Abstract
Graves’ ophthalmopathy (GO) is the commonest extra-thyroidal manifestation of Graves’ disease (GD). Associations between interleukin-related (IL) gene polymorphisms and GO have been reported in different populations. We aim to confirm such associations by conducting a meta-analysis. Totally 382 publications were retrieved in MEDLINE and EMBASE up to 25/2/2015. After removing the duplicates and assessing the studies, we retrieved 16 studies that met the selection criteria for meta-analysis, involving 12 polymorphisms in 8 IL-related genes, and 1650 GO cases and 2909 GD controls. The summary odds ratio (OR) and 95% confidence intervals (CI) were estimated. We found one polymorphism in IL1A (rs1800587, c.-889C>T) showing a suggestive association with GO in the meta-analysis (allelic model [T vs. C]: OR = 1.62, 95% CI: 1.00–2.62, P = 0.050, I2 = 53.7%; recessive model [TT vs. TC + CC]: OR = 2.39, 95% CI: 1.07–5.37, P = 0.039, I2 = 23.6%; heterozygous model [TC vs. CC]: OR = 1.52, 95% CI: 1.04–2.22, P = 0.034, I2 = 37.0%). No association with GO was detected for the other 7 genes (IL1B, IL1RA, IL4, IL6, IL12B, IL13 and IL23R). Our results thus indicate that IL1A is likely to be a genetic biomarker for GO. Further studies with larger sample sizes are warranted to confirm the associations of IL1A and other IL-related genes with GO.
Graves’ ophthalmopathy (GO), also known as thyroid-associated orbitopathy (TAO), is the commonest extra-thyroidal manifestation of Graves’ disease (GD), present in reportedly 25–50% of cases with GD1,2. It is also the commonest adult orbital disorder worldwide3. GO is characterized by lid retraction, lid lag, swelling and erythema of conjunctiva and periocular tissues, restrictive strabismus and proptosis4. Around 3–5% of GO patients develop sight-threatening complications, such as globe subluxation, corneal ulceration due to exposure keratopathy and optic neuropathy, which may result in irreversible visual impairment or even blindness if not treated promptly and properly3.
GO is a complex disease with interactive genetic and environmental factors5. Although the pathological mechanisms of GO are not completely understood6, cytokines, especially interleukins (ILs), are evidently involved4. Enlarged extraocular muscles and expansion of orbital adipose tissues7 were shown histologically with infiltration of activated T cells, B cells and macrophages8,9,10. Interleukins IL1RA11, IL1B12, IL412, IL612 and IL1012 were detected in the affected tissues of GO patients. In GO patients, higher levels of ILs were found in orbital tissues (IL1B13, IL714, IL813 and IL1013), tears (IL714) and serum (IL1RA11, IL615,16 and sIL-6R16). In addition, the expressions of IL1B and IL6 mRNA in the orbital adipose tissues were positively correlated with the radiological orbital volume in GO patients12. It has been speculated that Th1 lymphocytes and associated Th1-like cytokines (IL1B, IL2, IL12, INFG and TNFA) predominate in17, and promote the inflammatory, active phase of GO, while the Th2 family of cytokines (IL4, IL5 and IL10)18 affect the later fibrotic, inactive phases of the disease19,20.
Emerging studies have shown positive associations of polymorphisms in the IL-related genes (including ILs, interleukin receptors and receptor antagonists) with GO. Over 50 genetic polymorphisms of 17 IL-related genes, namely IL1A21,22, IL1B21,22,23,24, IL1R21, IL1RA21,25,26,27, IL228, IL329, IL429,30,31, IL529, IL628,32, IL833, IL929, IL1031, IL12B28,34,35, IL1329,30,36,37,38, IL1839, IL2140 and IL23R41,42,43, were reported in GO among different populations. However, the associations of these polymorphisms were inconsistent across different studies. For example, a single-nucleotide polymorphism (SNP), rs16944, in the IL1B gene was significantly associated with GO in a Chinese cohort from mainland China22, but not in Taiwan Chinese24, Caucasians23 or Iranian21. Also, the IL23R SNPs rs2201841 and rs10889677 were associated with GO in Caucasians41 but not in Japanese42,43. Therefore, we conducted a systematic review and meta-analysis to summarize the associations of reported IL-related genes with GO.
Results
Characteristics of identified studies
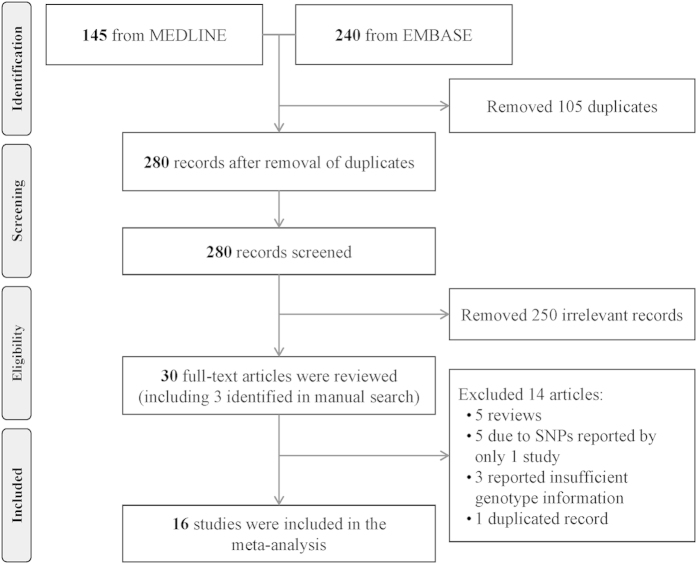
In the literature search, a total of 385 records, published between May 1, 1989 and February 25, 2015 were retrieved from the EMBASE and MEDLINE database. Among them, 105 were duplicated records. From the remaining 280 articles, we found 27 to be relevant according to our study criteria1,21,22,23,24,25,26,27,28,29,31,32,33,34,35,38,39,40,41,42,43,44,45,46,47,48,49. We further manually screened the reference lists and identified another 3 relevant studies30,36,37. Therefore, 30 articles were studied. We excluded 5 reviews1,44,47,48,49, 3 studies in which there were no sufficient genotypic or allelic data after communication with the authors38,45,46, and 1 study with duplicated samples43. Also excluded were 5 other studies, in which the SNPs were reported only once in the literature and not eligible for meta-analysis27,33,35,39,40. Finally, 16 studies investigating totally 12 genetic variations in 8 IL-related genes were included into this meta-analysis (Fig. 1).
Figure 1. Flow diagram of study selection process.
The 16 studies involved a total of 1,650 GO patients and 2,909 GD controls (GD without GO) recruited from Caucasian23,25,26,32,36,41, Chinese22,24,29,30, Japanese34,37,42 and Iranian21,28,31 populations. The sample sizes of GO patients ranged from 4426 to 20024, and GD controls from 2825 to 56922. The diagnostic criteria for GD were stated in all included studies except one23. In 12 studies, GO was classified according to the NOSPECS criteria (Supplementary Table 1) and only GO with NOSPECS Class 2 (or 3) and above were included21,22,23,24,26,28,29,31,32,34,36,37. The other 4 studies did not report the definition of GO25,30,41,42 (Table 1). Moreover, 9 studies reported the results of tests for Hardy-Weinberg equilibrium (HWE) in controls21,22,23,28,29,30,31,34,37, and there are 5 studies testing both genetic and non-genetic risk factors (e.g. gender, family history and smoking)21,24,30,32,37.
Table 1. Characteristics of studies included in the meta-analysis.
| No. | Study (year) | Country | Ethnicity | Definition |
Sample size(Cases/controls) | Gene | Test for HWE | Adjusted factor | |
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | ||||||||
| 1 | Cuddihy RM (1996) | USA | Caucasians | GO | GD without GO | 98/28 | IL1RA | Not reported | Not reported |
| 2 | Muhlberg T (1998) | Germany | Caucasians | GO | GD without GO | 44/100 | IL1RA | Not reported | Not reported |
| 3 | Bednarczuk T (2003) | Poland | Caucasians | GO | GD without GO | 93/168 | IL13 | Not reported | Not reported |
| 4 | Bednarczuk T (2004) | Poland | Caucasians | GO | GD without GO | 108/171 | IL6 | Not reported | Age, sex, smoking status |
| 5 | Yang Y (2005) | China | Chinese | GO | GD without GO | 98/89 | IL4 and IL13 | In HWE | Age of onset |
| 6 | Hiromatsu Y (2005) | Japan | Japanese | GO | GD without GO | 98/212 | IL13 | In HWE | Smoking status |
| 7 | Hiromatsu Y (2006) | Japan | Japanese | GO | GD without GO | 103/226 | IL12B | In HWE | Not reported |
| 8 | Huber AK (2008) | USA | Caucasians | GO | GD without GO | 103/111 | IL23R | Not reported | Not reported |
| 9 | Ban Y (2009) | Japan | Japanese | GO | GD without GO | 100/190 | IL 23R | Not reported | Not reported |
| 10 | Lacka K (2009) | Poland | Caucasians | GOa | GD with GOb | 75/42 | IL1B | In HWE | Not reported |
| 11 | Khalilzadeh O (2009) | Iran | Iranian | GO | GD without GO | 50/57 | IL1A, IL1B, IL1R, and IL1RA | In HWE | Family history of GD, age of onset, duration of GD |
| 12 | Anvari M (2010) | Iran | Iranian | GO | GD without GO | 50/57 | IL2, IL6, and IL12B | In HWE | Not reported |
| 13 | Liu N (2010) | China | Chinese | GO | GD without GOc | 190/569 | IL1A and IL1B | In HWE | Not reported |
| 14 | Zhu W (2010) | China | Chinese | GO | GD without GOc | 190/561 | IL3, IL4, IL5, IL9, and IL13 | In HWE | Not reported |
| 15 | Liu YH (2010) | China | Chinese | GO | GD without GO | 200/271 | IL1B | Not reported | Age, sex, smoking status |
| 16 | Khalilzadeh O (2010) | Iran | Iranian | GO | GD without GO | 50/57 | IL4 and IL10 | In HWE | Not reported |
GD: Grave’s Disease; GO: Grave’s Ophthalmopathy; HWE: Hardy Weinberg equilibrium; IL1A: Interleukin 1 Alpha; IL1B: Interleukin 1 Beta; IL1R: Interleukin 1 Receptor; IL1RA: Interleukin 1 Receptor Antagonist; IL12B: Interleukin 12 Beta; IL23R: Interleukin 23 Receptor; No.: number.
aGD with GO of NOSPECS class III or greater from the onset.
bGD with GO developed during 6 months to 7 years from GD onset.
cGD without GO of NOSPECS 0 and 1.
dNOSPECS classification system for the severity of GO (Supplementary Table 1).
Genetic associations of IL-related genes with GO
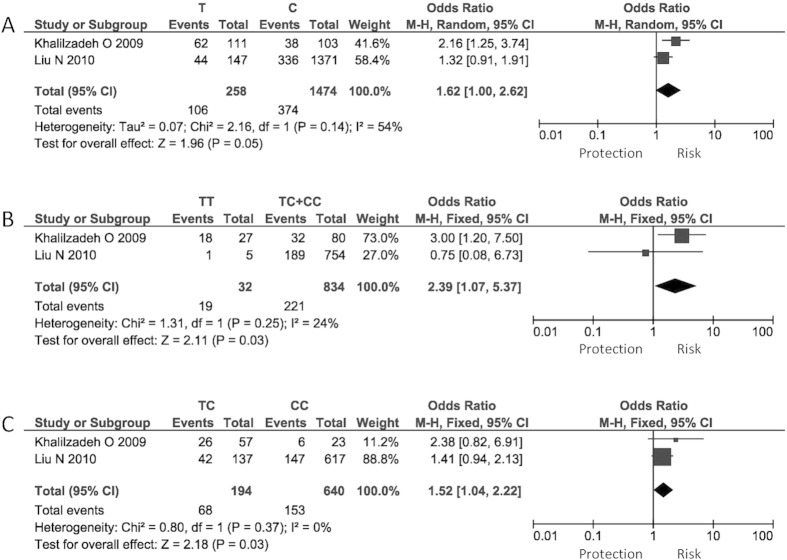
We meta-analyzed 12 variations in 8 IL-related genes, including IL1A (rs1800587), IL1B (rs1143634 and rs16944), IL1RA (A2, two copies of an 86-bp tandem repeat in intron2), IL4 (rs2070874), IL6 (rs1800795), IL12B (rs3212227), IL13 (rs1800925 and c.-2044G>A) and IL23R (rs10889677, rs2201841 and rs7530511). The number of studies on each variation ranged from 2 to 4. Only 1 SNP, IL1A rs1800587, showed a marginally significant association with GO in allelic (T vs. C, summary odds ratio [OR] = 1.62, P = 0.050, I2 = 53.7%), recessive (TT vs. TC + CC, OR = 2.39, P = 0.039, I2 = 23.6%) and heterozygous (TC vs. CC, OR = 1.52, P = 0.034, I2 = 0) models. This SNP was reported in 2 studies, involving a total of 240 GO patients and 626 GD controls (Table 2; Fig. 2). However, the associations were not significant after Bonferroni correction (P > 0.01).
Table 2. Meta-analysis of interleukin-related gene polymorphisms in Graves’ ophthalmopathy.
| Gene | Polymorphism | No. ofcohorts | Ethnicity | Geneticmodel | Total allele orgenotype counts |
FEM or REMa |
Heterogeneity |
Egger’stest (P) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | OR (95% CI) | P | P (Q) | I2 (%) | ||||||
| IL1A | rs1800587 | 2 | Chinese, Iranian | T vs. C | 480 | 1252 | 1.62 (1.00–2.62) | 0.050 | 0.14 | 53.7 | n.a. |
| TT vs. TC + CC | 240 | 626 | 2.39 (1.07–5.37) | 0.039 | 0.25 | 23.6 | n.a. | ||||
| TT + TC vs. CC | 240 | 626 | 1.80 (0.86–3.78) | 0.12 | 0.15 | 51.7 | n.a. | ||||
| TT vs. CC | 172 | 500 | 2.66 (0.41–17.22) | 0.31 | 0.13 | 57.0 | n.a. | ||||
| TC vs. CC | 221 | 613 | 1.52 (1.04–2.22) | 0.034 | 0.37 | 0.0 | n.a. | ||||
| IL1B | rs1143634 | 3 | Chinese, Caucasians, Iranian, | T vs. C | 650 | 736 | 0.93 (0.62–1.40) | 0.72 | 0.85 | 0.0 | 0.61 |
| TT vs. TC + CC | 325 | 368 | 1.02 (0.62–1.67) | 0.94 | 0.73 | 0.0 | 0.75 | ||||
| TT + TC vs. CC | 325 | 368 | 0.37 (0.10–1.32) | 0.13 | 0.85 | 0.0 | 0.82 | ||||
| TT vs. CC | 89 | 63 | 0.38 (0.10–1.40) | 0.15 | 0.92 | 0.0 | 0.85 | ||||
| TC vs. CC | 247 | 308 | 0.33 (0.09–1.22) | 0.10 | 0.72 | 0.0 | 0.84 | ||||
| IL1B | rs16944 | 4 | Chinese, Caucasians, Iranian | C vs. T | 1030 | 1880 | 1.09 (0.93–1.28) | 0.29 | 0.69 | 0.0 | 0.89 |
| CC vs. CT + TT | 515 | 940 | 1.11 (0.87–1.40) | 0.41 | 0.97 | 0.0 | 0.68 | ||||
| CC + CT vs. TT | 515 | 940 | 1.13 (0.83–1.54) | 0.45 | 0.15 | 38.9 | 0.95 | ||||
| CC vs. TT | 261 | 455 | 1.18 (0.83–1.67) | 0.36 | 0.31 | 7.2 | 0.85 | ||||
| CT vs. TT | 333 | 650 | 1.10 (0.79–1.52) | 0.58 | 0.11 | 46.2 | 0.90 | ||||
| IL1RA | A2/non-A2b | 2 | Caucasians | A2 vs. Non-A2 | 284 | 256 | 1.45 (0.89–2.35) | 0.13 | 0.68 | 0.0 | n.a. |
| IL4 | rs2070874 | 3 | Chinese, Iranian | T vs. C | 333 | 692 | 0.93 (0.74–1.17) | 0.52 | 0.30 | 22.3 | 0.37 |
| TT vs. TC + CC | 333 | 692 | 0.88 (0.66–1.18) | 0.39 | 0.44 | 7.5 | 0.26 | ||||
| TT + TC vs. CC | 333 | 692 | 0.95 (0.54–1.65) | 0.84 | 0.16 | 0.0 | 0.060 | ||||
| TT vs. CC | 212 | 457 | 1.16 (0.54–2.47) | 0.71 | 0.18 | 41.3 | 0.1 | ||||
| TC vs. CC | 154 | 284 | 0.97 (0.55–1.72) | 0.92 | 0.15 | 0.0 | 0.050 | ||||
| IL6 | rs1800795 | 2 | Caucasians, Iranian | C vs. G | 316 | 456 | 1.29 (0.66–2.54) | 0.45 | 0.040 | 77.3 | n.a. |
| CC vs. CG + GG | 158 | 228 | 1.48 (0.65–3.37) | 0.36 | 0.12 | 58.3 | n.a. | ||||
| CC + CG vs. GG | 158 | 228 | 1.53 (0.41–5.78) | 0.53 | 0.040 | 76.5 | n.a. | ||||
| CC vs. GG | 78 | 107 | 2.04 (0.36–11.64) | 0.42 | 0.020 | 80.9 | n.a. | ||||
| CG vs. GG | 119 | 184 | 1.34 (0.43–4.20) | 0.62 | 0.080 | 67.2 | n.a. | ||||
| IL12B | rs3212227 | 2 | Japanese, Iranian | A vs. C | 306 | 566 | 0.60 (0.26–1.39) | 0.23 | 0.010 | 85.4 | n.a. |
| AA vs. AC + CC | 153 | 283 | 0.46 (0.13–1.66) | 0.24 | 0.020 | 82.5 | n.a. | ||||
| AA + AC vs. CC | 153 | 283 | 0.57 (0.20–1.59) | 0.28 | 0.070 | 68.8 | n.a. | ||||
| AA vs. CC | 74 | 147 | 0.35 (0.06–2.04) | 0.24 | 0.010 | 84.2 | n.a. | ||||
| AC vs. CC | 123 | 202 | 0.81 (0.49–1.32) | 0.39 | 0.30 | 7.0 | n.a. | ||||
| IL13 | rs1800925 | 3 | Chinese, Japanese, Caucasians | C vs. T | 566 | 904 | 1.08 (0.82–1.41) | 0.59 | 0.27 | 29.5 | 0.17 |
| CC vs. CT + TT | 283 | 452 | 1.14 (0.83–1.58) | 0.42 | 0.51 | 0.0 | 0.70 | ||||
| CC + CT vs. TT | 283 | 452 | 0.88 (0.45–1.73) | 0.71 | 0.27 | 30.2 | 0.12 | ||||
| CC vs. TT | 200 | 301 | 0.92 (0.46–1.84) | 0.82 | 0.26 | 31.5 | 0.11 | ||||
| CT vs. TT | 99 | 175 | 0.84 (0.41–1.71) | 0.63 | 0.36 | 12.2 | 0.19 | ||||
| IL13 | c.-2044G>A | 2 | Japanese, Caucasians | G vs. A | 382 | 760 | 1.02 (0.77–1.36) | 0.87 | 0.31 | 2.3 | n.a. |
| GG vs. GA + AA | 191 | 380 | 0.99 (0.70–1.41) | 0.97 | 0.56 | 0.0 | n.a. | ||||
| GG + GA vs. AA | 191 | 380 | 1.14 (0.55–2.34) | 0.73 | 0.18 | 45.5 | n.a. | ||||
| GG vs. AA | 117 | 237 | 1.12 (0.54–2.35) | 0.76 | 0.17 | 46.4 | n.a. | ||||
| GA vs. AA | 86 | 171 | 1.18 (0.55–2.52) | 0.67 | 0.22 | 34.9 | n.a. | ||||
| IL23R | rs10889677 | 2 | Caucasians, Japanese | C vs. A | 400 | 604 | 1.15 (0.46–2.86) | 0.76 | 0.0018 | 89.7 | n.a. |
| CC vs. CA + AA | 200 | 302 | 1.40 (0.46–4.21) | 0.55 | 0.040 | 77.1 | n.a. | ||||
| CC + CA vs. AA | 200 | 302 | 0.93 (0.37–2.32) | 0.87 | 0.12 | 58.5 | n.a. | ||||
| CC vs. AA | 132 | 163 | 1.20 (0.31–4.66) | 0.80 | 0.070 | 70.1 | n.a. | ||||
| CA vs. AA | 129 | 238 | 0.72 (0.44–1.15) | 0.17 | 0.44 | 0.0 | n.a. | ||||
| IL23R | rs2201841 | 2 | Caucasians, Japanese | A vs. G | 402 | 606 | 1.20 (0.55–2.63) | 0.65 | 0.010 | 86.3 | n.a. |
| AA vs. AG + GG | 201 | 303 | 1.60 (0.70–3.66) | 0.26 | 0.10 | 64.0 | n.a. | ||||
| AA + AG vs. GG | 201 | 303 | 0.80 (0.51–1.25) | 0.33 | 0.23 | 30.2 | n.a. | ||||
| AA vs. GG | 133 | 163 | 1.17 (0.58–2.36) | 0.65 | 0.19 | 41.5 | n.a. | ||||
| AG vs. GG | 127 | 238 | 0.72 (0.45–1.16) | 0.18 | 0.62 | 0.0 | n.a. | ||||
| IL23R | rs7530511 | 2 | Caucasians, Japanese | C vs. T | 396 | 580 | 0.68 (0.39–1.17) | 0.16 | 0.41 | 0.0 | n.a. |
| CC vs. CT + TT | 198 | 290 | 0.64 (0.35–1.18) | 0.15 | 0.43 | 0.0 | n.a. | ||||
| CC + CT vs. TT | 198 | 290 | 0.64 (0.12–3.32) | 0.60 | 0.90 | 0.0 | n.a. | ||||
| CC vs. TT | 168 | 268 | 0.59 (0.11–3.09) | 0.53 | 0.93 | 0.0 | n.a. | ||||
| CT vs. TT | 33 | 24 | 0.98 (0.17–5.50) | 0.98 | 0.80 | 0.0 | n.a. | ||||
CI: Confidence interval; FEM: fixed effects model; GD: Grave’s Disease; GO: Graves’ Ophthalmopathy; IL: Interleukin; IL1A: Interleukin 1 Alpha; IL1B: Interleukin 1 Beta; IL1RA: Interleukin 1 Receptor Antagonist; IL12B: Interleukin 12 Beta; IL23R: Interleukin 23 Receptor; No.: number; OR: Odds ratio; REM: random effects model.
aIf the P value for Q-statistic was <0.10 or the I2 value ≥ 50%, a random-effects model was used, otherwise a fixed-effects model was adopted.
bA2 = 2 repeats of a 86-bp segment; non-A2 = other number of repeats of a 86-bp segment.
Figure 2. Association of IL1A rs1800587 with GO allelic, recessive and heterozygous models.
(A) Allelic model; (B) Recessive model; (C) Heterozygous model.
The other 11 variations in 7 genes did not show a significant association with GO in any inheritance models (P > 0.05; Table 2). Among the insignificant polymorphisms, IL1B rs1143634, IL1RA A2/non-A2 and IL23R rs7530511 showed no heterogeneity among studies (I2 = 0), whilst IL1B rs16944 (I2 ≤ 46.2%), IL4 rs2070874 (I2 ≤ 41.3%), IL6 rs1800795 (I2 ≤ 80.9%), IL12B rs3212227 (I2 ≤ 85.4%), IL13 rs1800925 (I2 ≤ 31.5%) and c.-2044G>A (I2 ≤ 46.4%), and IL23R rs10889677 (I2 ≤ 89.7%) and rs2201841 (I2 ≤ 86.3%) showed moderate to high heterogeneities (Table 2).
To explain the heterogeneity, we performed subgroup analysis by ethnicity. Due to the limited number of studies, we only tested the associations of 2 SNPs (IL1B rs16944 and IL4 rs2070874) in Chinese. However, these 2 SNPs did not show significant association with GO (P > 0.05), with low to moderate heterogeneities (Supplementary Table 2).
Assessment of potential biases and sensitivity analysis
Lacka et al. compared a subgroup of patients with GD associated with GO from the onset and a subgroup contained patients in whom GO developed from 6 months to 7 years from the onset of GD23. To avoid selection bias, we conducted sensitivity analysis by excluding this study and keeping only patients with GD without GO as controls from the meta-analysis of IL1B rs16944 and rs1143634. The associations remained insignificant (Supplementary Table 3). In the quality assessment of studies using the Newcastle Ottawa Scale (NOS), all of the studies were assigned 7 or more stars, indicating low risk of introducing biases. Therefore, no study was excluded from the meta-analysis due to poor quality (Supplementary Table 4). Moreover, for SNPs reported in 3 or more studies (i.e., IL1B rs1143634 and rs16944, IL4 rs2070874 and IL13 rs1800925), sensitivity analyses were performed by sequentially omitting one study at a time. The insignificant associations remained unchanged (P > 0.05; data not shown). There was no significant publication bias detected by the funnel plots (data not shown) and Egger’s test (Table 2 and Supplementary Table 3).
Discussion
This study has, for the first time, summarized the associations of IL-related genes with GO. Among the 11 reported genes, we performed meta-analyses on 12 polymorphisms in 8 genes. Unexpectedly, we found only one SNP, IL1A rs1800587 (c.-889C>T), being marginally associated with GO. No significant association was detected for SNPs in the other 7 genes (IL1B, IL1RA, IL4, IL6, IL12B, IL13 and IL23R), among which IL1B rs1143634, IL1RA A2/non-A2 and IL23R rs7530511 showed no heterogeneity across the study populations.
The IL1A SNP rs1800587 showed a suggestive association with no to moderate heterogeneities in different genetic models. This SNP was reported in 2 studies21,22. Although a significant association was reported only in one study21, the effect of the risk allele T pointed to the same direction in the both studies (OR = 2.1621 and OR = 1.3222). The heterogeneity could be due to the relatively small sample size in the study of Khalilzadeh et al. (about 50 cases and 50 controls), the ethnic differences in linkage disequilibrium structures, and the differences in the minor allele frequencies (MAF) of rs1800587 (T) between Iranian (about 43.0%)21 and Chinese (about 10%)22. Notably, however, since the P values did not survive the Bonferroni correction for multiple testing, the genetic association of the IL1A SNP with GO has yet to be confirmed in further studies with larger sample sizes.
IL1A, a major member of the IL1 superfamily, is the prototype pro-inflammatory and a potent pleiotropic cytokine involved in acute or chronic inflammation50. Associations between IL1A and GO were demonstrated in biochemical, histological, immunological and genetics studies. There were significant differences in the serum IL1A levels between controls and GO patients, and for the latter, before and after corticosteroid, corticosteroid with orbital irradiation, or decompression51. IL1A immunoreactivity was detected in the orbital tissues, their fibroblast cultures and supernatants from 5 out of 6 GO patients, but absent in those derived from 5 normal individuals52. An in vitro study demonstrated the induction of intercellular adhesion molecule 1 (ICAM-1), endothelial leukocyte adhesion molecule 1 (ELAM-1) and vascular cell adhesion molecule 1 (VCAM-1), which promote T cell chemotaxis upon the exposure of endothelial cells generated from retrobulbar tissues to IL1A53. The proliferation of orbital fibroblasts from GO patients was stimulated by IL1A, which has no effect on normal orbital fibroblasts54. Transcription of prostaglandin endoperoxidase H synthase-2 (an inflammatory cyclooxygenase that produces prostaglandin E2 and contributes to orbital inflammation in GO55) in orbital fibroblasts by leukoregulin (a product of activated T lymphocytes) was found to be mediated through an intermediate induction of IL1A56. In our meta-analysis, we found the IL1A SNP rs1800587 as a potential susceptibility genetic marker for GO, confirming the involvement of IL1A in the disease. In fact, the IL1A SNP rs1800587, located in the 5’ untranslated region (c.-889C>T), had been associated with autoimmune diseases including ankylosing spondylitis57, systemic lupus erythematosus58, psoriatic arthritis59 and Behcet’s disease60. IL1A and its SNP rs1800587 could thus play a role in the pathogenesis of autoimmune diseases including GO.
Except for IL1A, SNPs in other reported genes did not show a significant association with GO in our meta-analysis. Three SNPs showed no association with GO in all of the tested populations (P > 0.05) with no heterogeneity, including IL1B rs1143634 (Caucasians, Iranian and Chinese), IL1RA A2/non-A2 (Caucasians) and IL23R rs7530511 (Caucasians and Japanese). They are not likely to be genetic markers for GO. Another 6 insignificant SNPs (IL1B rs16944, IL4 rs2070874, IL6 rs1800795, IL13 rs1800925 and c.-2044G>A, and IL23R rs10889677) also lacked significant association in any of the studies with mild to high heterogeneities. In contrast, IL12B rs321222728 and IL23R rs220184141 showed significant associations with GO in Iranian and Caucasians, respectively, but not in Japanese34,42. In our meta-analysis, no significant association was found for these 2 SNPs by using the random-effect model, with moderate to high heterogeneities. Of note, the IL23R SNP rs2201841 was also significantly associated with other autoimmune diseases, such as Crohn’s Disease61,62 and rheumatoid arthritis62. Therefore, further replication of these 2 SNPs, IL12B rs3212227 and IL23R rs2201841, in GO among different populations are warranted.
Two IL1B SNPs, rs1143634 and rs16944, showed no association with GO. The summary results of the IL1B gene in our meta-analysis were inconsistent with that in the study of Liu et al.22. In this study, we used patients with GD but without GO as controls, with a view to assess the effects of the gene SNPs on GO in a background of GD. In contrast, Liu’s group compared GO patients with healthy subjects and detected a significant association22. Thus, our results cannot be compared directly with that of Liu et al. Further studies are needed to confirm if the IL1B SNPs are genuine markers differentiating GO patients from normal subjects.
IL1RA acts as a competitive inhibitor of IL1A and IL1B and blocks IL1-mediated cellular activities63, such as IL-1-induced glycosaminoglycan production by cultured human orbital fibroblasts64. IL1RA could also block the induction of prostaglandin endoperoxidase H synthase-2 by leukoregulin56. A study had shown that upon cytokines exposure, markedly lower level of IL1RA expression was found in cultured orbital fibroblasts of GO patients as compared to the normal orbital fibroblasts65. Although IL1RA A2/non-A2 was not significant in our meta-analysis, another IL1RA SNP (c.11100C>T) had shown positive association in Iranians21. Follow-up studies on the IL1RA polymorphisms are needed to confirm the role of IL1RA in GO in specific population such as Iranians.
IL4 is a potent Th2 cytokine which stimulates proliferation of IgE- and IgG- secreting B cells and the expression of HLA class II antigens via STAT666 against Th1 inflammatory response30. It has also been detected in orbital fat tissues of GO patients12. The promoter SNP rs2070874, which has transcriptional activity30,67, did not show a significant association with GO in our meta-analysis. However, significant associations of other IL4 SNPs, including c.-1098T>G and c.-33C>T, with GO have been reported in Iranians31. Therefore, these two IL4 polymorphisms should be tested in future studies. IL13 and IL4 have similar biological functions68. IL13 is an anti-inflammatory cytokine that regulates IgE synthesis68,69 and the maturation of B cells30. However, there was no significant association detected in Chinese (rs1800925)30, Japanese (rs1800925 and c.-2044G>A)37 and Caucasians (rs1800925 and c.-2044G>A)36. Consistently, we did not detect associations of these two IL13 SNPs with GO.
The IL6 SNP rs1800795 is associated with multiple autoimmune diseases, including systemic-onset juvenile chronic arthritis70, type I diabetes mellitus71, rheumatoid arthritis72 and Sjogren’s syndrome73. When compared with healthy controls, serum IL6 levels were significantly higher in GD and GO patients, especially in active GO patients74. However, only one SNP rs1800795 in IL6 was eligible for the meta-analysis, and it showed a lack of significant association.
This meta-analysis also reveals several limitations in the existing genetic studies of GO. First, the small number of published genetic studies on IL-related genes in GO limited the power of determining the associations, especially among different ethnic groups. Second, GO may not develop concurrently with GD. Classifying GO based on a cross-sectional assessment of observer-dependent signs and subject-dependent symptoms may therefore introduce bias. Third, as the pathogenesis of GO and GD is multifactorial, it would be more informative to test genetic, environmental (e.g. smoking), hormonal (e.g. fluctuation of thyroid function) and antigenic (thyroid related autoantibodies and use of radioactive iodine) factors, and their interactions in the study population. However, few genetic studies on IL-related genes and GO provided such information.
In conclusion, in this systematic review and meta-analysis of the association of IL-related genes with GO, we identified IL1A rs1800587 as the only SNP that is potentially associated with GO. Since the overall number of studies is small, further studies with larger sample sizes are needed to confirm IL1A rs1800587 as a genetic biomarker for GO, and also verify the roles of other IL-related genes in the disease.
Methods
Searching Strategy
We performed the literature search in the EMBASE and MEDLINE databases via the Ovid platform using structured search strategies. We identified citations recorded during the period starting from May 1, 1989 to February 25, 2015. Boolean logic and search terms with controlled vocabularies (i.e. Medical Subject Heading terms) were used: (Graves’ ophthalmopathy OR thyroid associated ophthalmopathy) AND interleukins (Supplementary Table 5). Moreover, we manually scanned the reference lists of the identified articles, reviews, and meta-analyses to include all potentially relevant articles. No language filters were applied in the literature search.
Inclusion and Exclusion Criteria
A study was included if it fulfilled the following criteria: (1) original case-control study on the associations of IL-related genes polymorphisms with GO; (2) cases were patients with GO defined by clinical signs of GO or NOSPECS Class 2 or 3 and above; (3) controls were patients with Graves’ disease without GO (no clinical signs of GO or NOSPECS Class 0 or 1); (4) study subjects were unrelated individuals from clearly defined populations; (5) allele or genotype counts or frequencies in both case and control groups were provided (or existing data allow their calculation). Animal studies, case reports, reviews, abstracts, conference proceedings, editorials and studies with incomplete data were excluded.
Literature Review and Data Extraction
Two investigators (W.K.H. and S.S.R.) screened and reviewed all studies independently. Disagreement was resolved by thorough discussion with a third investigator (L.J.C.) until consensus was reached. A customized data form was used to extract the data, which included the first author, year of publication, country of study, ethnicity, definition of cases and controls, sample size in case and control groups, gene and polymorphisms studied, allelic and genotypic counts, and result of the test for HWE in the control group. Two reviewers (W.K.H. and S.S.R.) extracted the data independently. Disagreement was resolved by consensus among the investigators. If the allele counts were not reported, we calculated them from the genotype data. If genotype counts were missing, we estimated the data using allele frequencies (if available) and sample sizes, assuming no deviation from HWE unless otherwise reported75. If there was no extractable genetic information in an eligible study, we communicated with the authors for the data. Allele counts of the eligible SNPs for meta-analysis were summarized in Supplementary Table 6.
Statistical Analysis
Meta-analysis for each gene polymorphism was performed if it was reported in 2 or more studies. The genetic association was assessed using different genetic models, including allelic (A vs. a), dominant (AA+Aa vs. aa), recessive (aa vs. AA+Aa) and codominant (homozygous: AA vs. aa; heterozygous: AA vs. Aa) models. The strength of association was evaluated using the summary odds ratios and 95% confidence intervals of each gene polymorphism. Heterogeneity was tested by the Q-statistic and the I2 value76. The Q-statistic was considered significant when P < 0.10. The I2 values indicated no (0–24.9%), low (25–49.9%), moderate (50–74.9%) or high (75–100%) inter-study heterogeneity76,77. If the P value for the Q-statistic was <0.10 or the I2 value ≥ 50%, a random-effect model was used, otherwise a fixed-effect model was adopted78. In the assessment of data quality, we first examined the HWE in the control groups. If HWE was not reported, we tested it using the control group data with the Chi-square test. Also, we adopted the Newcastle Ottawa Scale (accessed via http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp) to evaluate the quality of case-control studies (Appendix 1). A star was given to each study if one requirement in the NOS from 3 dimensions (selection, comparability and exposure) was met. The maximum number of stars that can be assigned to one study was 9. A study obtaining ≤6 stars was considered as of high risk in introducing bias79. We then conducted a sensitivity analysis to confirm the associations by sequentially omitting each of the studies one at a time, studies deviated from HWE, or studies of suboptimal quality80. Furthermore, the Funnel plots and Egger’s test were performed to assess potential biases (e.g. publication bias)81,82. The presence of bias was suggested when the P value of the Egger’s test was <0.05. All statistical analyses were performed using the R software for statistical computing (v3.0.0, http://cran.r-project.org/). Of note, since we tested the genetic association using 5 genetic models, the risk of type I error might be increased; therefore, we corrected the P values for association using the Bonferroni method. Thus, a P value of less than 0.010 (0.05/5) was considered statistically significant, where 5 is the number of genetic models being tested.
Additional Information
How to cite this article: Wong, K. H. et al. Genetic Associations of Interleukin-related Genes with Graves’ Ophthalmopathy: a Systematic Review and Meta-analysis. Sci. Rep. 5, 16672; doi: 10.1038/srep16672 (2015).
Supplementary Material
Acknowledgments
We express our gratitude to all participants in this study.
Footnotes
Author Contributions W.K.H., S.S.R., K.L.C. and L.J.C. conceived the study idea and participated in the study design. W.K.H. and S.S.R. conducted literature search, reviewed and extracted data and carried out the statistical analysis. A.L.Y. and C.P.P. provided critical comments for the conduct of the study. W.K.H. drafted the article. S.S.R., K.L.C., C.P.P. and L.J.C. revised the article. All authors read and approved the final manuscript.
References
- Bahn R. S. & Heufelder A. E. Pathogenesis of Graves’ ophthalmopathy. N Engl J Med 329, 1468–1475 (1993). [DOI] [PubMed] [Google Scholar]
- Tanda M. L. et al. Prevalence and natural history of Graves’ orbitopathy in a large series of patients with newly diagnosed graves’ hyperthyroidism seen at a single center. J Clin Endocrinol Metab 98, 1443–1449 (2013). [DOI] [PubMed] [Google Scholar]
- Wiersinga W. M. & Bartalena L. Epidemiology and prevention of Graves’ ophthalmopathy. Thyroid 12, 855–860 (2002). [DOI] [PubMed] [Google Scholar]
- Bahn R. S. Graves’ ophthalmopathy. N Engl J Med 362, 726–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomer Y. & Davies T. F. Searching for the autoimmune thyroid disease susceptibility genes: from gene mapping to gene function. Endocr Rev 24, 694–717 (2003). [DOI] [PubMed] [Google Scholar]
- Smith T. J. Pathogenesis of Graves’ orbitopathy: a 2010 update. J Endocrinol Invest 33, 414–421 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromatsu Y. et al. Role of magnetic resonance imaging in thyroid-associated ophthalmopathy: its predictive value for therapeutic outcome of immunosuppressive therapy. Thyroid 2, 299–305 (1992). [DOI] [PubMed] [Google Scholar]
- Weetman A. P., Cohen S., Gatter K. C., Fells P. & Shine B. Immunohistochemical analysis of the retrobulbar tissues in Graves’ ophthalmopathy. Clin Exp Immunol 75, 222–227 (1989). [PMC free article] [PubMed] [Google Scholar]
- Kahaly G., Hansen C., Felke B. & Dienes H. P. Immunohistochemical staining of retrobulbar adipose tissue in Graves’ ophthalmopathy. Clin Immunol Immunopathol 73, 53–62 (1994). [DOI] [PubMed] [Google Scholar]
- Hiromatsu Y. et al. Human histocompatibility leukocyte antigen-DR and heat shock protein-70 expression in eye muscle tissue in thyroid-associated ophthalmopathy. J Clin Endocrinol Metab 80, 685–691 (1995). [DOI] [PubMed] [Google Scholar]
- Li B. & Smith T. J. Divergent expression of IL-1 receptor antagonists in CD34+ fibrocytes and orbital fibroblasts in thyroid-associated ophthalmopathy: contribution of fibrocytes to orbital inflammation. J Clin Endocrinol Metab 98, 2783–2790 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromatsu Y. et al. Cytokine profiles in eye muscle tissue and orbital fat tissue from patients with thyroid-associated ophthalmopathy. J Clin Endocrinol Metab 85, 1194–1199 (2000). [DOI] [PubMed] [Google Scholar]
- Kumar S. & Bahn R. S. Relative overexpression of macrophage-derived cytokines in orbital adipose tissue from patients with graves’ ophthalmopathy. J Clin Endocrinol Metab 88, 4246–4250 (2003). [DOI] [PubMed] [Google Scholar]
- Cai K. & Wei R. Interleukin-7 expression in tears and orbital tissues of patients with Graves’ ophthalmopathy. Endocrine 44, 140–144 (2013). [DOI] [PubMed] [Google Scholar]
- Molnar I. & Balazs C. High circulating IL-6 level in Graves’ ophthalmopathy. Autoimmunity 25, 91–96 (1997). [DOI] [PubMed] [Google Scholar]
- Salvi M. et al. Serum concentrations of proinflammatory cytokines in Graves’ disease: effect of treatment, thyroid function, ophthalmopathy and cigarette smoking. Eur J Endocrinol 143, 197–202 (2000). [DOI] [PubMed] [Google Scholar]
- de Carli M. et al. Cytolytic T cells with Th1-like cytokine profile predominate in retroorbital lymphocytic infiltrates of Graves’ ophthalmopathy. J Clin Endocrinol Metab 77, 1120–1124 (1993). [DOI] [PubMed] [Google Scholar]
- Natt N. & Bahn R. S. Cytokines in the evolution of Graves’ ophthalmopathy. Autoimmunity 26, 129–136 (1997). [DOI] [PubMed] [Google Scholar]
- Han R. & Smith T. J. T helper type 1 and type 2 cytokines exert divergent influence on the induction of prostaglandin E2 and hyaluronan synthesis by interleukin-1beta in orbital fibroblasts: implications for the pathogenesis of thyroid-associated ophthalmopathy. Endocrinology 147, 13–19 (2006). [DOI] [PubMed] [Google Scholar]
- Naik V. et al. Biologic therapeutics in thyroid-associated ophthalmopathy: translating disease mechanism into therapy. Thyroid 18, 967–971 (2008). [DOI] [PubMed] [Google Scholar]
- Khalilzadeh O. et al. Graves’ ophthalmopathy and gene polymorphisms in interleukin-1alpha, interleukin-1beta, interleukin-1 receptor and interleukin-1 receptor antagonist. Clin Experiment Ophthalmol 37, 614–619 (2009). [DOI] [PubMed] [Google Scholar]
- Liu N. et al. The association of interleukin-1alpha and interleukin-1beta polymorphisms with the risk of Graves’ disease in a case-control study and meta-analysis. Hum Immunol 71, 397–401 (2010). [DOI] [PubMed] [Google Scholar]
- Lacka K. et al. Interleukin-1beta gene (IL-1beta) polymorphisms (SNP −511 and SNP +3953) in thyroid-associated ophthalmopathy (TAO) among the Polish population. Curr Eye Res 34, 215–220 (2009). [DOI] [PubMed] [Google Scholar]
- Liu Y. H. et al. Association of interleukin-1beta (IL1B) polymorphisms with Graves’ ophthalmopathy in Taiwan Chinese patients. Invest Ophthalmol Vis Sci 51, 6238–6246 (2010). [DOI] [PubMed] [Google Scholar]
- Cuddihy R. M. & Bahn R. S. Lack of an association between alleles of interleukin-1 alpha and interleukin-1 receptor antagonist genes and Graves’ disease in a North American Caucasian population. J Clin Endocrinol Metab 81, 4476–4478 (1996). [DOI] [PubMed] [Google Scholar]
- Muhlberg T. et al. Lack of association of Graves’ disease with the A2 allele of the interleukin-1 receptor antagonist gene in a white European population. Eur J Endocrinol 138, 686–690 (1998). [DOI] [PubMed] [Google Scholar]
- Blakemore A. I. F., Watson P. F., Weetman A. P. & Duff G. W. Association of Graves’ disease with an allele of the interleukin-1 receptor antagonist gene. J Clin Endocrinol Metab 80, 111–115 (1995). [DOI] [PubMed] [Google Scholar]
- Anvari M. et al. Genetic susceptibility to Graves’ ophthalmopathy: the role of polymorphisms in proinflammatory cytokine genes. Eye 24, 1058–1063 (2010). [DOI] [PubMed] [Google Scholar]
- Zhu W. et al. Association analysis of polymorphisms in IL-3, IL-4, IL-5, IL-9, and IL-13 with Graves’ disease. J Endocrinol Invest 33, 751–755 (2010). [DOI] [PubMed] [Google Scholar]
- Yang Y. et al. Association study between the IL4, IL13, IRF1 and UGRP1 genes in chromosomal 5q31 region and Chinese Graves’ disease. J Hum Genet 50, 574–582 (2005). [DOI] [PubMed] [Google Scholar]
- Khalilzadeh O. et al. Genetic susceptibility to Graves’ ophthalmopathy: The role of polymorphisms in anti-inflammatory cytokine genes. Ophthalmic Genet 31, 215–220 (2010). [DOI] [PubMed] [Google Scholar]
- Bednarczuk T. et al. Association of G-174C polymorphism of the interleukin-6 gene promoter with Graves’ ophthalmopathy. Autoimmunity 37, 223–226 (2004). [DOI] [PubMed] [Google Scholar]
- Gu L. Q. et al. Association studies of interleukin-8 gene in Graves’ disease and Graves’ ophthalmopathy. Endocrine 36, 452–456 (2009). [DOI] [PubMed] [Google Scholar]
- Hiromatsu Y. et al. Interleukin-12B gene polymorphism does not confer susceptibility to Graves’ ophthalmopathy in Japanese population. Endocr J 53, 753–759 (2006). [DOI] [PubMed] [Google Scholar]
- Liu Y. H. et al. Association of IL12B polymorphisms with susceptibility to Graves ophthalmopathy in a Taiwan Chinese population. J Biomed Sci 19, 97 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarczuk T. et al. Interleukin-13 gene polymorphisms in patients with Graves’ disease. Clin Endocrinol (Oxf) 59, 519–525 (2003). [DOI] [PubMed] [Google Scholar]
- Hiromatsu Y. et al. Interleukin-13 gene polymorphisms confer the susceptibility of Japanese populations to Graves’ disease. J Clin Endocrinol Metab 90, 296–301 (2005). [DOI] [PubMed] [Google Scholar]
- Chong K. K. L. et al. Association of CTLA-4 and IL-13 gene polymorphisms with Graves’ disease and ophthalmopathy in Chinese children. Invest Ophthalmol Vis Sci 49, 2409–2415 (2008). [DOI] [PubMed] [Google Scholar]
- Mukai T. et al. Lack of association of interleukin-18 gene polymorphisms with susceptibility of Japanese populations to Graves’ disease or Graves’ ophthalmopathy. Thyroid 16, 243–248 (2006). [DOI] [PubMed] [Google Scholar]
- Jia H. Y. et al. Association between interleukin 21 and Graves’ disease. Genet Mol Res 10, 3338–3346 (2011). [DOI] [PubMed] [Google Scholar]
- Huber A. K., Jacobson E. M., Jazdzewski K., Concepcion E. S. & Tomer Y. Interleukin (IL)-23 receptor is a major susceptibility gene for Graves’ ophthalmopathy: the IL-23/T-helper 17 axis extends to thyroid autoimmunity. J Clin Endocrinol Metab 93, 1077–1081 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban Y. et al. Association studies of the IL-23R gene in autoimmune thyroid disease in the Japanese population. Autoimmunity 42, 126–130 (2009). [DOI] [PubMed] [Google Scholar]
- Tozaki T. et al. Association studies of the IL-23R gene in autoimmune thyroid disease in the Japanese population. Autoimmunity 42, 126–130 (2009). [DOI] [PubMed] [Google Scholar]
- Chng C. L., Seah L. L. & Khoo D. H. Ethnic differences in the clinical presentation of Graves’ ophthalmopathy. Best Pract Res Clin Endocrinol Metab 26, 249–258 (2012). [DOI] [PubMed] [Google Scholar]
- Hiromatsu Y., Kaku H., Miyake I., Murayama S. & Soejima E. Role of cytokines in the pathogenesis of thyroid-associated ophthalmopathy. Thyroid 12, 217–221 (2002). [DOI] [PubMed] [Google Scholar]
- Niyazoglu M. et al. Association of PARP-1, NF-kappaB, NF-kappaBIA and IL-6, IL-1beta and TNF-alpha with Graves Disease and Graves Ophthalmopathy. Gene 547, 226–232 (2014). [DOI] [PubMed] [Google Scholar]
- Bednarczuk T., Gopinath B., Ploski R. & Wall J. R. Susceptibility genes in Graves’ ophthalmopathy: searching for a needle in a haystack? Clin Endocrinol (Oxf) 67, 3–19 (2007). [DOI] [PubMed] [Google Scholar]
- Khalilzadeh O., Noshad S., Rashidi A. & Amirzargar A. Graves’ Ophthalmopathy: A review of immunogenetics. Curr Genomics 12, 564–575 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaii F., McCoy A. N. & Smith T. J. Cytokines as villains and potential therapeutic targets in thyroid-associated ophthalmopathy: From bench to bedside. Expert Rev Ophthalmol 9, 227–234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello C. A. Interleukin-1 and interleukin-1 antagonism. Blood 77, 1627–1652 (1991). [PubMed] [Google Scholar]
- Laban-Guceva N., Bogoev M. & Antova M. Serum concentrations of interleukin (IL-)1alpha, 1beta, 6 and tumor necrosis factor (TNF-) alpha in patients with thyroid eye disease (TED). Med Arh 61, 203–206 (2007). [PubMed] [Google Scholar]
- Heufelder A. E. & Bahn R. S. Detection and localization of cytokine immunoreactivity in retro-ocular connective tissue in Graves’ ophthalmopathy. Eur J Clin Invest 23, 10–17 (1993). [DOI] [PubMed] [Google Scholar]
- Heufelder A. E. & Scriba P. C. Characterization of adhesion receptors on cultured microvascular endothelial cells derived from the retroorbital connective tissue of patients with Graves’ ophthalmopathy. Eur J Endocrinol 134, 51–60 (1996). [DOI] [PubMed] [Google Scholar]
- Heufelder A. E. & Bahn R. S. Modulation of Graves’ orbital fibroblast proliferation by cytokines and glucocorticoid receptor agonists. Invest Ophthalmol Vis Sci 35, 120–127 (1994). [PubMed] [Google Scholar]
- Smith T. J. The putative role of prostaglandin endoperoxide H synthase-2 in the pathogenesis of thyroid-associated orbitopathy. Exp Clin Endocrinol Diabetes 107 Suppl 5, S160–163 (1999). [DOI] [PubMed] [Google Scholar]
- Cao H. J. & Smith T. J. Leukoregulin upregulation of prostaglandin endoperoxide H synthase-2 expression in human orbital fibroblasts. Am J Physiol 277, C1075–1085 (1999). [DOI] [PubMed] [Google Scholar]
- Lea W. I. & Lee Y. H. The associations between interleukin-1 polymorphisms and susceptibility to ankylosing spondylitis: a meta-analysis. Joint Bone Spine 79, 370–374 (2012). [DOI] [PubMed] [Google Scholar]
- Parks C. G. et al. Systemic lupus erythematosus and genetic variation in the interleukin 1 gene cluster: a population based study in the southeastern United States. Ann Rheum Dis 63, 91–94 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran J. S. et al. Interleukin 1alpha, interleukin 1beta and interleukin 1 receptor gene polymorphisms in psoriatic arthritis. Rheumatology (Oxford) 43, 22–26 (2004). [DOI] [PubMed] [Google Scholar]
- Alayli G. et al. T helper 1 type cytokines polymorphisms: association with susceptibility to Behcet’s disease. Clin Rheumatol 26, 1299–1305 (2007). [DOI] [PubMed] [Google Scholar]
- Duerr R. H. et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farago B. et al. Functional variants of interleukin-23 receptor gene confer risk for rheumatoid arthritis but not for systemic sclerosis. Ann Rheum Dis 67, 248–250 (2008). [DOI] [PubMed] [Google Scholar]
- Arend W. P. Interleukin-1 receptor antagonist. Adv Immunol 54, 167–227 (1993). [DOI] [PubMed] [Google Scholar]
- Tan G. H., Dutton C. M. & Bahn R. S. Interleukin-1 (IL-1) receptor antagonist and soluble IL-1 receptor inhibit IL-1-induced glycosaminoglycan production in cultured human orbital fibroblasts from patients with Graves’ ophthalmopathy. J Clin Endocrinol Metab 81, 449–452 (1996). [DOI] [PubMed] [Google Scholar]
- Muhlberg T., Heberling H. J., Joba W., Schworm H. D. & Heufelder A. E. Detection and modulation of interleukin-1 receptor antagonist messenger ribonucleic acid and immunoreactivity in Graves’ orbital fibroblasts. Invest Ophthalmol Vis Sci 38, 1018–1028 (1997). [PubMed] [Google Scholar]
- Kelso A. Cytokines: principles and prospects. Immunol Cell Biol 76, 300–317 (1998). [DOI] [PubMed] [Google Scholar]
- Rosenwasser L. J. et al. Promoter polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clin Exp Allergy 25 Suppl 2, 74–78; discussion 95–76 (1995). [DOI] [PubMed] [Google Scholar]
- Chomarat P. & Banchereau J. Interleukin-4 and interleukin-13: their similarities and discrepancies. Int Rev Immunol 17, 1–52 (1998). [DOI] [PubMed] [Google Scholar]
- Shirakawa I. et al. Atopy and asthma: genetic variants of IL-4 and IL-13 signalling. Immunol Today 21, 60–64 (2000). [DOI] [PubMed] [Google Scholar]
- Fishman D. et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest 102, 1369–1376 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahromi M. M., Millward B. A. & Demaine A. G. A polymorphism in the promoter region of the gene for interleukin-6 is associated with susceptibility to type 1 diabetes mellitus. J Interferon Cytokine Res 20, 885–888 (2000). [DOI] [PubMed] [Google Scholar]
- Pascual M. et al. IL-6 promoter polymorphisms in rheumatoid arthritis. Genes Immun 1, 338–340 (2000). [DOI] [PubMed] [Google Scholar]
- Hulkkonen J., Pertovaara M., Antonen J., Pasternack A. & Hurme M. Elevated interleukin-6 plasma levels are regulated by the promoter region polymorphism of the IL6 gene in primary Sjogren’s syndrome and correlate with the clinical manifestations of the disease. Rheumatology (Oxford) 40, 656–661 (2001). [DOI] [PubMed] [Google Scholar]
- Lv M. et al. [Role of Treg/Th17 cells and related cytokines in Graves’ ophthalmopathy]. Nan Fang Yi Ke Da Xue Xue Bao 34, 1809–1813 (2014). [PubMed] [Google Scholar]
- Allen N. C. et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet 40, 827–834 (2008). [DOI] [PubMed] [Google Scholar]
- Higgins J. P. & Thompson S. G. Quantifying heterogeneity in a meta-analysis. Stat Med 21, 1539–1558 (2002). [DOI] [PubMed] [Google Scholar]
- Higgins J. P., Thompson S. G., Deeks J. J. & Altman D. G. Measuring inconsistency in meta-analyses. BMJ 327, 557–560 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuritz S. J., Landis J. R. & Koch G. G. A general overview of Mantel-Haenszel methods: applications and recent developments. Annu Rev Public Health 9, 123–160 (1988). [DOI] [PubMed] [Google Scholar]
- Rong S. S., Peng Y., Liang Y. B., Cao D. & Jhanji V. Does cigarette smoking alter the risk of pterygium? A systematic review and meta-analysis. Invest Ophthalmol Vis Sci 55, 6235–6243 (2014). [DOI] [PubMed] [Google Scholar]
- Ma L. et al. Association of PEDF polymorphisms with age-related macular degeneration and polypoidal choroidal vasculopathy: a systematic review and meta-analysis. Sci Rep 5, 9497 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. L., Sutton A. J., Jones D. R., Abrams K. R. & Rushton L. Contour-enhanced meta-analysis funnel plots help distinguish publication bias from other causes of asymmetry. J Clin Epidemiol 61, 991–996 (2008). [DOI] [PubMed] [Google Scholar]
- Sterne J. A., Gavaghan D. & Egger M. Publication and related bias in meta-analysis: power of statistical tests and prevalence in the literature. J Clin Epidemiol 53, 1119–1129 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.