hoeben E, Borghys H, Looszova A, Bouche MP, van Velsen F, Baert L. 2010. Pharmacokinetics and disposition of rilpivirine (TMC278) nanosuspension as a long-acting injectable antiretroviral formulation. Antimicrob Agents Chemother 54:2042–2050. doi: 10.1128/AAC.01529-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson AG, Else LJ, Mesquita PM, Egan D, Back DJ, Karolia Z, Ringner-Nackter L, Higgs CJ, Herold BC, Gazzard BG, Boffito M. 2014. A compartmental pharmacokinetic evaluation of long-acting rilpivirine in HIV-negative volunteers for pre-exposure prophylaxis. Clin Pharmacol Ther 96:314–323. doi: 10.1038/clpt.2014.118. [DOI] [PubMed] [Google Scholar]
- 23.Spreen W, Williams P, Margolis D, Ford SL, Crauwels H, Lou Y, Gould E, Stevens M, Piscitelli S. 2014. Pharmacokinetics, safety, and tolerability with repeat doses of GSK1265744 and rilpivirine (TMC278) long-acting nanosuspensions in healthy adults. J Acquir Immune Defic Syndr 67:487–492. doi: 10.1097/QAI.0000000000000365. [DOI] [PubMed] [Google Scholar]
- 24.Verloes R, Deleu S, Niemeijer N, Crauwels H, Meyvisch P, Williams P. 2015. Safety, tolerability and pharmacokinetics of rilpivirine following administration of a long-acting formulation in healthy volunteers. HIV Med 16:477–484. doi: 10.1111/hiv.12247. [DOI] [PubMed] [Google Scholar]
- 25.Wensing AM, Calvez V, Gunthard HF, Johnson VA, Paredes R, Pillay D, Shafer RW, Richman DD. 2014. 2014 update of the drug resistance mutations in HIV-1. Top Antivir Med 22:642–650. [PMC free article] [PubMed] [Google Scholar]
- 26.Chirwa LI, Johnson JA, Niska RW, Segolodi TM, Henderson FL, Rose CE, Li JF, Thigpen MC, Matlhaba O, Paxton LA, Brooks JT. 2014. CD4(+) cell count, viral load, and drug resistance patterns among heterosexual breakthrough HIV infections in a study of oral preexposure prophylaxis. AIDS 28:223–226. doi: 10.1097/QAD.0000000000000102. [DOI] [PubMed] [Google Scholar]
- 27.Lehman DA, Baeten JM, McCoy CO, Weis JF, Peterson D, Mbara G, Donnell D, Thomas KK, Hendrix CW, Marzinke MA, Frenkel L, Ndase P, Mugo NR, Celum C, Overbaugh J, Matsen FA, Partners PrEP Study Team. 2015. Risk of drug resistance among persons acquiring HIV within a randomized clinical trial of single- or dual-agent preexposure prophylaxis. J Infect Dis 211:1211–1218. doi: 10.1093/infdis/jiu677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liegler T, Abdel-Mohsen M, Bentley LG, Atchison R, Schmidt T, Javier J, Mehrotra M, Eden C, Glidden DV, McMahan V, Anderson PL, Li P, Wong JK, Buchbinder S, Guanira JV, Grant RM, iPrEx Study Team. 2014. HIV-1 drug resistance in the iPrEx preexposure prophylaxis trial. J Infect Dis 210:1217–1227. doi: 10.1093/infdis/jiu233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ambrose Z, Boltz V, Palmer S, Coffin JM, Hughes SH, Kewalramani VN. 2004. In vitro characterization of a simian immunodeficiency virus-human immunodeficiency virus (HIV) chimera expressing HIV type 1 reverse transcriptase to study antiviral resistance in pigtail macaques. J Virol 78:13553–13561. doi: 10.1128/JVI.78.24.13553-13561.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ambrose Z, Palmer S, Boltz VF, Kearney M, Larsen K, Polacino P, Flanary L, Oswald K, Piatak M Jr, Smedley J, Shao W, Bischofberger N, Maldarelli F, Kimata JT, Mellors JW, Hu SL, Coffin JM, Lifson JD, KewalRamani VN. 2007. Suppression of viremia and evolution of human immunodeficiency virus type 1 drug resistance in a macaque model for antiretroviral therapy. J Virol 81:12145–12155. doi: 10.1128/JVI.01301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balzarini J, Weeger M, Camarasa MJ, De Clercq E, Uberla K. 1995. Sensitivity/resistance profile of a simian immunodeficiency virus containing the reverse transcriptase gene of human immunodeficiency virus type 1 (HIV-1) toward the HIV-1-specific non-nucleoside reverse transcriptase inhibitors. Biochem Biophys Res Commun 211:850–856. doi: 10.1006/bbrc.1995.1890. [DOI] [PubMed] [Google Scholar]
- 32.Uberla K, Stahl-Hennig C, Bottiger D, Matz-Rensing K, Kaup FJ, Li J, Haseltine WA, Fleckenstein B, Hunsmann G, Oberg B, Sodroski J. 1995. Animal model for the therapy of acquired immunodeficiency syndrome with reverse transcriptase inhibitors. Proc Natl Acad Sci U S A 92:8210–8214. doi: 10.1073/pnas.92.18.8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ambrose Z, Kline C, Polacino P, Hu SL. 2014. Dysregulation of multiple inflammatory molecules in lymph node and ileum of macaques during RT-SHIV infection with or without antiretroviral therapy. J Med Primatol 43:298–309. doi: 10.1111/jmp.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hofman MJ, Higgins J, Matthews TB, Pedersen NC, Tan C, Schinazi RF, North TW. 2004. Efavirenz therapy in rhesus macaques infected with a chimera of simian immunodeficiency virus containing reverse transcriptase from human immunodeficiency virus type 1. Antimicrob Agents Chemother 48:3483–3490. doi: 10.1128/AAC.48.9.3483-3490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caron M, Besson G, Etenna SL, Mintsa-Ndong A, Mourtas S, Radaelli A, Morghen Cde G, Loddo R, La Colla P, Antimisiaris SG, Kazanji M. 2010. Protective properties of non-nucleoside reverse transcriptase inhibitor (MC1220) incorporated into liposome against intravaginal challenge of rhesus macaques with RT-SHIV. Virology 405:225–233. doi: 10.1016/j.virol.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Aravantinou M, Singer R, Derby N, Calenda G, Mawson P, Abraham CJ, Menon R, Seidor S, Goldman D, Kenney J, Villegas G, Gettie A, Blanchard J, Lifson JD, Piatak M Jr, Fernandez-Romero JA, Zydowsky TM, Teleshova N, Robbiani M. 2012. The nonnucleoside reverse transcription inhibitor MIV-160 delivered from an intravaginal ring, but not from a carrageenan gel, protects against simian/human immunodeficiency virus-RT Infection. AIDS Res Hum Retroviruses 28:1467–1475. doi: 10.1089/aid.2012.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang Y, Tian B, Agy MB, Saifuddin M, Tsai CC. 2009. Macaca fascicularis are highly susceptible to an RT-SHIV following intravaginal inoculation: a new model for microbicide evaluation. J Med Primatol 38(Suppl 1):S39–S46. doi: 10.1111/j.1600-0684.2009.00374.x. [DOI] [PubMed] [Google Scholar]
- 38.Jiang Y, Tian B, Saifuddin M, Agy MB, Emau P, Cairns JS, Tsai CC. 2009. RT-SHIV, an infectious CCR5-tropic chimeric virus suitable for evaluating HIV reverse transcriptase inhibitors in macaque models. AIDS Res Ther 6:23. doi: 10.1186/1742-6405-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu M, Keele BF, Aravantinou M, Krawczyk N, Seidor S, Abraham CJ, Zhang S, Rodriguez A, Kizima L, Derby N, Jean-Pierre N, Mizenina O, Gettie A, Grasperge B, Blanchard J, Piatak MJ Jr, Lifson JD, Fernandez-Romero JA, Zydowsky TM, Robbiani M. 2014. Exposure to MIV-150 from a high-dose intravaginal ring results in limited emergence of drug resistance mutations in SHIV-RT infected rhesus macaques. PLoS One 9:e89300. doi: 10.1371/journal.pone.0089300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kearney M, Spindler J, Shao W, Maldarelli F, Palmer S, Hu SL, Lifson JD, KewalRamani VN, Mellors JW, Coffin JM, Ambrose Z. 2011. Genetic diversity of simian immunodeficiency virus encoding HIV-1 reverse transcriptase persists in macaques despite antiretroviral therapy. J Virol 85:1067–1076. doi: 10.1128/JVI.01701-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kenney J, Aravantinou M, Singer R, Hsu M, Rodriguez A, Kizima L, Abraham CJ, Menon R, Seidor S, Chudolij A, Gettie A, Blanchard J, Lifson JD, Piatak M Jr, Fernandez-Romero JA, Zydowsky TM, Robbiani M. 2011. An antiretroviral/zinc combination gel provides 24 hours of complete protection against vaginal SHIV infection in macaques. PLoS One 6:e15835. doi: 10.1371/journal.pone.0015835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.North TW, Van Rompay KK, Higgins J, Matthews TB, Wadford DA, Pedersen NC, Schinazi RF. 2005. Suppression of virus load by highly active antiretroviral therapy in rhesus macaques infected with a recombinant simian immunodeficiency virus containing reverse transcriptase from human immunodeficiency virus type 1. J Virol 79:7349–7354. doi: 10.1128/JVI.79.12.7349-7354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.North TW, Higgins J, Deere JD, Hayes TL, Villalobos A, Adamson L, Shacklett BL, Schinazi RF, Luciw PA. 2010. Viral sanctuaries during highly active antiretroviral therapy in a nonhuman primate model for AIDS. J Virol 84:2913–2922. doi: 10.1128/JVI.02356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shao W, Kearney M, Maldarelli F, Mellors JW, Stephens RM, Lifson JD, KewalRamani VN, Ambrose Z, Coffin JM, Palmer SE. 2009. RT-SHIV subpopulation dynamics in infected macaques during anti-HIV therapy. Retrovirology 6:101. doi: 10.1186/1742-4690-6-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singer R, Derby N, Rodriguez A, Kizima L, Kenney J, Aravantinou M, Chudolij A, Gettie A, Blanchard J, Lifson JD, Piatak M Jr, Fernandez-Romero JA, Zydowsky TM, Robbiani M. 2011. The nonnucleoside reverse transcriptase inhibitor MIV-150 in carrageenan gel prevents rectal transmission of simian/human immunodeficiency virus infection in macaques. J Virol 85:5504–5512. doi: 10.1128/JVI.02422-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singer R, Mawson P, Derby N, Rodriguez A, Kizima L, Menon R, Goldman D, Kenney J, Aravantinou M, Seidor S, Gettie A, Blanchard J, Piatak M Jr, Lifson JD, Fernandez-Romero JA, Robbiani M, Zydowsky TM. 2012. An intravaginal ring that releases the NNRTI MIV-150 reduces SHIV transmission in macaques. Sci Transl Med 4:150ra123. doi: 10.1126/scitranslmed.3003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turville SG, Aravantinou M, Miller T, Kenney J, Teitelbaum A, Hu L, Chudolij A, Zydowsky TM, Piatak M Jr, Bess JW Jr, Lifson JD, Blanchard J, Gettie A, Robbiani M. 2008. Efficacy of Carraguard-based microbicides in vivo despite variable in vitro activity. PLoS One 3:e3162. doi: 10.1371/journal.pone.0003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wadford DA, Kauffman RC, Deere JD, Aoki ST, Stanton RA, Higgins J, Van Rompay KK, Villalobos A, Nettles JH, Schinazi RF, Pedersen NC, North TW. 2014. Variation of human immunodeficiency virus type-1 reverse transcriptase within the simian immunodeficiency virus genome of RT-SHIV. PLoS One 9:e86997. doi: 10.1371/journal.pone.0086997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol 74:8358–8367. doi: 10.1128/JVI.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cecilia D, KewalRamani VN, O'Leary J, Volsky B, Nyambi P, Burda S, Xu S, Littman DR, Zolla-Pazner S. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J Virol 72:6988–6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi C, Mellors JW. 1997. A recombinant retroviral system for rapid in vivo analysis of human immunodeficiency virus type 1 susceptibility to reverse transcriptase inhibitors. Antimicrob Agents Chemother 41:2781–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kimpton J, Emerman M. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol 66:2232–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cline AN, Bess JW Jr, Piatak M Jr, Lifson JD. 2005. Highly sensitive SIV plasma viral load assay: practival considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J Med Primatol 34:303–312. doi: 10.1111/j.1600-0684.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 54.Kline C, Ndjomou J, Franks T, Kiser R, Coalter V, Smedley J, Piatak M Jr, Mellors JW, Lifson JD, Ambrose Z. 2013. Persistence of viral reservoirs in multiple tissues after antiretroviral therapy suppression in a macaque RT-SHIV model. PLoS One 8:e84275. doi: 10.1371/journal.pone.0084275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Azijn H, Tirry I, Vingerhoets J, de Bethune MP, Kraus G, Boven K, Jochmans D, Van Craenenbroeck E, Picchio G, Rimsky LT. 2010. TMC278, a next-generation nonnucleoside reverse transcriptase inhibitor (NNRTI), active against wild-type and NNRTI-resistant HIV-1. Antimicrob Agents Chemother 54:718–727. doi: 10.1128/AAC.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Janssen PA, Lewi PJ, Arnold E, Daeyaert F, de Jonge M, Heeres J, Koymans L, Vinkers M, Guillemont J, Pasquier E, Kukla M, Ludovici D, Andries K, de Bethune MP, Pauwels R, Das K, Clark AD Jr, Frenkel YV, Hughes SH, Medaer B, De Knaep F, Bohets H, De Clerck F, Lampo A, Williams P, Stoffels P. 2005. In search of a novel anti-HIV drug: multidisciplinary coordination in the discovery of 4-[[4-[[4-[(1E)-2-cyanoethenyl]-2,6-dimethylphenyl]amino]-2-pyrimidinyl]amino]benzonitrile (R278474, rilpivirine). J Med Chem 48:1901–1909. doi: 10.1021/jm040840e. [DOI] [PubMed] [Google Scholar]
- 57.Sluis-Cremer N, Huber KD, Brumme CJ, Harrigan PR. 2014. Competitive fitness assays indicate that the E138A substitution in HIV-1 reverse transcriptase decreases in vitro susceptibility to emtricitabine. Antimicrob Agents Chemother 58:2430–2433. doi: 10.1128/AAC.02114-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yukl SA, Gianella S, Sinclair E, Epling L, Li Q, Duan L, Choi AL, Girling V, Ho T, Li P, Fujimoto K, Lampiris H, Hare CB, Pandori M, Haase AT, Gunthard HF, Fischer M, Shergill AK, McQuaid K, Havlir DV, Wong JK. 2010. Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. J Infect Dis 202:1553–1561. doi: 10.1086/656722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mansoor LE, Abdool Karim Q, Yende-Zuma N, MacQueen KM, Baxter C, Madlala BT, Grobler A, Abdool Karim SS. 2014. Adherence in the CAPRISA 004 tenofovir gel microbicide trial. AIDS Behav 18:811–819. doi: 10.1007/s10461-014-0751-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marrazzo JM, Ramjee G, Richardson BA, Gomez K, Mgodi N, Nair G, Palanee T, Nakabiito C, van der Straten A, Noguchi L, Hendrix CW, Dai JY, Ganesh S, Mkhize B, Taljaard M, Parikh UM, Piper J, Masse B, Grossman C, Rooney J, Schwartz JL, Watts H, Marzinke MA, Hillier SL, McGowan IM, Chirenje ZM, VOICE Study Team. 2015. Tenofovir-based preexposure prophylaxis for HIV infection among African women. N Engl J Med 372:509–518. doi: 10.1056/NEJMoa1402269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ton Q, Frenkel L. 2013. HIV drug resistance in mothers and infants following use of antiretrovirals to prevent mother-to-child transmission. Curr HIV Res 11:126–136. doi: 10.2174/1570162X11311020005. [DOI] [PubMed] [Google Scholar]
- 62.Boltz VF, Ambrose Z, Kearney MF, Shao W, Kewalramani VN, Maldarelli F, Mellors JW, Coffin JM. 2012. Ultrasensitive allele-specific PCR reveals rare preexisting drug-resistant variants and a large replicating virus population in macaques infected with a simian immunodeficiency virus containing human immunodeficiency virus reverse transcriptase. J Virol 86:12525–12530. doi: 10.1128/JVI.01963-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bacheler L, Jeffrey S, Hanna G, D'Aquila R, Wallace L, Logue K, Cordova B, Hertogs K, Larder B, Buckery R, Baker D, Gallagher K, Scarnati H, Tritch R, Rizzo C. 2001. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J Virol 75:4999–5008. doi: 10.1128/JVI.75.11.4999-5008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson BC, Pauly GT, Rai G, Patel D, Bauman JD, Baker HL, Das K, Schneider JP, Maloney DJ, Arnold E, Thomas CJ, Hughes SH. 2012. A comparison of the ability of rilpivirine (TMC278) and selected analogues to inhibit clinically relevant HIV-1 reverse transcriptase mutants. Retrovirology 9:99. doi: 10.1186/1742-4690-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Asahchop EL, Wainberg MA, Oliveira M, Xu H, Brenner BG, Moisi D, Ibanescu IR, Tremblay C. 2013. Distinct resistance patterns to etravirine and rilpivirine in viruses containing nonnucleoside reverse transcriptase inhibitor mutations at baseline. AIDS 27:879–887. doi: 10.1097/QAD.0b013e32835d9f6d. [DOI] [PubMed] [Google Scholar]
- 66.Rhee SY, Blanco JL, Jordan MR, Taylor J, Lemey P, Varghese V, Hamers RL, Bertagnolio S, Rinke de Wit TF, Aghokeng AF, Albert J, Avi R, Avila-Rios S, Bessong PO, Brooks JI, Boucher CA, Brumme ZL, Busch MP, Bussmann H, Chaix ML, Chin BS, D'Aquin TT, De Gascun CF, Derache A, Descamps D, Deshpande AK, Djoko CF, Eshleman SH, Fleury H, Frange P, Fujisaki S, Harrigan PR, Hattori J, Holguin A, Hunt GM, Ichimura H, Kaleebu P, Katzenstein D, Kiertiburanakul S, Kim JH, Kim SS, Li Y, Lutsar I, Morris L, Ndembi N, Ng KP, Paranjape RS, Peeters M, Poljak M, Price MA, Ragonnet-Cronin ML, Reyes-Terán G, Rolland M, Sirivichayakul S, Smith DM, Soares MA, Soriano VV, Ssemwanga D, Stanojevic M, Stefani MA, Sugiura W, Sungkanuparph S, Tanuri A, Tee KK, Truong HH, van de Vijver DA, Vidal N, Yang C, Yang R, Yebra G, Ioannidis JP, Vandamme AM, Shafer RW. 2015. Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: an individual-patient- and sequence-level meta-analysis. PLoS Med 12:e1001810. doi: 10.1371/journal.pmed.1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Penrose KJ, Parikh UM, Hamanishi KA, Panousis C, Else L, Back D, Boffito M, Jackson A, Mellors JW. 2014. Selection of rilpivirine resistant HIV-1 in a seroconverter on long-acting rilpivirine (TMC278) from the lowest dose arm of the SSAT 040 trial. AIDS Res Hum Retroviruses 30(S1):A69. doi: 10.1089/aid.2014.5127.abstract. [DOI] [Google Scholar]
- 68.Porter DP, Kulkarni R, Fralich T, Miller MD, White KL. 2015. 96-week resistance analyses of the STaR study: rilpivirine/emtricitabine/tenofovir DF versus efavirenz/emtricitabine/tenofovir DF in antiretroviral-naive, HIV-1-infected subjects. HIV Clin Trials 16:30–38. doi: 10.1179/1528433614Z.0000000009. [DOI] [PubMed] [Google Scholar]
- 69.Ren J, Nichols CE, Stamp A, Chamberlain PP, Ferris R, Weaver KL, Short SA, Stammers DK. 2006. Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138. FEBS J 273:3850–3860. doi: 10.1111/j.1742-4658.2006.05392.x. [DOI] [PubMed] [Google Scholar]
- 70.Kulkarni R, Babaoglu K, Lansdon EB, Rimsky L, Van Eygen V, Picchio G, Svarovskaia E, Miller MD, White KL. 2012. The HIV-1 reverse transcriptase M184I mutation enhances the E138K-associated resistance to rilpivirine and decreases viral fitness. J Acquir Immune Defic Syndr 59:47–54. doi: 10.1097/QAI.0b013e31823aca74. [DOI] [PubMed] [Google Scholar]
- 71.McCallum M, Oliveira M, Ibanescu RI, Kramer VG, Moisi D, Asahchop EL, Brenner BG, Harrigan PR, Xu H, Wainberg MA. 2013. Basis for early and preferential selection of the E138K mutation in HIV-1 reverse transcriptase. Antimicrob Agents Chemother 57:4681–4688. doi: 10.1128/AAC.01029-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu HT, Colby-Germinario SP, Asahchop EL, Oliveira M, McCallum M, Schader SM, Han Y, Quan Y, Sarafianos SG, Wainberg MA. 2013. Effect of mutations at position E138 in HIV-1 reverse transcriptase and their interactions with the M184I mutation on defining patterns of resistance to nonnucleoside reverse transcriptase inhibitors rilpivirine and etravirine. Antimicrob Agents Chemother 57:3100–3109. doi: 10.1128/AAC.00348-13. [DOI] [PMC free article] [PubMed] [Google Scholar]