Abstract
Enterobacter cloacae complex (ECC), an opportunistic pathogen causing numerous infections in hospitalized patients worldwide, is able to resist β-lactams mainly by producing the AmpC β-lactamase enzyme. AmpC expression is highly inducible in the presence of some β-lactams, but the underlying genetic regulation, which is intricately linked to peptidoglycan recycling, is still poorly understood. In this study, we constructed different mutant strains that were affected in genes encoding enzymes suspected to be involved in this pathway. As expected, the inactivation of ampC, ampR (which encodes the regulator protein of ampC), and ampG (encoding a permease) abolished β-lactam resistance. Reverse transcription-quantitative PCR (qRT-PCR) experiments combined with phenotypic studies showed that cefotaxime (at high concentrations) and cefoxitin induced the expression of ampC in different ways: one involving NagZ (a N-acetyl-β-d-glucosaminidase) and another independent of NagZ. Unlike the model established for Pseudomonas aeruginosa, inactivation of DacB (also known as PBP4) was not responsible for a constitutive ampC overexpression in ECC, whereas it caused AmpC-mediated high-level β-lactam resistance, suggesting a post-transcriptional regulation mechanism. Global transcriptomic analysis by transcriptome sequencing (RNA-seq) of a dacB deletion mutant confirmed these results. Lastly, analysis of 37 ECC clinical isolates showed that amino acid changes in the AmpD sequence were likely the most crucial event involved in the development of high-level β-lactam resistance in vivo as opposed to P. aeruginosa where dacB mutations have been commonly found. These findings bring new elements for a better understanding of β-lactam resistance in ECC, which is essential for the identification of novel potential drug targets.
INTRODUCTION
Species of the Enterobacter cloacae complex (ECC) are widely distributed in nature and are part of the commensal microbiota of the human gastrointestinal tract. For 2 decades, they have emerged as major human pathogens (1, 2). Indeed, they have become one of the leading causes of hospital-acquired infections worldwide, accounting for around 5% to 10% of intensive care unit (ICU) infections (1, 2). The ECC shows a genomic heterogeneity with 13 clusters and currently comprises six different species: Enterobacter asburiae, Enterobacter cloacae, Enterobacter hormaechei, Enterobacter kobei, Enterobacter ludwigii, and Enterobacter nimipressuralis (3). ECC species are highly adapted to the hospital environment and are able to contaminate various medical devices. Because of its huge ability to rapidly develop multiple antimicrobial resistances, therapeutic failures are commonly observed (1, 2).
It is well known that the ECC is intrinsically resistant to ampicillin, amoxicillin-clavulanate, and first- and second-generation cephalosporins due to a low-level but inducible expression of a chromosomal ampC gene encoding a β-lactamase (1, 4). The production of this cephalosporinase is highly inducible in the presence of strong β-lactam inducers such as imipenem, cefoxitin, and clavulanate (4). The chromosomal β-lactamase induction mechanism is complex and involves three major gene products intimately linked to the peptidoglycan (PG) recycling pathway: AmpR (a LysR-type transcriptional regulator), AmpD (a cytosolic amidase), and AmpG (an inner membrane permease) (5–12).
In the current model of Gram-negative bacteria, during normal growth, muropeptides from PG degradation are removed from the cell wall and transported via the AmpG permease into the cytoplasm where they are cleaved by AmpD to generate free peptides. To be recycled back into the cell wall synthesis, they are converted into UDP-pentapeptides. These interact with AmpR creating a conformation that represses the transcription of ampC (4, 11, 12). Jacobs et al. suggested that the pentapeptide may be the AmpR ligand, since the murein precursor decreases AmpR-mediated transcriptional activation in vitro (13). Under inducing conditions, there is an accumulation of muropeptides in the cytoplasm, and AmpD is unable to efficiently process the high levels of cell wall fragments. Therefore, the muropeptides (inducing peptides) are thought to displace the UDP-pentapeptides (repressing peptides) from AmpR, converting it into a transcriptional activator of ampC expression (11, 12). In Pseudomonas aeruginosa, several works showed that AmpR is a global transcriptional regulator. It is a potential membrane-associated dimer that regulates genes involved in virulence, quorum sensing, and stress response (14–18). In addition, three AmpD enzymes have been found in P. aeruginosa. The cytoplasmic AmpD protease appear to be involved in cell wall recycling events and the antibiotic resistance pathway, whereas the periplasmic AmpDh2 and AmpDh3 enzymes exhibited marginal activities (19). Mechanisms of AmpC regulation have been extensively studied in P. aeruginosa, and several studies demonstrated that other genes intimately linked to the cell wall recycling system are involved in the regulation of ampC expression. Thus, ampE (coding for an inner membrane-bound sensory transducer), dacB (coding for a d-alanyl-d-alanine carboxypeptidase also known as PBP4), and nagZ (coding for an N-acetyl-β-d-glucosaminidase) have been shown to play an important role in the regulatory network of ampC in P. aeruginosa (20–26). In contrast, the role of these different proteins on AmpC-mediated β-lactam resistance in ECC is not known, and regulatory pathways still remain to be fully elucidated.
Among ECC clinical isolates, high-level resistance to β-lactams (especially third-generation cephalosporins) is due to ampC constitutive overexpression (a phenomenon called derepression), mainly resulting from ampD mutations and much less commonly from ampR mutations (27–32). Alterations responsible for AmpD inactivation or decreased ampD expression lead to a permanent increase in concentrations of inducing muropeptides into the cytoplasm, which convert AmpR into a transcriptional activator conformation (33, 34). This development of resistance in Enterobacter spp. is a major concern since it appears among ca. 10% to 20% of patients treated with broad-spectrum cephalosporins (35–37). Also, once selected, AmpC overproduction is stable, and approximately 30% to 40% of ECC isolates are currently resistant to third-generation cephalosporins worldwide (38, 39).
The aim of this study was then to investigate in detail the regulation mechanisms of AmpC-mediated β-lactam resistance in ECC. First, in the genome of E. cloacae, we identified all of the gene products putatively involved in peptidoglycan recycling based on a P. aeruginosa model. Second, we constructed corresponding deletion mutants and tested their β-lactam resistance profiles and their impact on ampC expression. Third, we screened a collection of ECC clinical isolates for mutations putatively involved in acquisition of β-lactam resistance in vivo. Our results revealed that the model described for P. aeruginosa was not completely relevant for ECC and gave a better overview of regulatory mechanisms underlying β-lactam resistance in Enterobacteriaceae.
(A preliminary report of this work was presented at the 54th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC, 9 to 12 September 2014 [40].)
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. The reference strain used for the construction of knockout mutant strains was E. cloacae subsp. cloacae ATCC 13047 (ECL13047), of which the genome is completely sequenced and annotated (GenBank accession numbers NC_014121, NC_014107, and NC_014108) (41). The 37 independent clinical isolates were recovered from diverse infection sites between 2013 and 2014 (CHU, Caen, France). E. cloacae strains were cultured by shaking at 37°C in Luria-Bertani (LB) medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype/relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strain | ||
| E. cloacae subsp. cloacae ATCC 13047 | Reference strain completely sequenced and annotated | 41 |
| ECLΔampC | 13047 derivative with deletion of ampC (ECL_00553) | This study |
| ECLΔampR | 13047 derivative with deletion of ampR (ECL_00554) | This study |
| ECLΔampD | 13047 derivative with deletion of ampD (ECL_00906) | This study |
| ECLΔampE | 13047 derivative with deletion of ampE (ECL_00907) | This study |
| ECLΔampG | 13047 derivative with deletion of ampG (ECL_01191) | This study |
| ECLΔampH | 13047 derivative with deletion of ampH (ECL_01132) | This study |
| ECLΔdacB | 13047 derivative with deletion of dacB (ECL_04561) | This study |
| ECLΔdacBΔampC | 13047 derivative with deletion of dacB and ampC | This study |
| ECLΔdacBΔampR | 13047 derivative with deletion of dacB and ampR | This study |
| ECLΔnagZ | 13047 derivative with deletion of nagZ (ECL_02529) | This study |
| ECLΔ03254 | 13047 derivative with deletion of ECL_03254 | This study |
| ECLΔ03253 | 13047 derivative with deletion of ECL_03253 | This study |
| Plasmid | ||
| pKOBEG | Recombination vector, phage λ recγβα operon under the control of the pBAD promoter, Cmr | 42 |
| pKD4 | Plasmid containing an FRT-flanked kanamycin cassette, Kanr | 43 |
| pCP20_Gm | FLP-mediated recombination vector, Genr | 44 |
Cmr, chloramphenicol resistant; Genr, gentamicin resistant; Kanr, kanamycin resistant.
Antimicrobial susceptibility testing.
The MIC values of different β-lactams (amoxicillin, AMX; piperacillin-tazobactam, PTZ; cefoxitin, FOX; cefotaxime, CTX; ceftazidime, CAZ; cefepime, FEP; and imipenem, IPM) were determined on Mueller-Hinton agar using Etest strips (bioMérieux, Marcy-l'Etoile, France) in three independent experiments according to the manufacturer's instructions.
AmpC β-lactamase activity assay.
AmpC activity was assessed using the nitrocefin hydrolysis assay as previously described (26). E. cloacae cultures were grown in LB medium until reaching an optical density at 600 nm (OD600) of 0.5. At this time point, 50 μg/ml FOX or 25 μg/ml CTX was added for 2 h. During these treatments, no bactericidal effect was observed. One sample without antibiotic was used as a control. Nitrocefin hydrolysis was measured every minute for 15 min at room temperature by absorbance at 486 nm. AmpC activity was calculated using a nitrocefin extinction coefficient of 20,500 M−1 cm−1. Each assay was independently performed at least three times. Statistical significance was assessed using a two-tailed Student t test, and a P value of ≤0.05 was considered statistically significant.
Construction of knockout deletion mutants.
Disruption of the selected genes putatively implicated in the regulation of ampC was performed using the method described by Datsenko and Wanner with some modifications, using the plasmid pKOBEG (containing a gene for chloramphenicol resistance selection and a gene encoding a recombinase) (42, 43). Briefly, the plasmid pKOBEG was introduced into the ECL13047 strain by electroporation, and transformants were selected on Luria-Bertani (LB) agar with chloramphenicol (25 μg/ml) after incubation for 24 h at 30°C. A selectable kanamycin resistance gene was amplified by PCR using the pKD4 plasmid as a DNA template (42). The primers used included 5′ extensions with homology for the candidate genes and are listed in Table S1 in the supplemental material. The PCR product was introduced into the ECL13047/pKOBEG cells by electroporation, and after homologous recombination the disruption of the candidate gene was obtained. Selected clones were cured for the pKOBEG plasmid following a heat shock. In order to obtain deletion mutants after double crossover, the strains were transformed with the pCP20_Gm plasmid (44), which is able to express the FLP nuclease that recognizes the flippase recognition target (FRT) sequences present on either side of the kanamycin resistance gene.
PCR, sequencing, and quantification of gene expression.
Genomic DNA from the ECL13047 strain and clinical isolates were extracted using the QIAamp DNA minikit (Qiagen, Courtaboeuf, France). Different genes (hsp60 for species-level distinction [3], ampR, ampC, and dacB) were amplified by PCR with specific primers (see Table S1 in the supplemental material), and the purified PCR products were sequenced with the same sets of primers in both directions (GATC Biotech, Constance, Germany).
The levels of expression of ampC, ampR, and ECL_03254 were determined by real-time reverse transcription-PCR (RT-PCR) using specific primers (see Table S1 in the supplemental material). Bacterial cells were harvested 2 h after reaching an OD600 of 0.5, during which CTX (25 μg/ml) or FOX (50 μg/ml) was added for the induction experiments. Total RNAs were extracted from ECL13047 and its derivative mutants using the Direct-zol RNA miniprep kit (Zymo Research, Irvine, CA). Residual chromosomal DNA was removed by treating samples with the Turbo DNA-free kit (Life Technologies, Saint-Aubin, France). Samples were quantified using the BioSpec-nano spectrophotometer (Shimadzu, Noisiel, France), and the integrity was assessed using the Agilent 2100 bioanalyzer according to the manufacturer's instructions. cDNA was synthesized from total RNA (∼25 ng) using the QuantiFast SYBR green RT-PCR kit (Qiagen), and transcript levels were determined by the ΔΔ threshold cycle (ΔΔCt) method using the rpoB gene as a housekeeping control gene (see Table S1). Each experiment was performed in triplicate.
Transcriptomic analysis by RNA-seq.
Total RNAs were extracted from ECL13047 and ECLΔdacB (cultured to the late-exponential growth phase) in duplicate as mentioned above. Before library preparation, DNase-treated samples were depleted from 23S, 16S, and 5S rRNAs using the Ribo-Zero rRNA removal kit (Gram-negative bacteria) (Epicentre, Madison, WI) according to the manufacturer's instructions. To evaluate the degree of rRNA depletion, the samples were analyzed using the Agilent 2100 bioanalyzer. cDNA libraries were prepared with the strand-specific Nextflex rapid directional transcriptome sequencing (RNA-Seq) kit (dUTP-based) v2, and sequencing was performed using an Illumina HiSeq 2500 instrument with the paired-end (2 × 100 bp) multiplexing protocol (ProfileXpert-LCMT, Lyon, France).
For bioinformatic analysis, reads were mapped against the genomic sequence of ECL13047 (GenBank accession number NC_014121) using the CLC Genomics Workbench software v5.1 (Qiagen). Calculation of fold change (FC) values and statistical analysis were performed using the DESeq R package (45). Genes with an expression log2 FC superior to 2 or inferior to −2 were considered induced or repressed, respectively, and statistical significance was retained in the case of a P value of ≤0.05. For visualization, each gene was plotted according to its mean expression value and the differential expression (MA plot).
RESULTS AND DISCUSSION
Role and regulation of the two ampC-annotated genes in ECL13047.
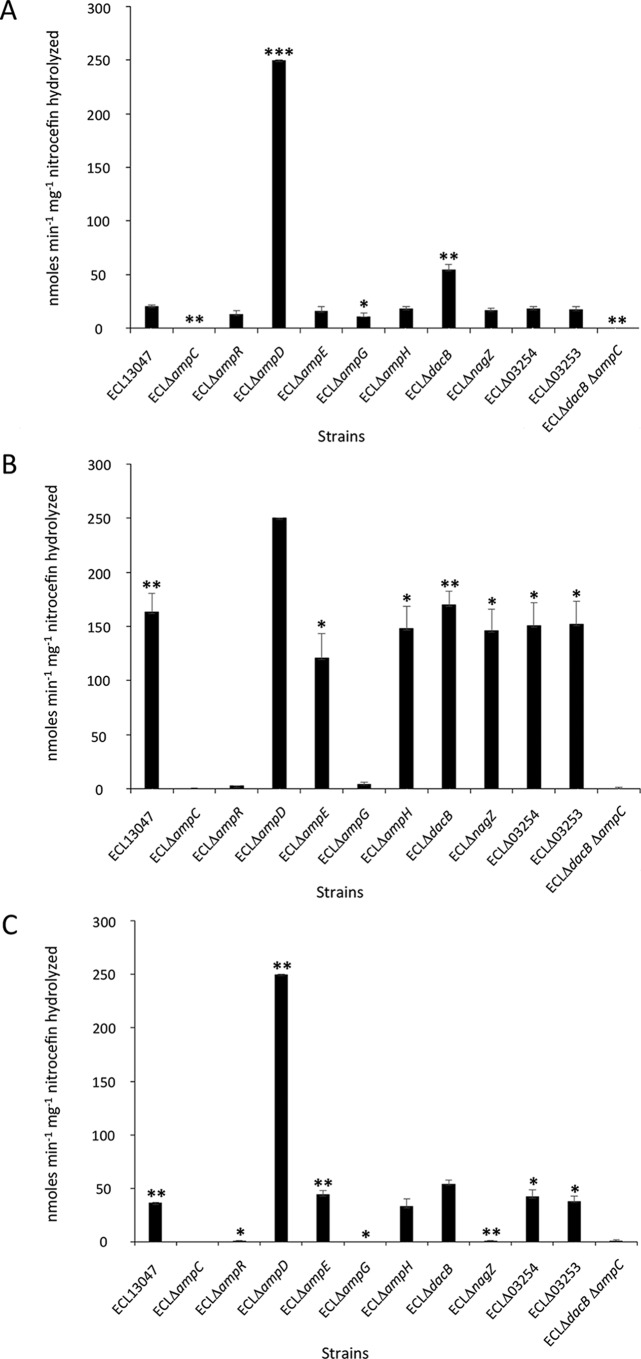
As expected, the wild-type ECL13047 strain was highly resistant to AMX and FOX (MICs, ≥256 μg/ml) but remained susceptible to the other β-lactams tested (Table 2). The AmpC enzyme affinity is indeed much higher to aminopenicillins and FOX than to third-generation cephalosporins and carbapenems (4). In parallel, the expression of the ampC gene in ECL13047 was induced 22- and 79-fold in the presence of FOX (50 μg/ml) and of high-concentration CTX (25 μg/ml), respectively. The β-lactamase activity of AmpC was also statistically significantly increased in the presence of FOX or CTX (Fig. 1). FOX and CTX at high concentrations should be considered β-lactam inducers of E. cloacae ampC expression. Because the induction by CTX was performed during a short period (2 h), this result was very unlikely due to a selection of derepressed mutants. Moreover, significant decreases in the MICs of AMX (≥128-fold), FOX (≥64-fold), and CTX and CAZ (4-fold) seen in the ampC deletion mutant correlated with the absence of β-lactamase activity (Table 2 and Fig. 1), confirming the crucial role of ampC in β-lactam resistance in E. cloacae (4). Detailed analysis of the E. cloacae genome sequence revealed the existence of another gene (ECL_03254) annotated as encoding a class C β-lactamase based on the presence of the conserved motif (CubicO group peptidase, beta-lactamase class C family, COG1680). Interestingly, the level of transcription of ECL_03254 was not modified in the presence of FOX or CTX (data not shown), while the mutant lacking ECL_03254 had the same profile of β-lactam resistance as the wild-type strain. In addition, the absence of ECL_03254 did not have any effect on basal or induced ampC expression and AmpC β-lactamase activity (Table 2 and Fig. 1). Altogether, these results demonstrate the minimal impact of such a protein in β-lactam resistance in ECC and its negligible role in regulation and induction pathways of ampC.
TABLE 2.
MICs and basal and induced ampC expression in wild-type E. cloacae ATCC 13047 and its derivative mutants
| Strain | MIC (μg/ml) |
Relative mRNA level of ampCa |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMX | PTZ | FOX | CTX | CAZ | FEP | IPM | Basal | FOX inducedb | CTX inducedb | |
| Wild-type 13047 | ≥256 | 4 | ≥256 | 0.5 | 1 | 0.03 | 0.5 | 1 | 21.6 ± 4.2 | 79.2 ± 4.5 |
| ECLΔampC | 2 | 4 | 4 | 0.12 | 0.25 | 0.03 | 0.25 | ≤0.01 | ≤0.01 | ≤0.01 |
| ECLΔampR | 4 | 4 | 4 | 0.12 | 0.25 | 0.03 | 0.25 | 1.5 ± 1.0 | 0.67 ± 0.1 | 2.9 ± 0.3 |
| ECLΔ03254 | ≥256 | 4 | ≥256 | 0.5 | 1 | 0.03 | 0.5 | 1.1 ± 0.3 | 16.6 ± 3.6 | 60.4 ± 5.0 |
| ECLΔ03253 | ≥256 | 4 | ≥256 | 0.5 | 1 | 0.03 | 0.5 | 1.1 ± 0.2 | 11.2 ± 1.6 | 46.3 ± 10.9 |
| ECLΔampD | ≥256 | 32 | ≥256 | 32 | 32 | 1 | 0.5 | 369 ± 45 | 707 ± 17 | 600 ± 106 |
| ECLΔampE | ≥256 | 4 | ≥256 | 0.5 | 1 | 0.03 | 0.5 | 1.2 ± 0.1 | 11.2 ± 1.4 | 145 ± 22 |
| ECLΔampG | 4 | 2 | 4 | 0.12 | 0.25 | 0.03 | 0.25 | 1.0 ± 0.2 | 1.9 ± 0.9 | 0.5 ± 0.2 |
| ECLΔdacB | ≥256 | 16 | ≥256 | 16 | 16 | 0.25 | 0.25 | 1.6 ± 0.8 | 7.0 ± 2.2 | 17.4 ± 8.5 |
| ECLΔdacBΔampC | 2 | 4 | 4 | 0.12 | 0.25 | 0.03 | 0.25 | ≤0.01 | ≤0.01 | ≤0.01 |
| ECLΔampH | ≥256 | 4 | ≥256 | 0.5 | 1 | 0.03 | 0.5 | 1.1 ± 0.7 | 6.0 ± 2.9 | 77.0 ± 18.2 |
| ECLΔnagZ | 16 | 2 | 256 | 0.25 | 0.5 | 0.03 | 0.25 | 1.1 ± 0.2 | 37.8 ± 14.1 | 2.1 ± 0.3 |
Relative amount of ampC mRNA compared to the wild-type 13047 basal level ± standard deviation; significant changes are indicated in bold.
Induction experiments carried out with 50 μg/ml of FOX or 25 μg/ml of CTX.
FIG 1.
AmpC activity (measured in nanomoles per minute per milligram nitrocefin hydrolyzed) of E. cloacae ATCC 13047 grown under basal conditions (A) and induced conditions, with the medium supplemented with 50 μg/ml of cefoxitin (B) or 25 μg/ml of CTX (C) for 2 h. Bars represent the means ± standard errors. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Expression of ampC is regulated by AmpR, a transcriptional regulator, coded by the ampR gene located next to ampC and divergently transcribed (5–7). The ECLΔampR mutant was more susceptible to β-lactams than the wild-type strain and lost the ability to induce the transcription of ampC (Table 2). Transcriptomic analysis also showed that ampR was constitutively transcribed in the wild-type strain and in all of the isogenic mutants (data not shown), confirming that the activity of AmpR depends on the interaction with coregulators rather than on an overexpression or underexpression of ampR. This is in accordance with the current model where, in the absence of β-lactams, AmpR represses ampC transcription, and when exposed to antibiotics, cells accumulate peptidoglycan catabolites (i.e., 1,6-anhydroMurNAc-peptides), changing AmpR to an activator of ampC transcription (33, 46, 47). In P. aeruginosa, AmpR plays a role in physiological processes and influences the expression of over 500 genes, including virulence-encoding genes and other transcriptional regulators (15, 16, 48). Then, AmpR appears as a global regulator, and its possible role in the virulence of ECC should be further investigated. Note that this regulation by AmpR is not a general mechanism in Enterobacteriaceae since some Escherichia coli strains do not possess an ampR gene (5–7). Interestingly, ECL_03254 was preceded by a gene (ECL_03253) that coded for a transcriptional regulator that also belonged to the LysR family. Because the transcription of ECL_03254 was significantly induced (8.4-fold) in ECLΔ3253, ECL_03253 likely acts as a repressor under standard growth conditions (data not shown). It is tempting to speculate that the protein encoded by ECL_03254 may be functional and that the role and substrate of this enzyme and the precise regulation of the corresponding gene remain to be elucidated.
ampD and ampG mutations also affect the ampC expression.
In order to verify that enzymes involved in PG recycling play a role in ampC regulation, mutant strains affected in ampD, ampE, and ampG were constructed. AmpD is a cytoplasmic N-acetylmuramyl-l-alanine amidase leading to the production of UDP-MurNAc-pentapeptides that change AmpR to a repressor of ampC transcription (13, 34, 49). This explains why, in the ECLΔampD mutant, the level of ampC expression was extremely high (between 369- and 707-fold more than in the wild type) even in the absence of β-lactams (Table 2). This was correlated with its high-level resistance to PTZ, CTX, and CAZ (MICs, 32 μg/ml) (Table 2). Based on what is already known about E. cloacae, this mutant likely accumulates peptidoglycan catabolites triggering the activation form of AmpR and the overexpression of ampC (7, 27, 28, 32, 50). Consequently, the high level of AmpC activity correlates with the high MICs observed (Fig. 1 and Table 2).
In E. cloacae, the ampE gene is part of the ampDE operon and encodes a cytoplasmic membrane protein thought to act as a β-lactam-reactive sensory transducer (7). The unique deletion of ampE did not reveal a significant difference in terms of ampC regulation and β-lactam resistance compared to the wild-type strain (Table 2). Nevertheless, transcomplementation experiments performed in P. aeruginosa suggest that AmpE may play an indirect role in resistance and that there are other unknown genes (likely located close to the ampDE operon) involved in AmpC overproduction (51).
AmpG is an inner membrane permease that transports PG catabolites involved in cell wall recycling (9, 10). We confirm that, in the absence of AmpG, the levels of ampC mRNA were not significantly modified by the addition of CTX or FOX, while ECLΔampG was more susceptible to all of the β-lactams tested, which is similar to the ECLΔampC and ECLΔampR mutants (Table 2). In accordance with the current model of regulation, this is explained by the fact that peptides are unable to enter the cytosol that leads to the absence of molecules interacting with AmpR and preventing the modulation of the ampC expression (9, 10).
These results, which combine resistance phenotypes, the level of ampC transcription, and AmpC activity, were in agreement with the current model linking PG recycling and β-lactam resistance in Gram-negative rods.
DacB and AmpH are two PBPs involved in the regulation of ampC expression.
DacB is a nonessential low-molecular mass penicillin-binding protein (PBP) (called PBP4) with d,d-carboxypeptidase and d,d-endopeptidase activities. In addition to its role in PG recycling, it is also involved in E. coli cell separation during division or bacterial morphology (52, 53). In the ECLΔdacB mutant strain, we showed that the level of ampC transcription did not significantly change in the absence of β-lactams. However, in the presence of FOX or CTX, the ampC transcription remained slightly induced, but this was 3- to 4-fold lesser than that observed in the wild-type strain (Table 2). Surprisingly, whereas no substantial increase of ampC expression was observed, a high-level resistance to PTZ, CTX, and CAZ (MICs, 16 μg/ml) was observed in the ECLΔdacB mutant (Table 2). It may be suggested that DacB regulates AmpC at a post-transcriptional level, since dacB deletion triggered a significant increase of AmpC β-lactamase activity without a strong upregulation of the ampC gene (Fig. 1 and Table 2). It has been shown that the production of AmpC in Serratia marcescens was controlled by the presence of a 5′-untranslated region (5′-UTR) stem-loop structure of the mRNA that may be related to RNA stability or translational efficiency (54). The 5′ end of the ampC transcript of E. cloacae did not show such specific organization, but it is conceivable that translational regulation may occur, either by interaction with a small trans-acting noncoding RNA or by site-specific ribosome stalling, which is a mechanism used for the control of translation of antibiotic resistance genes such as cat (chloramphenicol acetyltransferase) and erm (erythromycin ribosomal methylase) (55–57). This mechanism of regulation radically diverges from that described in the P. aeruginosa model, where the inactivation of PBP4 leads to a constitutive ampC overexpression (23). In this last species, the role of PBP4 in the ampC induction process is linked to the activation of the CreBC two-component system that, in turn, plays a crucial role in β-lactam resistance (58). Such a regulatory system has not been found in the genome of ECL13047, and this may explain why regulatory pathways of ampC expression are, in part, different from those of P. aeruginosa. Note that the protein coded by the dacB gene of E. cloacae shows 90% amino acid identity with that of E. coli K-12 but only shows 29% amino acid identity with that of P. aeruginosa; this may explain such differences. Mutations in dacB have been identified in high-level β-lactam-resistant clinical isolates of P. aeruginosa, and PBP4 was shown to be the main driver of the resistance (23). These data reveal the presence of alternative process in PG recycling and the existence of multiple pathways leading to the control of ampC expression. To assess the role of AmpC in the β-lactam resistance observed in ECLΔdacB, ampC was disrupted in the ΔdacB mutant background, showing that β-lactam susceptibility was restored to the levels observed for the single ECLΔampC mutant (Table 2). This confirms that high-level β-lactam resistance in the absence of dacB is mediated by AmpC.
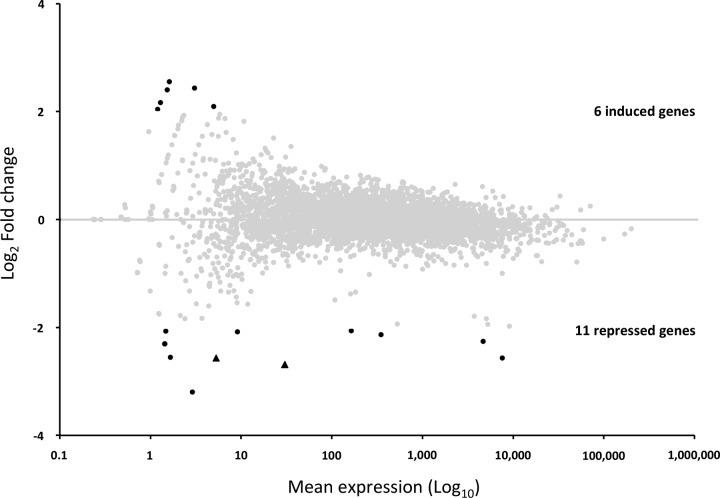
In order to verify whether DacB in E. cloacae was linked to a regulatory pathway able to modify the expression of some genes, we performed a global transcriptomic analysis by RNA-seq comparing the ΔdacB mutant with the wild-type strain (see Table S2 in the supplemental material). This comparison allowed us to identify only six genes of which expression was induced (but not statistically significantly) and 11 repressed genes (Fig. 2; see also Table S3 in the supplemental material). Of these 11 genes, only two exhibited statistically significantly decreased expression changes: ECL_01582 (FC, −6.4; P = 10−6) and ECL_01584 (FC, −5.9; P = 0.04) (see Table S3). The two genes coded for hypothetical proteins (135 and 52 amino acids long, respectively), and no homology was found with any other bacterial protein. Noteworthy, the levels of expression of ampC and ampR were not statistically significantly different (FC, 1.1) (see Table S3), which was also confirmed by reverse transcription-quantitative PCR (qRT-PCR) experiments (data not shown).
FIG 2.
MA plot representing the global gene expression analysis obtained by RNA-seq. Each dot represents one gene. The x axis reflects the mean expression level of a gene (baseMean in Table S3 in the supplemental material). The y axis reflects the differential expression of a gene (as log2 fold change) between the ECLΔdacB mutant and the wild-type ECL13047 strain (see Table S3). Plots corresponding to genes with no significant altered expression (i.e., log2 FC between −2 and 2) are faded. The two statistically differentially expressed genes (ECL_01582 and ECL_01584) are represented as triangles.
We also tested whether another PBP, AmpH, involved in the maturation and remodeling of PG may be part of the control of AmpC production in ECC. In E. coli, this low-molecular-mass PBP displays bifunctional d,d-endopeptidase and d,d-carboxypeptidase activity (59). The ECLΔampH mutant exhibited similar MICs of β-lactams compared to those of the wild-type strain (Table 2). However, we observed that the induction of ampC due to the addition of FOX was not as high as that measured for the ECL13047 wild-type strain (6-fold versus 22-fold, respectively) (Table 2). It may be proposed that AmpH has more affinity for cefoxitin and, despite a role in PG recycling, has a minor influence in ampC regulation.
FOX and CTX induce the transcription of ampC in two different ways.
Based on the P. aeruginosa and E. coli models, it is proposed that PG recycling also involves NagZ, which is a β-N-acetylglucosaminidase. This enzyme processes peptidoglycan degradation products in the cytoplasm, producing 1,6-anhydroMurNAc-peptides that may activate ampC transcription through its interaction with AmpR (60, 61). It is thus expected that the inactivation of nagZ should abolish the induction of ampC when β-lactams are present. In ECC, this appeared true with CTX since no transcriptional induction of ampC was observed in the ECLΔnagZ strain associated with a drastic reduction in the MIC of AMX (from ≥256 to 4 μg/ml) and, to a lesser extent, of PTZ, CTX, and CAZ (2-fold decrease) (Table 2). In P. aeruginosa, loss of nagZ reduces the capability to acquire resistance to β-lactams (62). However, the lack of ampC induction by the presence of CTX in the ECLΔnagZ mutant that we observed is very unlikely due to the selection of mutant strains because the antibiotic treatment only lasted 2 h. In contrast, when FOX was added to the culture of this mutant, the level of ampC mRNA increased 38-fold compared to the basal transcription without an antibiotic (Table 2). Note that the ECLΔnagZ mutant remained resistant to FOX (MIC, ≥256 μg/ml) (Table 2). Similar results were observed in P. aeruginosa, where nagZ inactivation had little effect on the induction of AmpC in the presence of FOX (62). Moreover, the nitrocefin hydrolysis activity data correlated well with the transcriptomic results (Fig. 1). This demonstrates that at least two different pathways exist for the overexpression of ampC: one stimulated by CTX and involving NagZ and another triggered by FOX and independent of NagZ. Shewanella oneidensis mutants lacking NagZ or AmpG are still able to induce the β-lactamase BlaA, which suggests a parallel signal transduction pathway independent of NagZ (61). Note that the blaA gene in S. oneidensis is inducible by ampicillin but this species does not have a neighbor gene coding for a transcriptional regulator such as ampR and that the nagZ mutation results in increased β-lactam resistance (61).
Prevalence of dacB mutations in β-lactam-resistant ECC clinical isolates.
Mutations in ampD and ampR genes have been shown to be responsible for AmpC overexpression and high-level β-lactam resistance in Enterobacter spp. for a long time (5–7, 63). However, the involvement of mutations in other genes (such as dacB) has never been investigated. To do that, screening of mutations in ampD, ampR, ampC, and dacB genes was done on a panel of 37 unrelated ECC clinical isolates (see Table S4 in the supplemental material). Determination of MICs showed that 31 (84%) exhibited constitutive AmpC overexpression (CAOP), while six (16%) presented a β-lactam susceptibility profile. Out of the 31 CAOP strains, 28 (90%) had at least one amino acid change in AmpD, seven of which resulted in premature termination of the protein by creation of a stop codon or by introducing a frameshift mutation (see Table S4). Some of these mutations have already been described in E. cloacae (50). Despite the existence of other amino acid substitutions in AmpR, AmpC, or DacB (without any sequence interruption), these data strongly argue for the crucial role of AmpD alterations in the development of high-level β-lactam resistance in E. cloacae. For the three CAOP strains devoid of AmpD mutation (strains 6, 20, and 30), it is tempting to speculate that the modifications observed in AmpR, AmpC, and/or DacB may play an important role in resistant phenotypes (see Table S4). In this context, the unique mutation in the DacB sequence was retrieved in strain 30. However, this strain showed MIC values significantly lower than the those of the other CAOP strains. These results were not in accordance with those reported for P. aeruginosa, where dacB mutations seem to be much more common (23).
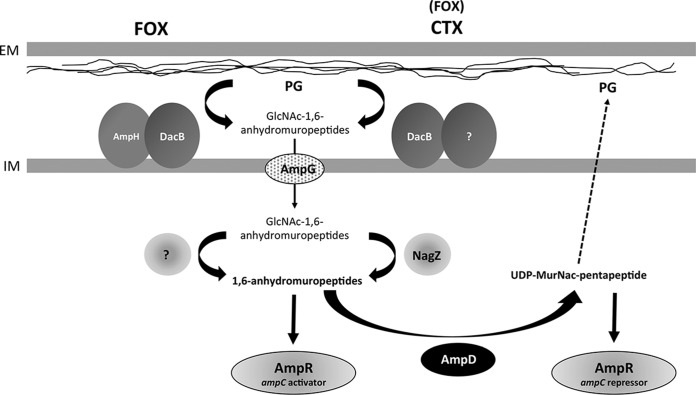
Taken altogether, our data, which were obtained with the different mutant strains affected in genes involved in PG recycling, allowed us to propose a model in which the presence of a β-lactam leads to the expression of the AmpC β-lactamase (Fig. 3). CTX (at high concentration) and FOX were able to significantly enhance the ampC expression. Growth of E. cloacae in the presence of β-lactams increases the production of PG degradation products (1,6-anhydroMurNAc-peptides), which enter the cytoplasm via the AmpG permease and which ultimately lead to the accumulation of MurNAc-tripeptide. NagZ, which catalyzes the production of the MurNAc-tripeptide, appeared important when CTX was present, whereas another mechanism, as yet unknown, is involved in the ampC induction in the presence of FOX. MurNAc-tripeptide may interact with the AmpR regulator, changing its activity from a negative to a transcriptional activator of ampC. In the absence of β-lactams, the MurNAc-tripeptide can be reintegrated into the peptidoglycan synthesis thanks to AmpD activity. Note that mutations in AmpD, which conduces to an increase of AmpC production and consequently β-lactam resistance, seems the most frequent mechanism of resistance in ECC clinical isolates. In addition, we have shown that DacB played an important role for β-lactam resistance in E. cloacae but also that AmpH intervened in ampC induction by FOX. This is the first work that describes the different steps of the regulation of AmpC expression in E. cloacae and that clearly diverges from those of E. coli (sometimes devoid of AmpR) or P. aeruginosa (where dacB inactivation leads to ampC overexpression). A better understanding of the molecular mechanism of resistance to β-lactams is crucial to better fight and prevent infections by this important opportunistic pathogen.
FIG 3.
Model of the transcriptional induction of ampC following exposure to cefoxitin (FOX) (left) or cefotaxime (CTX) (right) in E. cloacae. EM, external membrane; GlcNAC, N-acetylglucosamine; IM, inner membrane; MurNac, N-acetylmuramic acid.
Supplementary Material
ACKNOWLEDGMENTS
The technical assistance of Michel Auzou, Julie Guignot, and Isabelle Podglagen is gratefully acknowledged.
All authors read and approved the manuscript, and we declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01729-15.
REFERENCES
- 1.Sanders WE Jr, Sanders CC. 1997. Enterobacter spp.: pathogens poised to flourish at the turn of the century. Clin Microbiol Rev 10:220–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mezzatesta ML, Gona F, Stefani S. 2012. Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol 7:887–902. doi: 10.2217/fmb.12.61. [DOI] [PubMed] [Google Scholar]
- 3.Paauw A, Caspers MP, Schuren FH, Leverstein-van Hall MA, Deletoile A, Montijn RC, Verhoef J, Fluit AC. 2008. Genomic diversity within the Enterobacter cloacae complex. PLoS One 3(8):e3018. doi: 10.1371/journal.pone.0003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacoby GA. 2009. AmpC beta-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honore N, Nicolas MH, Cole ST. 1986. Inducible cephalosporinase production in clinical isolates of Enterobacter cloacae is controlled by a regulatory gene that has been deleted from Escherichia coli. EMBO J 5:3709–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolas MH, Honore N, Jarlier V, Philippon A, Cole ST. 1987. Molecular genetic analysis of cephalosporinase production and its role in beta-lactam resistance in clinical isolates of Enterobacter cloacae. Antimicrob Agents Chemother 31:295–299. doi: 10.1128/AAC.31.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honore N, Nicolas MH, Cole ST. 1989. Regulation of enterobacterial cephalosporinase production: the role of a membrane-bound sensory transducer. Mol Microbiol 3:1121–1130. doi: 10.1111/j.1365-2958.1989.tb00262.x. [DOI] [PubMed] [Google Scholar]
- 8.Holtje JV, Kopp U, Ursinus A, Wiedemann B. 1994. The negative regulator of beta-lactamase induction AmpD is a N-acetyl-anhydromuramyl-l-alanine amidase. FEMS Microbiol Lett 122:159–164. doi: 10.1111/j.1574-6968.1994.tb07159.x. [DOI] [PubMed] [Google Scholar]
- 9.Korfmann G, Sanders CC. 1989. ampG is essential for high-level expression of AmpC beta-lactamase in Enterobacter cloacae. Antimicrob Agents Chemother 33:1946–1951. doi: 10.1128/AAC.33.11.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindquist S, Weston-Hafer K, Schmidt H, Pul C, Korfmann G, Erickson J, Sanders C, Martin HH, Normark S. 1993. AmpG, a signal transducer in chromosomal beta-lactamase induction. Mol Microbiol 9:703–715. doi: 10.1111/j.1365-2958.1993.tb01731.x. [DOI] [PubMed] [Google Scholar]
- 11.Johnson JW, Fisher JF, Mobashery S. 2013. Bacterial cell-wall recycling. Ann N Y Acad Sci 1277:54–75. doi: 10.1111/j.1749-6632.2012.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong KF, Schneper L, Mathee K. 2010. Beta-lactam antibiotics: from antibiosis to resistance and bacteriology. APMIS 118:1–36. doi: 10.1111/j.1600-0463.2009.02563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs C, Frère JM, Normark S. 1997. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible beta-lactam resistance in gram-negative bacteria. Cell 88:823–832. doi: 10.1016/S0092-8674(00)81928-5. [DOI] [PubMed] [Google Scholar]
- 14.Caille O, Zincke D, Merighi M, Balasubramanian D, Kumari H, Kong KF, Silva-Herzog E, Narasimhan G, Schneper L, Lory S, Mathee K. 2014. Structural and functional characterization of Pseudomonas aeruginosa global regulator AmpR. J Bacteriol 196:3890–3902. doi: 10.1128/JB.01997-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumari H, Murugapiran SK, Balasubramanian D, Schneper L, Merighi M, Sarracino D, Lory S, Mathee K. 2014. LTQ-XL mass spectrometry proteome analysis expands the Pseudomonas aeruginosa AmpR regulon to include cyclic di-GMP phosphodiesterases and phosphoproteins, and identifies novel open reading frames. J Proteomics 96:328–342. doi: 10.1016/j.jprot.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balasubramanian D, Kumari H, Jaric M, Fernandez M, Turner KH, Dove SL, Narasimhan G, Lory S, Mathee K. 2014. Deep sequencing analyses expands the Pseudomonas aeruginosa AmpR regulon to include small RNA-mediated regulation of iron acquisition, heat shock and oxidative stress response. Nucleic Acids Res 42:979–998. doi: 10.1093/nar/gkt942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong KF, Jayawardena SR, Indulkar SD, Del Puerto A, Koh CL, Høiby N, Mathee K. 2005. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB beta-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob Agents Chemother 49:4567–4575. doi: 10.1128/AAC.49.11.4567-4575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balasubramanian D, Schneper L, Merighi M, Smith R, Narasimhan G, Lory S, Mathee K. 2012. The regulatory repertoire of Pseudomonas aeruginosa AmpC β-lactamase regulator AmpR includes virulence genes. PLoS One 7:e34067. doi: 10.1371/journal.pone.0034067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Lee M, Hesek D, Lastochkin E, Boggess B, Mobashery S. 2013. Reactions of the three AmpD enzymes of Pseudomonas aeruginosa. J Am Chem Soc 135:4950–4953. doi: 10.1021/ja400970n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asgarali A, Stubbs KA, Oliver A, Vocadlo DJ, Mark BL. 2009. Inactivation of the glycoside hydrolase NagZ attenuates antipseudomonal beta-lactam resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:2274–2282. doi: 10.1128/AAC.01617-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong KF, Aguila A, Schneper L, Mathee K. 2010. Pseudomonas aeruginosa beta-lactamase induction requires two permeases, AmpG and AmpP. BMC Microbiol 10:328. doi: 10.1186/1471-2180-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stubbs KA, Debowski AW, Mark BL, Stick RV, Vocadlo DJ. 2008. Synthesis and use of mechanism-based protein-profiling probes for retaining beta-d-glucosaminidases facilitate identification of Pseudomonas aeruginosa NagZ. J Am Chem Soc 130:327–335. doi: 10.1021/ja0763605. [DOI] [PubMed] [Google Scholar]
- 23.Moya B, Dotsch A, Juan C, Blazquez J, Zamorano L, Haussler S, Oliver A. 2009. Beta-lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog 5:e1000353. doi: 10.1371/journal.ppat.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zamorano L, Moya B, Juan C, Oliver A. 2010. Differential beta-lactam resistance response driven by ampD or dacB (PBP4) inactivation in genetically diverse Pseudomonas aeruginosa strains. J Antimicrob Chemother 65:1540–1542. doi: 10.1093/jac/dkq142. [DOI] [PubMed] [Google Scholar]
- 25.Zamorano L, Reeve TM, Juan C, Moya B, Cabot G, Vocadlo DJ, Mark BL, Oliver A. 2011. AmpG inactivation restores susceptibility of pan-beta-lactam-resistant Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother 55:1990–1996. doi: 10.1128/AAC.01688-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavallari JF, Lamers RP, Scheurwater EM, Matos AL, Burrows LL. 2013. Changes to its peptidoglycan-remodeling enzyme repertoire modulate beta-lactam resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:3078–3084. doi: 10.1128/AAC.00268-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopp U, Wiedemann B, Lindquist S, Normark S. 1993. Sequences of wild-type and mutant ampD genes of Citrobacter freundii and Enterobacter cloacae. Antimicrob Agents Chemother 37:224–228. doi: 10.1128/AAC.37.2.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ehrhardt AF, Sanders CC, Romero JR, Leser JS. 1996. Sequencing and analysis of four new Enterobacter ampD alleles. Antimicrob Agents Chemother 40:1953–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidtke AJ, Hanson DH. 2006. Model system to evaluate the effect of ampD mutations on AmpC-mediated β-lactam resistance. Antimicrob Agents Chemother 50:2030–2037. doi: 10.1128/AAC.01458-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanson ND, Sanders CC. 1999. Regulation of inducible AmpC beta-lactamase expression among Enterobacteriaceae. Curr Pharm Des 5:881–894. [PubMed] [Google Scholar]
- 31.Kuga A, Okamoto R, Inoue M. 2000. ampR gene mutations that greatly increase class C beta-lactamase activity in Enterobacter cloacae. Antimicrob Agents Chemother 44:561–567. doi: 10.1128/AAC.44.3.561-567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaneko K, Okamoto R, Nakano R, Kawakami S, Inoue M. 2005. Gene mutations responsible for overexpression of AmpC beta-lactamase in some clinical isolates of Enterobacter cloacae. J Clin Microbiol 43:2955–2958. doi: 10.1128/JCM.43.6.2955-2958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dietz H, Pfeifle D, Wiedemann B. 1997. The signal molecule for beta-lactamase induction in Enterobacter cloacae is the anhydromuramyl-pentapeptide. Antimicrob Agents Chemother 41:2113–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peter K, Korfmann G, Wiedemann B. 1988. Impact of the ampD gene and its product on beta-lactamase production in Enterobacter cloacae. Rev Infect Dis 10:800–805. doi: 10.1093/clinids/10.4.800. [DOI] [PubMed] [Google Scholar]
- 35.Chow JW, Fine MJ, Shlaes DM, Quinn JP, Hooper DC, Johnson MP, Ramphal R, Wagener MM, Miyashiro DK, Yu VL. 1991. Enterobacter bacteremia: clinical features and emergence of antibiotic resistance during therapy. Ann Intern Med 115:585–590. doi: 10.7326/0003-4819-115-8-585. [DOI] [PubMed] [Google Scholar]
- 36.Kaye KS, Cosgrove S, Harris A, Eliopoulos GM, Carmeli Y. 2001. Risk factors for emergence of resistance to broad-spectrum cephalosporins among Enterobacter spp. Antimicrob Agents Chemother 45:2628–2630. doi: 10.1128/AAC.45.9.2628-2630.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi SH, Lee JE, Park SJ, Choi SH, Lee SO, Jeong JY, Kim MN, Woo JH, Kim YS. 2008. Emergence of antibiotic resistance during therapy for infections caused by Enterobacteriaceae producing AmpC beta-lactamase: implications for antibiotic use. Antimicrob Agents Chemother 52:995–1000. doi: 10.1128/AAC.01083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrasevic AT, Dowzicky MJ. 2012. In vitro activity of tigecycline and comparators against Gram-negative pathogens isolated from blood in Europe (2004-2009). Int J Antimicrob Agents 39:115–123. doi: 10.1016/j.ijantimicag.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 39.Hoban DJ, Badal R, Bouchillon S, Hackel M, Kazmierczak K, Lascols C, Hawser S. 2014. In vitro susceptibility and distribution of beta-lactamases in Enterobacteriaceae causing intra-abdominal infections in North America 2010-2011. Diagn Microbiol Infect Dis 79:367–372. doi: 10.1016/j.diagmicrobio.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 40.Guérin F, Goudergues B, Giard J-C, Cattoir V. 2014. Revisiting the regulation mechanisms of Ampc-mediated beta-lactam resistance in Enterobacter cloacae, abstr C-1890. In Abstr 54th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC, 9 to 12 September 2014. [Google Scholar]
- 41.Ren Y, Ren Y, Zhou Z, Guo X, Li Y, Feng L, Wang L. 2010. Complete genome sequence of Enterobacter cloacae subsp. cloacae type strain ATCC 13047. J Bacteriol 192:2463–2464. doi: 10.1128/JB.00067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Derbise A, Lesic B, Dacheux D, Ghigo JM, Carniel E. 2003. A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol Med Microbiol 38:113–116. doi: 10.1016/S0928-8244(03)00181-0. [DOI] [PubMed] [Google Scholar]
- 43.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doublet B, Douard G, Targant H, Meunier D, Madec JY, Cloeckaert A. 2008. Antibiotic marker modifications of lambda Red and FLP helper plasmids, pKD46 and pCP20, for inactivation of chromosomal genes using PCR products in multidrug-resistant strains. J Microbiol Methods 75:359–361. doi: 10.1016/j.mimet.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 45.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiedemann B, Dietz H, Pfeifle D. 1998. Induction of beta-lactamase in Enterobacter cloacae. Clin Infect Dis 27(Suppl):S42–S47. doi: 10.1086/514921. [DOI] [PubMed] [Google Scholar]
- 47.Dietz H, Wiedemann B. 1996. The role of N-actylglucosaminyl-1,6 anhydro N-acetylmuramyl-l-alanyl-d-glutamyl-meso-diaminopimelic acid-d-alanine for the induction of beta-lactamase in Enterobacter cloacae. Zentralbl Bakteriol 284:207–217. doi: 10.1016/S0934-8840(96)80096-X. [DOI] [PubMed] [Google Scholar]
- 48.Balasubramanian D, Kumari H, Mathee K. 2015. Pseudomonas aeruginosa AmpR: an acute-chronic switch regulator. Pathog Dis 73:1–14. doi: 10.1111/2049-632X.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korfmann G, Sanders CC, Moland ES. 1991. Altered phenotypes associated with ampD mutations in Enterobacter cloacae. Antimicrob Agents Chemother 35:358–364. doi: 10.1128/AAC.35.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naas T, Massuard S, Garnier F, Nordmann P. 2001. AmpD is required for regulation of expression of NmcA, a carbapenem-hydrolyzing beta-lactamase of Enterobacter cloacae. Antimicrob Agents Chemother 45:2908–2915. doi: 10.1128/AAC.45.10.2908-2915.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Juan C, Macia MD, Gutierrez O, Vidal C, Perez JL, Oliver A. 2005. Molecular mechanisms of beta-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother 49:4733–4738. doi: 10.1128/AAC.49.11.4733-4738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghosh AS, Chowdhury C, Nelson DE. 2008. Physiological functions of d-alanine carboxypeptidases in Escherichia coli. Trends Microbiol 16:309–317. doi: 10.1016/j.tim.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 53.Meberg BM, Paulson AL, Priyadarshini R, Young KD. 2004. Endopeptidase penicillin-binding proteins 4 and 7 play auxiliary roles in determining uniform morphology of Escherichia coli. J Bacteriol 186:8326–8336. doi: 10.1128/JB.186.24.8326-8336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mahlen SD, Morrow SS, Abdalhamid B, Hanson ND. 2003. Analyses of ampC gene expression in Serratia marcescens reveal new regulatory properties. J Antimicrob Chemother 51:791–802. doi: 10.1093/jac/dkg133. [DOI] [PubMed] [Google Scholar]
- 55.Lovett PS, Rogers EJ. 1996. Ribosome regulation by the nascent peptide. Microbiol Rev 60:366–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vazquez-Laslop N, Thum C, Mankin AS. 2008. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell 30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 57.Yao S, Blaustein JB, Bechhofer DH. 2008. Erythromycin-induced ribosome stalling and RNase J1-mediated mRNA processing in Bacillus subtilis. Mol Microbiol 69:1439–1449. doi: 10.1111/j.1365-2958.2008.06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zamorano L, Moya B, Juan C, Mulet X, Blazquez J, Oliver A. 2014. The Pseudomonas aeruginosa CreBC two-component system plays a major role in the response to beta-lactams, fitness, biofilm growth, and global regulation. Antimicrob Agents Chemother 58:5084–5095. doi: 10.1128/AAC.02556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gonzalez-Leiza SM, de Pedro MA, Ayala JA. 2011. AmpH, a bifunctional dd-endopeptidase and dd-carboxypeptidase of Escherichia coli. J Bacteriol 193:6887–6894. doi: 10.1128/JB.05764-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park JT, Uehara T. 2008. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol Mol Biol. Rev 72:211–227. doi: 10.1128/MMBR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yin J, Mao Y, Ju L, Jin M, Sun Y, Jin S, Gao H. 2014. Distinct roles of major peptidoglycan recycling enzymes in beta-lactamase production in Shewanella oneidensis. Antimicrob Agents Chemother 58:6536–6543. doi: 10.1128/AAC.03238-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zamorano L, Reeve TM, Deng L, Juan C, Moyá B, Cabot G, Vocadlo DJ, Mark BL, Oliver A. 2010. NagZ inactivation prevents and reverts beta-lactam resistance, driven by AmpD and PBP 4 mutations, in Pseudomonas aeruginosa. Antimicrob Agents Chemother 54:3557–3563. doi: 10.1128/AAC.00385-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lindberg F, Normark S. 1987. Common mechanism of ampC beta-lactamase induction in enterobacteria: regulation of the cloned Enterobacter cloacae P99 beta-lactamase gene. J Bacteriol 169:758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.