Abstract
Immunological memory, which protects organisms from re-infection, is a hallmark of the mammalian adaptive immune system and the underlying principle of vaccination. In early life, however, mice and other mammals are deficient at generating memory CD8+ T cells, which protect organisms from intracellular pathogens. The molecular basis that differentiates adult and neonatal CD8+ T cells is unknown. MicroRNAs (miRNAs) are both developmentally regulated and required for normal adult CD8+ T cell functions. We used next-generation sequencing to identify mouse miRNAs that are differentially regulated in adult and neonatal CD8+ T cells, which may contribute to the impaired development of neonatal memory cells. The miRNA profiles of adult and neonatal cells were surprisingly similar during infection; however, we observed large differences prior to infection. In particular, miR-29 and miR-130 have significant differential expression between adult and neonatal cells before infection. Importantly, using RNA-Seq, we detected reciprocal changes in expression of messenger RNA targets for both miR-29 and miR-130. Moreover, targets that we validated include Eomes and Tbx21, key genes that regulate the formation of memory CD8+ T cells. Notably, age-dependent changes in miR-29 and miR-130 are conserved in human CD8+ T cells, further suggesting that these developmental differences are biologically relevant. Together, these results demonstrate that miR-29 and miR-130 are likely important regulators of memory CD8+ T cell formation and suggest that neonatal cells are committed to a short-lived effector cell fate prior to infection.
Keywords: microRNA regulation, adaptive immunity, development
THE majority of mammalian messenger RNAs (mRNAs) contain conserved target sites that bind microRNAs (miRNAs) (Friedman et al. 2009). Such target sites are typically located within the 3′ untranslated regions (3′ UTRs) of mRNAs, and binding of a miRNA to a target site predominantly causes the target’s accelerated decay and resulting protein repression (Bartel 2009; Fabian et al. 2010; Guo et al. 2010; Eichhorn et al. 2014). Because mammals have hundreds of miRNAs, each of which can have hundreds of targets, most regulatory pathways incorporate miRNAs (Kim and Nam 2006). One prominent example of miRNAs impacting cellular processes is found in the immune system, where it is clear that different immune cells require specific miRNAs to develop and function (Xiao and Rajewsky 2009; Dooley et al. 2013; Kroesen et al. 2015; Liang et al. 2015).
In the adaptive immune system, CD8+ T cells are responsible for recognizing and killing cells infected with viruses and other intracellular pathogens (Butz and Bevan 1998; Williams and Bevan 2007; Joshi and Kaech 2008; Kaech and Cui 2012). Hematopoietic stem cells migrate to the thymus, where they then undergo positive and negative selection to form CD8+ T cells (Starr et al. 2003; Schwarz and Bhandoola 2006), which then egress from the thymus and are capable of migrating to sites of infection (Weinreich and Hogquist 2008). CD8+ T cells express different T-cell receptor (TCR) isoforms, thus enabling the CD8+ T cell repertoire to recognize specific antigens. Upon stimulation by a specific and complementary antigen-presenting cell, a naive CD8+ T cell, which is one that has not previously been activated in response to infection, responds by proliferating and differentiating into cytotoxic effector cells that kill infected cells using proteases and cytolytic proteins (Harty et al. 2000). Effector cells are composed of short-lived effector cells (SLECs), which terminally differentiate and undergo apoptosis post-infection, and memory precursor effector cells (MPECs), which can transition into long-lived memory cells that are capable of robustly responding to secondary infection (Kaech et al. 2002; Joshi et al. 2007; Sarkar et al. 2008; Cruz-Guilloty et al. 2009; Banerjee et al. 2010; Yang et al. 2011).
During thymic maturation, CD8+ T cells require the miRNA biogenesis protein Dicer (Muljo et al. 2005), and many miRNAs undergo dynamic regulation (Neilson et al. 2007). Dicer is also required for in vivo activation of mature CD8+ T cells after infection (Zhang and Bevan 2010); in the absence of Dicer, CD8+ T cells neither proliferate nor migrate to sites of infection. Interestingly, Dicer-deficient CD8+ T cells respond more rapidly to in vitro stimulation than do wild type (Zhang and Bevan 2010; Trifari et al. 2013), suggesting that miRNAs have both activating and inhibitory effects on infection response. Additionally, many miRNAs are differentially expressed during effector and memory cell differentiation (Wu et al. 2007; Almanza et al. 2010; Trifari et al. 2013), some of which have known roles in generating effector cells (Wu et al. 2012; Gracias et al. 2013; Khan et al. 2013; Tsai et al. 2013). Importantly, many additional miRNAs with dynamic expression in CD8+ T cells do not have known roles, suggesting that they may also contribute to CD8+ T cell differentiation (Dooley et al. 2013; Kroesen et al. 2015; Liang et al. 2015).
Investigations into roles for miRNAs in lymphocytes have almost exclusively focused on adult cells, although expression of certain miRNAs changes across the life span. For example, miR-181 expression peaks in neonatal CD4+ T cells (Palin et al. 2013) and then declines with progressing age, which is important because miR-181 contributes to immune reactivity upon stimulation (Li et al. 2012). These data suggest that immune system changes during ontogeny might also be under control of miRNAs. In contrast to CD4+ T cells, nothing is known about the expression patterns and roles of miRNAs in neonatal CD8+ T cells.
Neonatal CD8+ T cells respond to infection profoundly differently than do adult cells (Campion et al. 2002; Adkins et al. 2003, 2004; Opiela et al. 2009; Rudd et al. 2013); specifically, neonatal cells proliferate more rapidly in response to primary infection and fail to generate memory cells, thus impairing their response to a secondary infection from the same pathogen (Smith et al. 2014). This inability to generate memory cells has important consequences for neonatal health and immunization. Remarkably little is known, however, about the underlying molecular bases that differentiate neonatal and adult CD8+ T cells. Because miRNAs are developmentally regulated (Stefani and Slack 2008; Sayed and Abdellatif 2011; Ebert and Sharp 2012) and are required for the adult CD8+ T cell response, we investigated developmental regulation of miRNAs in CD8+ T cells, motivated to discover if differences in both miRNA expression and regulation by specific miRNAs contribute to the important differences in how differently aged animals respond to primary infection from pathogens. To do so, we examined if miRNAs are differentially regulated between adult and neonatal CD8+ T cells, both before and during primary infection. In parallel, we also defined the mRNA transcriptome in the same sets of cells to identify primary targets of miRNAs as well as other mRNAs that have altered expression in different-aged CD8+ T cells. Surprisingly, our results demonstrate that differential gene regulation between adults and neonates is most pronounced prior to immunological challenge, suggesting that the developmentally regulated miRNAs do not regulate the outcome of effector cells at the peak of the infection response. Instead, these miRNAs may set the activation threshold prior to infection, causing the neonatal CD8+ T cells to differentiate more rapidly into effector cells, thus biasing them toward a terminally differentiated SLEC fate. Multiple lines of evidence implicate miR-29 and miR-130, in particular, as critical regulators of adult and neonatal CD8+ T cells’ differential infection responses.
Materials and Methods
Human samples
De-identified whole-adult (18–55 years of age) and cord blood (39–41 weeks gestation) samples, each from three healthy donors, were obtained from New York Blood Center and National Disease Research Interchange, respectively. Mononuclear cells were isolated using Ficoll-paque Plus (GE Healthcare); CD8+ T cells were then isolated using a Naive CD8+ T Cell Isolation Kit (Miltenyi Biotec).
Mice
gBT-I TCR transgenic mice (TCRαβ specific for SSIEFARL peptide from HSV-1 glycoprotein B498-505) were provided by Janko Nikolich-Zugich (University of Arizona, Tucson, AZ). Ly5.2 mice (8–12 wk) were purchased from the National Cancer Institute (Frederick, MD). Rag−/− OT-I mice were purchased from Taconic (Germantown, NY) and were crossed to C57Bl/6 mice from the National Cancer Institute, and F1 progeny were used. All pups (male and female) were used for newborn samples (1 day); only male neonatal pups (6–7 days) and adults (2–4 months) were used. Mice were maintained under pathogen-free conditions at Cornell University College of Veterinary Medicine. All experiments were performed in strict accordance with recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health; protocols were approved by the Institutional Animal Care and Use Committee (permit no. 2011-0090). All efforts were made to minimize animal suffering.
Naive cells
To isolate naive CD8+ T cells, spleens were harvested from uninfected mice of indicated age groups. Single-cell suspensions were prepared by mechanical dissociation of spleens through cell strainers. To obtain required numbers of cells, multiple newborn and neonatal spleens were pooled into single samples. CD8+ cells were purified using positive magnetic selection with CD8+ microbeads (Miltenyi Biotec). Following purification, samples were tested for purity, retaining only samples with >90% purity for subsequent sequencing.
Adoptive transfers
Adoptive transfers and infections were performed as described (Smith et al. 2014). Briefly, CD8+ T cells were isolated from congenically marked adult and neonatal mice, mixed at a 1:1 ratio, and 2 × 104 cells were transferred intravenously into Ly5.2 recipients. The next day, recipients were intravenously infected with Lm-gB (5 × 103 CFU). Effector CD8+ T cells from recipient spleens were isolated with anti-CD8 beads (Miltenyi Biotec) and then sorted via congenic marker expression on a FACS Aria (BD Biosciences).
Thymus isolation
To prepare thymic samples, thymi were collected from mice of indicated ages. Single-cell suspensions were prepared by mechanical dissociation through a cell strainer. Samples were depleted of CD4+ cells by incubating with anti-CD4-biotin followed by magnetic isolation with streptavidin microbeads (Miltenyi Biotec). To specifically isolate single positive CD8+ T cells, CD4− CD8+ Va2+ Vb8+ cells were FACS-sorted to >90% purity with an Aria instrument (BD). Sorted cells were placed in Trizol (Ambion) for extraction of RNA.
Antibodies and flow cytometry
Monoclonal antibodies anti-CD8α (53-6.7), anti-CD4 (GK1.5), anti-CD45.1 (A20), anti-CD45.2 (104), anti-CD90.1/Thy1.1 (OX-7), anti-KLRG1 (2F1), anti-CD127 (A7R34), anti-T-bet (eBio4B10), anti-Eomes (Dan11mag), anti-IL6ST (KGP-130), anti-CD25 (PC61), anti-CD69 (1H.2F3), and anti-CD62L (mel14) were purchased from commercial sources (Biolegend, eBioscience, or Invitrogen). Surface staining was performed by labeling cells for 30 min. Intracellular staining was performed using the Foxp3 staining buffer set (eBioscience). Flow cytometric data were acquired using a FACS LSRII with DiVa software (BD Biosciences) and analyzed with FlowJo (Tree Star).
RNA sequencing
RNA was isolated using Trizol (Life Technologies). One hundred nanograms of total RNA was used to generate small RNA libraries using the NEBNext Small RNA Library kit (NEB) or the TruSeq Small RNA Prep Kit (Illumina). miRNA expression was found with MirDeep2 (Friedländer et al. 2012, mm9, miRBase version 21), and differential expression was found with EdgeR (Robinson et al. 2010). Approximately 100 ng of total RNA was used to generate mRNA-Seq libraries using the RNA Sample Preparation Kit v2 (Illumina). Reads were aligned to the genome with Tophat (Trapnell et al. 2009, mm9), and differential expression was found with CuffDiff (Trapnell et al. 2013).
3′-Seq
3′-Seq was performed as described (Lianoglou et al. 2013). Briefly, total RNA was poly(A)-selected using a biotinylated poly(T) primer. After complementary DNA synthesis, the RNA was nick-translated and then the ends were cleaned up to allow adapter ligation and PCR amplification. Reads were aligned to the genome with Tophat (Trapnell et al. 2009, mm9), and reads whose 3′ ends were near a putative 3′ UTR polyadenylation sequence were quantified.
microRNA targeting
Strong miRNA targets were defined as expressed genes with at least one conserved miRNA target site and a TargetScan (version 6.2) context+ score ≤0.2 (Garcia et al. 2011). The background set consisted of any expressed gene that had a conserved miRNA target site with a context+ score ≤0.2 to any broadly conserved miRNA (Friedman et al. 2009), excluding all potential targets to the miRNA being tested. Differential targeting between miRNA targets and background were determined using two-sided Kolmogorov–Smirnov tests.
Luciferase assays
HEK293 cells were cotransfected with luciferase reporter plasmids and synthetic miRNAs (IDT) and harvested 24 hr later. Firefly and Renilla luciferase activities were measured using Dual-Luciferase Reporter Assay system (Promega) with a Veritas Microplate Luminometer (Turner).
Clustering gene expression data
We clustered expressed genes that were significantly differentially expressed and had at least a twofold difference in expression between adults and neonates in at least one sample [naive, 5, 7, or 15 days post-infection (dpi)]. Those genes were grouped into five clusters using the partitioning around the medoids method (Reynolds et al. 2006) in R.
Enrichment statistics
Gene sets were downloaded from the Immunologic Signatures collection at the Molecular Signatures Database (MSDB) (Subramanian et al. 2005). We found the number of genes from each data set that belonged to a cluster. For each cluster, we found the number of genes that were located in both the cluster and the data set (b), the total number of genes present in that cluster (n), the number of genes in the MSDB data set (B), and the total number of genes that had been clustered (N). Enrichment was calculated as (b/n)/(B/N). One-sided Fisher exact tests were used to measure significance.
More details regarding the methods are available in Supporting Information, File S1. Data in this publication have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (Edgar et al. 2002) under GSE65923.
Data availability
Plasmids are available upon request. Supporting information includes naive cell phenotyping (Figure S1), replicate miRNA expression data (Figure S2), miRNA expression data from CD8+ T cells with a different TCR (Figure S3), replicate mRNA expression data (Figure S4), miRNA targeting (Figure S5), protein expression data (Figure S6), 3′ Seq data (Figure S7), mRNA expression clustering (Figure S8), sorting strategy for thymic cells (Figure S9), mRNA expression of specific effector cell genes (Figure S10), detailed methods (File S1), miRNA expression values (File S2), differences in miRNA family expression (File S3), expression values for miRNA targets (File S4), 3′ UTR isoforms (File S5), mRNA expression values (File S6), and genes present in clusters (File S7). Gene expression data are available at GEO with the accession number: GSE65923.
Results
Neonatal effector CD8+ T cells are predominantly short-lived effectors
Naive CD8+ T cells respond to infection by proliferating and differentiating into cytotoxic effectors that kill infected cells. We previously demonstrated that neonates fail to effectively generate memory CD8+ T cells during primary infection due to cell-intrinsic differences (Smith et al. 2014), but the underlying molecular basis remained unknown. Here, we globally identified miRNAs present in neonatal and adult CD8+ T cells, reasoning that one or more miRNAs might contribute to these differences. To focus on cell-intrinsic differences, we eliminated extrinsic differences between adult and neonatal CD8+ T cells in two ways. First, we used CD8+ T cells from adult and neonatal gBT-I mice, which all contain the same TCR (transgenic for HSV-1 glycoprotein B498–505, gB), thus ensuring that any differences observed were not due to neonatal animals’ smaller TCR repertoire (Rudd et al. 2013). Second, we employed an adoptive co-transfer strategy, in which we co-transferred equal numbers of congenic gBT-I CD8+ T cells from adults and neonates into adult recipients, thus ensuring that the adult and neonatal cells were in the same environment. After the co-transfer, the adult recipients were systemically infected with Listeria monocytogenes expressing gB (Lm-gB). Donor cells were isolated at 5, 7, and 15 dpi using congenic markers (Figure 1A); this strategy allowed us to capture key events during the CD8+ T cell response to primary infection: expansion, peak response, and contraction, respectively. Consistent with previous findings (Smith et al. 2014), neonatal cells responded vigorously during expansion but were outnumbered by their adult counterparts on days 7 and 15 (Figure 1B). The neonatal effector cell population was predominantly composed of SLECs (KLRG1hiIL7Rlo), whereas adults generated many more MPECs (KLRG1loIL7Rhi), a difference that was observed at all three time points (Figure 1, C and D).
Figure 1.
Phenotypic differences between adult and neonatal CD8+ T cells. (A) Schematic of experimental strategy. Congenically marked adult (CD45.2+Thy1.2+) and neonatal (CD45.2+Thy1.1+) donor gBT-I CD8+ T cells (1 × 104) were transferred intravenously into wild-type congenically marked adult recipients (CD45.1+Thy1.2+), which were infected with L. monocytogenes expressing gB peptide. At 5, 7, and 15 dpi, spleens were harvested and donor cells sorted by flow cytometry based on congenic marker expression. Sorted cells were used for all downstream cellular and molecular assays performed on effector cells. (B–D) Distinguishing and quantifying both adult vs. neonatal and SLEC vs. MPEC CD8+ T cells in recipient animals. (B) Representative contour plots showing percentages (±SD) of adult and neonatal donor cells at 5, 7, and 15 dpi. The amount of Thy1.1 produced by CD45.2+ donor cells was used to differentiate adult (Thy1.1−) and neonatal (Thy1.2+) cells. (C) Representative contour plots of KLRG1 vs. IL7R for adult and neonatal donor cells at 5, 7, and 15 dpi. (D) Statistical analysis of SLEC (KLRGhiIL7Rlo) and MPEC (KLRG1loIL7Rhi) populations at 5, 7, and 15 dpi. Significance was determined by paired t-test; *P < 0.0001. Data are representative of two experiments (n = 8–12 mice/group). (E) Representative histograms show expression of SELL, IL2RA, CD69, KLRG1, and IL7R of adult and neonatal CD8+ cells after positive selection.
To establish that uninfected neonatal CD8+ T cells possess characteristics of naive cells equivalent to those of adults, we monitored several proteins whose expression patterns differentiate between naive and effector cells. We first found that adult and neonatal naive cells have equivalently high levels of the naive cell marker SELL/CD62L, which controls lymph homing (Figure 1E). Additionally, we found that adult and neonatal naive CD8+ T cells have similarly low levels of markers associated with activation (IL2RA/CD25 and CD69, Figure 1E). Finally, we measured KLRG1 and IL7R expression to determine if expression differences seen during infection are present prior to infection; however, adult and neonatal cells were similarly KLRG1loIL7Rhi (Figure 1E). Together, these results demonstrate that neonatal CD8+ T cells have naive characteristics prior to infection. Importantly, we also determined that our method for isolating CD8+ T cells for the adoptive cotransfer did not activate the cells, as demonstrated by cells before and after selection having equivalent expression of SELL, IL2RA, CD69, KLRG1, and Il7R (Figure S1).
Neonatal and adult CD8+ T cells from naive mice have pronounced differences in miRNA expression
MicroRNAs have been implicated in controlling adult naive CD8+ T cell differentiation into SLECs or MPECs (Liang et al. 2015); thus, we hypothesized that differential miRNA expression between adult and neonatal effector CD8+ T cells contributes to the lack of MPECs in neonates. We used high-throughput sequencing to profile miRNAs present in adult and neonatal CD8+ T cells at 5 and 7 dpi (as shown in Figure 1A).
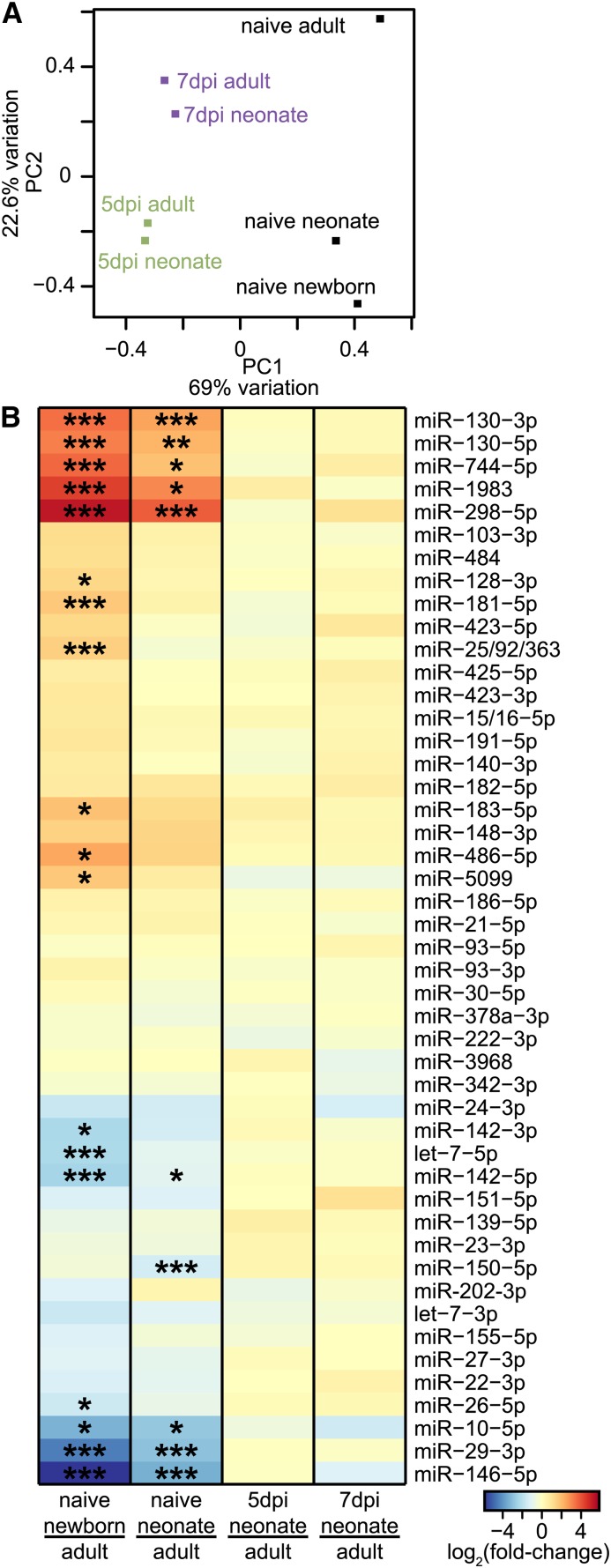
CD8+ T cells had 47 miRNA families that were well expressed [>1000 reads per million (RPM)] under at least one condition (File S2). MicroRNA families consist of miRNAs with identical or near identical targeting properties (Lewis et al. 2003). When we compared the miRNA profiles across all samples, we were surprised to find highly similar patterns of miRNA expression between neonatal and adult effector cells at both 5 and 7 dpi. Such similarities were observed using principal component analysis (PCA) (Figure 2A) and fold-change differences in expression (Pearson r > 0.99, P < 10−15, Figure 2B). Moreover, we found no miRNAs with statistically significant differential expression between adult and neonatal effector cells at either time point (Figure 2B and File S3). Interestingly, despite the pronounced difference in the SLEC:MPEC ratio between adults and neonates (Figure 1D), their highly concordant miRNA profiles suggested that SLECs and MPECs themselves possess extremely similar miRNA profiles.
Figure 2.
Differential microRNA expression in adults and neonates. (A) PCA using RPM values of well-expressed miRNAs. Adult and neonatal naive samples consist of two highly correlated pooled biological replicates (Figure S2); naive newborn and 5- and 7-dpi samples each consist of one replicate. The percentage of the overall variation accounted for by principal components 1 (x-axis) and 2 (y-axis) is indicated for each axis. (B) Fold-change difference in expression between adults and neonates was found for miRNA families in naive and 5- and 7-dpi CD8+ T cells, as well as between naive adult and newborn cells. Reads for miRNAs corresponding to the same family were summed. Significance was determined by edgeR exact test with multiple test correction (Benjamini–Hochberg; *P < 0.05, **P < 0.005, ***P < 0.0005). Small RNA expression data were replicated with independent biological samples, using an alternative method of sequencing library construction (see Figure S2).
We next profiled miRNAs from purified naive CD8+ T cells. In contrast to effector cells, miRNA expression differed greatly between naive neonatal and adult cells (Figure 2, A and B), including significant differential expression of 10 of the 47 well-expressed miRNA families. Many of the differentially expressed miRNAs have been implicated in CD8+ T cell processes. For example, miR-130 is upregulated in adult CD8+ T cells in early infection and is associated with T-cell activation (Zhang and Bevan 2010); it is also upregulated in neonatal naive CD8+ T cells (Figure 2B). Conversely, miR-29, miR-146, and miR-150, which are downregulated in naive neonatal CD8+ T cells (Figure 2B), are implicated as negative regulators of CD8+ T-cell infection response (Ma et al. 2011; Yang et al. 2012; Trifari et al. 2013). These results identify differential miRNA expression in naive cells as a possible basis for the altered behavior of neonatal and adult CD8+ T cells after infection.
To further explore developmental differences in miRNA expression, we profiled naive CD8+ T cells from newborn mice (1 day old). Differences in the global miRNA profile between newborn and adult naive cells were highly similar to differences that we observed between neonates and adults; such differences, however, were more pronounced in newborn mice than in neonates (Figure 2, A and B, P < 0.05). Together, these results show that the naive miRNA profile is largely age-dependent in CD8+ T cells.
To confirm that the age-dependent differences in miRNA expression that we observed (Figure 2) are not specific to a particular CD8+ TCR transgenic mouse line (gBT-I mice), we repeated our miRNA profiling in adult, neonatal, and newborn naive CD8+ T cells isolated from another transgenic T-cell mouse line (OT-I mice), which expresses a CD8+ TCR specific to the OVA peptide SIINFEKL. Importantly, the age-dependent miRNA expression differences observed in gBT-I CD8+ T cells were recapitulated well in OT-I CD8+ T cells (Figure S3), demonstrating that these differences in naive CD8+ T cell miRNA expression are robust across two different mouse genetic backgrounds.
Identification of conserved expression differences in developmentally regulated miRNAs
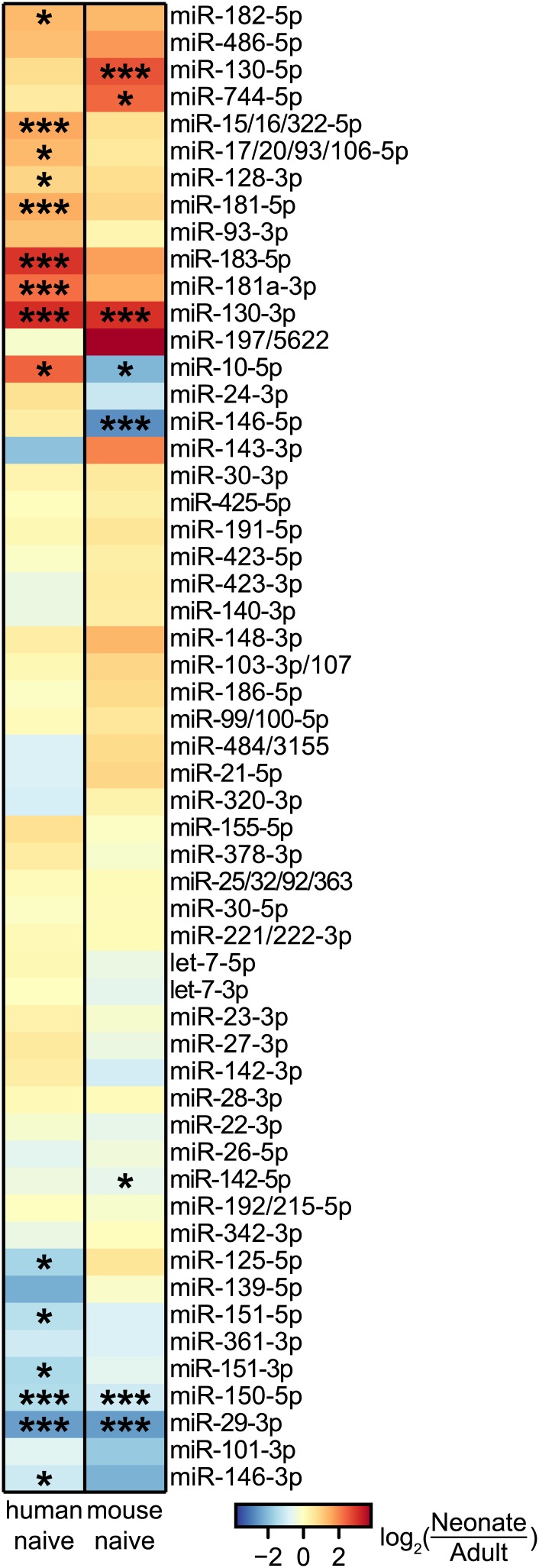
To determine if age-related miRNA expression differences are conserved in humans, we profiled miRNAs present in naive human CD8+ T cells from adult peripheral blood and from cord blood, which is developmentally similar to murine neonatal blood (Adkins et al. 2004). Fold-change expression differences between adult and neonatal cells correlated significantly between mice and humans for the 55 miRNA families that were well expressed in either one or both species (Figure 3). Of the 10 miRNAs with significant differential expression between adult and neonatal CD8+ T cells in mice, three had equivalent expression differences in humans: miR-29, miR-130, and miR-150. These particular miRNAs, therefore, are most likely to have conserved biological functions in CD8+ T cells.
Figure 3.
MicroRNA expression in naive human cells. Fold-change differences in expression were calculated comparing adults and neonates for miRNAs well expressed in humans and/or mice. Mice samples consist of two pooled biological replicates; human samples consist of three pooled biological replicates. *P < 0.05, ***P < 0.0005.
Targets of miR-29 and miR-130 differ in expression between adults and young mice
MicroRNAs affect phenotypes via repression of mRNA targets, yet changes in miRNA expression do not necessarily cause differential target regulation because only miRNAs expressed at high levels are able to mediate a detectable impact upon the transcriptome. Moreover, our understanding of miRNAs is insufficient to predict the consequences of modest changes in their expression levels (Bartel 2009; Mukherji et al. 2011). We therefore measured the effects of well-expressed miRNAs in differently aged naive CD8+ T cells by comparing the changes in expression for targets of well-expressed miRNAs between adults and neonates/newborns using RNA-Seq. For each sample, we sequenced at least two biological replicates, which were highly correlated for all samples (Figure S4).
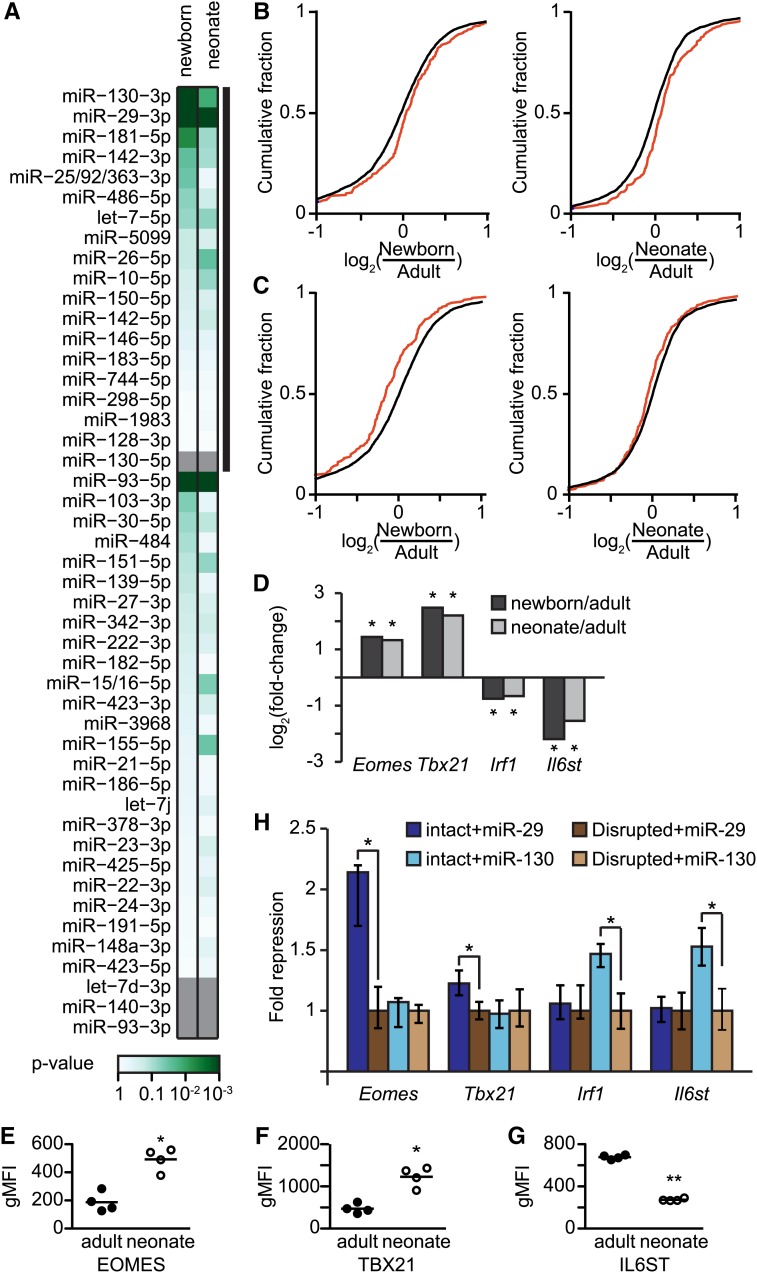
We predicted targets using TargetScan for each of the miRNA families that are well expressed in murine CD8+ T cells (Friedman et al. 2009; Garcia et al. 2011). To determine if a miRNA’s targets had altered expression in naive CD8+ T cells, we found the expression difference for those targets, individually comparing both newborns and neonates to adults, and then compared those values to a background set of genes targeted by other miRNAs. Only four miRNA families had significant differential targeting between adults and newborns, and three of those were also different between adults and neonates (P < 0.05, Figure 4A). Three of the miRNAs exhibiting differential targeting were themselves significantly differentially expressed; as expected, target sets had reciprocal expression to their cognate miRNAs. Notably, two of these miRNAs, miR-29 and miR-130, correspond to those with conserved expression differences in human adult and neonatal CD8+ T cells (Figure 3).
Figure 4.
miRNA targets. (A) Significant differences in miRNA target expression. Fold-change differences in expression between adults and neonates (or newborns) were found for predicted targets of each miRNA expressed in CD8+ T cells, which were then compared to fold changes in expression of a background set. Differentially expressed miRNAs are indicated with a thick black line, and miRNAs without predicted targets are shown in gray. Significance was determined by Kolmogorov–Smirnov tests. All samples consist of two pooled biological replicates; see also File S4. (B and C) Cumulative fold-change differences in expression of newborns (left) and neonates (right) compared to adults is shown for background genes (black) and targets (red) of miR-29 (B, P < 10−4) and miR-130 (C, P < 0.05). Expression values for targets are the mean Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values of two pooled biological replicates. (D) Mean differences in expression for neonate or newborn FPKM value compared to the adult value. Benjamini–Hochberg corrected significant differential expression was determined using CuffDiff. *P < 0.05 (E–G) Differences in protein expression between naive adult and neonatal samples as determined by flow cytometry for EOMES (E), TBX21 (F), and IL6ST (G). Significance determined by unpaired t-test, *P < 0.01, **P < 0.0001; see also Figure S6. (H) Fold repression of putative miRNA targets. The luciferase activity of reporter constructs containing intact miRNA target sites was normalized to otherwise identical reporters with disrupted target sites in the presence of miR-29 or miR-130. Reporter data plotted as fold repression (y-axis) of reporters with intact sites relative to those with disrupted sites. Data are represented as the geometric mean ± 33% of the spread of the data; n = 12–18; significance was determined by a two-sided Wilcoxon rank-sum test; *P < 0.0005.
The miRNA miR-29 is downregulated in newborn and neonatal CD8+ T cells, while its mRNA targets are more highly expressed in those cells (Figure 4B). Conversely, miR-130 is upregulated in newborn and neonatal CD8+ T cells, while its targets are more lowly expressed in those cells (Figure 4C). These targeting differences were robust to analyses using different background gene sets (Figure S5). Importantly, targets of both miRNAs that were predicted to be stronger exhibited more pronounced differences in their expression between adults and neonates when compared to those predicted to be weaker, strongly suggesting that differential expression of these miRNAs is directly controlling their targets’ expression (Figure S5).
Predicted targets are repressed by miR-29 and miR-130
We next investigated the differentially expressed targets of miR-29 and miR-130 (File S4) that had established roles in adult CD8+ T cell function. Notably, targets of miR-29 include Eomes and Tbx21 (T-bet), both of which encode transcription factors that upregulate genes required for effector function (Intlekofer et al. 2005; Banerjee et al. 2010; Kaech and Cui 2012). The transcripts of Eomes and Tbx21 were significantly upregulated in naive neonates and newborns, as compared to adults (Figure 4D); importantly, we confirmed this upregulation for both proteins in neonates (Figure 4, E and F; Figure S6). Targets of miR-130 include the transcription factor Irf1, which is required for CD8+ T cell development in the thymus (Penninger et al. 1997), and the chemokine receptor Il6st, which has been suggested to protect effector cells from apoptosis at the resolution of the CD8+ T cell response (Castellino and Germain 2007). Both Irf1 and Il6st transcripts are downregulated in newborn and neonatal mice; although we were unable to test IRF1 protein levels, we confirmed this downregulation for the IL6ST protein in neonates (Figure 4, D and G; Figure S6).
Because miRNA target prediction is imperfect, we used reporter assays to investigate whether targets were correctly predicted. Portions of the putative targets’ 3′ UTRs containing the miRNA target sites were placed downstream of a luciferase reporter; the target site was disrupted via mutagenesis to generate a paired control incapable of repression by the cognate miRNA. After cotransfecting each reporter with either miR-29 or miR-130, we compared differences in luciferase activity between the intact and disrupted target sites. Target sites in Eomes and Tbx21 were specifically repressed by miR-29, whereas sites in Irf1 and Il6st were specifically repressed by miR-130 (Figure 4H). These results, together with in vivo mRNA and protein quantifications (Figure 4, D–G), indicate that these genes are likely true targets of their respective miRNAs.
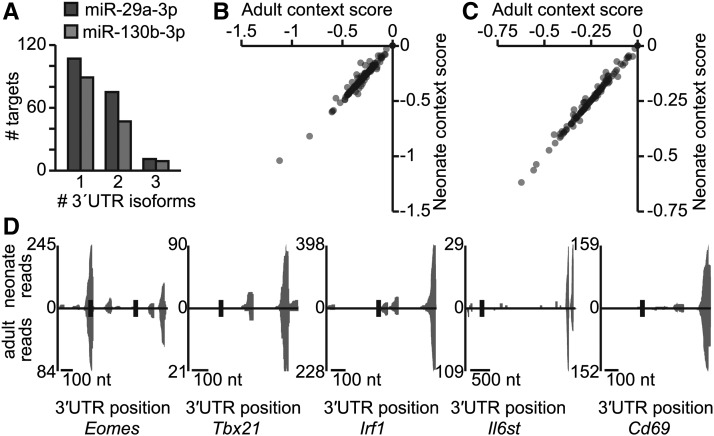
Mammalian 3′ UTRs are often alternatively processed to generate multiple isoforms to regulate gene expression (Mayr and Bartel 2009; Lianoglou et al. 2013), in part by changing the available miRNA target sites (Sandberg et al. 2008; Nam et al. 2014). We used 3′-Seq (Lianoglou et al. 2013), a technique that globally sequences the 3′ termini of mRNAs, to investigate alternative 3′ UTR processing in naive adult and neonatal CD8+ T cells. Most transcripts primarily express one 3′ UTR isoform; some genes (33%) express multiple isoforms, suggesting that different miRNA target sites might be present (Figure 5A). To determine if 3′ UTR isoform usage altered miR-29 and miR-130 targeting, we repeated our targeting analysis using the isoforms present in naive cells (File S5). Most predictions were unaffected, and adults and neonates almost always (∼90%) use the same 3′ UTR isoform in naive CD8+ T cells, resulting in the same miRNA target site availability for miR-29 and miR-130 (Figure 5, B and C; Figure S7). Specifically, we found that isoforms for Eomes, Tbx21, Irf1, and Il6st contain the expected miRNA targets sites, and importantly, isoform preference was equivalent in adults and neonates; Cd69 was also included as a likely target of miR-130 (Zhang and Bevan 2010). We found two Eomes 3′-UTR isoforms, one containing a single miR-29 target site and the other a pair of sites; however, relative usage of the isoforms was unchanged between adult and neonatal CD8+ T cells (Figure 5D). These results indicate that the differential miRNA targeting observed between adult and neonatal naive CD8+ T cells is not complicated by 3′-UTR isoform usage and further indicate that Eomes, Tbx21, Irf1, and Il6st are likely regulated by miR-29 or miR-130 in vivo.
Figure 5.
3′ UTR isoforms in naive CD8+ T cells. (A) The number of 3′-UTR isoforms expressed in adult and neonatal naive cells for miR-29 and miR-130 targets, as determined by 3′ Seq. The naive adult data are derived from two pooled biological replicates; the naive neonate has one replicate; see also File S5. (B and C) Isoform-weighted context scores for predicted targets of miR-29 (B) and miR-130 (C), comparing adult and neonatal samples (Pearson correlation coefficients > 0.98; P < 10−15 for targets of both miR-29 and miR-130). Context scores were weighted by the percentage of isoforms that contain each miRNA target; see also Figure S7. (D) Gene models shown with miRNA target sites in black, and density of 3′ Seq reads corresponding to 3′ termini of different isoforms.
Global differences in adult and neonatal CD8+ T-cell transcriptomes are pronounced in naive cells
Our results identify miR-29 and miR-130 as developmentally regulated miRNAs in differently aged naive CD8+ T cells in both mice and humans. Importantly, their targets include genes whose functions are known to affect differentiation into SLECs and MPECs. MicroRNAs exert their cumulative effects not only by repression of their direct targets, but also from resulting downstream expression differences due to target repression, creating indirect miRNA targets. Thus, the overall age-related transcriptome differences in CD8+ T cells are derived from direct and indirect effects of miRNAs, together with additional differences not attributed to miRNAs. We therefore systematically investigated age-related expression differences in CD8+ T-cell transcriptomes of adults, neonates, and newborns.
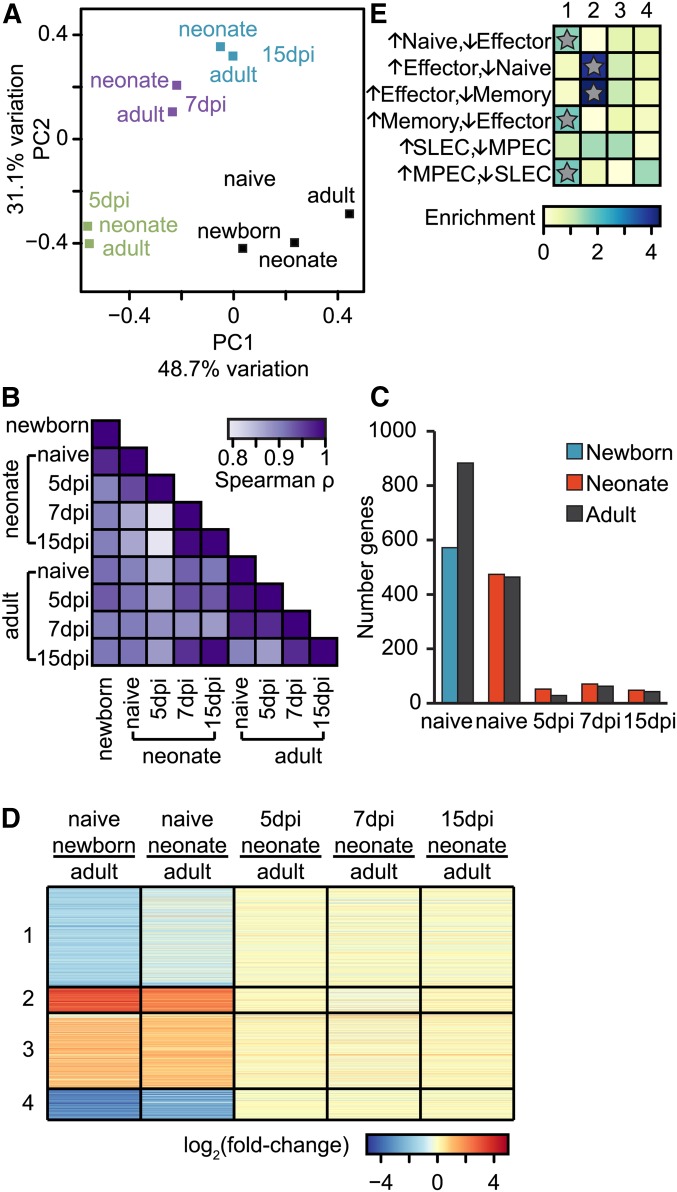
Global transcriptome differences were visualized using PCA (Figure 6A) and pairwise similarity between samples (Figure 6B), both of which indicate large transcriptomic differences in response to infection but smaller differences between adults and neonates at each time point; for naive cells, neonatal gene expression is more similar to newborns than to adults. Additionally, nearly 10 times more genes were significantly differentially expressed between adults and neonates in naive cells than in any stage of effector cells. Moreover, newborn naive CD8+ T cells have more differentially expressed genes with adults than do neonates (Figure 6C and File S6). Thus, similar to the age-dependent changes that we observed in miRNA expression (Figure 2A), we found that the CD8+ T-cell transcriptome also exhibited the most pronounced age-dependent changes in naive cells, rather than effector cells at any stage of the primary immune response.
Figure 6.
Differential gene expression in adult and neonatal CD8+ T cells. (A) PCA comparing naive and effector CD8+ T cells from different developmental stages. PCA, and all subsequent analyses (B–F), were performed on genes with an FPKM > 1 in at least one sample. The mean FPKM value was found for each sample, and PCA was performed on log-transformed values. The percentage of the overall variation accounted for by principal components 1 (x-axis) and 2 (y-axis) is indicated for each axis. (B) Color-coded pair-wise Spearman rank correlation coefficients comparing transcriptomes between different samples; P < 10−15 for all comparisons. (C) Number of genes that exhibited differences in expression that is at least twofold and significant (Benjamini–Hochberg corrected P-value <0.05) between pairs of samples. For naive and 5-, 7-, and 15-dpi samples, we compared adults and neonates; for naive, we additionally compared adults and newborns. See also File S6. (D) Clustering of co-regulated genes in newborn, neonatal, and adult CD8+ T-cell transcriptomes. Fold-change differences for all genes represented in B were calculated between adults and neonates or newborns. Clustering was performed to identify genes with similar changes in expression throughout infection; fold change for each gene is plotted in each sample, and genes are shown in their clusters; see also File S7. (E) Genes in each cluster were compared to genes that define naive, effector, memory, MPEC, or SLEC cells. Enrichment was calculated as the number of genes in each cluster as compared to the number expected. Significance was determined by Fisher exact tests; *P < 10−4.
Genes with similar changes in expression are often co-regulated and tend to possess similar biological functions (D’haeseleer 2005). Using unsupervised clustering, we found four groups of genes that have differential expression between adult and either neonatal or newborn CD8+ T cells (Figure 6D and File S7). The more modest differences between adult and neonatal effector cells were masked by the large expression differences in naive cells. Clusters 1 and 4 contain genes that are upregulated in adult CD8+ T cells, whereas clusters 2 and 3 show upregulation in the newborns and neonatal cells. To potentially detect expression differences in effector cells, we omitted the newborn expression data while performing the clustering; the differences in naive cells were still far larger than those at 5, 7, or 15 dpi (Figure S8). This clustering analysis further demonstrates that the age-related differences in gene expression in naive cells are far more extreme than those detected in effector cells. Taken together with our miRNA profiling, these analyses strongly suggest that the differences that exist between adult and neonatal CD8+ effector T cells are likely established in naive cells prior to infection and differentiation into effector cells.
To gain perspective on the possible biological functions of co-regulated clusters, we cross-referenced each cluster with curated gene sets that define and differentiate SLECs, MPECs, naive, effector, and memory cells (Kaech et al. 2002; Subramanian et al. 2005; Joshi et al. 2007). We found that both clusters 1 and 2 significantly overlap with certain of the curated gene sets. Cluster 1, which is downregulated in naive neonates and newborns, is significantly enriched for genes that distinguish naive from effector cells, memory from effector cells, and MPECs from SLECs. Taken together, these results suggest that, compared to naive adults, cells from both neonates and newborns have a lower expression of genes that defines naive and memory cells. Cluster 2, which is upregulated in naive neonates and newborns, is enriched in genes that distinguish effector cells from both naive and memory cells (Figure 6E). Importantly, for both clusters 1 and 2, the observed differences in gene expression were present only in naive cells, and not in effector cells. Together, these results demonstrate that, prior to infection, neonatal and newborn CD8+ T cell transcriptomes already begin to resemble those of effector cells, suggesting that this effector-like gene expression lowers the activation threshold of naive neonatal CD8+ T cells.
MicroRNA profiles in the thymus resemble neonatal naive cells
The underlying causes of the age-related differences in CD8+ T-cell miRNA expression that we have described are likely complex and due to developmental differences between adults, neonates, and newborns. CD8+ T cells are derived from hematopoietic stem cells (HSC) that undergo differentiation in the thymus. After exiting the thymus, CD8+ T cells are not yet fully mature; they undergo additional development for 3 weeks after their thymic emigration (Berzins et al. 1998). Stem cell origin is one possible basis for differential expression of miRNAs: CD8+ T cells of neonates and newborns are derived from fetal liver HSCs, whereas those of adults are derived from bone marrow HSCs (Jotereau et al. 1987; Foss et al. 2001). Alternatively, differences in cellular maturity might contribute to differential expression of miRNAs; neonatal and newborn naive cells are composed of less mature cells that have recently exited the thymus, whereas ∼80% of adult cells have undergone complete post-thymic maturation (Hale et al. 2006; Fink 2013). Importantly, these models are not mutually exclusive; moreover, the extent to which each model contributes may differ for specific miRNAs. To distinguish between these models, we compared the miRNA profiles of naive CD8+ T cells (located in the spleen) to those of immature adult, neonatal, and newborn CD8+ T cells found in the thymus (Figure S9). If age-dependent differences in miRNA expression are caused by HSC origin, adult and newborn thymic CD8+ T cells will have miRNA differences that mirror those differences observed in adult and neonatal/newborn splenic CD8+ T cells. It is worth noting that the miRNA profiles of neonatal thymic cells may be less informative as they are likely composed of both HSC populations. Alternatively, if the differences in miRNA profiles are instead due to time spent in the periphery, where additional post-thymic maturation occurs, neonatal and newborn splenic miRNA expression will more closely resemble that of thymic samples from all ages, which together will be distinct from miRNA expression found in the adult spleen.
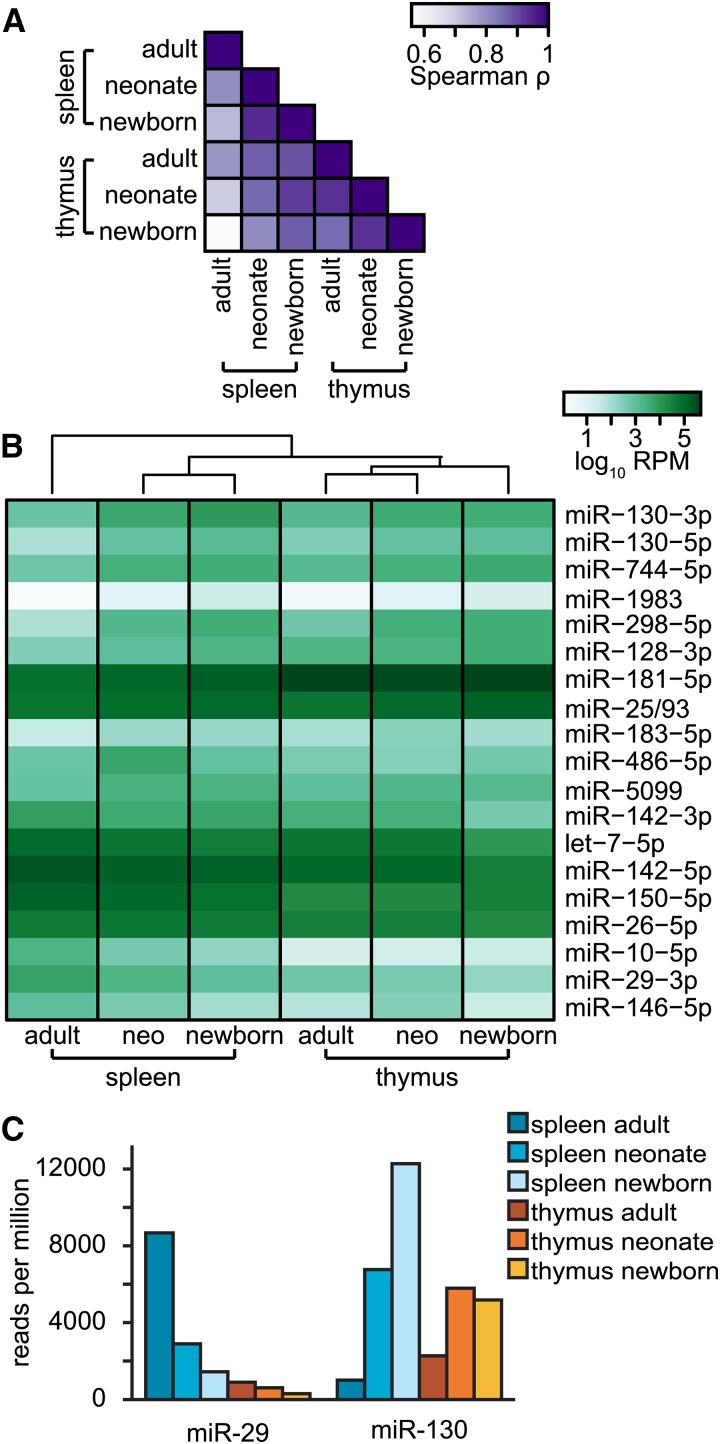
We first globally compared the expression of all well-expressed miRNAs by using pair-wise similarities between the samples (Figure 7A). When we compared miRNA profiles between thymus-derived samples, the profiles differed according to the age of the animal, suggesting that HSC origin contributes, at least to a degree, to differential regulation of miRNAs. Overall, however, the thymus profiles from all samples more closely resemble splenic neonatal and newborn profiles than that of the splenic adult. This result implies that the major determinant of the overall miRNA profile corresponds to the amount of time that CD8+ T cells have spent out of the thymus. Notably, these same patterns were also evident when we focused on only the expression of miRNAs identified previously as differentially expressed in naive cells (Figure 2B): patterns in thymic samples were more similar to those of neonatal and newborn splenic samples than to the profile found in the adult spleen, which possesses the most distinct profile of all the samples (Figure 7B).
Figure 7.
MicroRNA expression in immature and mature CD8+ T cells. (A) Color-coded pair-wise Pearson correlation coefficients comparing miRNA RPM values for adult and neonatal CD8+ T cells derived from the spleen and thymus. All adult samples and neonatal thymus samples are derived from two pooled biological replicates; the remaining samples consist of one biological replicate. See also Figure S9 and File S2. (B) Expression, in RPM, is shown for miRNAs that are significantly differentially expressed between naive adult cells as compared to neonatal and newborn cells (in the same order as Figure 2B). The samples were ordered via Euclidean clustering. (C) RPM values for miR-29 and miR-130 expression levels are shown for all samples.
Importantly, some miRNAs were expressed relatively uniformly in the thymus, independent of age, whereas others varied by age (Figure 7B); this result indicated that changes in expression of different miRNAs do not all share a common basis. Because our previous data implicate miR-29 and miR-130 as miRNAs associated with age-dependent changes in CD8+ T-cell phenotypes, we specifically examined their expression in the thymus-isolated and spleen-isolated naive CD8+ T cells (Figure 7C). Levels of miR-29 in the immature thymus-derived cells are low, regardless of the age of the animal and thus HSC lineage, indicating that control of miR-29 expression is likely primarily determined by maturation after egress from the thymus. Specifically, levels of miR-29 are highest in the adult spleen, which contains CD8+ T cells that are more mature than any of the other conditions that we examined. For miR-130, the upregulation in neonates and newborns when compared to adults is mirrored in the thymus, which implicates HSC origin as a major factor controlling levels of miR-130. Levels of miR-130, however, were different when the two adult samples were compared: miR-130 only reaches its lowest levels in the adult spleen, which indicates that the maturation status of CD8+ T cells also correlates with control of miR-130. These data imply that both HSC origin and post-thymic maturation influence control of miR-130 levels. Together, these data demonstrate that one model alone does not explain differences in splenic adult and neonatal immunity. While miRNA expression in thymic adult CD8+ T cells closely resembles that found in thymic neonates and newborns, indicating that post-thymic cellular maturation is a major determinant of miRNA profiles in CD8+ T cells, the expression of some miRNAs appears to be due also to HSC origin.
Discussion
Here, we show that adult and neonatal CD8+ T cells have highly similar gene expression profiles during primary infection despite large phenotypic differences. Instead, the greatest differences between adults and neonates are found in naive cells in mRNA and especially miRNA expression profiles. Downstream targets of the differentially expressed miRNAs miR-29 and miR-130 include the chemokine receptor Il6st and the transcription factors Irf1, Eomes, and Tbx21. These results suggest that regulatory differences in naive cells underlie neonates’ inability to create memory cells during infection, and they identify miR-29 and miR-130 as potential factors that contribute to establishment of these differences.
After primary microbial challenge, naive adult CD8+ T cells differentiate into SLECs and MPECs, whereas neonates primarily differentiate into SLECs. We were therefore surprised that the adult and neonatal effector miRNA profiles were more similar to each other than to cells at differing infection stages. These results suggest that miRNA expression differences previously seen between SLECs and MPECs (Khan et al. 2013) were relatively minor, especially when compared to the far greater differences that we observed between adult and neonatal/newborn naive cells. Another possibility is that SLECs produce vastly more miRNAs than MPECs, or vice versa, and thus the miRNAs from one cell type might dominate the overall profile. Regardless, phenotypic differences in adult and neonatal immunity are not explained by miRNA expression during infection.
MicroRNAs miR-29 and miR-130 are of particular interest because gBT-I mice, OT-I mice, and humans show consistent expression differences between adults and neonatal naive CD8+ T cells. miR-29 is highly expressed in adult naive cells; importantly, downregulation of miR-29 upon activation is necessary for efficient cell killing, and mice that lack miR-29 are more resistant to infection than wild type (Ma et al. 2011). Thus, naive neonates’ lowered miR-29 expression resembles that of adults post-activation, suggesting that neonatal naive cells more easily undergo activation, which primes them for effector functions. Conversely, miR-130 is upregulated shortly after activation in adult cells, and its expression correlates with the cells’ ability to exit lymphoid organs (Shiow et al. 2006; Zhang and Bevan 2010). Naive neonatal expression of miR-130, therefore, also resembles that of adult effector cells. Increased neonatal miR-130 in naive cells might allow the cells to more quickly reach sites of inflammation after activation, thereby promoting SLEC differentiation (Joshi et al. 2007; Sarkar et al. 2008; Gerlach et al. 2010; Jung et al. 2010). Thus, changes in miR-29 and miR-130 expression in naive neonates are consistent with rapid acquisition of effector functions and may allow neonates to compensate for smaller repertoires of CD8+ T cells in early life (Rudd et al. 2011, 2013).
We demonstrated that miR-29 can directly repress Tbx21 and Eomes, which encode partially redundant transcription factors that upregulate genes needed during infection, such as Ifng, Cxcr3, and Il2rb (Intlekofer et al. 2005; Banerjee et al. 2010), each of which is significantly upregulated in naive neonatal cells (Figure S10). Expression of Tbx21 and Eomes is low in adult naive cells, and their upregulation during infection is driven by external cell signaling (Takemoto et al. 2006; Joshi et al. 2007; Cruz-Guilloty et al. 2009; Pipkin et al. 2010; Rao et al. 2010, 2012; Kaech and Cui 2012). Therefore, lowered expression of miR-29 in neonatal naive cells likely directly results in elevated expression of Tbx21 and Eomes, which facilitates upregulation of effector-function proteins prior to infection. Thus, upregulation of Tbx21 and Eomes may be one specific mechanism by which decreased miR-29 expression in naive neonatal cells promotes a SLEC fate.
Importantly, our data suggest that age per se does not control expression of miR-29 and miR-130 (Figure 7). Instead, the miRNA profile of adult thymic CD8+ T cells is similar to that of neonates. In mice, the fetal thymus is colonized by at least two different layers of HSCs. The first, which is derived from the liver, colonizes the thymus around midgestation (Jotereau et al. 1987) and gives rise to neonatal and newborn CD8+ T cells. The second layer of hematopoietic stem cells originates from the bone marrow, and these cells seed the thymus just before birth, eventually replacing the first-layer cells and producing adult CD8+ T cells (Foss et al. 2001). Our data suggest that the alternative HSC origins contribute, to a degree, to differences in miRNAs expressed in neonates and adults, in particular contributing to differences observed for miR-130. By virtue of age, all neonatal and newborn CD8+ T cells have recently exited the thymus. In adults, by contrast, such recent thymic emigrants make up only ∼20% of the mature CD8+ T population, and they are known to preferentially differentiate as SLECs rather than as MPECs (Makaroff et al. 2009; Fink 2013). Our data identify the maturity of CD8+ T cells as the major factor determining the miRNA profile, fully explaining differential levels of miR-29 and contributing to control of miR-130. To further understand the contribution of these models, it would be informative to profile miRNAs found in adult recent thymic emigrants.
Considering the large differences in naive miRNA profiles, it is interesting to consider whether miRNAs could serve as biomarkers to predict vaccine success or infection outcomes in neonates and infants. For example, because miR-29 is enriched in adult naive cells whereas miR-130 is enriched in neonatal naive cells, the ratio of miR-29 to miR-130 in naive CD8+ T cells could provide important information regarding the proportion of adult- and young-derived CD8+ T cells, which, in turn, could be used to predict if the starting population of CD8+ T cells can differentiate into long-lived memory cells. This information could be particularly useful in the clinical setting, where there appears to be extensive heterogeneity in the response to infections and vaccines during early life (Green et al. 1994; Hsu et al. 1996; Poland 1998; Nair et al. 2007; Gaucher et al. 2008; Poland et al. 2011).
Supplementary Material
Acknowledgments
We acknowledge the use of tissues procured by the National Disease Research Interchange with support from National Institutes of Health (NIH) grant 2 U42 OD011158. Sequencing was performed at the Cornell Biotechnology Resource Center Genomics Facility with support from NIH grant 1S10OD010693 and Cornell University. This work was supported by NIH grants R01AI105265 and R01AI110613 (to B.D.R.); grants R01GM105668 and Core B of P50HD076210 (to A.G.); the Cornell Center for Vertebrate Genomics (B.D.R. and A.G.); and National Science Foundation grant DGE-1144153 (to E.M.W.). Cytometry core is supported in part by the Empire State Stem Cell Fund, the New York State Department of Health (NYS-DOH), contract #C026718. Opinions expressed are solely the authors; they do not necessarily reflect the view of Empire State Stem Cell Board, the NYS-DOH, or New York State.
Footnotes
Communicating editor: A. Gasch
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.179176/-/DC1.
Data in this publication have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus under accession no. GSE65923.
Literature Cited
- Adkins B., Williamson T., Guevara P., Bu Y., 2003. Murine neonatal lymphocytes show rapid early cell cycle entry and cell division. J. Immunol. 170: 4548–4556. [DOI] [PubMed] [Google Scholar]
- Adkins B., Leclerc C., Marshall-Clarke S., 2004. Neonatal adaptive immunity comes of age. Nat. Rev. Immunol. 4: 553–564. [DOI] [PubMed] [Google Scholar]
- Almanza G., Fernandez A., Volinia S., Cortez-Gonzalez X., Croce C. M., et al. , 2010. Selected microRNAs define cell fate determination of murine central memory CD8 T cells. PLoS One 5: e11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Gordon S. M., Intlekofer A. M., Paley M. A., Mooney E. C., et al. , 2010. Cutting edge: the transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J. Immunol. 185: 4988–4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P., 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berzins S. P., Boyd R. L., Miller J. F. P., 1998. The role of the thymus and tecent thymic migrants in the maintenance of the adult peripheral lymphocyte pool. J. Exp. Med. 187: 1839–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butz E. A., Bevan M. J., 1998. Massive expansion of antigen-specific CD8+ T cells during an acute virus infection. Immunity 8: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campion A. L., Bourgeois C., Lambolez F., Martin B., Léaument S., et al. , 2002. Naive T cells proliferate strongly in neonatal mice in response to self-peptide/self-MHC complexes. Proc. Natl. Acad. Sci. USA 99: 4538–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellino F., Germain R. N., 2007. Chemokine-guided CD4+ T cell help enhances generation of IL-6RαhighIL-7Rαhigh prememory CD8+ T cells. J. Immunol. 178: 778–787. [DOI] [PubMed] [Google Scholar]
- Cruz-Guilloty F., Pipkin M. E., Djuretic I. M., Levanon D., Lotem J., et al. , 2009. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 206: 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’haeseleer P., 2005 How does gene expression clustering work? Nat. Biotechnol. 23: 1499–1501. [DOI] [PubMed]
- Dooley J., Linterman M. A., Liston A., 2013. MicroRNA regulation of T-cell development. Immunol. Rev. 253: 53–64. [DOI] [PubMed] [Google Scholar]
- Ebert M. S., Sharp P. A., 2012. Roles for microRNAs in conferring robustness to biological processes. Cell 149: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E., 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn S. W., Guo H., McGeary S. E., Rodriguez-Mias R. A., Shin C., et al. , 2014. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 56: 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian M. R., Sonenberg N., Filipowicz W., 2010. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79: 351–379. [DOI] [PubMed] [Google Scholar]
- Fink P. J., 2013. The biology of recent thymic emigrants. Annu. Rev. Immunol. 31: 31–50. [DOI] [PubMed] [Google Scholar]
- Foss D. L., Donskoy E., Goldschneider I., 2001. The importation of hematogenous precursors by the thymus is a gated phenomenon in normal adult mice. J. Exp. Med. 193: 365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer M. R., Mackowiak S. D., Li N., Chen W., Rajewsky N., 2012. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40: 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R. C., Farh K. K.-H., Burge C. B., Bartel D. P., 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D. M., Baek D., Shin C., Bell G. W., Grimson A., et al. , 2011. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 18: 1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher D., Therrien R., Kettaf N., Angermann B. R., Boucher G., et al. , 2008. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J. Exp. Med. 205: 3119–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach C., van Heijst J. W. J., Swart E., Sie D., Armstrong N., et al. , 2010. One naive T cell, multiple fates in CD8+ T cell differentiation. J. Exp. Med. 207: 1235–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracias D. T., Stelekati E., Hope J. L., Boesteanu A. C., Doering T. A., et al. , 2013. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat. Immunol. 14: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. S., Shohat T., Lerman Y., Cohen D., Slepon R., et al. , 1994. Sex differences in the humoral antibody response to live measles vaccine in young adults. Int. J. Epidemiol. 23: 1078–1081. [DOI] [PubMed] [Google Scholar]
- Guo H., Ingolia N. T., Weissman J. S., Bartel D. P., 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale J. S., Boursalian T. E., Turk G. L., Fink P. J., 2006. Thymic output in aged mice. Proc. Natl. Acad. Sci. USA 103: 8447–8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty J. T., Tvinnereim A. R., White D. W., 2000. CD8+ T cell effector mechanisms in resistance to infection. Annu. Rev. Immunol. 18: 275–308. [DOI] [PubMed] [Google Scholar]
- Hsu L.-C., Lin S.-R., Hsu H.-M., Chao W.-H., Hsieh J.-T., et al. , 1996. Ethnic differences in immune responses to hepatitis B vaccine. Am. J. Epidemiol. 143: 718–724. [DOI] [PubMed] [Google Scholar]
- Intlekofer A. M., Takemoto N., Wherry E. J., Longworth S. A., Northrup J. T., et al. , 2005. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6: 1236–1244. [DOI] [PubMed] [Google Scholar]
- Joshi N. S., Kaech S. M., 2008. Effector CD8 T cell development: a balancing act between memory cell potential and terminal differentiation. J. Immunol. 180: 1309–1315. [DOI] [PubMed] [Google Scholar]
- Joshi N. S., Cui W., Chandele A., Lee H. K., Urso D. R., et al. , 2007. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27: 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jotereau F., Heuze F., Salomon-Vie V., Gascan H., 1987 Cell kinetics in the fetal mouse thymus: precursor cell input, proliferation, and emigration. J. Immunol. 138: 1026–1030. [PubMed] [Google Scholar]
- Jung Y. W., Rutishauser R. L., Joshi N. S., Haberman A. M., Kaech S. M., 2010. Differential localization of effector and memory CD8 T cell subsets in lymphoid organs during acute viral infection. J. Immunol. 185: 5315–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S. M., Cui W., 2012. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12: 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S. M., Hemby S., Kersh E., Ahmed R., 2002. Molecular and functional profiling of memory CD8 T cell differentiation. Cell 111: 837–851. [DOI] [PubMed] [Google Scholar]
- Khan A. A., Penny L. A., Yuzefpolskiy Y., Sarkar S., Kalia V., 2013. MicroRNA-17∼92 regulates effector and memory CD8 T-cell fates by modulating proliferation in response to infections. Blood 121: 4473–4483. [DOI] [PubMed] [Google Scholar]
- Kim V. N., Nam J.-W., 2006. Genomics of microRNA. Trends Genet. 22: 165–173. [DOI] [PubMed] [Google Scholar]
- Kroesen B.-J., Teteloshvili N., Smigielska-Czepiel K., Brouwer E., Boots A. M. H., et al. , 2015. Immuno-miRs: critical regulators of T-cell development, function and ageing. Immunology 144: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. P., Shih I., Jones-Rhoades M. W., Bartel D. P., Burge C. B., 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798. [DOI] [PubMed] [Google Scholar]
- Li G., Yu M., Lee W.-W., Tsang M., Krishnan E., et al. , 2012. Decline in miR-181a expression with age impairs T cell receptor sensitivity by increasing DUSP6 activity. Nat. Med. 18: 1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Pan H.-F., Ye D.-Q., 2015. microRNAs function in CD8+T cell biology. J. Leukoc. Biol. 97: 487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianoglou S., Garg V., Yang J. L., Leslie C. S., Mayr C., 2013. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 27: 2380–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F., Xu S., Liu X., Zhang Q., Xu X., et al. , 2011. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat. Immunol. 12: 861–869. [DOI] [PubMed] [Google Scholar]
- Makaroff L. E., Hendricks D. W., Niec R. E., Fink P. J., 2009. Postthymic maturation influences the CD8 T cell response to antigen. Proc. Natl. Acad. Sci. USA 106: 4799–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C., Bartel D. P., 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji S., Ebert M. S., Zheng G. X. Y., Tsang J. S., Sharp P. A., et al. , 2011. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 43: 854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo S. A., Ansel K. M., Kanellopoulou C., Livingston D. M., Rao A., et al. , 2005. Aberrant T cell differentiation in the absence of Dicer. J. Exp. Med. 202: 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair N., Gans H., Lew-Yasukawa L., Long-Wagar A. C., Arvin A., et al. , 2007. Age-dependent differences in IgG isotype and avidity induced by measles vaccine received during the first year of life. J. Infect. Dis. 196: 1339–1345. [DOI] [PubMed] [Google Scholar]
- Nam J.-W., Rissland O. S., Koppstein D., Abreu-Goodger C., Jan C. H., et al. , 2014. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol. Cell 53: 1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson J. R., Zheng G. X. Y., Burge C. B., Sharp P. A., 2007. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 21: 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opiela S. J., Koru-Sengul T., Adkins B., 2009. Murine neonatal recent thymic emigrants are phenotypically and functionally distinct from adult recent thymic emigrants. Blood 113: 5635–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palin A. C., Ramachandran V., Acharya S., Lewis D. B., 2013. Human neonatal naive CD4+ T cells have enhanced activation-dependent signaling regulated by the microRNA miR-181a. J. Immunol. 190: 2682–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninger J. M., Sirard C., Mittrücker H. W., Chidgey A., Kozieradzki I., et al. , 1997. The interferon regulatory transcription factor IRF-1 controls positive and negative selection of CD8+ thymocytes. Immunity 7: 243–254. [DOI] [PubMed] [Google Scholar]
- Pipkin M. E., Sacks J. A., Cruz-Guilloty F., Lichtenheld M. G., Bevan M. J., et al. , 2010. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 32: 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland G. A., 1998. Variability in immune response to pathogens: using measles vaccine to probe immunogenetic determinants of response. Am. J. Hum. Genet. 62: 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland G. A., Kennedy R. B., Ovsyannikova I. G., 2011. Vaccinomics and personalized vaccinology: Is science leading us toward a new path of directed vaccine development and discovery? PLoS Pathog. 7: e1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R. R., Li Q., Odunsi K., Shrikant P. A., 2010. The mTOR Kinase determines effector vs. memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and eomesodermin. Immunity 32: 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R. R., Li Q., Bupp M. R. G., Shrikant P. A., 2012. Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8+ T cell differentiation. Immunity 36: 374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. P., Richards G., de la Iglesia B., Rayward-Smith V. J., 2006. Clustering rules: a comparison of partioning and hierarchical clustering algorithms. J. Math. Model. Algor. 5: 475–504. [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd B. D., Venturi V., Davenport M. P., Nikolich-Zugich J., 2011. Evolution of the antigen-specific CD8+ TCR repertoire across the life span: evidence for clonal homogenization of the old TCR repertoire. J. Immunol. 186: 2056–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd B. D., Venturi V., Smith N. L., Nzingha K., Goldberg E. L., et al. , 2013. Acute neonatal infections “lock-in” a suboptimal CD8+ T cell repertoire with impaired recall responses. PLoS Pathog. 9: e1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R., Neilson J. R., Sarma A., Sharp P. A., Burge C. B., 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Kalia V., Haining W. N., Konieczny B. T., Subramaniam S., et al. , 2008. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205: 625–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D., Abdellatif M., 2011. MicroRNAs in development and disease. Physiol. Rev. 91: 827–887. [DOI] [PubMed] [Google Scholar]
- Schwarz B. A., Bhandoola A., 2006. Trafficking from the bone marrow to the thymus: a prerequisite for thymopoiesis. Immunol. Rev. 209: 47–57. [DOI] [PubMed] [Google Scholar]
- Shiow L. R., Rosen D. B., Brdičková N., Xu Y., An J., et al. , 2006. CD69 acts downstream of interferon-α/β to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature 440: 540–544. [DOI] [PubMed] [Google Scholar]
- Smith N. L., Wissink E., Wang J., Pinello J. F., Davenport M. P., et al. , 2014. Rapid proliferation and differentiation impairs the development of memory CD8+ T cells in early life. J. Immunol. 193: 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T. K., Jameson S. C., Hogquist K. A., 2003. Positive and negative selection of T cells. Annu. Rev. Immunol. 21: 139–176. [DOI] [PubMed] [Google Scholar]
- Stefani G., Slack F. J., 2008. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 9: 219–230. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., et al. , 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto N., Intlekofer A. M., Northrup J. T., Wherry E. J., Reiner S. L., 2006. Cutting edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J. Immunol. 177: 7515–7519. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L., et al. , 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifari S., Pipkin M. E., Bandukwala H. S., Äijö T., Bassein J., et al. , 2013. MicroRNA-directed program of cytotoxic CD8+ T-cell differentiation. Proc. Natl. Acad. Sci. USA 110: 18608–18613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.-Y., Allie S. R., Zhang W., Usherwood E. J., 2013. MicroRNA miR-155 affects antiviral effector and effector memory CD8 T cell differentiation. J. Virol. 87: 2348–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M. A., Hogquist K. A., 2008. Thymic emigration: when and how T cells leave home. J. Immunol. 181: 2265–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M. A., Bevan M. J., 2007. Effector and memory CTL differentiation. Annu. Rev. Immunol. 25: 171–192. [DOI] [PubMed] [Google Scholar]
- Wu H., Neilson J. R., Kumar P., Manocha M., Shankar P., et al. , 2007. miRNA profiling of naïve, effector and memory CD8 T cells. PLoS One 2: e1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Wieland A., Araki K., Davis C. W., Ye L., et al. , 2012. Temporal expression of microRNA cluster miR-17–92 regulates effector and memory CD8+ T-cell differentiation. Proc. Natl. Acad. Sci. USA 109: 9965–9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Rajewsky K., 2009. MicroRNA control in the immune system: basic principles. Cell 136: 26–36. [DOI] [PubMed] [Google Scholar]
- Yang C. Y., Best J. A., Knell J., Yang E., Sheridan A. D., et al. , 2011. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat. Immunol. 12: 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Boldin M. P., Yu Y., Liu C. S., Ea C.-K., et al. , 2012. miR-146a controls the resolution of T cell responses in mice. J. Exp. Med. 209: 1655–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Bevan M. J., 2010. Dicer controls CD8+ T-cell activation, migration, and survival. Proc. Natl. Acad. Sci. USA 107: 21629–21634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plasmids are available upon request. Supporting information includes naive cell phenotyping (Figure S1), replicate miRNA expression data (Figure S2), miRNA expression data from CD8+ T cells with a different TCR (Figure S3), replicate mRNA expression data (Figure S4), miRNA targeting (Figure S5), protein expression data (Figure S6), 3′ Seq data (Figure S7), mRNA expression clustering (Figure S8), sorting strategy for thymic cells (Figure S9), mRNA expression of specific effector cell genes (Figure S10), detailed methods (File S1), miRNA expression values (File S2), differences in miRNA family expression (File S3), expression values for miRNA targets (File S4), 3′ UTR isoforms (File S5), mRNA expression values (File S6), and genes present in clusters (File S7). Gene expression data are available at GEO with the accession number: GSE65923.