Abstract
Emerging resistance to “last-resort” polymyxin antibiotics in Gram-negative bacteria is a significant threat to public health. We identified the Acinetobacter baumannii NaxD deacetylase as a critical mediator of lipid A modification resulting in polymyxin resistance and demonstrated that naxD is regulated by the sensor kinase PmrB. This represents the first description of a specific PmrB-regulated gene contributing to polymyxin resistance in A. baumannii and highlights NaxD as a putative drug target to reverse polymyxin resistance.
TEXT
Multidrug antibiotic resistance is a rapidly growing threat to global public health. In the United States alone, the Centers for Disease Control and Prevention estimate that at least 2 million infections are caused by antibiotic-resistant pathogens each year, leading to roughly 25,000 deaths (1). A growing number of bacterial isolates are now resistant to most or all classes of antibiotics, rendering some infections untreatable. Polymyxin antibiotics, including polymyxin B and colistin (polymyxin E), are increasingly used as the “last resort” to treat highly drug-resistant Gram-negative bacterial pathogens. These cationic antimicrobial peptide antibiotics bind to the negatively charged lipid A moiety of lipopolysaccharide (LPS) through electrostatic interactions that lead to the disruption of the outer and inner bacterial membranes (2–4). Unfortunately, polymyxin resistance has emerged and is increasing globally (2, 5, 6), undermining the utility of these “last-line” antimicrobials. This clinical challenge is exemplified by Acinetobacter baumannii, a Gram-negative bacterium that accounts for up to 12,000 infections per year in the United States, 63% of which are multidrug resistant and some of which are polymyxin resistant (1).
Polymyxin resistance can emerge in A. baumannii via modifications to lipid A (4, 5). In particular, modifications of the highly anionic lipid A phosphate groups with positively charged moieties, such as phosphoethanolamine (pEtN), lead to increased bacterial surface charge and repulsion of cationic polymyxins (2, 7). In Acinetobacter spp., polymyxin resistance is often controlled by the PmrA/PmrB (PmrAB) two-component regulatory system, in which PmrB is a membrane-localized sensor kinase that phosphorylates and activates PmrA, the cytosolic DNA-binding response regulator. Point mutation(s) in PmrB (commonly within the histidine kinase domain) and/or constitutive expression of pmrAB has been observed in polymyxin-resistant A. baumannii strains (8–10). A. baumannii lipid A has been shown to be modified with pEtN (9, 10, 11), and recently, Pelletier et al. (11) first described a galactosamine (GalN) modification in polymyxin-resistant strains that have activating mutations in PmrB. However, it has remained unclear how PmrB exerts its effects on lipid A modification and polymyxin resistance since PmrB-regulated genes mediating these functions have not yet been identified in A. baumannii.
We previously identified NaxD, a deacetylase belonging to the YdjC superfamily, as being required for the GalN modification of Francisella tularensis lipid A (12). Specifically, NaxD deacetylates N-acetylgalactosamine (linked to the lipid carrier undecaprenyl phosphate) to galactosamine, a step required for the subsequent addition of galactosamine to lipid A. Protein sequence analysis revealed a YdjC-like hypothetical protein with 32% identity to F. tularensis NaxD in the A. baumannii ATCC 17978 (17978 strain) genome (annotated as A1S_2623; see Fig. S1 in the supplemental material). As a first step to determine whether naxD/A1S_2623 might be a gene involved in lipid A modification in A. baumannii, we measured naxD expression by quantitative real-time PCR (qRT-PCR) in the polymyxin-susceptible A. baumannii 17978 strain and a polymyxin-resistant derivative (R2 [10]). We observed that naxD was highly overexpressed in R2, compared to its expression level in the parental wild-type A. baumannii 17978 strain (Fig. 1). Since R2 has an activating mutation in PmrB (10), we tested whether PmrB was responsible for the overexpression of naxD. Indeed, naxD expression was reduced to a baseline level in an isogenic pmrB deletion mutant of R2 and restored to the wild-type level in the R2 pmrB-complemented strain, indicating that naxD expression was dependent on PmrB. Furthermore, the promoter region of naxD contains a consensus PmrA binding site (13–16) (see Fig. S2 in the supplemental material). These data clearly demonstrate PmrB-mediated regulation of naxD in A. baumannii.
FIG 1.
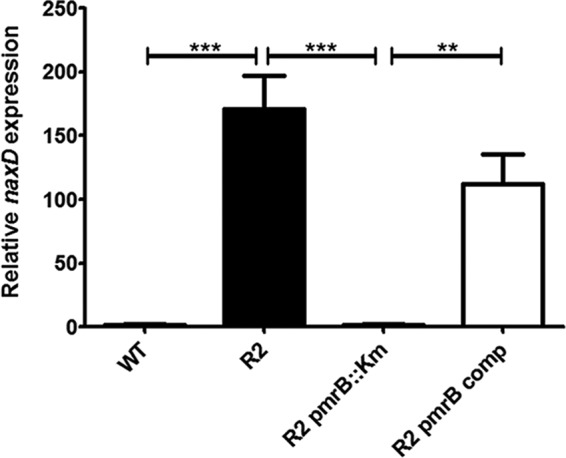
PmrB-dependent overexpression of naxD in a polymyxin-resistant strain of A. baumannii. The wild-type polymyxin-sensitive strain 17978 (WT), the polymyxin-resistant derivative R2, the isogenic R2 pmrB deletion mutant, and the pmrB-complemented (R2 pmrB comp) strains were grown in lysogeny broth (LB) supplemented with 1 mM MgCl2 to an optical density at 600 nm of 0.6. RNA was harvested and used for quantitative real-time PCR analysis of naxD expression in relation to the housekeeping 16S rRNA. **, P < 0.005; ***, P < 0.0005.
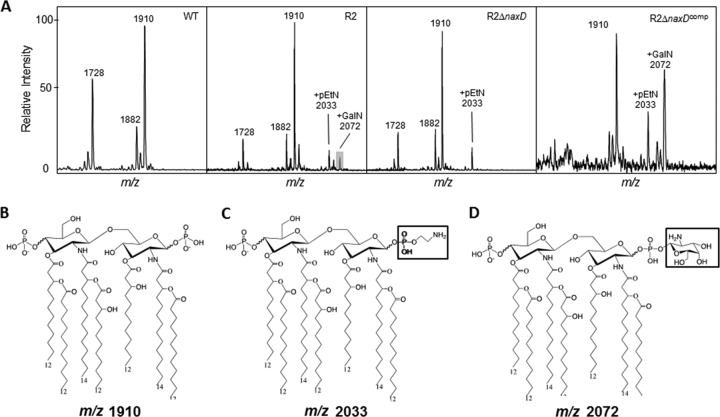
We next set out to directly determine whether NaxD plays a role in A. baumannii lipid A modification by generating a naxD deletion mutant in the R2 strain and comparing the lipid A profiles of the parental and mutant strains by mass spectrometry. Lipid A was isolated as described previously (11) and analyzed using a Bruker Microflex matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrophotometer in the negative ion mode (11). Mass spectra of lipid A isolated from the polymyxin-resistant R2 strain revealed pEtN (m/z 2,033; Δm/z 123) and GalN (m/z 2,072; Δm/z 161) additions to the terminal phosphates of the hexa-acylated lipid A structure (m/z 1,910) (Fig. 2). This was in contrast to lipid A isolated from the parental polymyxin-susceptible 17978 strain (Fig. 2), which did not contain either pEtN or GalN additions, confirming that these lipid A modifications are associated with polymyxin resistance in A. baumannii as previously reported (11). However, while the pEtN modification was not affected, the GalN modification was absent in the lipid A from the naxD deletion mutant. Furthermore, the GalN modification was restored when the naxD mutant was complemented with naxD in trans (Fig. 2). These data indicate that naxD plays a critical role in the modification of lipid A with GalN in A. baumannii.
FIG 2.
NaxD is required for the galactosamine modification of A. baumannii lipid A. (A) Quantification of lipids in the wild-type polymyxin-susceptible A. baumannii 17978 (WT) strain, the polymyxin-resistant derivative R2, the isogenic naxD deletion mutant strain (R2ΔnaxD), and the naxD-complemented (R2 ΔnaxDcomp) strain. (B to D) The peaks corresponding to bis-phosphorylated hepta-acylated lipid A (m/z 1,910) (B), to that with phosphoethanolamine (m/z 2,033) (C), and to that with galactosamine (m/z 2,072) (D) are present in R2, but the galactosamine is absent in the R2ΔnaxD mutant. For each spectrum, the m/z range between 1,600 and 2,300 is shown. Note that phosphate additions can be at either the 1 or the 4′ position. Ion peaks m/z 1,728 and m/z 1,882 correspond to bis-phosphorylated hepta-acylated lipid A (m/z 1,910) with the loss of a C12 fatty acid and C2H4, respectively.
We next tested whether NaxD, and the GalN lipid A modification that it controls, was required for A. baumannii polymyxin resistance. Briefly, overnight cultures were grown from frozen stock in lysogeny broth (LB) (BD Biosciences, Sparks, MD) at 37°C with aeration and then diluted to a final concentration of 107 CFU/ml in 30% LB. Bacteria were treated with polymyxin B (7 μg/ml) (USB Affymetrix, OH) or colistin sulfate (10 μg/ml) (referred to as “colistin” here; Sigma-Aldrich, St. Louis, MO) and incubated at 37°C with aeration for 2 h. The parental wild-type 17978 strain was highly susceptible to both polymyxin and colistin, while the R2 strain was resistant (Fig. 3). This resistance was partially dependent on NaxD, since the viability of the naxD deletion mutant was significantly reduced after polymyxin B or colistin treatment compared to the R2 strain (Fig. 3). Furthermore, this susceptibility was reversed in the naxD-complemented strain. These data demonstrate the important contribution of NaxD to A. baumannii polymyxin resistance and are the first description of a specific PmrB-regulated gene that mediates polymyxin resistance in this clinically important nosocomial pathogen.
FIG 3.
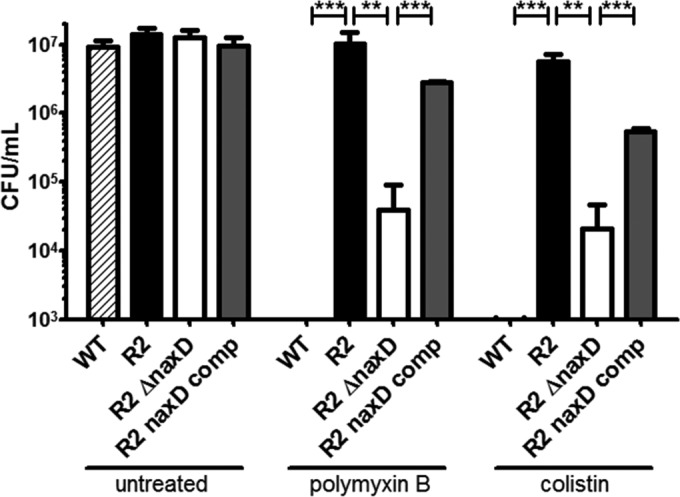
A. baumannii NaxD is required for polymyxin resistance. The A. baumannii wild-type strain, polymyxin-susceptible A. baumannii strain 17978 (WT), polymyxin-resistant strain R2, the R2ΔnaxD mutant, and the naxD-complemented strain (R2ΔnaxD comp) were incubated with 7 μg/ml polymyxin B or 10 μg/ml colistin for 2 h, and CFU were enumerated after plating. **, P < 0.005; ***, P < 0.0005.
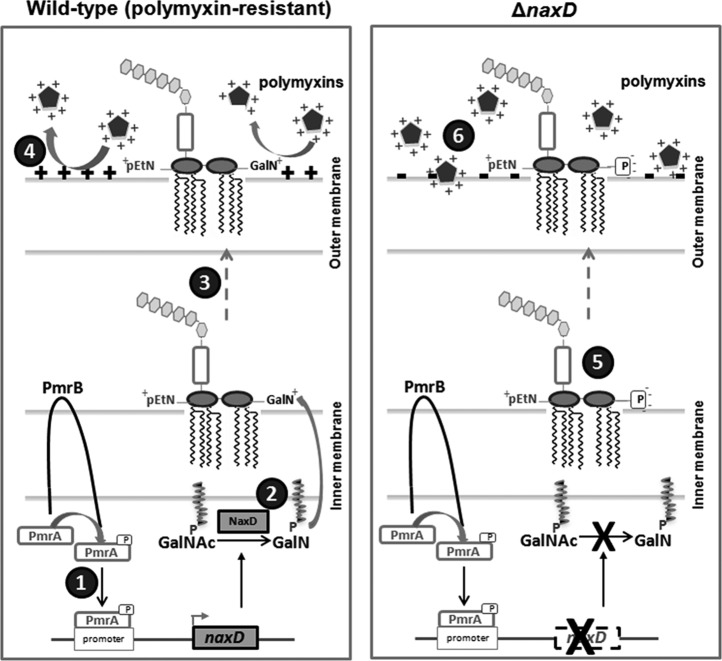
Taken together, the data presented here demonstrate that an activating mutation in PmrB can lead to overexpression of naxD. NaxD then mediates modification of A. baumannii lipid A with positively charged GalN, thus lowering the affinity for cationic antibiotics and leading to increased resistance (Fig. 4). In addition to the NaxD-dependent GalN modification, the NaxD-independent pEtN modification is also likely important for this resistance (9, 10, 11). This work reveals the first identification of a two-component regulatory system controlling naxD expression and highlights NaxD as a potential drug target for inhibitors that reverse polymyxin resistance. Since NaxD is conserved in several pathogenic Gram-negative bacteria (see Fig. S1 in the supplemental material), such an approach could have significant utility in combating antibiotic-resistant bacteria.
FIG 4.
Model of the role of A. baumannii NaxD in resistance to polymyxins. Schematics of polymyxin-resistant wild-type A. baumannii and naxD (ΔnaxD) mutant bacteria are shown. (1) PmrB phosphorylates and activates PmrA upon sensing of environmental stimuli (or when rendered constitutively active due to an amino acid mutation, often in the kinase domain). PmrA subsequently binds the naxD promoter, facilitating transcription. (2) NaxD deacetylates N-acetylgalactosamine (GalNAc) to galactosamine (GalN), which is attached to a lipid carrier and transported from the cytosolic to the periplasmic side of the inner membrane. GalN is subsequently added to the 1 position phosphate of lipid A. The lipid A is also modified with phosphoethanolamine (pEtN) through a NaxD-independent process. (3) Lipopolysaccharide (LPS), composed of lipid A, core, and O antigen, is then transported to the outer leaflet of the outer membrane. (4) GalN and pEtN are positively charged, and these modifications to lipid A increase the surface charge of the bacteria such that positively charged polymyxin antibiotics (pentagons) are repelled. (5) In naxD mutant bacteria, LPS lacks the lipid A GalN modification. (6) Therefore, the bacterial surface charge is decreased, allowing polymyxins to interact with and disrupt the bacterial membranes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Robert Hancock (University of British Columbia, Vancouver, Canada) for generously providing the A. baumannii R2, R2 pmrB::Km, and R2 pmrB-complemented strains. We thank Emily Crispell, Hannah Ratner, and Edgar Sherman for critical reading of the manuscript.
This work was supported by NIH grant AI098800 and a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease fellowship to D.S.W.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00515-15.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 2.Falagas ME, Rafailidis PI, Matthaiou DK. 2010. Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist Updat 13:132–138. doi: 10.1016/j.drup.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Vaara M. 2010. Polymyxins and their novel derivatives. Curr Opin Microbiol 13:574–581. doi: 10.1016/j.mib.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai Y, Chai D, Wang R, Liang B, Bai N. 2012. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J Antimicrob Chemother 67:1607–1615. doi: 10.1093/jac/dks084. [DOI] [PubMed] [Google Scholar]
- 6.Neonakis IK, Samonis G, Messaritakis H, Baritaki S, Georgiladakis A, Maraki S, Spandidos DA. 2010. Resistance status and evolution trends of Klebsiella pneumoniae isolates in a university hospital in Greece: ineffectiveness of carbapenems and increasing resistance to colistin. Chemotherapy 56:448–452. doi: 10.1159/000320943. [DOI] [PubMed] [Google Scholar]
- 7.Gunn JS. 2008. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol 16:284–290. doi: 10.1016/j.tim.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother 53:3628–3634. doi: 10.1128/AAC.00284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother 55:3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arroyo LA, Herrera CM, Fernandez L, Hankins JV, Trent MS, Hancock RE. 2011. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob Agents Chemother 55:3743–3751. doi: 10.1128/AAC.00256-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelletier MR, Casella LG, Jones JW, Adams MD, Zurawski DV, Hazlett KR, Doi Y, Ernst RK. 2013. Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob Agents Chemother 57:4831–4840. doi: 10.1128/AAC.00865-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llewellyn AC, Zhao J, Song F, Parvathareddy J, Xu Q, Napier BA, Laroui H, Merlin D, Bina JE, Cotter PA, Miller MA, Raetz CR, Weiss DS. 2012. NaxD is a deacetylase required for lipid A modification and Francisella pathogenesis. Mol Microbiol 86:611–627. doi: 10.1111/mmi.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamayo R, Ryan SS, McCoy AJ, Gunn JS. 2002. Identification and genetic characterization of PmrA-regulated genes and genes involved in polymyxin B resistance in Salmonella enterica serovar typhimurium. Infect Immun 70:6770–6778. doi: 10.1128/IAI.70.12.6770-6778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aguirre A, Lejona S, Vescovi EG, Soncini FC. 2000. Phosphorylated PmrA interacts with the promoter region of ugd in Salmonella enterica serovar typhimurium. J Bacteriol 182:3874–3876. doi: 10.1128/JB.182.13.3874-3876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchal K, De Keersmaecker S, Monsieurs P, van Boxel N, Lemmens K, Thijs G, Vanderleyden J, De Moor B. 2004. In silico identification and experimental validation of PmrAB targets in Salmonella typhimurium by regulatory motif detection. Genome Biol 5:R9. doi: 10.1186/gb-2004-5-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McPhee JB, Lewenza S, Hancock RE. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol 50:205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.