Abstract
Simeprevir (TMC435) is a once-daily, single-pill, oral hepatitis C virus (HCV) NS3 protease inhibitor approved for the treatment of chronic HCV infection. Phenotypic characterization of baseline isolates and isolates from HCV genotype 1-infected patients failing with a simeprevir-based regimen was performed using chimeric replicons carrying patient-derived NS3 protease sequences. Cutoff values differentiating between full susceptibility to simeprevir (≤2.0-fold reduction in simeprevir activity) and low-level versus high-level resistance (≥50-fold reduction in simeprevir activity) were determined. The median simeprevir fold change in the 50% effective concentration (FC) of pretreatment genotype 1a isolates, with and without Q80K, and genotype 1b isolates was 11, 0.9, and 0.4, respectively. Naturally occurring NS3 polymorphisms that reduced simeprevir activity, other than Q80K, were uncommon in the simeprevir studies and generally conferred low-level resistance in vitro. Although the proportion of patients with failure differed by HCV geno/subtype and/or presence of baseline Q80K, the level of simeprevir resistance observed at failure was similarly high irrespective of type of failure, HCV genotype 1 subtype, and presence or absence of baseline Q80K. At the end of the study, simeprevir activity against isolates that lost the emerging amino acid substitution returned to pretreatment values. Activity of simeprevir against clinical isolates and site-directed mutant replicons harboring the corresponding single or double amino acid substitutions correlated well, showing that simeprevir resistance can be attributed to these substitutions. In conclusion, pretreatment NS3 isolates were generally fully susceptible (FC, ≤2.0) or conferred low-level resistance to simeprevir in vitro (FC, >2.0 and <50). Treatment failure with a simeprevir-based regimen was associated with emergence of high-level-resistance variants (FC, ≥50).
INTRODUCTION
Currently, multiple direct-acting antiviral agents (DAAs) with different mechanisms of action are approved, and this has revolutionized the treatment of chronic hepatitis C virus (HCV) infection (1). Simeprevir (TMC435) is a one-pill, once-daily, oral HCV NS3/4A protease inhibitor approved for the treatment of chronic hepatitis C infection. In clinical studies, simeprevir 150 mg in combination with peginterferon and ribavirin (PegIFN/RBV) significantly improved sustained virologic response (SVR) rates in treatment-naive and treatment-experienced patients with chronic HCV genotype 1 infection versus PegIFN/RBV alone and enabled a shorter, 24-week overall treatment duration in treatment-naive patients and prior relapsers (2–4). Simeprevir in combination with sofosbuvir given for 12 or 24 weeks with or without RBV resulted in high SVR rates in traditionally difficult-to-cure HCV genotype 1-infected patients (5).
The HCV NS5B polymerase has low fidelity, which, combined with the high replication rate of the virus, results in high genetic variability (6). Naturally occurring variants with DAA-resistant amino acid substitutions have been described for NS3 protease, NS5A protein, and the NS5B polymerase region and may affect treatment outcome (7, 8). During DAA treatment, resistant mutations can emerge in the gene encoding the protein targeted by the drug in patients not achieving SVR. For simeprevir, the amino acid substitutions identified in patients failing treatment with simeprevir plus PegIFN/RBV were mainly located at NS3 positions 80, 122, 155, and/or 168 (9). These emerging substitutions were no longer detected in a substantial proportion of patients after treatment was stopped, suggesting that the substitutions reduce the fitness of the virus in the absence of drug pressure (9).
Viral resistance analysis is commonly used during development programs of antivirals to characterize the resistance profile of the drug. Resistance analysis includes sequencing of the viral target gene and phenotypic assessment of drug susceptibility, which together provide complementary information on the presence or emergence of amino acid substitutions affecting the activity of the antiviral. For the treatment of viral infections such as human immunodeficiency virus (HIV) infection and, to some extent, influenza and hepatitis B virus infections, drug-resistance testing has proved to be a useful tool in the management of patients (10–12).
In this study, the activity of simeprevir against chimeric replicons carrying NS3 sequences derived from clinical isolates of HCV genotype 1-infected patients enrolled in phase 1 to phase 3 clinical studies is described. The relationship between the presence of amino acid substitutions in clinical isolates and the in vitro susceptibility of the isolates to simeprevir was investigated, and cutoff values were determined to differentiate clinical isolates fully susceptible to simeprevir from isolates with low-level or high-level resistance to simeprevir.
MATERIALS AND METHODS
Sample selection.
Isolates collected pretreatment, at the time of failure, at the end of the study, and/or at other time points during the study of HCV genotype 1-infected patients naive to HCV NS3/4A protease inhibitors who received simeprevir alone (clinical studies TMC435-C101 [13] and -C201 [14]) or who were treated with simeprevir in combination with PegIFN/RBV (clinical studies TMC-C201, -C205 [15], -C206 [16], -C208 [3], -C216 [2], and HPC3007 [4]) were selected for phenotypic analysis. In addition, 4 pretreatment isolates from 4 patients enrolled in the placebo arm of clinical study TMC435-C201 were analyzed.
Results were available for a total of 522 clinical isolates, and results from 465 clinical isolates from 241 prior protease inhibitor treatment-naive HCV genotype 1-infected patients (142 genotype 1a, 97 genotype 1b, and 2 genotype 1/other) were included in this analyses (Table 1). Of the 465 clinical isolates, 224 were obtained at baseline and/or screening (i.e., pretreatment).
TABLE 1.
Overview of clinical isolates derived from HCV genotype 1-infected patients analyzed in the chimeric replicon assaya
| Parameter | PBO | Simeprevir monotherapy |
Simeprevir plus PegIFN/RBV |
Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C101b | C201b,c,d,e | C201b,c,d,e,f | C205d,e | C206e,f | C208e | C216e | HPC3007e | |||
| ITT (n) | 6 | 28 | 60 | 309 | 396 | 264 | 257 | 260 | 1,580 | |
| Patients with phenotyping data (n) | 4 | 6 | 13 | 33 | 61 | 34 | 34 | 28 | 28 | 241 |
| Genotype 1a | 3 | 4 | 4 | 17 | 38 | 18 | 23 | 13 | 22 | 142 |
| Genotype 1b | 1 | 2 | 8 | 16 | 23 | 15 | 11 | 15 | 6 | 97 |
| Genotype 1/otherg | 1 | 1 | 2 | |||||||
| Total clinical isolates analyzed | 4 | 18 | 24 | 57 | 109 | 73 | 63 | 59 | 58 | 465 |
| Pretreatmenth | 4 | 5 | 12 | 29 | 59 | 30 | 32 | 26 | 27 | 224 |
| Postbaseline | 13 | 12 | 28 | 50 | 43 | 31 | 33 | 31 | 241 | |
HCV, hepatitis C virus; ITT, intent-to-treat population (patients who received at least 1 dose of simeprevir); PBO, placebo; PegIFN, peginterferon; RBV, ribavirin. C101, C201, etc., refer to the clinical trials described in Materials and Methods.
Simeprevir 200 mg.
Simeprevir 25 mg.
Simeprevir 75 mg.
Simeprevir 150 mg.
Simeprevir 100 mg.
Includes 1 patient with genotype 1e and 1i, respectively.
If no baseline sample was available for analysis, the screening sample was used.
All studies were conducted in full compliance with the Declaration of Helsinki and Good Clinical Practice guidelines. All patients provided written informed consent before participating in any study-related activity.
HCV NS3 sequence analysis.
The NS3 protease domain was sequenced using standard Sanger population sequencing (17). Briefly, viral RNA was isolated from plasma and the HCV NS3/4A protease region was amplified using a nested reverse-transcription PCR assay. The resulting DNA was purified and sequenced directly using Sanger population sequencing.
Phenotypic characterization using transient replicon assay.
The 2 transient replicon assay approaches used in this study were described earlier (18, 19). In brief, for analysis of the individual effect of NS3 amino acid substitutions on simeprevir activity in vitro, the NS3 mutations of interest were engineered in a genotype 1a or genotype 1b replicon backbone via site-directed mutagenesis (19). For analysis of patient-derived NS3 protease sequences, sequences from the NS3 protease domain (amino acid 7 to 192) derived from patient isolates were amplified and inserted into a replication-deficient HCV genotype 1b replicon backbone generating chimeric replicons (18). The NS3 region of each newly generated chimeric replicon was sequenced. For each site-directed mutant (SDM) and chimeric replicon, the in vitro susceptibility to simeprevir was determined in a transient replicon assay and expressed as the 50% effective concentration (EC50). Results from the chimeric replicons were excluded from the analyses when the EC90/EC50 ratio was >5. This ratio was used to exclude isolates with biphasic dose-response curves to avoid misinterpretation of the EC50 value due to the presence of minor variants (20–22). The simeprevir EC50 values for the SDMs and chimeric replicons were compared with the wild-type HCV genotype 1b EC50 value, and the ratios between both EC50 values were expressed as the fold change in the EC50 (FC) values. FC values for SDMs and individual clinical isolates were calculated as median values across experimental replicates. For analyses in which results from individual isolates were grouped, the median of the FC values for the individual isolates was used.
Statistical correlation analysis of isolates and SDMs.
The activity of simeprevir against clinical isolates and SDMs carrying corresponding amino acid substitutions at NS3 positions 43, 80, 122, 155, 156, 168, and/or I/V170T was compared. The intraclass correlation coefficient (ICC) between the SDM and patient isolate FC values is quantified using a linear mixed-effects model on the log-transformed FC values, with the three-way interaction among genotype 1a and 1b, source indicator (SDM versus isolate), and amino acid substitution type as covariates, and the HCV geno/subtype included as a clustering factor. Subsequently, the ICC is obtained as the ratio of the between-cluster variance and the total variance. The uncertainty on the ICC is quantified using the delta approach for the standard error and Fisher's variance-stabilizing transformation, from which the 95% confidence intervals can readily be obtained. For more detail, see Vangeneugden et al. (23).
RESULTS
Biological cutoff.
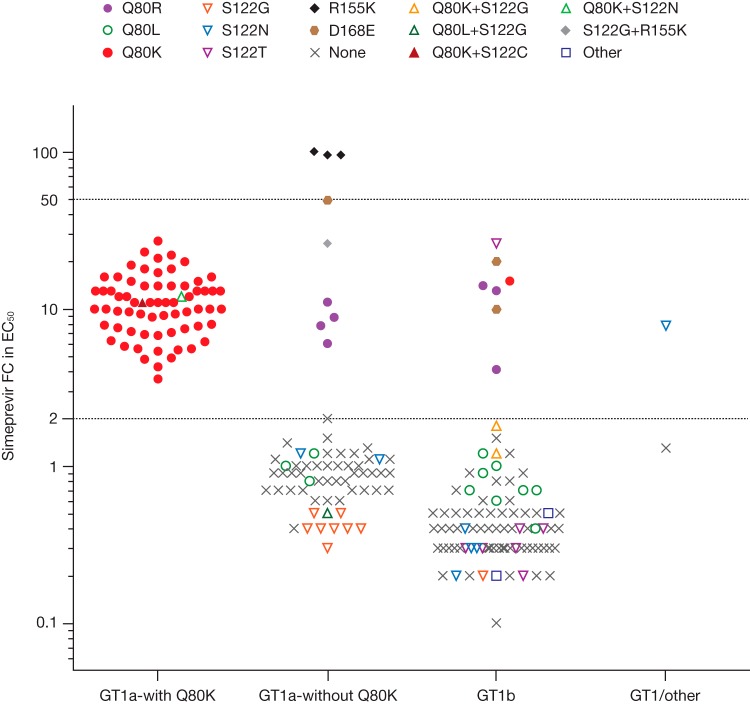
To differentiate isolates susceptible to simeprevir from those with reduced simeprevir susceptibility, a biological cutoff FC value was determined. This cutoff FC value was based on the natural distribution of the simeprevir FC values for chimeric replicons containing NS3 sequences from clinical isolates obtained pretreatment with no amino acid substitution at NS3 positions 43, 80, 122, 155, 156, and/or 168. Amino acid substitutions at these positions are known to reduce simeprevir activity in vitro and were found to emerge during in vitro selection experiments (19). Forty-four of 134 genotype 1a, 58 of 88 genotype 1b, and 1 of 2 genotype 1/other pretreatment clinical isolates had no baseline polymorphism at any of the aforementioned positions (Fig. 1). The median simeprevir FC values against these genotype1a, 1b, and 1/other clinical isolates were 0.9 (range, 0.4 to 2.0), 0.4 (range, 0.1 to 1.5), and 1.3 (not applicable), respectively (Table 2). Because the highest observed FC value was 2.0, this was considered the biological cutoff. Hence, FC values of >2.0 were considered indicative of reduced activity of simeprevir in vitro. Analysis of simeprevir activity against wild-type genotype 1a replicons and genotype 1b replicons carrying amino acid substitutions in the NS5A or NS5B region, which were not expected to affect simeprevir activity (NS5A positions 28, 30, 31, and 93 [n = 6] and NS5B positions 96, 142, 282, 316, 414, 415, 423, 448, 495, 496, and 559 [n = 20]), showed FC values of ≤2.1, supporting this biological cutoff value (see Table S1 in the supplemental material).
FIG 1.
Activity of simeprevir against chimeric replicons carrying NS3 sequences derived from pretreatment isolates of hepatitis C virus (HCV) genotype (GT) 1-infected patients by baseline polymorphism (NS3 positions 43, 80, 122, 155, 156, and/or 168). Each data point represents the median simeprevir fold-change (FC) value in 50% effective concentration (EC50) calculated from 1 to 5 repeat transient chimeric replicon experiments of the same clinical isolate (the majority [168 of 224] of clinical isolates depicted in this graph were tested ≥3 times).
TABLE 2.
Activity of simeprevir against chimeric replicons carrying NS3 sequences derived from pretreatment isolates of HCV genotype 1-infected patientsa
| Genotype | N | Simeprevir FC in EC50b |
||
|---|---|---|---|---|
| Median | Q1–Q3 | Range (min. to max.) | ||
| Genotype 1a | 134 | 5.9 | 0.9–12 | 0.3–100 |
| With Q80K | 67 | 11 | 7.6–14 | 3.6–27 |
| Without Q80K | 67 | 0.9 | 0.7–1.2 | 0.3–100 |
| No polymorphisms at any of NS3 positions 43, 80, 122, 155, 156, 168 | 44 | 0.9 | 0.7–1.1 | 0.4–2.0 |
| Genotype 1b | 88 | 0.4 | 0.3–0.7 | 0.1–26 |
| No polymorphisms at any of NS3 positions 43, 80, 122, 155, 156, 168 | 58 | 0.4 | 0.3–0.5 | 0.1–1.5 |
| Genotype 1/otherc | 2 | 4.6 | NA | 1.3–7.8 |
| No polymorphisms at any of NS3 positions 43, 80, 122, 155, 156, 168 | 1 | 1.3 | NA | NA |
EC50, 50% effective concentration; FC, fold change; HCV, hepatitis C virus; N, number of isolates analyzed; NA, not applicable; Q1–Q3, range of values between quartiles 1 and 3 (only provided if N ≥ 3).
Compared with reference genotype 1b wild-type replicon.
One was infected with genotype 1e and one with genotype 1i.
Activity of simeprevir against clinical isolates obtained pretreatment.
The activity of simeprevir against chimeric replicons carrying NS3 sequences obtained pretreatment was analyzed (Fig. 1; Table 2).
The median simeprevir FC value for genotype 1a isolates was 5.9 and for genotype 1b isolates was 0.4. The median simeprevir FC was 11 for the genotype 1a isolates with Q80K and 0.9 for genotype 1a isolates without Q80K. Similar median simeprevir FC values were obtained for genotype 1a isolates with Q80K polymorphism from North America (FC, 11), Europe (FC, 12), and Australia/New Zealand (FC, 7.2) (see Table S2 in the supplemental material).
Pretreatment isolates carrying polymorphisms at NS3 positions 43, 80, 122, 155, 156, and/or 168 with reduced susceptibility to simeprevir, other than Q80K, included isolates with NS3 polymorphisms Q80R (median FC, 8.3 [genotype 1a; n = 4] and 13 [genotype 1b; n = 3]), R155K (FC, 95 [genotype 1a; n = 3]), S122G plus R155K (FC, 26 [genotype 1a; n = 1]), and D168E (FC, 49 [genotype 1a; n = 1] and 15 [genotype 1b; n = 2]). No reduction in simeprevir activity (FC, ≤2.0) was observed for pretreatment genotype 1a and 1b isolates that carried polymorphisms Q80L and S122G or N. Simeprevir remained fully active against 6 of 7 isolates carrying S122T pretreatment (FC, ≤2.0); the reason for the higher FC value (FC, 26) of the 1 remaining isolate is not understood, since no additional amino acid substitutions were identified by population sequencing. Of note, subsequent deep sequencing analyses of another pretreatment isolate from this patient did not identify additional amino acid substitutions. Three pretreatment genotype 1b isolates with Q80K were tested; 1 had a single Q80K, which was associated with a 15-fold reduction in simeprevir activity, while the other 2 isolates carrying Q80K and S122G did not impact simeprevir activity (FC, ≤2.0).
Two pretreatment isolates with a genotype 1 subtype other than 1a or 1b were analyzed in the chimeric replicon assay (Table 2). The isolate from a genotype 1e-infected patient (NS3 amino acid substitutions V36L, T54S, S122N, I132V, and I170V, when considering an extended list of amino acid positions) resulted in a 7.8-fold reduction in simeprevir activity, while the genotype 1i isolate (no baseline polymorphisms) displayed no reduction (FC, ≤2.0). With the exception of I132V, which reduced simeprevir activity 2.3-fold when tested as an SDM in a genotype 1a replicon backbone, none of the other polymorphisms in the genotype 1e isolate reduced simeprevir activity in vitro.
Cutoff FC value differentiating low-level and high-level resistance to simeprevir.
To determine an FC cutoff value for differentiating amino acid substitutions with low-level resistance to simeprevir from those with high-level resistance, the clinical isolates obtained at the time of failure (i.e., on-treatment failure or viral relapse) in the 150 mg simeprevir dosing groups of clinical studies TMC435-C205, -C206, -C208, -C216, and HPC3007 were tested in the chimeric replicon assay. In addition, the emerging amino acid substitutions observed in patients who failed simeprevir treatment were tested as SDMs in a genotype 1a or 1b replicon background. The threshold for high-level resistance was defined as the lowest simeprevir FC value observed with SDMs or clinical isolates carrying amino acid substitutions commonly observed at time of failure.
The lowest simeprevir FC values in this analysis were observed for genotype 1a patients without baseline Q80K and a single emerging R155K, with a median simeprevir FC value of 73 (n = 11; range, 54 to 222). The amino acid substitution R155K resulted in a median FC value of 88 (range, 60 to 116) when tested as an SDM in a genotype 1a replicon backbone (see Table S3 in the supplemental material). Given the lower end of the range (54) for the FC against the clinical isolates, a simeprevir FC value of 50 was selected to differentiate isolates or SDMs with low-level resistance from those with high-level resistance to simeprevir.
Activity of simeprevir against isolates obtained at time of failure and end of study.
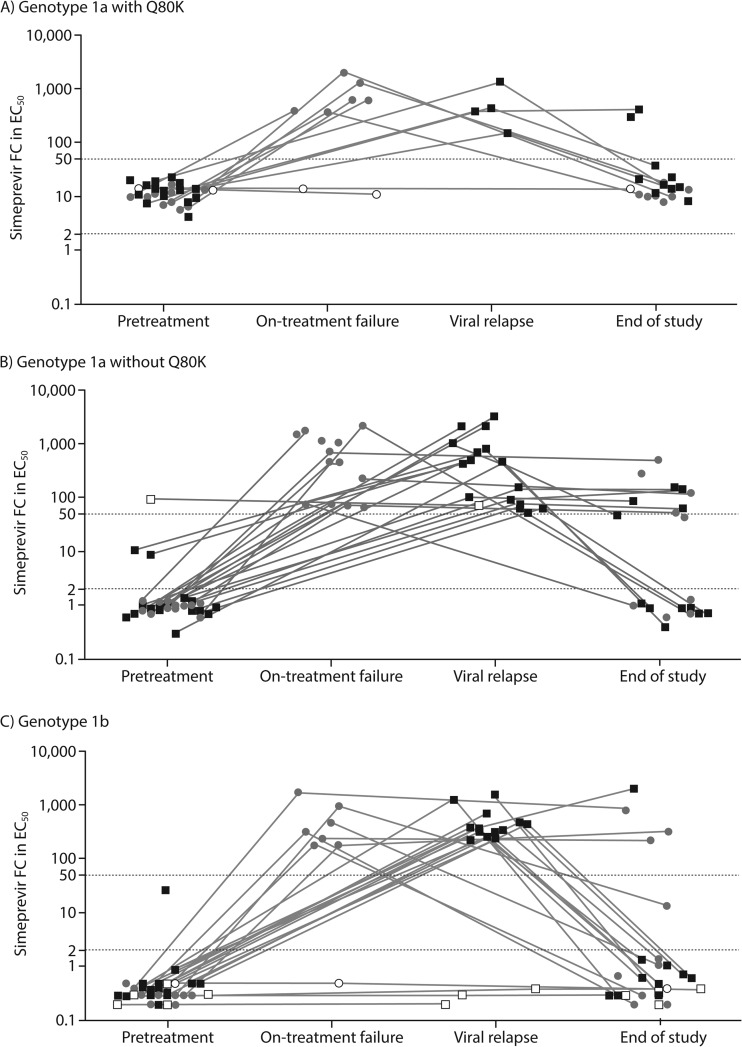
The activity of simeprevir was assessed against NS3 isolates obtained pretreatment, at the time of failure, and at the end of the study from patients with treatment failure (i.e., on-treatment failure or viral relapse) who were enrolled in the 150 mg simeprevir plus PegIFN/RBV groups of clinical studies TMC435-C205, -C206, -C208, -C216, and HPC3007 (Fig. 2; see also Table S4 in the supplemental material).
FIG 2.
Activity of simeprevir against chimeric replicons carrying NS3 sequences derived from isolates obtained pretreatment, at the time of failure (i.e., on-treatment failure and viral relapse), and at the end of the study from HCV genotype 1-infected patients treated with simeprevir 150 mg plus PegIFN/RBV who did not achieve sustained virologic response, by HCV geno/subtype. Gray circles, on-treatment failure, i.e., patients with confirmed detectable HCV RNA at the end of treatment; black squares, viral relapse, i.e., patients with undetectable HCV RNA at the end of treatment and last HCV RNA measurement during follow-up of ≥25 IU/ml; filled and open symbols, patients with and without emerging amino acid substitutions at the time of failure; end of study, last available time point with phenotype data available during follow-up. Gray lines connect pretreatment, time of failure, and/or end of study phenotype data obtained from individual patients. Each data point represents the median simeprevir FC in EC50 calculated from 1 to 6 repeat transient chimeric replicon experiments of the same clinical isolate (the majority [180 of 221] of clinical isolates shown in this graph were tested ≥3 times). EC50, 50% effective concentration; FC, fold change; HCV, hepatitis C virus.
Phenotypic data at the time of failure were available for 60 of 182 150-mg-simeprevir-treated patients with treatment failure and emerging amino acid substitutions at NS3 positions 80, 122, 155, 168, and/or I/V170T and for 7 of 17 patients with failure without any emerging amino acid substitution at these positions (9). An increase in simeprevir FC values at the time of failure compared with pretreatment was observed in most patients with treatment failure, consistent with sequencing results showing emerging amino acid substitutions in these patients. Overall, median simeprevir FC values increased from 1.0 (pretreatment) to 437 (on-treatment failure) and 359 (viral relapse). The median simeprevir FC of genotype 1a isolates with Q80K increased from 12 (pretreatment) to 499 (on-treatment failure) and 411 (viral relapse), while for genotype 1a isolates without Q80K, the median FC value increased from 1.0 (pretreatment) to 449 (on-treatment failure) and 441 (viral relapse). For genotype 1b isolates, median simeprevir FC values increased from 0.3 at pretreatment to 274 (on-treatment failure) and 321 (viral relapse).
Isolates were obtained at the time of failure from 3 genotype 1a- and 4 genotype 1b-infected patients with treatment failure and no emerging amino acid substitutions at the time of failure. The 3 genotype 1a isolates carried pretreatment polymorphisms known to reduce simeprevir activity in vitro: R155K (1 isolate) and Q80K (2 isolates). The 4 genotype 1b isolates did not have baseline polymorphisms known to reduce simeprevir susceptibility in vitro. Consistent with the absence of emerging amino acid substitutions at the time of failure in these patients, no increase in simeprevir FC values between the baseline and time-of-failure isolates was noted.
Evaluation of the evolution of viral variants after treatment failure until the last available time point of the study (end of study) showed that NS3 sequences returned to the pretreatment sequences in the majority of patients with emerging amino acid substitutions at the time of failure (9). Paired pretreatment and end-of-study phenotype data were available for 36 patients with failure for whom the NS3 sequence returned to the pretreatment sequence at the end of the study. The simeprevir FC values returned to pretreatment levels in all of these isolates consistent with the loss of the emerging amino acid substitution (Fig. 2; see Table S4 in the supplemental material). Data pairing time-of-failure to end-of-study FC values were available for 6 patients who changed to a different mutation profile at the end of study and showed a 66-fold decrease in median simeprevir FC values from 839 at the time of failure. All 6 patients had double (n = 5) or triple (n = 1) mutant variants emerging at the time of failure but “lost” one of the amino acid substitutions at the end of the study, resulting in a decreased simeprevir FC value. Similar simeprevir FC values were observed in patients with an unchanged mutation profile at the end of the study compared with the profile at the time of failure (n = 9; median FC values 222 versus 221, respectively).
Activity of simeprevir against clinical isolates and their corresponding SDMs.
To assess if the simeprevir FC values against clinical isolates were attributable to the effect of the amino acid substitutions present in these isolates, HCV genotype 1 clinical isolates carrying the same substitutions at NS3 positions 43, 80, 122, 155, 156, and/or 168 were grouped. The simeprevir FC value of each isolate of the group was compared with the values for the SDMs carrying the corresponding amino acid substitutions. In addition, clinical isolates and SDMs carrying I/V170T were analyzed, as it emerged in a few subjects (as a single substitution in 2 genotype 1a patients with Q80K and in combination with R155K in 3 genotype 1a patients without Q80K) at the time of failure during clinical studies with simeprevir (9). For 13 of the 23 identified groups of genotype 1a isolates, the corresponding SDM data in a genotype 1a replicon backbone were available. SDM data in a genotype 1b backbone were available for all 18 identified genotype 1b groups of isolates and for the 10 genotype 1a groups for which no corresponding genotype 1a SDM data were available. For 7 mutation profiles, only 1 individual clinical isolate was available (see Table S5 in the supplemental material).
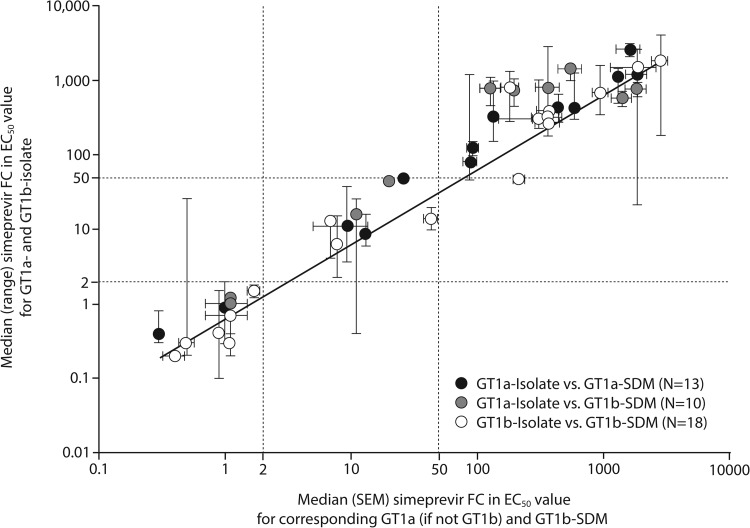
An overall correlation between the simeprevir FC values for the groups of isolates by genotype 1a and 1b and the simeprevir FC values for the corresponding SDMs in a genotype 1a backbone (if available for comparison with genotype 1a isolates) or genotype 1b backbone was observed, with an intraclass correlation coefficient (ICC) of 0.699 (95% confidence interval, 0.659 to 0.736) (Fig. 3). Correlation analysis of only those genotype 1a and 1b isolates for which a corresponding SDM in the genotype 1a or 1b backbone was available showed ICC values of 0.704 and 0.705, respectively (data not shown).
FIG 3.
Correlation of the median simeprevir FC values for HCV genotype 1a and genotype 1b isolates carrying amino acid substitutions at NS3 positions 80, 122, 155, 156, 168, and/or I/V170T, with the median simeprevir FC values for the corresponding SDMs in a genotype 1a or genotype 1b replicon backbone. Black/gray and white symbols, genotype 1a and genotype 1b isolates/SDMs, respectively. Simeprevir FC values for individual clinical isolates were calculated as median values across 1 to 7 replicates (the majority [341 of 435] of isolates were tested ≥3 times). Clinical isolates containing the same single, double, triple, or no amino acid substitutions at NS3 positions 43, 80, 122, 155, 156, 168, and/or 170 were grouped, and the median FC value for each group was determined (each group contained 1 to 95 individual isolates; for details, see Table S5 in the supplemental material). Simeprevir FC values for SDMs carrying corresponding simeprevir resistance-associated amino acid substitutions were calculated as median values across 1 to 24 experimental replicates (the majority [38 of 41] of SDMs were tested ≥3 times). EC50, 50% effective concentration; GT, genotype; FC, fold change; HCV, hepatitis C virus; SDM-1a or -1b, site-directed mutant assessed in an HCV genotype 1a or genotype 1b replicon backbone, respectively; SEM, standard error of the mean.
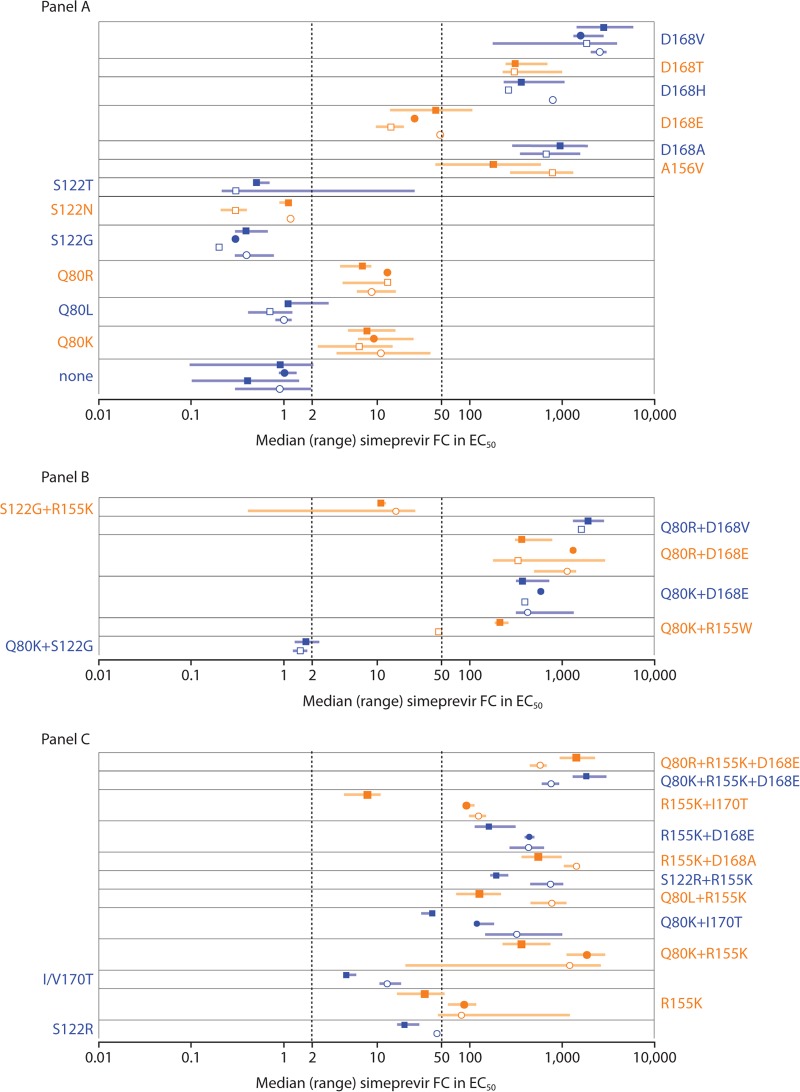
Consistent with the overall correlation, the range of simeprevir FC values for the genotype 1a and 1b isolates corresponded well with the range of simeprevir FC values obtained with the genotype 1a and 1b SDMs carrying the corresponding amino acid substitutions (Fig. 4).
FIG 4.
Activity of simeprevir against HCV genotype 1a and genotype 1b NS3 isolates carrying no, single (A), double, and triple (B) amino acid substitutions at NS3 positions 80, 122, 155, 156, 168, and/or I/V170T and against corresponding SDMs in genotype 1a and genotype 1b replicon backbones. (C) Data for isolates and SDMs with differences in simeprevir susceptibility by HCV geno/subtype. Open circles (○) and boxes (□), median simeprevir FC values of genotype 1a and 1b isolates, respectively; filled circles (●) and boxes (■), median simeprevir FC values of SDMs in genotype 1a and 1b replicon backbones, respectively. Error bars represent the range. Simeprevir FC values for individual clinical isolates were calculated as median values across 1 to 7 replicates (the majority of isolates [341 of 435] were tested ≥3 times). Clinical isolates containing the same single, double, triple, or no amino acid substitutions at NS3 positions 43, 80, 122, 155, 156, 168, and/or 170 were grouped, and the median FC value for each group was determined (each isolate group contained 1 to 95 individual isolates; for details, see Table S5 in the supplemental material). Simeprevir FC values for SDMs carrying corresponding simeprevir resistance-associated amino acid substitutions were calculated as median values across 1 to 24 experimental replicates (the majority of SDMs [38 of 41] were tested ≥3 times). EC50, 50% effective concentration; FC, fold change; HCV, hepatitis C virus; SDM, site-directed mutant.
Simeprevir FC values observed for clinical isolates with single substitutions Q80L, S122G, S122N, or S122T were ≤2.0, indicating that these isolates remained fully susceptible to simeprevir. Isolates and SDMs with NS3 substitutions Q80K, Q80R, S122R, or D168E conferred low-level resistance to simeprevir (FC, >2.0 and <50), whereas clinical isolates and SDMs with NS3 substitutions R155K (genotype 1a), A156V, D168A, D168H, D168T, or D168V conferred high-level resistance (FC, ≥50) (Fig. 4A). The highest simeprevir FC values for clinical isolates and SDMs carrying single NS3 substitutions were observed for genotype 1a and 1b clinical isolates carrying a single D168V (median FC values, 2,605 and 1,865, respectively). None of the isolates carried a single I/V170T; however, when analyzed as an SDM in genotype 1a and 1b backbones, I/V170T reduced simeprevir in vitro activity, with FC values of 13 and 4.7, respectively.
Most isolates with double and triple amino acid substitutions at NS3 positions 80, 122, 155, 168, and/or 170 conferred high-level resistance to simeprevir (FC, ≥50), in line with the simeprevir FC values observed for the corresponding SDMs (Fig. 4B). Interestingly, the addition of S122G to Q80K or R155K resulted in lower simeprevir FC values compared with the values observed when Q80K or R155K was present alone (Fig. 4).
Of note, the simeprevir FC value for clinical isolates carrying R155K, or combinations of R155K or I/V170T with amino acid substitutions at NS3 positions 80, 122, and/or 168, differed substantially when the same substitutions were analyzed as SDMs in a genotype 1a or 1b replicon backbone. Examples include Q80K plus R155K and R155K plus I170T, for which the median simeprevir FC values for clinical isolates were comparable to the FC value observed for the corresponding SDM in a genotype 1a replicon (1,210 versus 1,850 and 125 versus 92, respectively) but higher than the value observed for the SDM in a genotype 1b replicon backbone (364 and 7.9 for Q80K plus R155K and R155K plus I170T, respectively) (Fig. 4C).
DISCUSSION
In this study, the susceptibility to simeprevir of isolates collected from HCV genotype 1-infected patients naive to HCV NS3/4A protease inhibitors who were enrolled in phase 1/2a (TMC435-C101 and -C201), phase 2b (TMC435-C205 and -C206), and phase 3 (TMC435-C208, -C216, and HPC3007) studies was analyzed in a transient chimeric replicon phenotyping assay.
To aid the interpretation of HIV phenotyping assays, biological cutoff values are determined to account for the natural phenotypic variability among isolates from treatment-naive individuals (24). Here, the in vitro biological cutoff for simeprevir was based on the natural distribution of the simeprevir FC values for clinical isolates derived from patients naive to HCV NS3/4A protease inhibitors with no amino acid substitution at NS3 positions 43, 80, 122, 155, 156, and/or 168 (19). Below the biological cutoff (FC, ≤2.0), simeprevir was considered fully active against the clinical isolate or SDM in vitro. The second cutoff value established marked the border between low-level and high-level resistance to simeprevir and was set at FC, ≥50. This value was based on the simeprevir FC values observed with SDMs and clinical isolates carrying amino acid substitutions commonly observed at the time of failure from patients who failed a regimen of 150 mg simeprevir plus PegIFN/RBV. Although determined differently, mainly through in vitro selection experiments and early dose-finding clinical trials, also for other DAAs, such as the NS5A inhibitor daclatasvir and the NS3 protease inhibitors boceprevir and telaprevir, cutoff values have been described that separate amino acid substitutions that confer low-level and high-level resistance (25–30). For example, the cutoff values determined for telaprevir to separate amino acid substitutions conferring low-level resistance from substitutions conferring high-level resistance were FCs of >3 and ≤25 and of >25, respectively (27, 29). The FC values obtained in isolates from patients failing telaprevir-based therapy were generally lower than the FC values observed in isolates from patients failing simeprevir-based therapy (29). This difference might be attributable to the higher ratio of plasma exposure (minimum concentration [Cmin]) to the in vitro EC50 for 150 mg simeprevir once daily (mean Cmin, 1,936 ng/ml [2,581 nM]; in vitro replicon EC50, 9.4 nM [genotype 1b] [31]) compared with 750 mg telaprevir 3 times daily (mean Cmin, 2,030 ng/ml [2,990 nM]; in vitro replicon EC50, 354 nM [genotype 1b] [32]), suggesting that viral variants conferring higher levels of resistance are needed to overcome simeprevir drug pressure compared with that of telaprevir.
Baseline NS3 polymorphisms that reduced simeprevir activity, other than Q80K, were uncommon and usually conferred low-level resistance in vitro (FC, >2.0 and <50). The median FC values of simeprevir against pretreatment genotype 1a isolates without Q80K and against genotype 1b isolates were comparable to that of the wild-type genotype 1b replicon, while the presence of Q80K in genotype 1a resulted in a median 11-fold reduction in simeprevir activity. Although the effect of Q80K on simeprevir in vitro activity is limited, its presence may facilitate the emergence of additional amino acid substitutions, resulting in a higher failure rate in patients treated with simeprevir plus PegIFN/RBV (9, 33, 34).
Failure of treatment with simeprevir plus PegIFN/RBV was associated with high-level resistance to simeprevir (FC, ≥50), with a median FC value of ∼400 due to the emergence of amino acid substitutions at NS3 positions 80, 122, 155, 168, and/or I/V170T. In contrast to patients failing with telaprevir- or boceprevir-based treatments (29, 35), similar FC values were observed for isolates obtained from patients experiencing on-treatment failure and patients experiencing viral relapse. Although the proportion of patients with treatment failure differed by HCV geno/subtype and/or presence of baseline Q80K (2–4, 9), there was no difference in the median FC values observed at the time of failure between patients infected with genotype 1a with or without Q80K and patients infected with genotype 1b. Of note, given the low-level resistance conferred by baseline polymorphism Q80K, the emergence of one additional low-level resistance substitution, such as D168E or I/V170T, was sufficient to result in high-level resistance. In contrast, the few patients with high-level R155K at baseline who failed treatment had no emerging amino acid substitutions at the time of failure. Consistent with sequence data showing a loss of emerging resistance variants over time, simeprevir FC values at the end of the clinical study returned to pretreatment values in all isolates in which the emerging substitutions became undetectable by population sequencing.
The activity of simeprevir against patient-derived NS3 protease sequences showed good correlation with the activity against the SDMs harboring the corresponding single, double, or triple substitutions (ICC, 0.699), showing that the presence of the amino acid substitutions identified in the clinical isolates caused the resistance to simeprevir. These data suggest no benefit of phenotyping using chimeric replicon assays over population sequence-based methods and SDM analysis for the detection and measurement of simeprevir resistance. For some isolates and SDMs, such as those carrying R155K or combinations of R155K or I/V170T with substitutions at NS3 positions 80, 122, and/or 168, the simeprevir FC value depended on the replicon backbone used. Differences in FC values of amino acid substitutions when analyzed in genotype 1a or 1b backbone have also been observed for NS5A inhibitors (25).
A potential limitation of this study is that we used population-based sequencing, which has limited sensitivity. In addition, results from 57 isolates were not included in this analysis because their EC90/EC50 ratio was >5. This was done to exclude data from isolates with biphasic simeprevir dose-response curves, which may lead to a misinterpretation of EC50 values due to the presence of minor variants, as described previously (22). A subanalysis of these 57 isolates showed that the majority carried an amino acid mixture at the position of interest, i.e., wild-type and resistant amino acid, detectable by either population sequencing (20 isolates) or deep sequencing (13/20 isolates with no amino acid mixture identified by population sequencing and deep sequencing data available), confirming that for these types of isolates, assessment of the EC90 can increase the sensitivity of the phenotyping assay, as previously reported (20–22). In addition, in isolates with an EC90/EC50 ratio of ≤5, only a major variant was generally observed (data not shown). Importantly, however, this subanalysis confirmed the results from the analyses presented here showing that the resistant substitutions present (as mixture) at the NS3 position of interest conferred the resistance observed.
In conclusion, a relationship was found between the reduced susceptibility conferred by NS3 amino acid substitutions detected by population sequencing and the simeprevir FC value obtained from NS3 clinical isolates in a transient replicon phenotyping assay. At treatment failure, simeprevir in vitro activity was decreased considerably compared with activity pretreatment, which was consistent with the emergence of simeprevir-resistant variants at NS3 positions 80, 122, 155, 168, and/or I/V170T.
Supplementary Material
ACKNOWLEDGMENTS
Special thanks to the patients participating in these studies and their families. Thanks also to all colleagues from Janssen who contributed to this work.
Editorial support was provided by Gill Becker and Chrissie Kouremenou of Complete Medical Communications and funded by Janssen Research & Development.
Sequencing and phenotypic analysis of patient isolates was performed at Janssen Diagnostics BVBA (Beerse, Belgium).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01444-15.
REFERENCES
- 1.Pawlotsky JM. 2014. New hepatitis C virus (HCV) drugs and the hope for a cure: concepts in anti-HCV drug development. Semin Liver Dis 34:22–29. doi: 10.1055/s-0034-1371007. [DOI] [PubMed] [Google Scholar]
- 2.Manns M, Marcellin P, Poordad F, de Araujo ES, Buti M, Horsmans Y, Janczewska E, Villamil F, Scott J, Peeters M, Lenz O, Ouwerkerk-Mahadevan S, De La Rosa G, Kalmeijer R, Sinha R, Beumont-Mauviel M. 2014. Simeprevir with pegylated interferon alfa 2a or 2b plus ribavirin in treatment-naive patients with chronic hepatitis C virus genotype 1 infection (QUEST-2): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet 384:414–426. doi: 10.1016/S0140-6736(14)60538-9. [DOI] [PubMed] [Google Scholar]
- 3.Jacobson IM, Dore GJ, Foster GR, Fried MW, Radu M, Rafalsky VV, Moroz L, Craxi A, Peeters M, Lenz O, Ouwerkerk-Mahadevan S, De La Rosa G, Kalmeijer R, Scott J, Sinha R, Beumont-Mauviel M. 2014. Simeprevir with pegylated interferon alfa 2a plus ribavirin in treatment-naive patients with chronic hepatitis C virus genotype 1 infection (QUEST-1): a phase 3, randomised, double-blind, placebo-controlled trial. Lancet 384:403–413. doi: 10.1016/S0140-6736(14)60494-3. [DOI] [PubMed] [Google Scholar]
- 4.Forns X, Lawitz E, Zeuzem S, Gane E, Bronowicki JP, Andreone P, Horban A, Brown A, Peeters M, Lenz O, Ouwerkerk-Mahadevan S, Scott J, De La Rosa G, Kalmeijer R, Sinha R, Beumont-Mauviel M. 2014. Simeprevir with peginterferon and ribavirin leads to high rates of SVR in patients with HCV genotype 1 who relapsed after previous therapy: a phase 3 trial. Gastroenterology 146:1669-1679.e3. doi: 10.1053/j.gastro.2014.02.051. [DOI] [PubMed] [Google Scholar]
- 5.Lawitz E, Sulkowski MS, Ghalib R, Rodriguez-Torres M, Younossi ZM, Corregidor A, DeJesus E, Pearlman B, Rabinovitz M, Gitlin N, Lim JK, Pockros PJ, Scott JD, Fevery B, Lambrecht T, Ouwerkerk-Mahadevan S, Callewaert K, Symonds WT, Picchio G, Lindsay KL, Beumont M, Jacobson IM. 2014. Simeprevir plus sofosbuvir, with or without ribavirin, to treat chronic infection with hepatitis C virus genotype 1 in non-responders to pegylated interferon and ribavirin and treatment-naive patients: the COSMOS randomised study. Lancet 384:1756–1765. doi: 10.1016/S0140-6736(14)61036-9 (Erratum, Lancet 384:1748.). [DOI] [PubMed] [Google Scholar]
- 6.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 7.Bartels DJ, Sullivan JC, Zhang EZ, Tigges AM, Dorrian JL, De Meyer S, Takemoto D, Dondero E, Kwong AD, Picchio G, Kieffer TL. 2013. Hepatitis C virus variants with decreased sensitivity to direct-acting antivirals (DAAs) were rarely observed in DAA-naive patients prior to treatment. J Virol 87:1544–1553. doi: 10.1128/JVI.02294-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alves R, Queiroz AT, Pessoa MG, da Silva EF, Mazo DF, Carrilho FJ, Carvalho-Filho RJ, de Carvalho IM. 2013. The presence of resistance mutations to protease and polymerase inhibitors in hepatitis C virus sequences from the Los Alamos databank. J Viral Hepat 20:414–421. doi: 10.1111/jvh.12051. [DOI] [PubMed] [Google Scholar]
- 9.Lenz O, Verbinnen T, Fevery B, Tambuyzer L, Vijgen L, Peeters M, Buelens A, Ceulemans H, Beumont M, Picchio G, De Meyer S. 2015. Virology analyses of HCV isolates from genotype 1-infected patients treated with simeprevir plus peginterferon/ribavirin in phase IIb/III studies. J Hepatol 62:1008–1014. doi: 10.1016/j.jhep.2014.11.032. [DOI] [PubMed] [Google Scholar]
- 10.Pozo F, Lina B, Andrade HR, Enouf V, Kossyvakis A, Broberg E, Daniels R, Lackenby A, Meijer A, Community Network of Reference Laboratories for Human Influenza in Europe. 2013. Guidance for clinical and public health laboratories testing for influenza virus antiviral drug susceptibility in Europe. J Clin Virol 57:5–12. doi: 10.1016/j.jcv.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Durantel D, Brunelle M-N, Gros E, Carrouée-Durantel S, Pichoud C, Villet S, Trepo C, Zoulim F. 2005. Resistance of human hepatitis B virus to reverse transcriptase inhibitors: from genotypic to phenotypic testing. J Clin Virol 34(Suppl 1):S34–S43. doi: 10.1016/S1386-6532(05)80008-3. [DOI] [PubMed] [Google Scholar]
- 12.Vandamme AM, Camacho RJ, Ceccherini-Silberstein F, de Luca A, Palmisano L, Paraskevis D, Paredes R, Poljak M, Schmit JC, Soriano V, Walter H, Sönnerborg A, European HIV Drug Resistance Guidelines Panel. 2011. European recommendations for the clinical use of HIV drug resistance testing: 2011 update. AIDS Rev 13:77–108. http://www.aidsreviews.com/files/2011_13_2_077-108.pdf. [PubMed] [Google Scholar]
- 13.Reesink HW, Fanning GC, Farha KA, Weegink C, Van Vliet A, Van 't Klooster G, Lenz O, Aharchi F, Mariën K, Van Remoortere P, de Kock H, Broeckaert F, Meyvisch P, Van Beirendonck E, Simmen K, Verloes R. 2010. Rapid HCV-RNA decline with once daily TMC435: a phase I study in healthy volunteers and hepatitis C patients. Gastroenterology 138:913–921. doi: 10.1053/j.gastro.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 14.Manns M, Reesink H, Berg T, Dusheiko G, Flisiak R, Marcellin P, Moreno C, Lenz O, Meyvisch P, Peeters M, Sekar V, Simmen K, Verloes R. 2011. Rapid viral response of once-daily TMC435 plus pegylated interferon/ribavirin in hepatitis C genotype-1 patients: a randomized trial. Antivir Ther 16:1021–1033. doi: 10.3851/IMP1894. [DOI] [PubMed] [Google Scholar]
- 15.Fried MW, Buti M, Dore GJ, Flisiak R, Ferenci P, Jacobson I, Marcellin P, Manns M, Nikitin I, Poordad F, Sherman M, Zeuzem S, Scott J, Gilles L, Lenz O, Peeters M, Sekar V, De Smedt G, Beumont-Mauviel M. 2013. Once-daily simeprevir (TMC435) with pegylated interferon and ribavirin in treatment-naive genotype 1 hepatitis C: the randomized PILLAR study. Hepatology 58:1918–1929. doi: 10.1002/hep.26641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeuzem S, Berg T, Gane E, Ferenci P, Foster GR, Fried MW, Hezode C, Hirschfield GM, Jacobson I, Nikitin I, Pockros PJ, Poordad F, Scott J, Lenz O, Peeters M, Sekar V, De Smedt G, Sinha R, Beumont-Mauviel M. 2014. Simeprevir increases rate of sustained virologic response among treatment-experienced patients with HCV genotype-1 infection: a phase IIb trial. Gastroenterology 146:430-441.e6. doi: 10.1053/j.gastro.2013.10.058. [DOI] [PubMed] [Google Scholar]
- 17.Koletzki D, Pattery T, Fevery B, Vanhooren L, Stuyver LJ. 2013. Amplification and sequencing of the hepatitis C virus NS3/4A protease and the NS5B polymerase regions for genotypic resistance detection of clinical isolates of subtypes 1a and 1b. Methods Mol Biol 1030:137–149. doi: 10.1007/978-1-62703-484-5_12. [DOI] [PubMed] [Google Scholar]
- 18.Vijgen L, Verbeeck J, Van Kerckhove B, Berke JM, Koletzki D, Fanning G, Lenz O. 2013. A cellular replicon-based phenotyping assay to determine susceptibility of hepatitis C virus clinical isolates to NS3/4A protease inhibitors. Methods Mol Biol 1030:105–117. doi: 10.1007/978-1-62703-484-5_9. [DOI] [PubMed] [Google Scholar]
- 19.Lenz O, Verbinnen T, Lin TI, Vijgen L, Cummings MD, Lindberg J, Berke JM, Dehertogh P, Fransen E, Scholliers A, Vermeiren K, Ivens T, Raboisson P, Edlund M, Storm S, Vrang L, de Kock H, Fanning GC, Simmen KA. 2010. In vitro resistance profile of the hepatitis C virus NS3/4A protease inhibitor TMC435. Antimicrob Agents Chemother 54:1878–1887. doi: 10.1128/AAC.01452-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi X, Bae A, Liu S, Yang H, Sun SC, Harris J, Delaney W, Miller M, Mo H. 2009. Development of a replicon-based phenotypic assay for assessing the drug susceptibilities of HCV NS3 protease genes from clinical isolates. Antiviral Res 81:166–173. doi: 10.1016/j.antiviral.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Sheaffer AK, Lee MS, Hernandez D, Chaniewski S, Yu F, Falk P, Friborg J, Zhai G, McPhee F. 2011. Development of a chimeric replicon system for phenotypic analysis of NS3 protease sequences from HCV clinical isolates. Antivir Ther 16:705–718. doi: 10.3851/IMP1825. [DOI] [PubMed] [Google Scholar]
- 22.Verbinnen T, Jacobs T, Vijgen L, Ceulemans H, Neyts J, Fanning G, Lenz O. 2012. Replication capacity of minority variants in viral populations can affect the assessment of resistance in HCV chimeric replicon phenotyping assays. J Antimicrob Chemother 67:2327–2337. doi: 10.1093/jac/dks234. [DOI] [PubMed] [Google Scholar]
- 23.Vangeneugden T, Laenen A, Geys H, Renard D, Molenberghs G. 2005. Applying concepts of generalizability theory on clinical trial data to investigate sources of variation and their impact on reliability. Biometrics 61:295–304. doi: 10.1111/j.0006-341X.2005.031040.x. [DOI] [PubMed] [Google Scholar]
- 24.Harrigan PR, Montaner JS, Wegner SA, Verbiest W, Miller V, Wood R, Larder BA. 2001. World-wide variation in HIV-1 phenotypic susceptibility in untreated individuals: biologically relevant values for resistance testing. AIDS 15:1671–1677. doi: 10.1097/00002030-200109070-00010. [DOI] [PubMed] [Google Scholar]
- 25.Fridell RA, Qiu D, Wang C, Valera L, Gao M. 2010. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob Agents Chemother 54:3641–3650. doi: 10.1128/AAC.00556-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fridell RA, Wang C, Sun JH, O'Boyle DR 2nd, Nower P, Valera L, Qiu D, Roberts S, Huang X, Kienzle B, Bifano M, Nettles RE, Gao M. 2011. Genotypic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: in vitro and in vivo correlations. Hepatology 54:1924–1935. doi: 10.1002/hep.24594. [DOI] [PubMed] [Google Scholar]
- 27.Sarrazin C, Kieffer TL, Bartels D, Hanzelka B, Müh U, Welker M, Wincheringer D, Zhou Y, Chu HM, Lin C, Weegink C, Reesink H, Zeuzem S, Kwong AD. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767–1777. doi: 10.1053/j.gastro.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 28.Susser S, Welsch C, Wang Y, Zettler M, Domingues FS, Karey U, Hughes E, Ralston R, Tong X, Herrmann E, Zeuzem S, Sarrazin C. 2009. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology 50:1709–1718. doi: 10.1002/hep.23192. [DOI] [PubMed] [Google Scholar]
- 29.Kieffer TL, De Meyer S, Bartels DJ, Sullivan JC, Zhang EZ, Tigges A, Dierynck I, Spanks J, Dorrian J, Jiang M, Adiwijaya B, Ghys A, Beumont M, Kauffman RS, Adda N, Jacobson IM, Sherman KE, Zeuzem S, Kwong AD, Picchio G. 2012. Hepatitis C viral evolution in genotype 1 treatment-naive and treatment-experienced patients receiving telaprevir-based therapy in clinical trials. PLoS One 7:e34372. doi: 10.1371/journal.pone.0034372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tong X, Chase R, Skelton A, Chen T, Wright-Minogue J, Malcolm BA. 2006. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antiviral Res 70:28–38. doi: 10.1016/j.antiviral.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Janssen Therapeutics. 2013. Olysio (simeprevir): prescribing information. Janssen Therapeutics, Titusville, NJ: https://www.olysio.com/shared/product/olysio/prescribing-information.pdf. [Google Scholar]
- 32.Janssen Cilag International NV. 2011. Incivo (telaprevir): summary of product characteristics. Janssen Cilag International NV, Beerse, Belgium: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Product_Information/human/002313/WC500115529.pdf. [Google Scholar]
- 33.Palanisamy N, Danielsson A, Kokkula C, Yin H, Bondeson K, Wesslén L, Duberg AS, Lennerstrand J. 2013. Implications of baseline polymorphisms for potential resistance to NS3 protease inhibitors in hepatitis C virus genotypes 1a, 2b and 3a. Antiviral Res 99:12–17. doi: 10.1016/j.antiviral.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 34.Janssen Cilag International NV. 2015. Olysio (simeprevir): summary of product characteristics. Janssen Cilag International NV, Beerse, Belgium; https://www.medicines.org.uk/emc/medicine/28888/SPC/OLYSIO+150mg+hard+capsules/. [Google Scholar]
- 35.Ogert RA, Howe JA, Vierling JM, Kwo PY, Lawitz EJ, McCone J, Schiff ER, Pound D, Davis MN, Gordon SC, Ravendhran N, Rossaro L, Jacobson IM, Ralston R, Chaudhri E, Qiu P, Pedicone LD, Brass CA, Albrecht JK, Barnard RJ, Hazuda DJ, Howe AY. 2013. Resistance-associated amino acid variants associated with boceprevir plus pegylated interferon-alpha2b and ribavirin in patients with chronic hepatitis C in the SPRINT-1 trial. Antivir Ther 18:387–397. doi: 10.3851/IMP2549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.