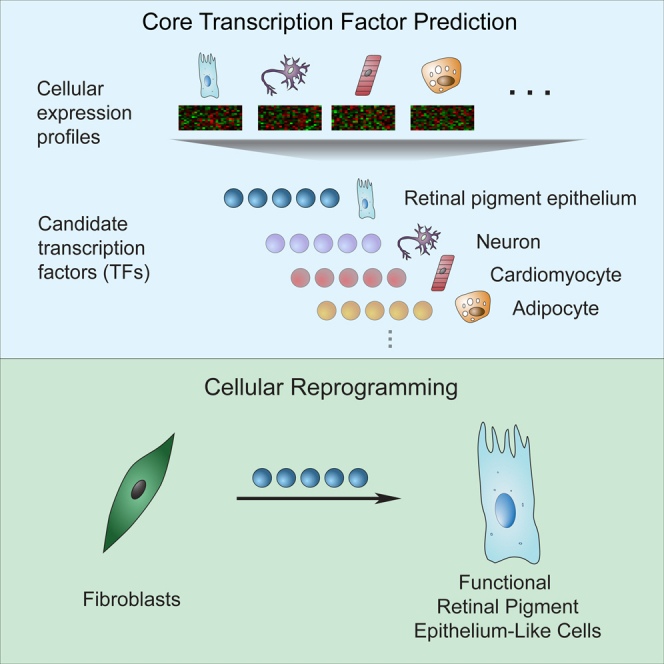
Summary
Hundreds of transcription factors (TFs) are expressed in each cell type, but cell identity can be induced through the activity of just a small number of core TFs. Systematic identification of these core TFs for a wide variety of cell types is currently lacking and would establish a foundation for understanding the transcriptional control of cell identity in development, disease, and cell-based therapy. Here, we describe a computational approach that generates an atlas of candidate core TFs for a broad spectrum of human cells. The potential impact of the atlas was demonstrated via cellular reprogramming efforts where candidate core TFs proved capable of converting human fibroblasts to retinal pigment epithelial-like cells. These results suggest that candidate core TFs from the atlas will prove a useful starting point for studying transcriptional control of cell identity and reprogramming in many human cell types.
Graphical Abstract
Highlights
-
•
Core transcription factors (TFs) are predicted for >200 cell types/tissues
-
•
Predicted TFs for retinal pigment epithelial (RPE) cells can reprogram fibroblasts
-
•
These reprogrammed RPE-like cells are functionally similar to primary RPE
-
•
The sets of predicted factors may facilitate studies of control of cell identity
Small sets of core transcription factors establish gene expression programs that determine cellular function. Young, Lee, and colleagues have generated an atlas of candidate core transcription factors for over 200 human cell types. Candidate core transcription factors proved capable of reprogramming cells, suggesting this atlas may facilitate studies of transcriptional regulation and reprogramming for clinically important cell types.
Introduction
Cell identity is controlled in large part by the action of transcription factors (TFs) that recognize and bind specific sequences in the genome and regulate gene expression. While approximately half of all TFs are expressed in any one cell type (Vaquerizas et al., 2009), a small number of core TFs are thought to be sufficient to establish control of the gene expression programs that define cell identity (Buganim et al., 2013, Graf and Enver, 2009, Morris and Daley, 2013, Sancho-Martinez et al., 2012, Vierbuchen and Wernig, 2012, Yamanaka, 2012). It would be valuable to identify these core TFs for all cell types; an atlas of candidate core regulators would complement the Encyclopedia of Regulatory DNA Elements (ENCODE) (Rivera and Ren, 2013, Stergachis et al., 2013), guide exploration of the principles of transcriptional regulatory networks, enable more systematic research into the mechanistic and global functions of these key regulators of cell identity, and facilitate advances in direct reprogramming for clinically relevant cell types (Henriques et al., 2013, Iwafuchi-Doi and Zaret, 2014, Soufi et al., 2012, Xie and Ren, 2013).
Core TFs that control individual cell identity have been identified previously, but systematic efforts to do so for most cell types have been relatively rare until recently. Early efforts focused on the experimental identification of genes that were differentially expressed in one cell type, compared to a small range of other cell types, and shown to have roles in controlling specific cell identities. Examples include MyoD1, which can convert fibroblasts to muscle cells upon overexpression in fibroblasts (Tapscott et al., 1988), and Oct4, whose loss results in the loss of the pluripotent cell population in the mammalian embryo (Nichols et al., 1998). More recently, cellular reprogramming experiments, where ectopic expression of TFs converts cells from one type to another, arose as a particularly stringent test of the ability of TFs to establish cell identity (Buganim et al., 2013, Graf and Enver, 2009, Morris and Daley, 2013, Sancho-Martinez et al., 2012, Vierbuchen and Wernig, 2012, Yamanaka, 2012). While powerful demonstrations of the role of TFs in control of cell identity, these experimental approaches are necessarily focused on specific cell types.
The development of genome-scale technologies has enabled more global attempts to predict candidate factors that control cell identity. Genome-wide gene expression and epigenome analysis across multiple cell types have been used to identify candidate core factors via computational methods (Cahan et al., 2014, Heinäniemi et al., 2013, Lang et al., 2014, Morris et al., 2014, Roost et al., 2015). While broad in scope, these studies assess their predictions using more easily scalable methods and typically do not assess whether predicted factors are sufficient to establish cell identity.
We describe here the identification of candidate core TFs across the largest collection of different human cell types to date. A computational approach was devised to systematically identify candidate core TFs for most known human cell types. Importantly, we demonstrate with ectopic expression experiments that these predictions can identify factors capable of converting cell identity, thus satisfying a stringent criterion that is not tested with most other approaches to identify key TFs. Expression of core factors identified for retinal pigment epithelial (RPE) cells was sufficient to reprogram human fibroblasts into RPE-like cells. These cells were functionally characterized for their similarity to RPE cells derived from healthy individuals and were shown to share many features—including morphology, gene expression, and the ability to perform canonical RPE processes—and to integrate into the host RPE layer in transplantation experiments. These results suggest that the atlas of candidate core TFs should be useful for reprogramming additional clinically important cell types and for systematically discovering the regulatory circuitries for these cells.
Results
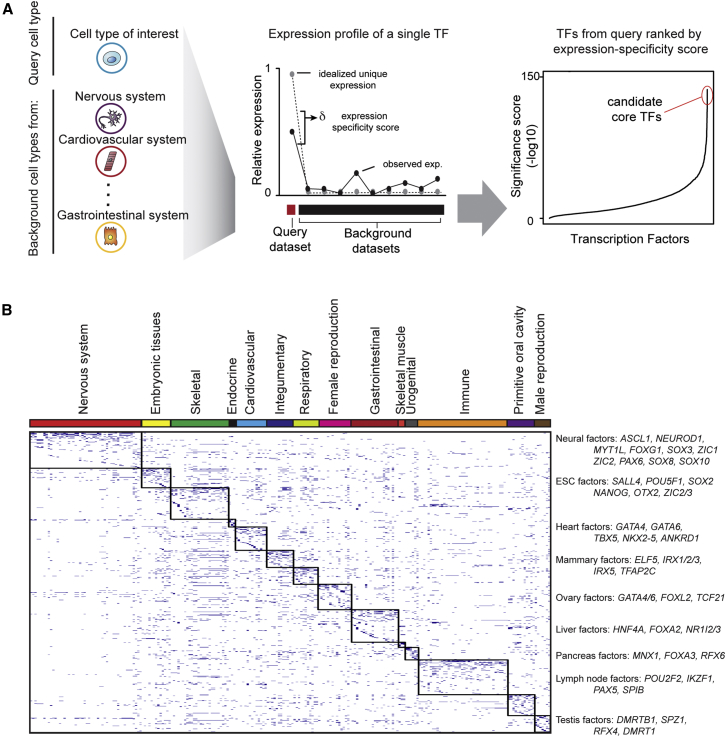
To identify candidate core TFs, we searched for TFs that fit two basic characteristics of known core TFs: the genes encoding known core TFs are typically expressed in a relatively cell-type-specific fashion and at relatively high levels (Figure 1A). The algorithm we developed quantifies both cell type specificity and the relative level of gene expression by using an entropy-based measure of Jensen-Shannon divergence (Cabili et al., 2011, Fuglede and Topsoe, 2004) to compare the expression of a TF in a cell type of interest (the query dataset) to the expression of that factor across a range of cell types that represent the diversity of human cell types and tissues (the background dataset) (Supplemental Information). The idealized case is when a TF is expressed at a relatively high level in the query dataset and not expressed in any other cell type in the background dataset. The algorithm generates a specificity score for each TF based on how well the actual expression data match with this idealized case and ranks each TF using a nonparametric rank-product approach to aggregate the results from multiple query datasets for a given cell type (Breitling et al., 2004).
Figure 1.
A General Approach to Identify Candidate Core TFs in Human Cells
(A) Computational approach used to identify candidate core TFs in human cells. Left panel: collection of gene expression profiles of a query cell type and representative cell types from the Human Body Index collection of expression data. Middle panel: expression profile of a single TF across a query dataset and a range of background datasets. The idealized case of expression level of a TF (gray circle, dashed line) is compared to the observed data to calculate the expression-specificity score of the TF. Right panel: plot depicting the distribution of significance scores of expression specificity for all TFs. Factors are arranged on the x axis in order of significance scores. Significance scores are indicated on the y axis. The highest scoring TFs are considered the best candidate core TFs and highlighted in the red circle.
(B) Representation of the collection of candidate core TFs for 233 tissue and cell types. Tissue and cell types are arranged on the x axis and ordered according to anatomical groups, represented by the colored bar at the top. Genes are arranged on the y axis. Blue dashes represent candidate core TFs in a cell type. Clusters of candidate core TFs in cell types representing an anatomical group are boxed. Representative genes are listed on the side.
This approach was used to score TFs for over 200 cell types/tissues collated from the Human Body Index collection of expression data, together with additional well-studied cell types (Figure 1B; Figure 2; Table S1; Supplemental Information). The complete atlas contains the scores for all TFs in all cell types, but for simplicity of additional analyses and manageability of experimental validation, we focused on the ten top-scoring TFs in each cell type as the primary candidate core TFs.
Figure 2.
Candidate Core TFs for 233 Tissue and Cell Types
Tissue and cell types were grouped into categories corresponding to different anatomical systems in the human body. Within each category, tissue and cell types were ordered using hierarchical clustering. The distance matrix was calculated by first rank-ordering the specificity scores for all TFs in each tissue and cell type within a category and then finding the Kendall tau correlation coefficient for each pairwise comparison of tissue and cell types within the category. For each individual tissue or cell type, the ten top-scoring candidate core TFs are listed.
503 different TFs were considered candidate core TFs for one or more cell types or tissues. As expected given our methodology, the candidate core TFs were expressed at higher levels than non-core TFs (Figure 3A), and individual factors were generally considered candidate core TFs in limited numbers of cell or tissue types (Figure 3B). DNA-binding domain analysis indicates that the candidate core TFs have a different distribution of DNA binding domains compared to other TFs, with relatively increased frequencies of domains frequently associated with developmental regulators (homeobox and helix-loop-helix) and relatively decreased frequencies of other domains such as SCAN, KRAB, and C2H2 zinc finger (Figure 3C). The candidate core TFs are generally well conserved, as orthologs typically exist for the factors through vertebrate and metazoan species (Figure 3D). The genes encoding candidate core TFs are generally associated with super-enhancers, transcriptional regulatory elements that are associated with genes that play important roles in cell identity (Hnisz et al., 2013, Parker et al., 2013, Whyte et al., 2013). Gene set enrichment analysis (GSEA) shows that super-enhancers of each cell type are enriched among the highest scoring TFs in the atlas, and this enrichment occurs in a cell-type-specific manner (Figure 3E). These data are consistent with the highest scoring TFs in the atlas having roles in control of cell identity. More extensive characterization and discussion of the set of candidate core TFs is included in the Supplemental Information and Figure S1.
Figure 3.
Characterization of Candidate Core TFs
(A) Box plots depicting the expression levels of candidate core TFs and non-core TFs. The significance of the difference between two groups was determined using a two-tailed Mann-Whitney test. For each plot, the top and bottom box edges mark the first and third quartiles, while the solid black line within the box marks the median. The top whisker line marks the largest data point that is within 1.5-fold of the interquartile range from the third quartile. The bottom whisker line marks the smallest data point that is within 1.5-fold of the interquartile range from the first quartile. Candidate core TFs are shown in gold. Non-core-TFs are shown in gray.
(B) Pie chart depicting the number of cell types in which a TF is considered as a candidate core TF.
(C) Bar chart representing the percentage of candidate core TFs and non-core TFs that are associated with different classes of DNA binding domains. The significance of the difference in distribution between candidate core TFs and non-core TFs across these categories is p < 0.003 and was determined using a chi-square test. The gray oval indicates the percentage of all TFs that are associated with the class of DNA binding domains as a point of comparison. Abbreviations for protein domains are: HOX, homeodomain; HLH, helix-loop-helix; BRLZ, basic region leucine zipper; HOLI, ligand binding domain of hormone receptor; ZnF_C4, c4 zinc finger in nuclear hormone receptors; HMG, high mobility group; ETS, erythroblast transformation specific; FH, forkhead; TBOX, T-box; POU, Pit-Oct-Unc; ZnF_GATA, zinc finger binding to DNA consensus sequence [AT]GATA[AG]; DWB, domain B in dwarfin family proteins; SANT, SWI3-ADA2-N-CoR-TFIIB DNA-binding domain; SCAN, SCAN domain; KRAB, Krueppel-associated box; ZnF_C2H2, zinc finger C2H2.
(D) Heatmap depicting the presence (blue) or absence (white) of orthologous genes in a species for each candidate core TF. The candidate core TFs are arranged as rows, and species are shown as columns. Species labels are colored using the following scheme: blue (primate), orange (mammal), purple (vertebrates), green (metazoa), and black (eukaryote). In the image, rows are clustered according to k-means clustering (k = 3).
(E) GSEA enrichment plots depicting the relationship between super-enhancer associated genes and high expression-specificity scores. Top panel: GSEA plot for genes associated with super-enhancers in CD4+ naive T cells and expression-specificity score. Enrichment score is plotted on the y axis. The x axis represents genes ordered by specificity score. The relationship when ordered by the expression specificity scores from CD4+ naive T cells is shown in blue. The relationship when ordered by the expression specificity scores from a non-matching cell type (embryonic stem cells) is shown in gray for comparison. p values for each are shown. Subsequent panels show similar relationships in different cell types. For each panel, the cell type is indicated. Super-enhancer associated genes are from that cell type. Blue curves represent the relationship when ordered by expression-specificity scores for that cell type. Gray curves represent the relationship when ordered by expression-specificity scores for a non-matching cell type (embryonic stem cells). p values for each are shown. E.S., enrichment score.
We examined whether factors from this atlas could induce a new cell identity as a stringent test of whether the atlas successfully identifies TFs that control cell identity. Ectopic expression of core TFs in fibroblasts can reprogram gene expression and produce cells with functional states similar to those that normally express those TFs (Buganim et al., 2013, Graf, 2011, Morris and Daley, 2013, Vierbuchen and Wernig, 2012, Yamanaka, 2012). Examination of the list of candidate core TFs predicted for embryonic stem cells shows good overlap with factors already used to reprogram murine or human fibroblasts to pluripotent stem cells (Table S2). Similar results are seen for several other cell types, including cardiomyocytes and hepatocytes (Table S2), and a comparison to a set of TFs that have been used for lineage reprogramming in human cells—summarized in Xu et al. (2015)—shows that roughly 70% of these lineage reprogramming factors are called candidate core TFs in the atlas (Table S3). To test factors from this atlas, RPE cells were chosen as the target cell type due to their growing relevance to cell therapy applications. Progressive degeneration of RPE cells is a major cause of age-related macular degeneration (AMD), and several clinical trials are currently assessing transplantation of RPE cells and stem-cell-derived RPE cells as a treatment for ocular disorders (Cyranoski, 2013, Cyranoski, 2014).
For ectopic expression experiments, nine of the top-scoring RPE core TF candidates—PAX6, LHX2, OTX2, SOX9, MITF, SIX3, ZNF92, GLIS3, and FOXD1—were selected and cloned into doxycycline-inducible lentiviral expression vectors (Figure 4A). Human foreskin fibroblasts (HFF) were then transduced with a cocktail of all nine factors. Colonies showing a “cobblestone”-like morphology characteristic of RPE cells were evident after 2 weeks of doxycycline induction (Figure 4A). Colonies were manually picked and further expanded into six independent RPE-like cell lines. Genotyping analysis showed that all six cell lines contained the PAX6, OTX2, MITF, SIX3, GLIS3, and FOXD1 expression constructs (Figure 4B).
Figure 4.
Ectopic Expression of RPE Candidate Core TFs Is Sufficient to Drive the Morphology and Gene Expression Program of Fibroblasts toward an RPE-like State
(A) Schematic outlining the ectopic expression of candidate core TFs in HFF. Lentiviral constructs were induced to express candidate core TFs with doxycycline (Dox). Scale bar, 50 μm.
(B) PCR and gel analysis of transgene integration for iRPE lines. Positive control (DNA of the constructs used to generate lentivirus) and negative control reactions are shown. Six different iRPE lines, labeled 1–6 are shown. Genes are indicated on the side.
(C) Immunostaining of iRPE-1 and iRPE-2 cells. Cells were immunostained with TJP1 (ZO-1). Scale bar 50 μm.
(D) Immunostaining imaging of RPE, iRPE-1, and iRPE-2 cells. Cells were immunostained for RPE cell markers CRALBP (green) and RPE65 (red) and with DAPI (blue). Scale bar, 50 μm.
(E) PCA comparing the gene expression profiles of iRPE cells to gene expression profiles of other cell types. Principal components (PC1–PC3) are shown on the x, y, and z axes. The expression profiles of HFF (black), iRPE cells (blue), RPE cells (light green), induced pluripotent stem (iPS)-RPE cells (green), iPS cells (red), and ES cells (orange red), and 106 additional cell types (gray) are shown.
(F) GSEA enrichment score of a previously published RPE signature gene set (Strunnikova et al., 2010) compared with genes differentially expressed between iRPE and fibroblasts. Genes are ranked along the x axis based on differential expression in iRPE cells versus fibroblasts, with more expressed in iRPE (red) to more expressed in fibroblasts (blue). Black tick marks indicate a gene from the RPE signature set. Enrichment score is shown on the y axis.
Two of the induced RPE (iRPE)-like cell lines, iRPE-1 and iRPE-2, were subjected to additional analysis. The iRPE cell lines exhibited characteristic expression of membrane-associated TJP1 (ZO-1), together with a “cobblestone” sheet morphology involving individual cells connected by tight junctions (Figure 4C), and maintained an RPE-like morphology in the presence of doxycycline for over 6 months (12 passages). Additionally, immunostaining indicates that the iRPE cells showed co-expression of CRALBP and RPE65 (Figure 4D), two well-known markers for RPE cells (Sparrow et al., 2010). Expression analysis shows that the candidate core TFs are expressed in both iRPE lines, and genes considered part of the RPE gene expression signature (Strunnikova et al., 2010) show substantial upregulation compared to fibroblasts (Table S4). Principal-component analysis (PCA) of genome-wide gene expression revealed that the two iRPE lines were as similar to primary RPE cells and induced pluripotent stem-cell-derived RPE cells as induced pluripotent stem cells are to embryonic stem cells (Figure 4E). Analysis of the genes differentially expressed between iRPE and fibroblasts shows that differentially expressed genes are enriched for genes considered part of the RPE gene expression signature (Figure 4F) (Strunnikova et al., 2010).
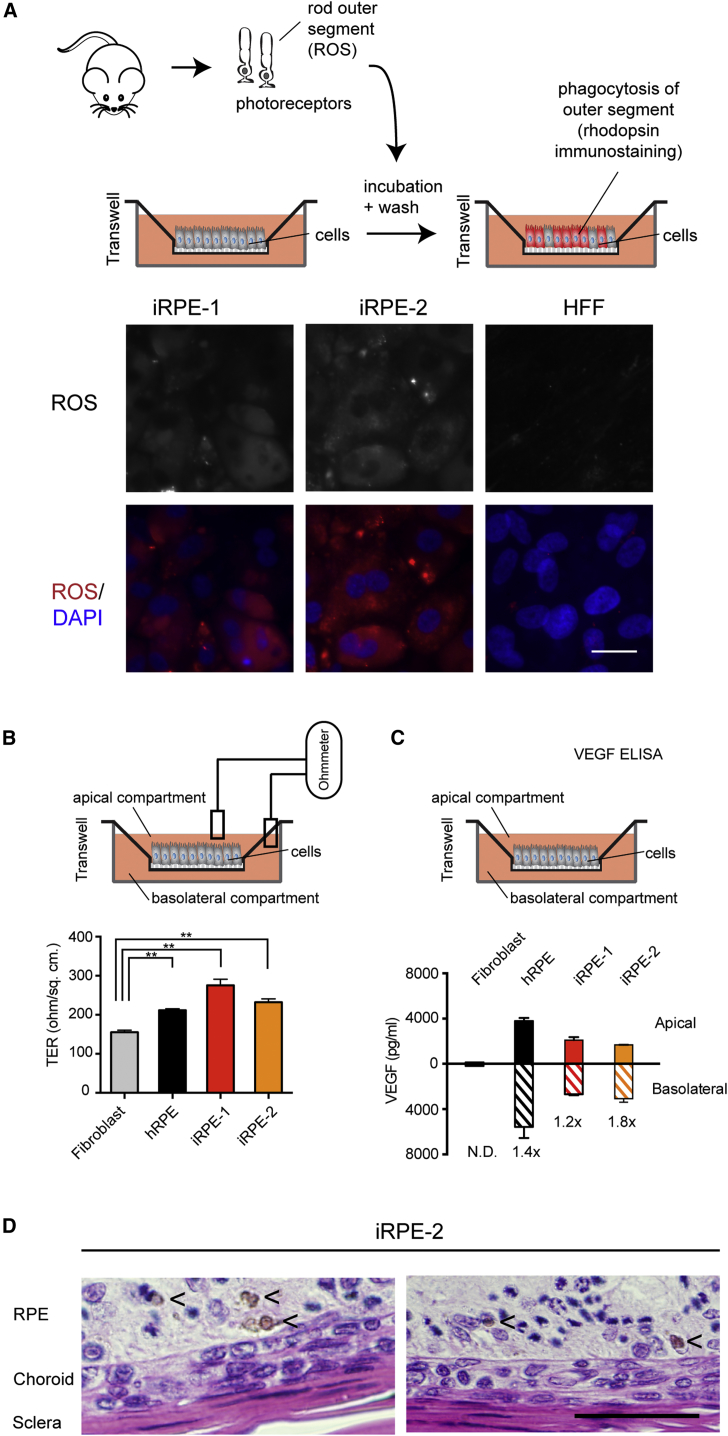
Ectopic expression of the RPE candidate core TFs results in cells that are functionally similar to RPE cells. RPE play crucial roles in the maintenance and function of retinal photoreceptors, including phagocytosis of shed outer segments of photoreceptors (Bok, 1993), transepithelial transport of nutrients and ions between the neural retina and the blood vessels (Strauss, 2005), and secretion of growth factors and hormones (Ford et al., 2011). For assaying phagocytosis, mouse rod outer segments (ROS) were incubated with iRPE cells or HFF cells. ROS incorporation was measured using an antibody against rhodopsin, which specifically recognizes a component of ROS. Both iRPE cell lines stained positive for rhodopsin, indicating binding and incorporation of ROS into the iRPE cells by phagocytosis (Figure 5A; Figure S2). To measure ion transport barrier function, we analyzed transepithelial electrical resistance (TER), which detects functional tight junctions (Stevenson et al., 1986). iRPE cells demonstrated effective barrier function that was significantly higher than fibroblasts and was as effective as that observed for RPE cells (Figure 5B). To evaluate secretion of growth factors, iRPE cells were examined for production of vascular endothelial growth factor (VEGF), which is released preferentially to the basolateral side of RPE cells to prevent endothelial cell apoptosis in the blood vessels (Saint-Geniez et al., 2009). No VEGF release was detected when fibroblasts were assayed (Figure 5C). The iRPE lines exhibit polarized secretion of VEGF similar to that produced by RPE cells (Figure 5C). Subretinal transplantation experiments showed that iRPE cells survive in vivo when transplanted in albino rats, and some integrate to the host RPE layer as pigmented cells (Figure 5D). Taken together, these results provide the most extensive characterization of iRPE to date and indicate that cells generated with the factors from our atlas are similar to RPE cells in terms of morphology, gene expression, and functionality.
Figure 5.
RPE-like Cells Have Functional Characteristics
(A) Schematic of the phagocytosis of ROS assay for iRPE function. Immunostaining for rhodopsin and DAPI are shown. The top row of images shows immunostaining for rhodopsin. The lower row of images shows the same fields with rhodopsin indicated in red and DAPI staining for DNA shown in blue. Scale bar, 25 μm.
(B) Schematic and results of TER assay for iRPE-1, iRPE-2, and hRPE cells (Salero et al., 2012). TER values for fibroblasts (gray), hRPE cells (black), iRPE-1 cells (red), and iRPE-2 cells (gold) are 155.2 ± 5 Ω/cm2, 211.4 ± 4 Ω/cm2, 275.6 ± 15 Ω/cm2, and 232.2 ± 8 Ω/cm2, respectively. TER was assayed in at least five biological replicates and is displayed as mean ± SD.
(C) Schematic and results for polarized release of VEGF assayed by ELISA. Values are shown for fibroblasts (nearly undetectable) and for hRPE (black), iRPE-1 (red), and iRPE-2 (gold), with the apical secretion values indicated with solid colors and the basolateral secretion values indicated with striped colors. The ratio of VEGF release (basolateral/apical) is shown below each bar. N.D., non detectable. ELISA was assayed in biological duplicates and is displayed as mean ± SD.
(D) Xenotransplant subretinal transplantations of wild-type albino Sprague-Dawley rats. H&E staining show pigmented donor cells iRPE-2 visible in the RPE layer. Single pigmented cells were identified in the host RPE layer in the doxycycline-treated group but not in the control iRPE group that did not receive doxycycline (data not shown). Pigmented cells are indicated with a “<” sign. Scale bar, 50 μm.
Discussion
The atlas of candidate core TFs presented here (Supplemental Information) provides a powerful starting point for studies of transcriptional regulation of cell identity and eventual applications for therapeutic purposes. The atlas itself is easily expanded with additional genome-wide expression data, which are relatively easy to obtain compared to other data types, especially for cell types that may be available in limited quantities. The approach is easy to implement and can be adapted to next-generation sequencing data as sufficient numbers and variety of datasets become available and may, thus, be generally useful for a wide range of researchers. The approach presented here capitalizes on basic principles of the expression level of known core TFs: relatively high expression and relatively cell-type-specific expression. Additional principles commonly associated with core TFs, such as autoregulation, binding in regulatory regions, or motif enrichment in regulatory regions may be useful to integrate in future versions.
The iRPE cells here represent the results of a stringent test for whether our approach successfully identifies TFs that can control cell identity. The factors here differ from, but overlap with, a set of factors previously used for RPE reprogramming (Zhang et al., 2014). Significantly, known oncogenic TFs such as MYC and signaling molecules such as activin A or retinoid acid together with Sonic hedgehog (SHH) were components of previous factor cocktails but are not required here. The iRPE cells generated here were characterized for morphology and gene expression and found to be largely similar to RPE. Moreover, the iRPE cells generated here were shown to have functional similarity to RPE cells and, thus, represent a step forward in the characterization of iRPE cells and our understanding of their potential use. These cells require continued expression of the transgenes, as withdrawal of doxycycline causes the cells to revert back toward a fibroblast morphology. This dependency is similarly observed for many other transdifferentiated cells (Buganim et al., 2012, Huang et al., 2011, Lujan et al., 2012, Sheng et al., 2012, Vierbuchen et al., 2010) and indicates that establishment of a fully self-sustaining RPE identity will require additional study. We anticipate that analysis of additional factors from our ranked list, as well as analysis of additional transdifferentiated and differentiated versions of RPE cells (Idelson et al., 2009, Kamao et al., 2014, Zhang et al., 2014), will prove useful in ultimately unraveling the complete transcriptional circuitry of RPE cells.
Multiple methods have been developed that can use high-throughput genomic data to identify factors critical for cell identity (Benayoun et al., 2014, Cahan et al., 2014, Davis and Eddy, 2013, Heinäniemi et al., 2013, Hwang et al., 2011, Lang et al., 2014, Morris et al., 2014, Roost et al., 2015, Zhou et al., 2011, Ziller et al., 2015). Many of these methods focus primarily on quantifying the differences between cell identities and less on the direct identification of factors controlling cell identity. Several of these approaches have experimentally verified that they are capable of identifying TFs important for cell identity, although none has demonstrated that the factors can establish cell identity to the extent shown here, possibly due to the extreme technical difficulty of these types of reprogramming experiments. Our expectation is that results for different methods of identifying candidate core TFs will eventually be compared and used in complementary fashions to gain insight on which TFs are critical for different cell types and which characteristics best define core TFs.
For the vast majority of human cell types, the core TFs and the transcriptional programs they control is poorly understood. Furthermore, much of disease-associated sequence variation occurs in transcriptional regulatory regions (Farh et al., 2015, Hnisz et al., 2013, Maurano et al., 2012), but the transcriptional mechanisms that lead to disease pathology are understood in only a few instances. The atlas of candidate core TFs described here should, therefore, facilitate future exploration of the functions of key regulators of cell identity, mapping of cellular regulatory circuitries, and investigation of disease-associated mechanisms.
Experimental Procedures
Additional details are provided in the Supplemental Experimental Procedures.
Identification of Candidate Core TFs
Briefly, an entropy-based measure of Jensen-Shannon divergence (Cabili et al., 2011) was adopted to identify candidate core TFs based on the relative level and cell type specificity of expression of a given factor in one cell type compared to a background dataset of diverse human cell and tissue types. Expression datasets used are provided in Table S5. Additional details are provided in the Supplemental Experimental Procedures.
Cell Culture
Human RPE cells were maintained in epithelial cell medium supplemented with 2% fetal bovine serum, 1× epithelial cell growth supplement, and 1× penicillin-streptomycin solution. HFF were maintained in DMEM supplemented with 15% Tet System Approved fetal bovine serum, 2 mM L-glutamine, and 100 U/ml penicillin-streptomycin.
Construction of Lentiviral Vectors
The Lenti-X Tet-On Advanced Inducible Expression System was used. Plasmids containing the full coding sequence of PAX6, OTX2, LHX2, MITF, SIX3, SOX9, GLIS3, FOXD1, or ZNF92 were obtained from Open Biosystems, Origene, or the Dana Farber/Harvard Cancer Center DNA Resource Core. Coding DNA sequences were amplified and cloned into the target vector via homologous recombination using the In-Fusion cloning system. Expression plasmids were transformed and maintained in STBL4 cells.
Viral Preparation and Transduction of HFF
For ectopic expression experiments, a human fetal fibroblast line expressing rtTA Advanced was generated by viral-mediated integration of an rtTA-expressing construct (from pLVX-Tet-On-Advanced). Cells were grown in 1 mg/ml geneticin for 2 weeks to select for cells harboring the construct.
For virus preparation, replication-incompetent lentiviral particles were packaged in 293T cells in the presence of the envelope (pMD2) and packaging (psPAX) plasmids. Viral supernatants from cultures 36, 48, 60, and 72 hr post-transfection were filtered through a 0.45-μM filter. High-titer virus preparations for all nine TFs were then added to HFF in the presence of 5 μg/ml of polybrene (day 1). A second transduction with virus for all nine factors was performed the next day (day 2). After 2 days, transduced HFF were split and transferred to iRPE medium (discussed later) (day 3). The following day, iRPE medium was supplemented with 2 mg/ml doxycycline (day 4). Medium was replaced every 3 days, and fresh doxycycline was added with every medium replacement.
iRPE Growth Conditions
iRPE lines were plated on Matrigel Basement Membrane Matrix-coated plates. iRPE cells were grown on Minimum Essential Medium (MEM) Eagle Alpha Modification containing 5% Tet System Approved fetal bovine serum, 1× N1 Medium Supplement, 1% sodium pyruvate, 2 mM L-glutamine, 1× MEM Non-Essential Amino Acids, 1 mg/ml Geneticin, 100 U/ml penicillin-streptomycin and THT (20 mg/l hydrocortisone, 250 mg/l taurine, 0.013 mg/l triiodothyronine). Media was supplemented with 2 mg/ml doxycycline. Cells were incubated at 37°C with 5% CO2 in a humidified incubator.
Genotyping
Cells were lysed, and genomic DNA was purified by treating samples with proteinase K, RNase A, and phenol-chloroform extraction. DNA was amplified using GoTaq Green Core Mix.
Immunostaining and Imaging
For immunostaining analysis, cells were grown in Corning transwell polyester membrane cell culture inserts for 8 weeks in iRPE medium supplemented with 2 mg/ml doxycycline. Medium was replaced every 3 days. Cells were then fixed and washed, and a 2-mm biopsy punch of the transwell membrane was transferred to a glass slide. Slides were incubated in blocking/permeabilizing solution, washed, treated with antibodies, and mounted. Slides were visualized under a fluorescence microscope (Zeiss Axio Observer D1).
Phagocytosis Assay
ROS were isolated following previously described protocols (Ryeom et al., 1996). Retinas were dissected immediately following sacrifice from 25 mice, ROS were isolated, and approximately 1.0 × 104 ROS were added to the supernatant of confluent cell cultures in transwells. All work was performed under protocols approved by the institutional animal care and use committee. The cells were then incubated for 2 hr at 37°C. Transwells were washed to remove all unbound ROS before fixation. Each transwell was fixed, immunostained for rhodopsin and DAPI, and visualized.
TER
iRPE and RPE cells (Salero et al., 2012) were grown in Corning transwell polyester membrane cell culture inserts for 8 weeks in iRPE medium supplemented with 2 mg/ml doxycycline. Medium was replaced every 3 days. Resistance was measured using the EVOM Epithelial Voltohmmeter.
VEGF-A Release
iRPE and RPE cells (Salero et al., 2012) were grown in Corning transwell polyester membrane cell culture inserts for 8 weeks in iRPE medium supplemented with 2 mg/ml doxycycline. RPE cells were collected from human cadaver donors for research purposes. The protocol was reviewed and found to be exempt category 4 by the Albany Medical College Institutional Review Board. Medium was replaced every 3 days with fresh doxycycline. Conditioned medium from apical and basal chambers of the same transwell insert was collected 24 hr following a complete medium change. VEGF-A protein secretion in conditioned medium was measured using a Human VEGF ELISA kit following the manufacturer’s suggested protocol (Life Technologies).
Transplantation
To study the ability of iRPE to integrate into the native retina, we have performed subretinal transplantations into the wild-type rat retina. All animal studies were designed and performed according to the Association for Research in Vision and Ophthalmology guidelines for the use of animals in ophthalmic and visual research and approved by the institutional animal care and use committee. Three-week-old albino Sprague-Dawley rats (Taconic Biosciences) were used in these experiments. One day before the surgery, all animals were switched to cyclosporine-A-supplemented water (210 mg/l) and remained on immunosuppressive treatment until the end of the study. One group of iRPE-transplanted animals also received doxycycline in the water.
For the surgery, animals were anesthetized by intraperitoneal injection of ketamine/xylazine. Topical proparacaine (anesthetic) and tropicamide (mydriatic agent) drops were applied.
The subretinal injection was performed in one eye per animal using a 50-μm beveled glass needle connected to a 10-μl Hamilton syringe through polyethylene tubing. The success of the injection and lack of complications (hemorrhage, retinotomy, leakage of cells into the vitreous) was assessed by fundus examination. Antibiotic ointment was applied to the eye for recovery.
Experimental groups were as follows: iRPE with doxycycline treatment (n = 5), iRPE without doxycycline treatment (n = 5), hRPE (n = 5) as positive control, vehicle injection (n = 5), and non-injected eyes (n = 5) as negative controls—five groups total.
Two weeks after the injection animals were euthanized by CO2 inhalation, eyes were enucleated and fixed in alcohol fixative (Excalibur Pathology), embedded in paraffin, and sectioned.
Author Contributions
A.C.D. contributed to the design of all experiments and performed reprogramming experiments. Z.P.F. developed the computational method used and provided all bioinformatics-based analyses. K.J.W. performed the phagocytosis assay. P.B. and M.J.Y. performed the in vivo transplantation experiments. M.A.C. performed iRPE colony selection. J.S.S. performed immunostaining and VEGF release experiments. E.C. provided maintenance of RPE and iRPE lines. C.C. performed the TER experiments. D.D. contributed to experimental design. N.M.H. generated the lentiviral constructs. R.J. and S.T. contributed to the conceptual development of the study. T.I.L. and R.A.Y. initially conceived the study and contributed to the design of experiments. A.C.D., Z.P.F., T.I.L., and R.A.Y. wrote the manuscript. M.A.C., K.J.W., R.J., and S.T. contributed critical comments.
Acknowledgments
We thank T. Volkert, J. Love, and S. Gupta at the Whitehead Genome Technologies Core; T. Blenkinsop, B. Lesch, A. Sigova, and S. Tsang for experimental assistance; Y. Bouganim, M. Mitalipova, F. Soldner, for helpful discussion; and G. Frampton and P. Thiru for curation of datasets. This work was supported by the NIH grants HG002668 (R.A.Y.) and CA146445 (R.A.Y. and T.I.L.) and a grant from the Skolkovo Foundation (R.A.Y. and R.J.). The authors declare competing financial interests: R.J. is a cofounder of Fate Therapeutics and an advisor to Stemgent, and R.A.Y. is a founder of Syros Pharmaceuticals.
Published: October 22, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, two figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2015.09.016.
Contributor Information
Tong Ihn Lee, Email: tlee@wi.mit.edu.
Richard A. Young, Email: young@wi.mit.edu.
Accession Numbers
The accession number for the raw and processed sequencing and microarray data reported in this paper is GEO: GSE64264 (http://www.ncbi.nlm.nih.gov/geo/).
Supplemental Information
References
- Benayoun B.A., Pollina E.A., Ucar D., Mahmoudi S., Karra K., Wong E.D., Devarajan K., Daugherty A.C., Kundaje A.B., Mancini E. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158:673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok D. The retinal pigment epithelium: a versatile partner in vision. J. Cell Sci. Suppl. 1993;17:189–195. doi: 10.1242/jcs.1993.supplement_17.27. [DOI] [PubMed] [Google Scholar]
- Breitling R., Armengaud P., Amtmann A., Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Buganim Y., Itskovich E., Hu Y.C., Cheng A.W., Ganz K., Sarkar S., Fu D., Welstead G.G., Page D.C., Jaenisch R. Direct reprogramming of fibroblasts into embryonic Sertoli-like cells by defined factors. Cell Stem Cell. 2012;11:373–386. doi: 10.1016/j.stem.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Faddah D.A., Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat. Rev. Genet. 2013;14:427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P., Li H., Morris S.A., Lummertz da Rocha E., Daley G.Q., Collins J.J. CellNet: network biology applied to stem cell engineering. Cell. 2014;158:903–915. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyranoski D. Stem cells cruise to clinic. Nature. 2013;494:413. doi: 10.1038/494413a. [DOI] [PubMed] [Google Scholar]
- Cyranoski D. Stem-cell method faces fresh questions. Nature. 2014;507:283. doi: 10.1038/507283a. [DOI] [PubMed] [Google Scholar]
- Davis F.P., Eddy S.R. Transcription factors that convert adult cell identity are differentially polycomb repressed. PLoS ONE. 2013;8:e63407. doi: 10.1371/journal.pone.0063407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh K.K., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J., Shishkin A.A. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford K.M., Saint-Geniez M., Walshe T., Zahr A., D’Amore P.A. Expression and role of VEGF in the adult retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 2011;52:9478–9487. doi: 10.1167/iovs.11-8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglede B., Topsoe F. IEEE; 2004. Jensen-Shannon Divergence and Hilbert space embedding. Proceedings of the 2004 International Symposium on Information Theory; p. p. 31. [Google Scholar]
- Graf T. Historical origins of transdifferentiation and reprogramming. Cell Stem Cell. 2011;9:504–516. doi: 10.1016/j.stem.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Graf T., Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Heinäniemi M., Nykter M., Kramer R., Wienecke-Baldacchino A., Sinkkonen L., Zhou J.X., Kreisberg R., Kauffman S.A., Huang S., Shmulevich I. Gene-pair expression signatures reveal lineage control. Nat. Methods. 2013;10:577–583. doi: 10.1038/nmeth.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques T., Gilchrist D.A., Nechaev S., Bern M., Muse G.W., Burkholder A., Fargo D.C., Adelman K. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol. Cell. 2013;52:517–528. doi: 10.1016/j.molcel.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., He Z., Ji S., Sun H., Xiang D., Liu C., Hu Y., Wang X., Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- Hwang P.I., Wu H.B., Wang C.D., Lin B.L., Chen C.T., Yuan S., Wu G., Li K.C. Tissue-specific gene expression templates for accurate molecular characterization of the normal physiological states of multiple human tissues with implication in development and cancer studies. BMC Genomics. 2011;12:439. doi: 10.1186/1471-2164-12-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idelson M., Alper R., Obolensky A., Ben-Shushan E., Hemo I., Yachimovich-Cohen N., Khaner H., Smith Y., Wiser O., Gropp M. Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell. 2009;5:396–408. doi: 10.1016/j.stem.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Iwafuchi-Doi M., Zaret K.S. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamao H., Mandai M., Okamoto S., Sakai N., Suga A., Sugita S., Kiryu J., Takahashi M. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Reports. 2014;2:205–218. doi: 10.1016/j.stemcr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A.H., Li H., Collins J.J., Mehta P. Epigenetic landscapes explain partially reprogrammed cells and identify key reprogramming genes. PLoS Comput. Biol. 2014;10:e1003734. doi: 10.1371/journal.pcbi.1003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan E., Chanda S., Ahlenius H., Südhof T.C., Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc. Natl. Acad. Sci. USA. 2012;109:2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S.A., Daley G.Q. A blueprint for engineering cell fate: current technologies to reprogram cell identity. Cell Res. 2013;23:33–48. doi: 10.1038/cr.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S.A., Cahan P., Li H., Zhao A.M., San Roman A.K., Shivdasani R.A., Collins J.J., Daley G.Q. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell. 2014;158:889–902. doi: 10.1016/j.cell.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Parker S.C., Stitzel M.L., Taylor D.L., Orozco J.M., Erdos M.R., Akiyama J.A., van Bueren K.L., Chines P.S., Narisu N., Black B.L., NISC Comparative Sequencing Program. National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program Authors. NISC Comparative Sequencing Program Authors Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. USA. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera C.M., Ren B. Mapping human epigenomes. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roost M.S., van Iperen L., Ariyurek Y., Buermans H.P., Arindrarto W., Devalla H.D., Passier R., Mummery C.L., Carlotti F., de Koning E.J. KeyGenes, a tool to probe tissue differentiation using a human fetal transcriptional atlas. Stem Cell Reports. 2015;4:1112–1124. doi: 10.1016/j.stemcr.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryeom S.W., Sparrow J.R., Silverstein R.L. CD36 participates in the phagocytosis of rod outer segments by retinal pigment epithelium. J. Cell Sci. 1996;109:387–395. doi: 10.1242/jcs.109.2.387. [DOI] [PubMed] [Google Scholar]
- Saint-Geniez M., Kurihara T., Sekiyama E., Maldonado A.E., D’Amore P.A. An essential role for RPE-derived soluble VEGF in the maintenance of the choriocapillaris. Proc. Natl. Acad. Sci. USA. 2009;106:18751–18756. doi: 10.1073/pnas.0905010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salero E., Blenkinsop T.A., Corneo B., Harris A., Rabin D., Stern J.H., Temple S. Adult human RPE can be activated into a multipotent stem cell that produces mesenchymal derivatives. Cell Stem Cell. 2012;10:88–95. doi: 10.1016/j.stem.2011.11.018. [DOI] [PubMed] [Google Scholar]
- Sancho-Martinez I., Baek S.H., Izpisua Belmonte J.C. Lineage conversion methodologies meet the reprogramming toolbox. Nat. Cell Biol. 2012;14:892–899. doi: 10.1038/ncb2567. [DOI] [PubMed] [Google Scholar]
- Sheng C., Zheng Q., Wu J., Xu Z., Wang L., Li W., Zhang H., Zhao X.Y., Liu L., Wang Z. Direct reprogramming of Sertoli cells into multipotent neural stem cells by defined factors. Cell Res. 2012;22:208–218. doi: 10.1038/cr.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A., Donahue G., Zaret K.S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow J.R., Hicks D., Hamel C.P. The retinal pigment epithelium in health and disease. Curr. Mol. Med. 2010;10:802–823. doi: 10.2174/156652410793937813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis A.B., Neph S., Reynolds A., Humbert R., Miller B., Paige S.L., Vernot B., Cheng J.B., Thurman R.E., Sandstrom R. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B.R., Siliciano J.D., Mooseker M.S., Goodenough D.A. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J. Cell Biol. 1986;103:755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- Strunnikova N.V., Maminishkis A., Barb J.J., Wang F., Zhi C., Sergeev Y., Chen W., Edwards A.O., Stambolian D., Abecasis G. Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum. Mol. Genet. 2010;19:2468–2486. doi: 10.1093/hmg/ddq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott S.J., Davis R.L., Thayer M.J., Cheng P.F., Weintraub H., Lassar A.B. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242:405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- Vaquerizas J.M., Kummerfeld S.K., Teichmann S.A., Luscombe N.M. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T., Wernig M. Molecular roadblocks for cellular reprogramming. Mol. Cell. 2012;47:827–838. doi: 10.1016/j.molcel.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Südhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Ren B. Developmental biology. Enhancing pluripotency and lineage specification. Science. 2013;341:245–247. doi: 10.1126/science.1236254. [DOI] [PubMed] [Google Scholar]
- Xu J., Du Y., Deng H. Direct lineage reprogramming: strategies, mechanisms, and applications. Cell Stem Cell. 2015;16:119–134. doi: 10.1016/j.stem.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10:678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Zhang K., Liu G.H., Yi F., Montserrat N., Hishida T., Esteban C.R., Izpisua Belmonte J.C. Direct conversion of human fibroblasts into retinal pigment epithelium-like cells by defined factors. Protein Cell. 2014;5:48–58. doi: 10.1007/s13238-013-0011-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.X., Brusch L., Huang S. Predicting pancreas cell fate decisions and reprogramming with a hierarchical multi-attractor model. PLoS ONE. 2011;6:e14752. doi: 10.1371/journal.pone.0014752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller M.J., Edri R., Yaffe Y., Donaghey J., Pop R., Mallard W., Issner R., Gifford C.A., Goren A., Xing J. Dissecting neural differentiation regulatory networks through epigenetic footprinting. Nature. 2015;518:355–359. doi: 10.1038/nature13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.