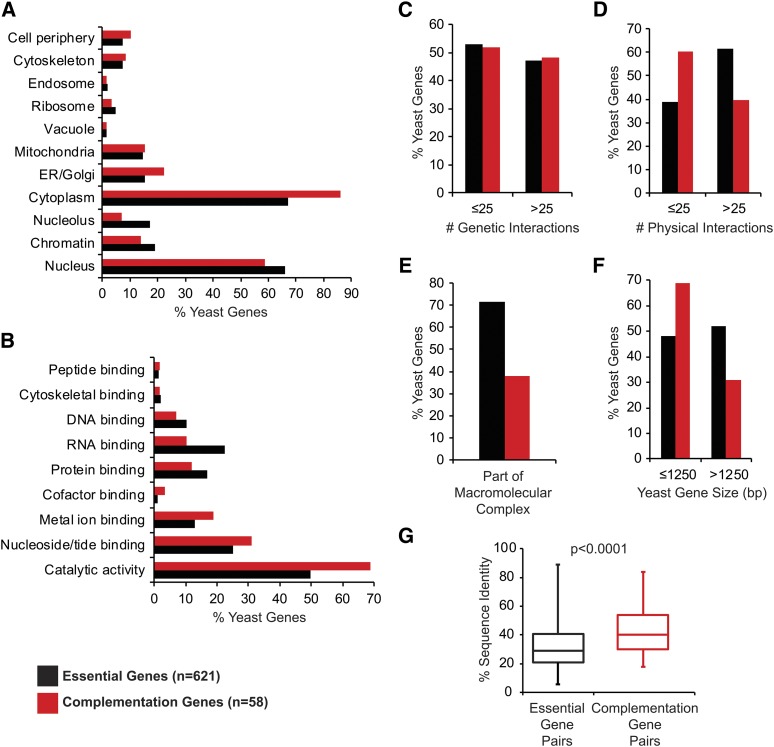
Figure 3.
Analyzing features of essential yeast genes that predict replaceability including (A) localization patterns, (B) molecular function, (C) no. of genetic interactions, (D) no. of physical interactions, (E) part of macromolecular complexes, (F) yeast gene size, and (G) human–yeast sequence identity. Localization data, Gene Ontology (GO) terms, no. of genetic/physical interactions, and gene size for each yeast gene were obtained from Yeastmine and each feature is represented as a proportion of the total number of genes input for each set (n = 621 for all essential genes included in both screens and n = 58 for the complementation genes). Overall, the complementation set was enriched for yeast proteins that localize to the cytoplasm (P = 8.18E-03), have catalytic activity (P = 2.28E-02), less physical interactions (P = 5.77E-03), are less likely to be part of macromolecular complexes (P = 3.36E-07), and have smaller gene size (P = 9.16E-03). For sequence identity, “essential gene pairs” refers to the 1076 human–yeast pairs included in this study corresponding to 621 yeast genes and “complementation gene pairs” refers to the 65 complementation pairs corresponding to 58 yeast genes. The box plot highlights the median and range of sequence identity for each set of gene pairs.