Abstract
Fruit flies of the genus Drosophila have been an attractive and effective genetic model organism since Thomas Hunt Morgan and colleagues made seminal discoveries with them a century ago. Work with Drosophila has enabled dramatic advances in cell and developmental biology, neurobiology and behavior, molecular biology, evolutionary and population genetics, and other fields. With more tissue types and observable behaviors than in other short-generation model organisms, and with vast genome data available for many species within the genus, the fly’s tractable complexity will continue to enable exciting opportunities to explore mechanisms of complex developmental programs, behaviors, and broader evolutionary questions. This primer describes the organism’s natural history, the features of sequenced genomes within the genus, the wide range of available genetic tools and online resources, the types of biological questions Drosophila can help address, and historical milestones.
Keywords: Drosophila, development, comparative genomics, model organism
Natural History
Drosophila are small flies in the order Diptera and family Drosophilidae. Commonly known as fruit or vinegar flies, they are often found on rotting fruit or other decaying matter (Powell 1997). Drosophila melanogaster, in the subgenus Sophophora, was first made famous at the beginning of the 20th century when the Morgan lab at Columbia University confirmed the chromosome theory of inheritance (Morgan 1910; Bridges 1916). Now studied by >1800 labs around the world, D. melanogaster is a powerful model organism. Flies are easily cultured in the lab and have many offspring and short generation times; in addition, they have a compact genome, are easy to manipulate genetically, and have many orthologous genes associated with human disease. In phylogenetic terms, the next-closest common invertebrate model, with an evolutionary divergence time of at least 600 million years (Adoutte et al. 2000), is the nematode Caenorhabditis elegans.
D. melanogaster is a human commensal species that inhabits all of the earth’s continents except Antarctica. Around 15,000 years ago, D. melanogaster migrated from its ancestral range in southern Sub-Saharan Africa to Europe and subsequently colonized much of the rest of the world, reaching the Americas in just the last few hundred years (David and Capy 1988; Lachaise et al. 1988). Humans are largely responsible for D. melanogaster migration in recent history, probably through the trade of fruit (David and Capy 1988; Markow and O’Grady 2007). Because the recent dispersion from its native home in tropical Sub-Saharan Africa required surviving in habitats with temperate climates, D. melanogaster are used to study adaptation to new environments (e.g., Schmidt et al. 2005).
There are >2000 described Drosophila species (Powell 1997; Markow and O’Grady 2006); however, the phylogenetic relationships between most of these species are not resolved and are debated among taxonomists (Dalton 2009; O’Grady and Markow 2009). Researchers study species across the genus Drosophila for everything from the size of their sperm (Pitnick et al. 1995) to their speciation history (Coyne and Orr 1998; Orr et al. 2007). Nearly two dozen Drosophila species have available genome sequences (more on this below), of which D. melanogaster was the first to be completed (Adams et al. 2000). Together, these species span a wide range of ecological habitats, life history characteristics, and evolutionary divergence times (Singh et al. 2009). This tractable complexity makes Drosophila a powerful model for comparative genomics studies on topics such as gene family evolution (Hahn et al. 2007), gene regulation (Stark et al. 2007), and ecological adaptation (Markow and O’Grady 2007).
Life Cycle
A major advantage of using D. melanogaster and related species as model systems is their particularly short life cycle, which allows for the rapid generation of large numbers of progeny to use in genetic crosses (Ashburner 1989). In D. melanogaster, the process of developing from a fertilized egg to adult (Figure 1) requires on average only 9–10 days at 25°; however, temperature can greatly influence the speed of this process, with flies cultured at 18° requiring ∼19 days from egg to adult. Upon fertilization, embryogenesis is completed in ∼24 hr, followed by three larval stages (termed first, second, and third instar) with a molting event at each stage transition. The first two instars each last on average 1 day, whereas the third instar typically requires 2 days. Thus, 5 days after fertilization, larval development is complete and the animals metamorphose within a hard, protective chitin-based pupal case (or puparium) that forms from the outer larval cuticle. The steroid hormone ecdysone is a central player in Drosophila metamorphosis, mediating gene expression shifts from the larval to adult fly pattern (Yamanaka et al. 2013). The animal remains in the pupal case for 4–5 days, during which most larval tissues break down and many adult structures develop from 19 imaginal discs present in the larvae. Imaginal discs are a collection of tissue-specific progenitor cells that develop during embryonic and larval stages and later give rise to most adult structures (such as eyes, legs, and wings) during pupal stages. Adult flies emerge from the pupal case in a process termed eclosion and become sexually mature in ∼8–12 hr, allowing the life cycle to repeat itself.
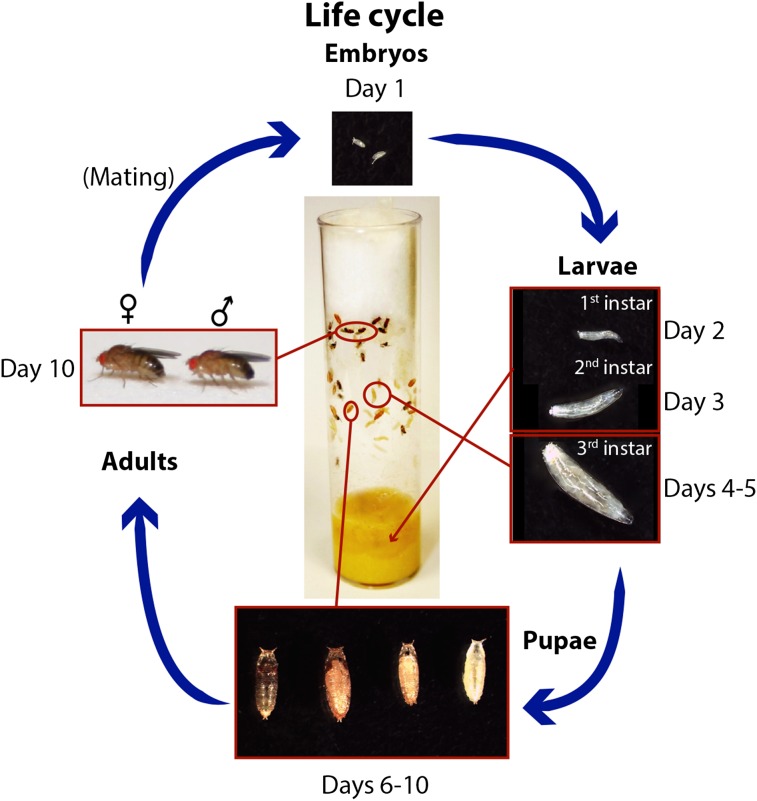
Figure 1.
Life cycle of D. melanogaster. D. melanogaster are cultured in vials with food in the bottom and a cotton, rayon, or foam plug at the top. The pictured vial shows each major stage of the life cycle, which is completed in 9–10 days when flies are maintained at 25°. Embryos hatch from the egg after ∼1 day and spend ∼4 days as larvae in the food. Around day 5, third instar larvae crawl out of the food to pupate on the side of the vial. During days 5–9, metamorphosis occurs, and the darkening wings within the pupal case indicate that maturation is nearly complete. Adult flies eclose from pupal cases around days 9–10.
Food and Husbandry
In the wild, fruit flies feed on yeast, bacteria, and plant matter within ripe or rotting fruit. In the lab, Drosophila media generally consists of a cornmeal/yeast/agar base supplemented with various carbohydrates and preservatives (see http://flystocks.bio.indiana.edu/Fly_Work/media-recipes/media-recipes.htm for recipe variants). The firmness of the food can be adjusted with varying amounts of agar depending on the health of the strain and level of larval activity. Ingredients are typically mixed with water, boiled, poured hot into vials or bottles, and then allowed to cool to create a solid plug of food at the bottom of the container. Some commercially available instant fly foods (with a potato-flake base) are simply mixed with water directly in the culture vial. Adult flies are transferred into the container (which is capped with a cotton or foam plug), where they lay eggs on the surface of the food. When eggs hatch into larvae, the larvae burrow into the food and progress through the instar stages until ready to undergo metamorphosis. As “wandering” third instar larvae, they crawl out of the food and pupate along the side of the container. Currently there are no efficient methods to freeze and recover adult flies or larvae, so the typical approach to maintain fly lines is continual transferring of adult flies to fresh media. It is recommended that flies are transferred to new food frequently (once every 2–3 weeks) to avoid bacterial/mold contamination and mite accumulation; however, this timing is commonly extended for strains that are not being actively used by maintaining them at 18°.
Sex Determination
While the presence of a Y chromosome is sufficient for male determination in mammals, it does not directly participate in sex determination in fruit flies. Instead sex determination in D. melanogaster is regulated by an X chromosome counting mechanism that “senses” the dosage of X chromosomes (Erickson and Quintero 2007). Thus, normal female flies are XX and males XY, but XO flies are males due to the decreased X dosage. In addition, each cell in the animal undergoes its own sex determination, with some cells continually monitoring X dosage. It is therefore possible to find gynandromorphs, in which part of the fly body has male characteristics and other parts female (for example, due to loss of one X chromosome in some cells within an otherwise XX female).
At the molecular level, sex determination in Drosophila is controlled by activation of the RNA processing gene Sex-lethal (Sxl) in females but not males (Bell et al. 1988; Salz and Erickson 2010; Verhulst et al. 2010). XX dosage in females results in early expression of Sxl, which then initiates a cascade of alternative splicing events that ultimately regulate differential splicing of the transcription factors doublesex (dsx) and fruitless (fru) (Figure 2). Sex-specific isoforms of Dsx and Fru then mediate expression of downstream effectors that govern sexual morphology and behavior (Baker et al. 1987; Demir and Dickson 2005). For example, female-specific isoforms promote expression of yolk proteins in the fat body of females but not males. Work is underway to identify specific genes regulated by sex-specific isoforms of Fru (Neville et al. 2014; Nojima et al. 2014; Vernes 2014) and Dsx (Clough et al. 2014; Luo and Baker 2015). For a review on sex determination in Drosophila, see Salz and Erickson (2010).
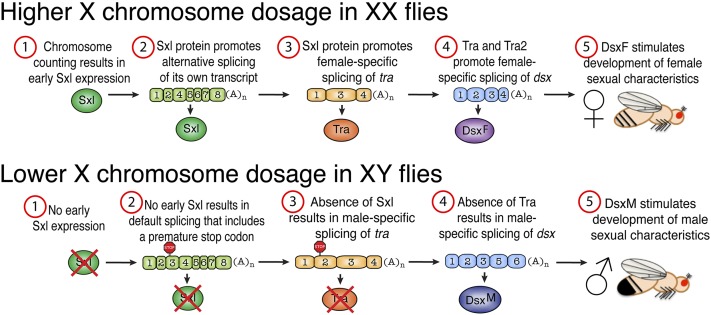
Figure 2.
Sex determination. The number of X chromosomes in D. melanogaster is determined by an X chromosome counting mechanism. In XX females, early expression of the RNA-processing gene Sex lethal (Sxl) later results in female-specific processing of its own transcript. Sxl then begins a cascade of alternative splicing events that ultimately result in generation of the female-specific isoform of Dsx (DsxF). Note that Fru is not shown here for clarity. In males, the absence of early Sxl expression results in default processing of Sxl and tra transcripts that contain an early stop codon. Dsx pre-mRNA is then processed for a male-specific isoform of Dsx (DsxM). These Dsx isoforms then promote expression of downstream genes that govern sex-specific decisions related to morphology and behavior.
Drosophila Genome Features
With its extraordinarily well-curated genome and large genetic toolkit (see below), D. melanogaster is a powerful genetic system. The entire D. melanogaster genome size is estimated at ∼180 Mb (Bosco et al. 2007), ∼120 Mb of which is euchromatin (Adams et al. 2000; Celniker et al. 2002). Of the four chromosome pairs (Figure 3A), the first is the sex chromosomes, including an acrocentric X chromosome and the submetacentric Y chromosome that is gene poor and almost entirely composed of heterochromatin (highly compacted, transcriptionally silent DNA, which is dense in repeats). The remaining three pairs are autosomes. Chromosomes 2 and 3 are large metacentric autosomes, whose left and right arms are sometimes referred to separately as 2L, 2R, 3L, and 3R. Chromosome 4 (the dot chromosome) is a very small autosome. The first physical maps of these chromosome arms were created by Calvin Bridges in Thomas Hunt Morgan’s group (Bridges 1935) based on polytene chromosomes, which are giant chromosomes (commonly isolated from salivary glands) usually consisting of >1000 endoreplicated DNA copies adhering in register. Adding a chemical dye to polytene chromosomes gives rise to unique banding patterns for each chromosomal region; these banded regions were numbered by Bridges and standardized as map coordinates that are still in use today. Before the D. melanogaster genome was sequenced, mapping of a DNA fragment to a genomic region was typically performed by in situ hybridization to polytene chromosomes.
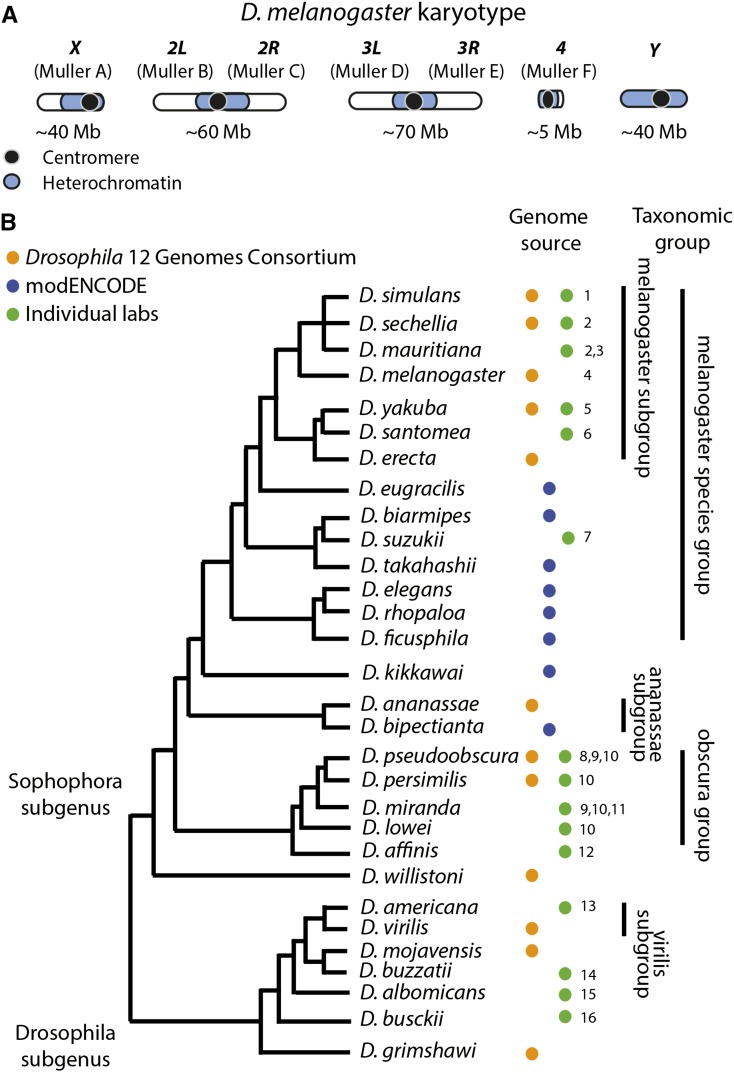
Figure 3.
Genome organization and phylogeny. (A) Organization of the Drosophila melanogaster genome. D. melanogaster has two metacentric autosome arms (chromosomes 2 and 3; Muller elements B and C and D and E), a small autosome (chromosome 4; Muller element F) and a pair of sex chromosomes (chromosome X—Muller element A—and chromosome Y). The approximate sizes and division of heterochromatin/euchromatin are shown. (B) Comparative genomics resources. Phylogeny of Drosophila species whose genomes were sequenced either by a large consortium (i.e., Drosophila 12 Genomes Consortium et al. 2007 or modENCODE https://www.hgsc.bcm.edu/drosophila-modencode-project) or other community or individual lab sequencing project. While likely not an exhaustive list, it highlights the power to do comparative genomics in Drosophila. The tree topology is derived from several sources (Drosophila 12 Genomes Consortium et al. 2007; Gao et al. 2007; Seetharam and Stuart 2013), as the phylogenetic relationships between some of these species are not well resolved. The references for each genome are as follows: (1) Hu et al. (2013); (2) Garrigan et al. (2012); (3) Nolte et al. (2013); (4) Adams et al. (2000); (5) Rogers et al. (2014); (6) http://genomics.princeton.edu/AndolfattoLab/Dsantomea_genome.html; (7) Chiu et al. (2013); (8) Richards et al. (2005); (9) Kulathinal et al. (2009); (10) McGaugh et al. (2012); (11) Zhou and Bachtrog (2012); (12) Palmieri et al. (2014); (13) Fonseca et al. (2013); (14) Guillen et al. (2014); (15) Zhou et al. (2012); and (16) Zhou and Bachtrog (2015).
The small genome size and longtime use as a model organism made D. melanogaster an appealing candidate genome to use as a proof of principle for whole genome shotgun sequencing and assembling of larger, more complex genomes (Adams et al. 2000). As a result, D. melanogaster was the second multicellular genome to be sequenced, after C. elegans (C. elegans Sequencing Consortium 1998). The first assembly was published in 2000 (Adams et al. 2000), and several iterations of assembly “finishing” were published subsequently (Celniker et al. 2002; Hoskins et al. 2007).
Initial genome annotation efforts identified ∼13,600 genes, ∼2500 of which had already been characterized (Adams et al. 2000; Misra et al. 2002). The current number of protein-coding genes predicted based on the latest genome assembly is 13,920 (R6.05; http://flybase.org/static_pages/docs/release_notes.html). Computationally predicted gene models are supported by evidence from sequenced transcripts (i.e., ESTs, cDNAs, and RNAseq data) and homology to known genes in GenBank (Misra et al. 2002). Each gene is assigned an annotation ID that begins with two letters—in D. melanogaster, “CG” for protein-coding genes and “CR” for nonprotein-coding genes—and followed by a series of numbers. Annotation IDs in other Drosophila species are identified by a different prefix (e.g., “GA” for D. pseudoobscura IDs and “GD” for D. simulans IDs). Each genomic feature in the D. melanogaster genome is assigned a unique FlyBase identifier: genes begin with “FBgn” and transcripts begin with “FBtr.” Genes typically are not assigned a descriptive name until they have been studied—those not yet studied are referred to by their “CG” number. Drosophila biologists have a tradition of assigning gene names based on their mutant phenotypes rather than their wild-type functions. For example, dunce mutants have a learning defect (Dudai et al. 1976), shaven baby mutants are missing the small hairs (trichomes) of the larval cuticle (Wieschaus et al. 1984), and tinman mutants lack a heart (Evans et al. 1995). Gene names and abbreviations are italicized but usually not capitalized, unless the allele for which they are named is dominant to wild type. Protein names and abbreviations are usually capitalized but not italicized.
Defining functional elements in the D. melanogaster genome
In 2007, the modENCODE project was launched with the goal of defining and describing all functional elements in the genomes of D. melanogaster and C. elegans (Celniker et al. 2009; modEncode Consortium et al. 2010). As a result of this large-scale collaboration, the fly community now has access to large datasets documenting many facets of Drosophila genetics, including transcription across the genome at different stages, transcription start and stop sites, alternative splicing patterns, promoter and other regulatory elements (and proteins that bind them), histone variants, and chromatin structure. Guidance in understanding and navigating available modENCODE data can be found in Boley et al. (2014).
modENCODE data indicate that the D. melanogaster transcriptome is much more complex than previously thought, suggesting an increased number of predicted protein-coding genes (14,692) along with 2872 noncoding genes, collectively covering 72% of the genome (Brown et al. 2014). More than half of D. melanogaster genes show alternative splicing, sometimes with differential promoter-driven use of alternative first exons, and 45% of genes encode more than one protein isoform. Many paralogous pairs of genes exist as a result of past gene duplication events, and in most cases the newer copy shows enriched expression in the testis at first, with gradually broadening expression over evolutionary time (Assis and Bachtrog 2013). Nearly 2000 long noncoding RNAs (lncRNAs) were identified in the modENCODE project, of which many overlap with protein-coding genes in an antisense orientation and likely are involved in regulation of the overlapping genes.
The broader view of genome-wide gene regulation obtained through modENCODE and other complementary efforts (de Graaf and van Steensel 2013) suggests (consistent with work in other organisms) that the three-dimensional folding of DNA in the nucleus brings together disparate regions of the genome into coregulated domains of genes showing similar expression patterns (modEncode Consortium et al. 2010). Binding of 84 transcription factors has been mapped at different developmental stages (Slattery et al. 2014). The map of transcription across the D. melanogaster genome was validated via comparison with transcriptional patterns in other Drosophila species (Chen et al. 2014).
Transposable elements
Transposable elements (TEs) are mobile genetic elements, first discovered in maize (McClintock 1950), that are common in Drosophila genomes. The two main classes of TEs are retrotransposons and DNA transposons (Craig et al. 2002). Retrotransposons move in the genome through an RNA intermediate using a “copy and paste” mechanism—DNA is transcribed to RNA, reverse transcribed, and inserted in new genomic location. DNA transposons move in the genome through a DNA intermediate using a “cut and paste” mechanism—transposon DNA is excised and moved to a new genomic location. In total there are ∼150 different TE families in D. melanogaster (Mackay et al. 2012), most of which are retrotransposons (Kaminker et al. 2002). These families, such as gypsy, copia, and minos, vary in their abundance and activity across Drosophila genomes (Drosophila 12 Genomes Consortium et al. 2007). One type of DNA transposon, the P element, was recently introduced to D. melanogaster from a distantly related species, D. willistoni (Daniels et al. 1990) and spread through D. melanogaster populations in a staggering <75 years (Bingham et al. 1982; Engels 1983; Kidwell 1983)! TEs are often deleterious, as they can insert into genes or cause chromosomal rearrangements. RNA interference mechanisms control the activity of TEs by silencing endogenous elements (Brennecke et al. 2007; Czech et al. 2008). These pathways involve 24–31 nucleotide Piwi-interacting RNAs (piRNAs) that associate with Piwi proteins (Brennecke et al. 2007) mainly in the germline, and ∼21 nucleotide endogenous siRNAs that associate with Dicer2 (Czech et al. 2008) in both the soma and germline. Not all TEs cause problems in the genome; rather, some TEs now serve an important biological role. For example, the activity of some retrotransposon families (Het-A, TART, and Tahre) maintains telomere length (George et al. 2006), a feature unique to Drosophila species. In the lab, researchers have developed ways to use TEs as tools for mutagenesis and transformation (see A Drosophila Genetic Toolkit below).
Conserved chromosome arms across Drosophila species
The gene content of chromosome arms tends to be conserved across Drosophila species, but the order and orientation of these arms are shuffled (Muller 1940). These conserved chromosome arms, lettered from A to F, are called Muller elements and correspond to the X, 2L, 2R, 3L, 3R, and 4 chromosome regions of D. melanogaster, respectively (Figure 3A). In general, the karyotype of each Drosophila species can be achieved by reorganizing these Muller elements. In Drosophila, chromosomal translocations involving the centromere and paracentric inversions (those not involving the centromere) are more common and fixed more often than pericentric inversions (those involving the centromere) (Sturtevant and Novitski 1941). The conservation of some Muller elements extends beyond Drosophila into other Dipterans such as botflies, house flies (Boyes and Vanbrink 1965), and even mosquitos (Bolshakov et al. 2002).
Drosophila as a model for comparative genomics
Since the publication of the D. melanogaster genome sequence (Adams et al. 2000), researchers have sequenced the genomes of over two dozen Drosophila species to varying levels of completion. The second sequenced Drosophila genome was D. pseudoobscura, a species with historical importance in evolutionary genetics (Richards et al. 2005). D. pseudoobscura is well studied for its abundance of chromosomal inversions (Sturtevant and Dobzhansky 1936) and the possibility that these genomic arrangements contribute to adaptation and the formation of new species (Noor et al. 2001; Navarro and Barton 2003). One motivating factor for sequencing D. pseudoobscura was to discover cis-regulatory elements; however, it turned out that there is little conservation between the noncoding regions of D. pseudoobscura and D. melanogaster (Richards et al. 2005). The Drosophila 12 Genomes Consortium involved a community-wide effort to sequence 10 additional species with a wide range of divergence times from D. melanogaster (Drosophila 12 Genomes Consortium et al. 2007). Of these sequenced species, the closest relatives to D. melanogaster are members of the simulans clade—D. sechellia and D. simulans—species that differ in ecology and life history characteristics despite diverging just 240,000 years ago (Garrigan et al. 2012). D. sechellia is endemic to the Seychelles Archipelago and specializes on a fruit toxic to other Drosophila species (Morinda citrifolia), whereas D. simulans is a geographically widespread and cosmopolitan species (Lachaise et al. 1988). The most distant relative to D. melanogaster is the Hawaiian “picture wing” species D. grimshawi. Hawaiian Drosophila are the most diverse group of drosophilids (Kaneshiro 1997) and are studied for their interesting morphological characteristics and speciation history (Carson 1975, 1982, 1997; Templeton 1981). Species with intermediate divergence times from D. melanogaster allowed for more efficient identification of conserved cis-regulatory elements and noncoding RNAs (Stark et al. 2007). One goal of the modENCODE project has been to sequence the genomes of eight additional species with next-generation technology (Celniker et al. 2009). Several more Drosophila species genomes are available through the efforts of individual labs (e.g., Garrigan et al. 2012; Zhou and Bachtrog 2012; Zhou et al. 2012; Nolte et al. 2013; Guillen et al. 2014) (Figure 3B). The large number of sequenced Drosophila genomes provides an important resource for comparative genomics and offers a powerful approach to the discovery of functional elements in genomes and their evolution (Singh et al. 2009), and ecological genomics (Markow 2015).
A Drosophila Genetic Toolkit
A strong attribute of the Drosophila experimental system is the ability to ask a wide variety of questions about the role of genes in the development and function of the organism, such as the following: Which genes are involved in the development of particular organs and tissues, or in a behavior, and what are the molecular pathways? Where is a certain gene expressed in the fly during development and/or later in the adult? What genes mediate basic cell biological events within specific cell types? Can controlled expression of a particular gene (or variants thereof) in developmental time and cellular location help elucidate that gene’s function? Ingenious genetic tools to address these questions in D. melanogaster have enabled dramatic advances in our understanding of basic cell biology and development. In addition, since most human genes associated with genetic disease have a Drosophila counterpart, genetic analysis in flies has led to important applications toward human health.
How can I identify the genes involved in my favorite developmental or behavioral process?
Finding genes associated with a developmental process or behavior can be accomplished in a variety of ways but most often is associated with a forward genetic screen in which mutagenized fly lines are examined for alterations in the phenotype of interest. Before we discuss how genetic screens are carried out, we need a brief overview of the basics of doing a genetic cross and some of the tricks of the trade.
Genetic crosses and balancer chromosomes:
Hallmarks of a model genetic organism are the ability to create stable inbred stocks carrying mutations or other experimental genome manipulations and the ability subsequently to cross these stocks to other stocks to generate desired genotypes. In D. melanogaster, multigenerational crossing schemes can be easily designed and carried out by isolating unmated or “virgin” females <8 hr old (since females who previously mated with siblings can store sperm for later use) and mixing them with males of defined genotypes. Males and females are distinguished primarily by abdominal pigmentation patterns (Figure 1) as well as genital structures and presence of “sex combs” on the front legs of males. A typical component of a Drosophila researcher’s day is the collecting of virgins upon arriving in the lab in the morning and leaving the lab at night. Accurate genetic crosses are enabled by visible “marker” mutations that allow selection of offspring that inherited one version or the other of a chromosome. Classic examples of these markers are yellow and white on the X chromosome (affecting body and eye color, respectively), Curly (Cy) on the second chromosome (affecting wing shape), and Stubble (Sb) on the third chromosome (affecting bristle length) (Greenspan 1997). For example, a geneticist may collect noncurly winged flies to obtain those that inherited the parent’s other copy of the second chromosome with a mutation of interest. Observant genetics students might wonder whether meiotic crossing over could be a big problem here—what if the mutation of interest had recombined onto the Curly chromosome during meiosis in the parent? To prevent such movement, the chromosomes with visible markers also contain multiple inversions to prevent viable recombinant offspring; thus the only viable offspring are those with one or the other of the parent’s two intact chromosomes (either with the mutation of interest or with the dominant marker). These engineered chromosomes with visible markers and multiple inversions are called “balancer chromosomes.” Balancer chromosomes have a third feature as well—recessive lethal mutations—to prevent mutations of interest from being selected out of an inbred population. For example, if you are studying mutation f that is homozygous lethal, you must propagate f via heterozygotes that have one wild-type and one mutant copy. Over time in a population, the f allele would gradually decrease in frequency since f/f homozygotes die while heterozygotes and wild-type homozygotes reproduce. If the f mutation is instead maintained with a balancer chromosome, the f allele persists since balancer homozygotes die, and heterozygotes are the only viable genotype. See Greenspan (1997) for a deeper examination of the logistics for setting up crosses and using balancer chromosomes.
Inducing mutations:
The first step in designing a genetic screen is choosing how you will disrupt the genome in hopes that particular genes affecting your process of interest are among the many genes randomly mutated. As with other model systems, the fly community has used chemical mutagens such as ethyl methane sulfonate (EMS) added to fly food or ionizing radiation such as X-rays to trigger everything from single base pair mutations to larger chromosomal changes (Greenspan 1997). As described earlier, the identification of transposable elements provided an additional tool to alter genetic sequences through the mobilization of genetic elements throughout the genome. The Gene Disruption Project has used different transposable elements to achieve broad coverage of insertions and to generate a variety of extremely useful types of insertions, although this has not completely saturated the genome due to the particular idiosyncrasies of the elements (Bellen et al. 2004, 2011; Venken et al. 2011). To date, the project has created >12,000 insertion lines that are available for public use in the stock collections. Current estimates suggest that only 30% of the coding sequences in the genome have been targeted using these traditional methods (Kondo 2014). These insertions have also been used in creative ways to create small deletions through imprecise transposon excision (Voelker et al. 1984), mutations in nearby genes through “local hopping” of the element (Tower et al. 1993), and defined sets of isogenic deletions that cover the genome through the excision of DNA between elements at defined insertion points (Parks et al. 2004; Ryder et al. 2004; Thibault et al. 2004). Most often researchers will use a combination of transposable element insertion libraries and chemical mutagenesis to provide broad mutational coverage of all the chromosomes in hopes of “saturating” or creating mutations in all the possible genes that may play a role in their favorite developmental or behavioral process.
A basic forward genetic screen:
The goal of a forward genetic screen is to identify genes involved in a particular developmental process, biochemical activity, or behavior, with no a priori model of which genes might be important. Success is often dependent on how efficiently a particular phenotype can be observed. A variety of innovative screening approaches have identified genes associated with many aspects of D. melanogaster biology (St Johnston 2002), with an early example being the Nobel Prize-winning embryonic lethal zygotic screen to dissect the basis of early development (Nusslein-Volhard and Wieschaus 1980). Their hypothesis was that mutations causing embryonic lethality likely affected genes that normally played a role in early development. Indeed, when they visualized embryonic morphology of the many mutants they isolated, the observed phenotypes suggested different levels of genetic and spatial organization in the developing embryo. Most mutagenesis screens start with mutagenizing males via chemicals or radiation and crossing them to females carrying balancer chromosomes (Figure 4). Single male offspring from this cross are outcrossed again, and male and female siblings within these individual lines are then crossed to start inbred lines that propagate different founding mutations. Within these lines, heterozygotes (with a balancer) propagate the stock, while homozygotes can be examined for recessive phenotypes of interest. Independent inbred lines with similar phenotypes can be crossed together in complementation tests to see if mutations happen to affect the same gene.
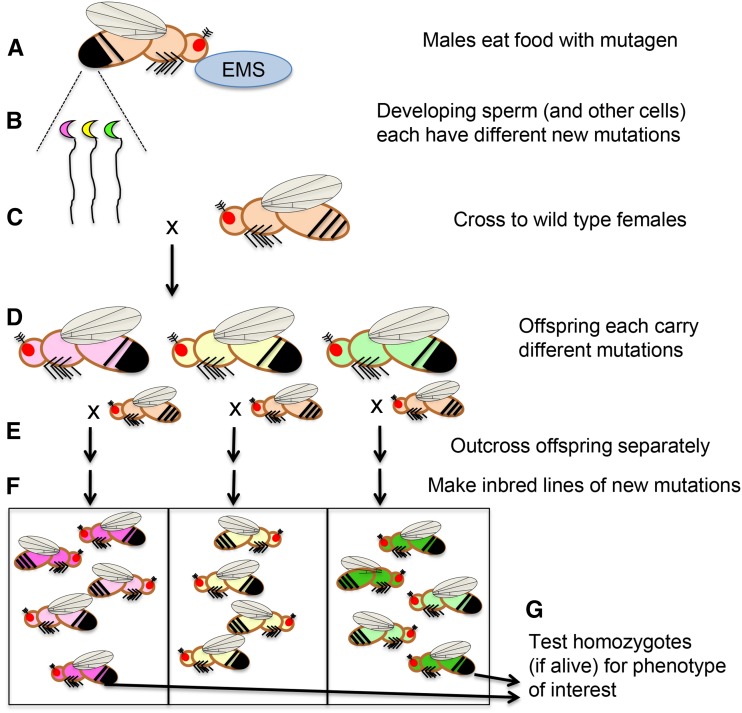
Figure 4.
Generalized scheme for a forward genetic screen using chemical mutagenesis. (A) Male flies eat food laced with ethane methyl sulfonate (EMS), an alkylating agent which typically causes point mutations. (B) Different mutations occur in each cell of the feeding flies, including sperm (indicated by pink, yellow, and green sperm cells). (C) Outcrossing the mutagenized flies to untreated females yields (D) offspring that each potentially have a different new mutations throughout their bodies, indicated schematically by body colors corresponding to the sperm cells above. (E) Outcrossing these flies individually and (F) inbreeding each set of offspring gives a population of flies for each new mutation. (G) Researchers then test homozygous flies (darker pink and green) for the phenotype of interest. In some cases, adult homozygotes are not viable (as in the yellow population) and so researchers interested in earlier developmental steps may examine embryos and larvae within these populations to find dying homozygotes.
An interesting recent application of genetic screening is the elucidation of the fly as a model for alcoholism and related behaviors (Singh and Heberlein 2000). The Heberlein lab invented an inebriometer—a tube containing ethanol vapor and multiple platforms from which flies can fall from level to level—to screen mutagenized flies and identify genes associated with alcohol sensitivity (Singh and Heberlein 2000). The barfly mutant showed decreased alcohol sensitivity, and the tipsy mutant showed increased sensitivity compared to wild type. These are further examples of the creative naming of D. melanogaster gene mutations based on the phenotypes they express. Another mutant identified by their lab, cheapdate, turned out to be an allele of the amnesiac gene, which encodes a PACAP-like neuropeptide and thus connects the cAMP pathway with ethanol sensitivity (Moore et al. 1998). The implications of this work for understanding effects of alcohol on the nervous system go beyond just the biology of the fly. Although the structures and networks associated with the fly and human brains are different, the molecular mechanisms uncovered in the fly associated with the neural response to alcohol and addictive behaviors are, at a basic level, shared with humans (Robinson and Atkinson 2013).
Enhancer and suppressor screens:
A forward genetic screen to find additional genes of interest can be performed starting from an already-mutant genetic background—the goal is to find new mutations that make worse (“enhance”) the preexisting mutant phenotype or that alleviate (“suppress”) that original phenotype. Often the new genes found in an enhancer or suppressor screen are directly involved in the biological phenomenon of interest but would not have had a significant enough effect to be found in the original forward genetic screen. In other cases, enhancers and suppressors that function through reduced dosage are genes that would cause lethality alone as homozygotes and thus perhaps would not be found in an original screen.
Mapping mutations identified in mutagenesis screens:
The next step is mapping a mutation to a chromosomal region to enable identification of candidate genes. Researchers commonly map genes using parallel complementation tests against a collection of known chromosomal deficiencies that collectively span the chromosome in question. A “deficiency kit” for a given chromosome can be obtained from the Bloomington Stock Center (Cook et al. 2010). An older approach is meiotic or recombination mapping with visible markers (Greenspan 1997). Many multiply marked chromosomes with recessive mutations are available at the Bloomington Drosophila Stock Center for this purpose, and a more recent approach using dominant markers is available (Sapiro et al. 2013). As sequencing technologies increase in speed and decrease in price, brute force sequencing will become a more common way to identify new mutations in inbred fly strains as well as in individual flies within a population.
I have identified a candidate gene possibly altered in an inbred line from a mutant screen—How can I make sure that that gene is truly associated with the effect I am observing? How can I test the effects of different versions of that gene?
An important next step in characterizing a new mutation found in a screen is testing whether a candidate gene you identified is truly associated with the phenotype you are observing, instead of being a random secondary occurrence. In the fly community, this is called doing a “rescue” experiment. The question you are asking is whether adding back a wild-type copy of the gene will reverse, or rescue, the mutant phenotype. (Once you have confirmed that the gene you found is the correct one, you can also add back different, manipulated versions of the gene to do structure–function studies.) The key innovations that allow us to answer these questions are P-element-mediated transformation as well as the Gal4/UAS bipartite expression system.
Transformation:
Much of D. melanogaster genetics was literally “transformed” with the identification and development of the P-element as a germ-line transformation vector. The P-element is a classic transposable element (Bingham et al. 1982; Rubin et al. 1982) that originally contained a gene encoding the transposase enzyme, which together with the inverted repeats found on the terminus of the DNA element permitted movement within the genome. Rubin and Spradling (1982) hypothesized that replacing the internal transposase gene with a gene of interest would produce an ideal system for inserting DNA into the fly genome. This P-element construct could then be co-injected into early (still syncytial) embryos along with an independent source of the transposase enzyme to insert the transposable element into the developing germline. The proof-of-principle experiments demonstrated heritable and stable transformation of the D. melanogaster germline using the wild-type rosy gene as a marker (Rubin and Spradling 1982). Later work developed the now classic wild-type white (w+) eye color marker gene that is used to identify transformants in a white mutant background (Klemenz et al. 1987). This discovery and the development of a variety of other elements was the genesis of much of the genetic technology that makes the fruit fly such a powerful model system. The entire spectrum of their uses is beyond the scope of this article (Venken and Bellen 2005, 2007) but their use as a mutagenesis tool and the design of the binary Gal4/UAS expression system are two of the most important stages in the development of the fly as a modern genetic system.
GAL4/UAS expression system:
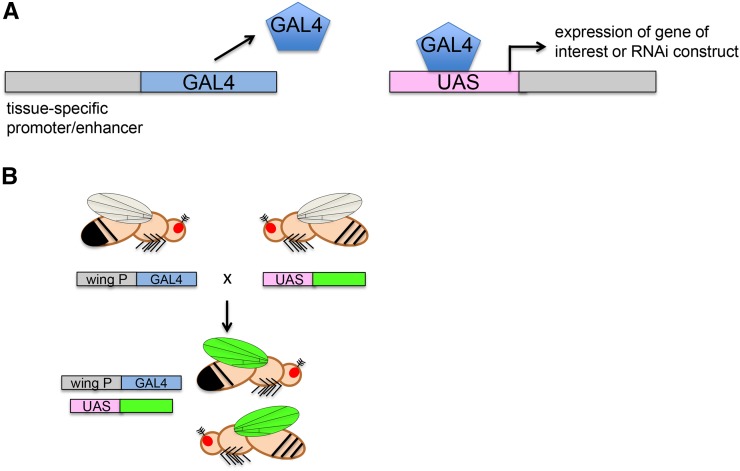
Brand and Perrimon (1993) took the P-element transformation vector and used it to create a gene expression system that would eventually allow for the expression of any gene of interest in any particular tissue within the fly. They cloned the yeast transcription factor GAL4 into a P-element vector and showed that one could place a defined promoter upstream of GAL4 or integrate the GAL4 element into the genome to use endogenous transcriptional enhancers to express GAL4 (an enhancer trap). To accompany this “driver” of gene expression, they created a corresponding P-element vector, pUAST, containing the upstream activating sequences (UAS) to which GAL4 protein can bind. These UAS sequences were connected to a general promoter and a cloning site to allow for the insertion of any gene of interest. This bipartite expression system could be used to drive the expression of a gene in a defined way (Figure 5), allowing researchers to perform rescue experiments and other manipulations (see below).
Figure 5.
GAL4/UAS system for modular expression of transgenes in specific tissues. To express a transgene or RNAi construct in a particular tissue, one needs flies carrying (A) a “driver” with a tissue-specific promoter/enhancer placed 5′ of the gene encoding the yeast GAL4 transcription factor (left) and (right) the gene of interest placed 3′ of the upstream activating sequence (UAS), which is activated by GAL4. (B) Transgenic flies carrying either of the two constructs alone (top) do not express the gene of interest, but when crossed into the same fly, the tissue-specific promoter (a wing promoter in this example) drives expression of GAL4, which turns on the gene of interest (here indicated by green) in the specified tissue. The system can also be used to express a hairpin RNA to knock down a gene in the target tissue.
Gene expression libraries that enabled the GAL4-UAS system:
The advent of transposable-element-based genetics in D. melanogaster spurred the development of reagents to identify promoter sequences driving cell-type-specific gene expression throughout development. Early screens aimed to find genes important for particular processes based on their expression patterns. Random genome-wide insertion of P-elements containing a cellular marker such as Beta-Galactosidase (LacZ) or GFP would be expressed if inserted just after an endogenous promoter (Bellen et al. 1989; Grossniklaus et al. 1989; Wilson et al. 1989). Later modifications inserted a transposable element with the GAL4 gene to create cell-type-specific GAL4 expression lines, which then allowed the manipulation of specific cell types using the GAL4/UAS system. These “enhancer-trap” lines were the first generation of expression tools that would eventually lead to the unprecedented specificity in the expression of particular transgenes. A second approach is to clone small enhancer regions from genes of interest to create GAL4 expression lines that replicate the expression pattern of that gene. Systematic approaches to the generation of enhancer specific expression of GAL4 has led to the creation of the Janelia Farm GAL4 collection that uses small defined regions of noncoding or intronic DNA to create expression patterns that are more restricted than previous generations—in some cases only one or two cells (Pfeiffer et al. 2008).
The GAL4-UAS system has been modified in various ways to improve utility (reviewed in Duffy 2002). One example of a challenge addressed is lack of control over final resting place of a transposable element as a transformation vector within the genome. An insertional event may disrupt an endogenous gene’s function, or the local transcriptional/chromatin environment may reduce expression of either the GAL4 transcription factor or the genes associated with the UAS elements. Unpredictable expression levels can add uncertainty regarding whether a rescue experiment, or other manipulations, are working as designed. To remedy this unpredictability, a new transformation system was developed using the serine recombinase protein from the PhiC31 bacteriophage, which recombines sequences using the attB and attP site-specific DNA sequences (Groth et al. 2004). A series of attP landing sites have been created at distinct points through out the genome providing molecularly defined positions for the high-efficiency creation of transgenic flies using transformation vectors with an incorporated attB sequence (Venken et al. 2006). These sites have also been characterized for the levels of expression of an inserted transgene, thus allowing for more finely tuned expression of a transgene as well as the comparison of different transgenes at the same chromosomal location (Ni et al. 2008).
I have identified many different genes using a mutant screen, but I suspect some specific additional genes may be involved in my biological process of interest, too. How do I test whether they are involved in this process?
We have already described the forward genetics approach of the mutant screen, in which you go from biological phenomenon to gene(s). In contrast, a reverse genetics approach is one in which you start with a gene and try to determine its function via direct genetic manipulation and assessment of phenotypes. A gene’s sequence or expression pattern may flag it as possibly interesting to people studying a particular process. How do researchers alter a gene if no chemically induced mutations exist or no transposable elements are inserted? Candidate gene experiments have led to new approaches to altering the genome of the fly. Initially researchers turned to homologous recombination processes similar to familiar mouse knock-out technology (Rong and Golic 2000). While this system achieved the desired outcomes, it was inefficient and not ideal for large-scale implementation throughout the genome. The two most promising approaches have been genome editing technology and RNA interference.
Genome editing:
The recent excitement about purposefully altering genomes has centered on genome-editing techniques that take advantage of particular types of nuclease enzymes. Genome-editing technologies have enabled significant progress toward the goal of a complete collection of mutant strains for every gene. Several different technologies have been successfully used in D. melanogaster, including zinc finger directed nucleases, and transcription activator like effector nucleases (TALENs) (Beumer and Carroll 2014). The most promising editing technology is the modified bacterial clustered regularly interspaced short palindromic repeat (CRISPR/Cas9) system. In its original form, the targeted endonuclease is used by bacteria as a defense against viruses and plasmids by combining an endonuclease and a targeting guide RNA. Modification of this guide RNA allows the targeting of particular genes to induce the endonuclease to produce a double strand break. This double strand break efficiently results in the creation of short insertions/deletions and large deletions in a gene through nonhomologous end joining repair (Gratz et al. 2013; Bassett and Liu 2014). Development of this technology in the fly has created an efficient mechanism by which the community can generate complete loss-of-function (or “null”) mutations in all of the ∼14,000 genes in the genome (Gratz et al. 2013; Bassett and Liu 2014). This technology also has the potential to “knock-in”/create specific mutations within a coding sequence to model the effects of particular genetic mutations, to probe the function of a particular protein domain, characterize splice sites, or fuse specific reporters (e.g., GFP, YFP, etc.) at precise locations within a gene.
RNA interference:
A second approach to creating genome-wide libraries to examine gene function has been the development of RNA interference in both transgenic animals and cell lines. Transgenic approaches use the GAL4/UAS system described earlier to express short inverted repeat RNA hairpins that target specific genes to reduce or “knock-down” the expression of the gene’s messenger RNA (mRNA) (Kennerdell and Carthew 2000). These are not permanent alterations of a gene’s coding sequence, but rather eliminate a gene’s function by reducing mRNA levels from that gene. The original versions of these systems used random insertions of transgenes carrying the UAS–RNA interference (RNAi) constructs specifically designed to target a particular candidate gene. These were inefficient in their ability to knock-down RNA expression of a particular gene to null levels and could produce lethality or sterility depending on the insertion site (Dietzl et al. 2007; Ni et al. 2008). In addition, work in cell culture suggested that the inverted repeat could produce off-target effects where multiple genes’ expression was altered by the system (Perrimon and Mathey-Prevot 2007). Some of these issues have been addressed with new libraries that use the phiC31a/AttP site-specific integration system (described above) to insert the UAS–RNAi transgene in a well-characterized location on the chromosome that ensures high levels of expression and an insertion in a position that does not affect other genes (Ni et al. 2008). These libraries at the Drosophila RNAi Screening Center and Vienna Drosophila Research Center can be used to screen almost 90% of the protein coding genes for phenotypes of interest. With the use of well-characterized GAL4 driver lines that are activated at specific stages or in specific cell types, researchers can elucidate the function of particular genes in cellular and developmental processes from embryo to adult by expressing the RNAi hairpin constructs (and thus knocking down the gene) at those developmental stages or in those cell types (Boutros et al. 2004; Armknecht et al. 2005; Mathey-Prevot and Perrimon 2006; Dietzl et al. 2007).
If a mutation produces lethality at some point in development, how do I examine a gene’s role in other stages of the life cycle? How do I determine in which cells the gene is exerting its effect?
Assessing the effects of a mutation on a particular developmental/cellular/behavioral process can be difficult for genes that play multiple roles at different stages of the life cycle. If a mutation prevents the completion of embryonic development (lethality) then analysis of the gene’s role in adult tissues can be preempted. In response to this challenge, geneticists have created tools that allow more fine-tuned analysis of gene function at any stage of development. For example, the previously discussed RNAi system allows researchers to knock down gene function at any stage of development or in specific cell types using the GAL4/UAS system; stage- or cell-specific GAL4 drivers can knock down a candidate gene or can drive a whole library of RNAi lines to do a genome-wide screen. If instead of RNAi knock-down, a fly biologist would like to analyze how a lethal mutation or other allele of a gene affects a tissue (or determine in which cell type a mutation has to be to exert an effect), they can use clonal analysis.
Clonal analysis:
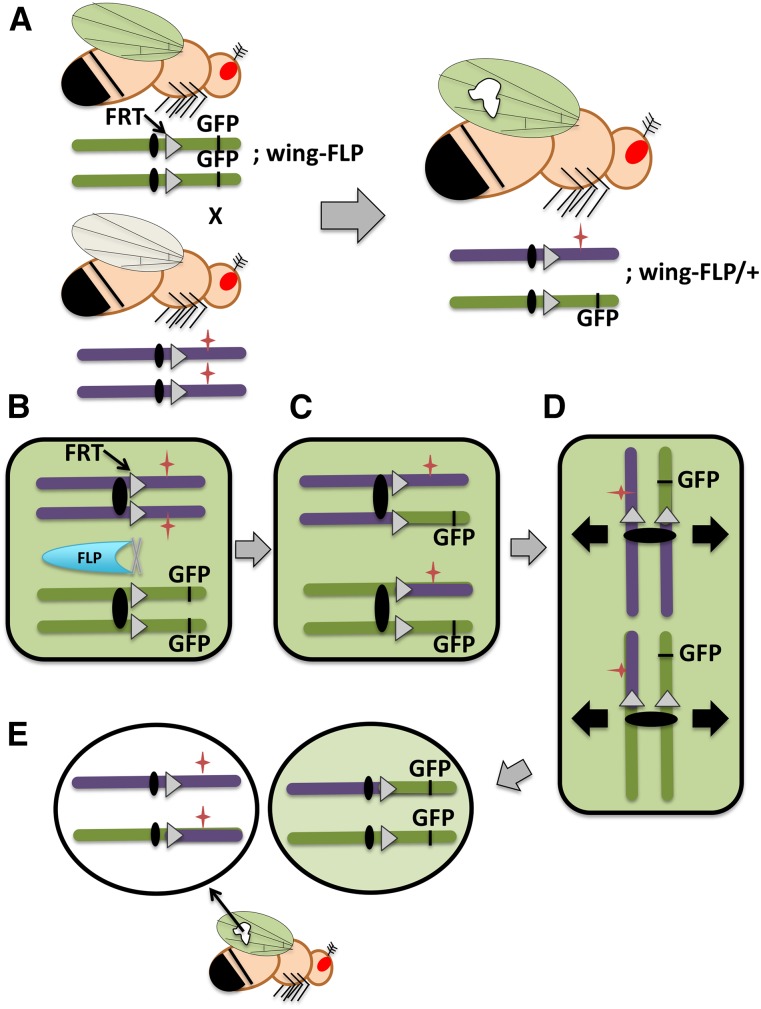
To characterize later roles of a gene essential for early development, one can produce mosaics that have homozygous mutant patches of cells (clones) in an otherwise heterozygous animal via mitotic recombination (Figure 6). While triggering mitotic recombination can be triggered by radiation, a more efficient approach hinges on genetic manipulation of flies to incorporate on their chromosomes the site-specific recombination components associated with the Saccharomyces cerevisiae 2-μm plasmid, the FLP recombinase and its site-specific recombination sites (FRTs) (Golic and Lindquist 1989). The original versions of this system placed FRT recombination sites on all of the arms of the D. melanogaster chromosomes to enable mitotic recombination between homologous chromosomes in the presence of the FLP recombinase. A researcher would recombine a mutation of interest onto a FRT chromosome, distal of the FRT site. In flies heterozygous for that mutation but with FRTs on both homologous chromosomes, FLP-triggered mitotic recombination would lead to patches of homozygous mutant cells, typically identified by a linked recessive marker or loss of a linked fluorescent gene product. The first FLP transgenic flies had the gene under the control of the hsp70 promoter to permit the induction of the FLP and thus recombination at specific time points during development with an increase of temperature (Golic and Lindquist 1989). In D. melanogaster this technique was originally developed to examine maternal contributions to development through the creation of germline clones in females, and then it was adapted for somatic clones to observe the behavior of mutant cells in a variety of tissues throughout development and in the adult (St Johnston 2002).
Figure 6.
Clonal analysis in somatic tissue. (A) Schematic diagram of a genetic cross used to create homozygous somatic clones of a mutation of interest within a heterozygous background. In this example, the clone is a patch of mutant cells within a wild-type wing. One parent is homozygous for a chromosome that carries the FLP recombinase recognition site, FRT (triangles) and a distal GFP marker that is being expressed under the control of a wing promoter. All of the cells of this fly’s wings will express GFP and appear green under a fluorescent microscope. This fly is also homozygous on another chromosome (not drawn) for the FLP recombinase gene, which is being expressed under the control of a wing promoter. The other parental fly is homozygous for a mutation of interest (red star) on the same FRT-carrying chromosome. Progeny from this cross will be transheterozygous for the GFP-marked chromosome and the mutant chromosome, and they will be heterozygous for FLP. During development, FLP-mediated mitotic recombination in the developing wing will produce patches of unmarked homozygous mutant cells (white patch). Panels B–E show the mechanics of clone production through mitotic recombination in the progeny. (B) Cells in mitotic G2 have replicated chromosomes with sister chromatids. In some wing cells, the FLP recombinase triggers recombination between FRT sites on nonsister chromatids, and (C) one copy each of the GFP marker and the mutation of interest will switch between homologous chromosomes. (D) One of two possible chromatid alignments at mitotic metaphase for the cell pictured in C. (The other alignment, not shown, leads to two heterozygous daughter cells.) The black arrows show the direction of sister chromatid separation during the completion of mitotic division. (E) Daughter cells produced upon completion of cell division for the cell pictured in D. One daughter cell is homozygous for the mutation of interest and will subsequently divide to give rise to a homozygous patch of cells, which can be identified for phenotypic analysis based on loss of the GFP marker. The other daughter cell is homozygous for the GFP marker and will blend into the surrounding heterozygous cells that also fluoresce.
Early in development of the embryo, the maternal contribution of RNA and protein from a heterozygous mother to a homozygous mutant embryo may mask an early developmental role for a gene. Germline clones in which homozygous mutant germ cells are created within a heterozygous female permits the characterization of earlier functional roles for such a gene. This technique takes advantage of the dominant ovoD mutation, which in females leads to atrophic ovaries that produce no eggs (Perrimon 1984). In females transheterozygous for ovoD and the mutation of interest on FRT-containing chromosomes, heat shock induces FLP-mediated mitotic recombination, allowing production of homozygous mutant clones free of the ovoD allele—these are the only cells that can continue through oogenesis to produce eggs. The phenotype of these cells, if any, can reveal a role for the gene of interest in germline and early development.
A variation of this process can produce somatic clones in any cell type at any stage of development. Rather than ovoD, the nonmutant chromosome contains a marker gene such as eye color (white+), body color (yellow+), or a fluorescent cell maker (GFP) to identify the nonlabeled clonal mutant cells in the labeled wild-type background (Perrimon 1998). As the FLP-FRT system has developed, a variety of promoters have been fused to the FLP recombinase gene to allow for more precise control of mitotic recombination during developmental time or in particular tissues. In addition, a more sophisticated system has been built, mosaic analysis with a repressible cell marker (MARCM), to allow for the analysis of individually marked mutant cells in an unlabeled heterozygous background (Lee and Luo 2001). This innovation has created a significant step forward in resolution, allowing for the examination and genetic manipulation of single mutant neurons within the adult brain to determine which neurons and which genes affect certain behaviors.
Drosophila cell lines:
While Drosophila is touted primarily as a fast genetic system for studying biological processes in a live animal, culturing of fruit fly cells can also be extremely advantageous for certain applications such as testing effects of gene knock-down at the subcellular level. Nearly 100 different D. melanogaster cell lines exist, with the S2 and Kc lines being the most commonly used (Echalier and Ohanessian 1969; Schneider 1972). These lines, originally derived by mechanical dissociation of embryos, became immortalized spontaneously and are likely of hematopoietic lineage. Other protocols established cell lines from protease-digested imaginal discs (Ui et al. 1987); however, a Drosophila cell line of true epithelial origin has yet to be derived. In addition, no method of forced immortalization exists (like retroviral infection in mammalian cells), and so establishing a new Drosophila cell line is dependent on spontaneous immortalization and can be somewhat variable (Baum and Cherbas 2008). As in cultured cells from other organisms, chromosome loss, duplication, and rearrangement can lead to altered karyotypes in some lines. D. melanogaster cell lines are commonly used to express and purify recombinant proteins when problems in protein activity or solubility are found in prokaryotic systems (Davis et al. 1993; Ikonomou et al. 2003). Transfection frequency in S2 cells is fairly high, making transgene analysis relatively straightforward (e.g., for immunoprecipitation or subcellular localization or proteins). Cell culture enables scaled-up biochemical approaches that can be difficult to perform on isolated tissues. Perhaps the most powerful approach using D. melanogaster cells is RNAi (Hannon 2002). While RNAi in mammalian cell culture typically involves transfection of a pool of short double-stranded RNAs (dsRNAs) matching the target gene, D. melanogaster S2 cells readily absorb dsRNA from the culture media, increasing the percentage of cells with knock-down (Echeverri and Perrimon 2006). Also, this response is not length dependent in flies, as dsRNA of 500–1000 bp can be added to cultured cells, with Dicer then chopping this into shorter pieces of dsRNA. Including such a large dsRNA increases the likelihood of effective knock-down. These properties make D. melanogaster S2 cells particularly amenable to high-throughput RNAi approaches (Armknecht et al. 2005; Perrimon and Mathey-Prevot 2007), and several RNAi libraries have been developed for this purpose.
Using the Power of Drosophila Genetics to Study Developmental and Cell Biology
Genetic approaches in D. melanogaster have proven highly successful in elucidating the mechanisms that regulate a wide variety of biological processes ranging from developmental pathways to cytoskeletal regulation to organelle trafficking. In this section, we describe key developmental steps and cell biological processes whose molecular mechanisms have been explored largely through genetic analysis. A resource for more information on these processes is Campos-Ortega and Hartenstein (1997).
Fertilization
Embryogenesis begins with fertilization of the oocyte. Upon mating, female flies store sperm for up to 2 weeks in specialized organs called seminal receptacles and spermathecae (Lefevre and Jonsson 1962). Sperm storage is thought to allow for the coordination of ovulation with sperm release and to reduce ecological costs associated with multiple matings (Wolfner 2003). After mating, females tend not to mate again for several days; however, sperm from more than one male can be stored at a given time. Sperm competition and sperm preference have thus been observed based on the genetics of both the male and female. Fertilization itself does not occur until the egg is ready to be laid. Mature eggs leave the ovaries and travel through the oviduct, during which time some of the stored sperm are released. As the egg passes through the oviduct, one or a few sperm enter the egg through a small, anterior opening in the chorion called the micropyle. Interestingly, fertilization in flies does not involve membrane fusion, but rather the sperm completely enters into the egg, with subsequent sperm plasma membrane breakdown occurring in the cytosol of the egg. Also, fertilization can only occur in what will become the anterior pole of the developing embryo.
Superficial cleavage and cellularization
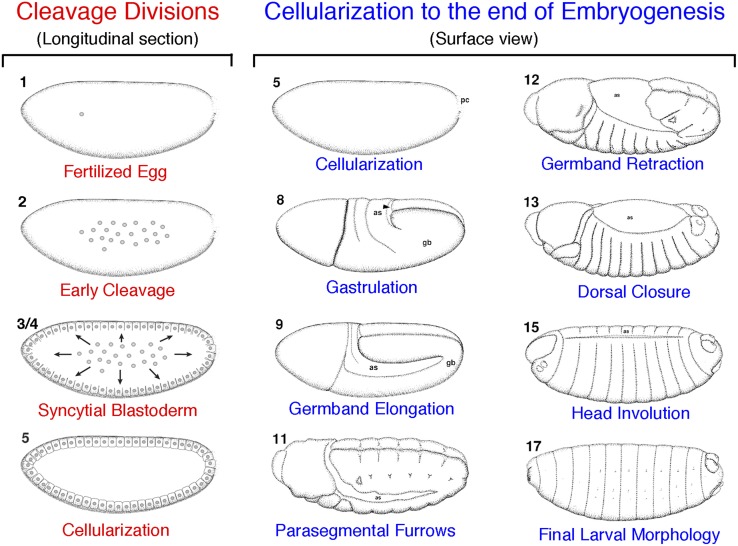
As in most insects, D. melanogaster development begins with nuclear divisions in a common cytoplasm with no new cellular membranes (thus it is a syncytium). The first nuclear divisions occur in the center of the egg and are coordinated such that all nuclei divide simultaneously in a cycle. After 10 division cycles, most of the nuclei migrate to the periphery where they become partially encapsulated by cytoskeletal proteins that create furrow canals. Bulk zygotic transcription initiates shortly thereafter and occurs prior to true cellularization. Cellularization marks the beginning of asynchronous cell divisions and occurs through invagination of the oocyte’s plasma membrane along the furrow canals. The cellular blastoderm, formed 3 hr postfertilization, now consists of a layer of ∼6000 cells lining the periphery of the zygote with yolk on the interior (Figure 7).
Figure 7.
Stages of embryonic development. D. melanogaster development begins in a syncytium characterized by nuclear divisions without cytokinesis (stage 2). After 10 synchronized rounds of division, nuclei migrate to the periphery where they become partially encapsulated by actin-based furrow canals (stage 3/4). True cellularization occurs in stage 5, followed by gastrulation (stage 8), which determines the three germ layers. Dramatic morphogenetic movements then reshape the body plan as cells from the posterior migrate toward the anterior in germband extension (stage 9) followed by later retraction to the posterior (germband retraction; stage 12). Epithelial cells then migrate toward the dorsal midline in dorsal closure (stage 13), and head structures begin to mature (head involution; stage 15). Finally the larva reaches its mature state (stage 17) and hatches from the eggshell. Images adapted from the Atlas of Drosophila Development (Hartenstein 1993) and used with permission. In each panel, anterior is to the right and dorsal is up.
Gastrulation/morphogenesis
Next, gastrulation serves to specify the three germ layers of the animal (mesoderm, endoderm, and ectoderm), and is characterized by cell fate decisions and shape changes that drive cells to move in sheets to different regions of the embryo. The mesoderm is specified along the ventral surface as cells fold inward along the ventral midline and pinch off to form a hollow tube immediately beneath the ventral ectoderm (Leptin 1995). A video illustrating these movements is at https://www.youtube.com/watch?v=ymRYxFYLsZ4. Similar cell-shape changes drive anterior and posterior midgut invaginations of cells fated to become the endoderm.
Following specification of the germ layers, dramatic movements then reshape the body plan. Convergent extension (a process whereby cells narrow along one axis and extend along the perpendicular axis) causes cells of the prospective trunk region (germband) to first extend toward the anterior of the animal and later retract to the posterior. Videos showing these movements are at https://www.youtube.com/watch?v=FChS4KU5jDM and https://www.youtube.com/watch?v=MefTPoeVQ3w. Several important processes begin while the germband is in the extended position including organogenesis, segmentation, and neurogenesis. As the germband retracts, epithelial tissue from the cellular blastoderm (amnioserosal cells) spreads to cover the dorsal surface. The lateral ectoderm from both sides of the embryo then migrates over the top of these amnioserosal cells in a process termed “dorsal closure.” Amnioserosal cells then degenerate and cuticle deposition begins. Now embryogenesis is nearly complete; the trachea fills with air, and the first muscle twitches can be observed before the first instar larva hatches from the eggshell (Figure 7). A useful resource illustrating these events and the development of particular organ systems is Volker Hartenstein’s Atlas of Drosophila Development (Hartenstein 1993), available online at http://www.sdbonline.org/sites/fly/atlas/00atlas.htm.
Anterior/posterior patterning
While the sperm entry site determines anterior/posterior polarity in some organisms such as C. elegans, this polarity is already specified in fly eggs before fertilization. In D. melanogaster, maternally contributed mRNAs are differentially localized within the embryo through anchoring to the cytoskeleton (van Eeden and St Johnston 1999; Lasko 2012). Translation of these mRNAs after fertilization results in protein gradients, with the highest concentration of protein being adjacent to its mRNA pool. For example, bicoid mRNA is localized to the anterior, and analysis of bicoid mutants revealed that it is critical for head and thorax formation (Berleth et al. 1988; Driever and Nusslein-Volhard 1988). In contrast, nanos localizes to the posterior and regulates abdominal segment formation (Gavis and Lehmann 1992). These protein gradients are then used to specify expression of a series of zygotic genes involved in segmentation and cell fate determination. There are three groups of segmentation genes (gap genes, pair-rule genes, and segment polarity genes), each of which is sequentially expressed and serves to further specify positioning within the embryo (St Johnston and Nusslein-Volhard 1992). Finally, after establishing identity within each segment, the homeotic selector genes (also called HOX genes) control the specification of tissues or organs in particular segments (Lewis 1978). The HOX genes in flies are all related transcription factors found in two complexes: the Antennapedia complex, whose five genes control the identity of the segments anterior to the midthorax and the Bithorax complex, whose three genes control the identity of the segments posterior to the midthorax. When HOX genes are mutant, a “homeotic” transformation occurs in which segmental identities are misspecified, thus resulting in replacement of one structure with another that is normal in form, but inappropriately positioned. For example, legs may develop in place of antennae in an Antennapedia mutant, or a second pair of wings may replace small dorsal appendages called halteres because of Ultrabithorax abnormalities. This work in Drosophila set the stage for our current understanding that HOX genes help determine segmental patterns in a way broadly conserved across widely diverged species; many human developmental disorders stem from aberrations in these genes.
Dorsal/ventral patterning
Like anterior/posterior patterning, the dorsal/ventral axis in D. melanogaster is established through protein gradients, but using a very different mechanism (Morisato and Anderson 1995). Genetic studies revealed that the ventral embryonic region is specified by the transcription factor Dorsal and its cytoplasmic anchor Cactus (thus embryos mutant for dorsal show only dorsalized structures). Dorsal mRNA is maternally provided to the oocyte, yet translation of the transcript does not occur until cellularization and occurs uniformly throughout the embryo. Complex signaling interactions then occur between the oocyte and follicle cells of the egg chamber to regulate nuclear localization of Dorsal protein only in cells fated to become the ventral surface (Neuman-Silberberg and Schupbach 1993). These signaling events involve a cascade of proteases that ultimately activate Pelle kinase, which phosphorylates Cactus, and stimulates Cactus destruction. Without its cytoplasmic anchor, Dorsal is now free to move into the nucleus and stimulate expression of genes specifying ventral fate. More information about embryonic patterning and axis determination in D. melanogaster can be found at http://www.ibiology.org/ibioseminars/development-stem-cells/eric-wieschaus-part-1.html and http://www.ibiology.org/ibioseminars/development-stem-cells/trudi-schupbach-part-1.html.
Imaginal discs, with eye development as an example:
Following embryogenesis, flies undergo three larval stages prior to pupation. These larvae feed, thereby building energy reserves, and they make (and specify) new cells in preparation for metamorphosis. As in most insects, adult structures in fruit flies arise from imaginal cells that are fated during embryogenesis. These imaginal cells form “disc”-shaped tissues, and each disc will ultimately become a unique structure in the adult animal, such as adult mouthparts, eyes/antennae, legs, wings, and genitalia. Each disc originates from a small cluster of cells in the embryo that invaginate to form a flattened sac of epithelium. Continued cell divisions expand the discs during embryonic and larval stages, until they ultimately develop during metamorphosis into adult structures. Imaginal discs can serve as useful systems to elucidate molecular events, and here we discuss eye development as an example of the process.
The adult D. melanogaster compound eye serves as a useful model system to elucidate a number of biological processes not only related to visual systems, but also general signaling pathways like the Ras pathway. The fruit fly eye is a highly ordered array of nearly 800 ommatidia, each functioning as an individual visual receptor. The ommatidium is composed of eight photoreceptor cells, with photoreceptors 1–6 positioned radially and photoreceptors 7 and 8 in the middle (with R7 positioned above R8). The photoreceptor cells are surrounded by two primary pigment cells, themselves surrounded by secondary pigment cells shared by adjacent ommatidia. Cone cells overlay the photoreceptors, and a bundle of eight neurons runs to the preoptic stalk.
Cells of the prospective eye are first fated in the embryo through expression of regulatory transcription factor families like eyeless. Initially ∼20 cells are fated to become eye cells; however, the disc grows through cell division to 10,000 cells by the third larval stage. Cell proliferation then ceases and differentiation begins, with BMP, Hedgehog, and EGFR signaling controlling differentiation of the photoreceptor cells and formation of the ommatidia. In the pupal stage, the disc tissue undergoes dramatic morphological changes to acquire its final shape, and it develops its light-sensing capabilities. Differentiation events sweep across the eye tissue in a seeming wave, at the crest of which is the so-called “morphogenetic furrow.” Photoreceptor cells acquire their rhabdomeres (organelles specialized for phototransduction), and differential expression of rhodopsins allows for some photoreceptors to be involved in motion detection and image construction, while others detect color differences.
The compound eye has served as a useful model system to elucidate a number of biological processes not only related to visual systems, but also general signaling pathways like the Ras pathway. In 1976, mutant flies specifically lacking photoreceptor 7 were reported (Harris et al. 1976), and cloning of the gene (named sevenless) revealed that it was a receptor tyrosine kinase (RTK) with homology to the EGF receptor. A subsequent enhancer screen using temperature-sensitive alleles of sevenless identified other components that cooperate with sevenless in this process, including bride of sevenless (boss), son of sevenless (sos), and surprisingly the oncogene Ras. Bride of Sevenless turned out to be the ligand for the pathway, and Son of Sevenless a Ras guanine exchange factor (GEF); thus these genetic studies in the fly eye helped elucidate the major players in Ras signaling.
Gametogenesis:
The development of egg cells in females (oogenesis) and sperm cells in males (spermatogenesis) involves not only the reductional chromosomal divisions of meiosis but also dramatic cellular reorganization and shape changes. In both the ovary and testis, germline stem cells at one end of the organ replenish their own populations while generating cells that undergo four mitotic divisions to make a cluster (“cyst”) of 16 still-diploid cells, each encompassed in a layer of somatic cells. In the ovary, one of the 16 diploid germ cells becomes the oocyte, eventually undergoing meiosis. The remaining 15 cells, nurse cells, are directly connected to the oocyte through actin-based tubes called ring canals (King 1970; Bastock and St Johnston 2008). These nurse cells are essentially RNA and protein factories, and late in oogenesis their actin cytoskeletons contract to squeeze their cytosolic contents into the oocyte. This “dumping” will provide the fertilized oocyte with the mRNA and proteins necessary to begin embryogenesis. (Consequently, a homozygous mutant zygote for a gene required early can develop up to a point as long as wild-type RNA and protein from the heterozygous mother lasts; on the other hand, a homozygous mutant mother’s embryos will be defective regardless of their own genotypes—this is called a maternal effect mutation.) The nurse cells then undergo apoptosis and are eliminated, while the follicle cells secrete both a vitelline membrane and outer chorion shell to protect the maturing egg. In the testis, all 16 diploid germline cells in a cyst undergo meiosis to form a cyst of 64 haploid round spermatids. These cells undergo dramatic reshaping of the cytoskeleton, mitochondria, membranes (Fabian and Brill 2012) to give rise to mature sperm with a condensed nucleus and a long tail containing a microtubule-based axoneme.
Gametogenesis in flies has served as a useful system to study stem cell specification and maintenance (Matunis et al. 2012), cell shape changes, and other events. Spermatocytes are a particularly advantageous system in which to study cytokinesis since the cells are large and easy to image and have a weak spindle assembly checkpoint (Cabernard 2012). Aspects of cytokinesis can therefore be studied in mutant cells with a spindle defect that would otherwise activate a checkpoint that precludes analysis of effects on cytokinesis. Genetic studies in Drosophila spermatocytes have revealed important mechanisms regulating cleavage furrow formation, aspects of contractile ring formation and function, as well as the role of membrane trafficking from internal compartment in cytokinesis (Giansanti and Fuller 2012).
Drosophila as a Genetic Model for Studying Neurobiology and Behavior
Genetic analysis of the fruit fly has helped the neuroscience community understand all levels of neural function from the development of a nervous system, neuronal function and plasticity at the molecular level, adult neural networks, and the neurobiology of complex behaviors (Bellen et al. 2010). Maybe most surprising to non-Drosophilists is the variety of complex behaviors exhibited by the fly that can be dissected to identify their underlying genetic and biological components. This approach to behavior, defined as neurogenetics, was pioneered in the laboratory of Seymour Benzer in the 1960s and has led to a bounty of discoveries that touch on the control of biological rhythms, sensory biology, learning and memory, sleep, aggressiveness, sexual behavior, and more (Vosshall 2007). Simple but powerful genetic screens underlie many of these findings. The field’s initial forays into biological circadian rhythms was through the identification of a series of period mutants that had altered daily cycles—too long, too short, or none at all (Konopka and Benzer 1971). The subsequent molecular cloning of the D. melanogaster period gene in 1984 led to identification of the transcriptional feedback loop of period/timeless and a continually growing set of interconnected pathways that play a role in behavioral rhythms (Bargiello et al. 1984; Zehring et al. 1984). The cloning of the period locus also represents the first time the gene responsible for a particular phenotype was identified using the newly developed P-element transformation technology (Bargiello et al. 1984; Zehring et al. 1984). Jonathan Weiner’s book Time, Love, Memory covers Benzer’s important contributions in detail and in a compelling way (Weiner 1999).
In fact, many of our genetic footholds in the field of neurobiology and behavior are the result of a simple behavioral assay paired with a genetic screen. The findings within the fly system have relevance to understanding of all of these processes through to higher vertebrate systems. Evolutionary conservation of gene functions have shown that much of what we have learned about behaviors such as circadian rhythms and memory is applicable to the more complex versions of these processes in mice and humans (Bellen et al. 2010). Furthermore, the conservation of these genetic processes has also provided significant insight into the underlying mechanisms of the human diseases and disorders that result from the alteration of typical neuronal function (Fortini and Bonini 2000; Muqit and Feany 2002). This fundamental connection between fly research and human biology is highlighted in the many examples of similar gene mutations giving rise to similar functional phenotypes. For example, shortened rhythm mutations in a per homolog in humans have been identified in individuals with a sleep syndrome (Toh et al. 2001).
The fly system has developed in parallel with significant technological advances in our ability to probe and manipulate neuronal systems, making it one of the best systems to move from gene discovery to the study of the development, function, and plasticity of neural networks. While the connectome of all 302 C. elegans adult neurons was determined by TEM many years ago (White et al. 1986), the efforts to map connectomes of more complicated adult nervous systems is being aided by the genetic and cell biological reagents available in the fly. Thus, the more complex nervous system of the brain has become more tractable with each passing decade. The Drosophila brain has ∼100,000 neurons, and the recent publication of single-cell resolution maps of the projections of 16,000 of them has pushed this field in new directions (Chiang et al. 2011). The map, paired with the ability to manipulate, ablate, and activate/repress individual neurons within a network, will allow researchers to ask increasingly sophisticated questions about the neural control of behavior and the higher cognitive calculations being made within networks (Kazama 2015). One important new technology is optogenetics, which allows the stimulation or repression of individual neurons using light of a specified wavelength (Fenno et al. 2011). Some early optogenetic work was done in the fly to address a variety of behavioral circuits including odorant learning and responses (Bellmann et al. 2010), proboscis extension (Gordon and Scott 2009), nocioception (Hwang et al. 2007), and the escape response (Zimmermann et al. 2009). The small size of the Drosophila brain has also been exploited to do single-cell recordings within defined networks either through technically challenging electrophysiological recordings or with calcium imaging on intact animals who are often performing a particular behavior task (Kazama 2015; Owald et al. 2015). Work over the past decade on the olfactory system in the fly has mapped neural connections associated with response to individual odorants. Electrophysiology and calcium imaging of animals in behavioral assays has indicated how the fly computes the response to directionally applied odors (Gaudry et al. 2013). The single-cell resolution in defined networks of neurons makes Drosophila one of the key systems for future dissection of the complex interplay between neural network computation and behavioral output.
Drosophila in Studies of Population and Evolutionary Genetics
The Drosophila community has been instrumental in combining evolutionary biology with other disciplines to understand the differences within and between species from molecular to phenotypic levels, yet another example of the tractable complexity of Drosophila. Below we highlight how work with Drosophila has enabled advances at the intersections of evolutionary biology with genetics and molecular biology, genome biology, and developmental biology.
Evolutionary genetics and molecular evolution
By the mid-20th century, the previously disparate fields of genetics and evolutionary biology (Fisher 1930; Haldane 1932; Dobzhansky 1937) came to be integrated into what is now known as the Modern Synthesis (Huxley 1942). Out of these ideas came much of what we understand about evolution today, including that populations evolve by natural selection acting on genetic variation. Early Drosophilists like Muller and Dobzhansky had an important influence on the Modern Synthesis and the fields of evolutionary and population genetics. The first studies of population variation were at the level of chromosomal inversion polymorphisms (Dobzhansky and Queal 1938) and the frequency of lethal mutations (Dubinin 1946; Greenberg and Crow 1960). Drosophilists also pioneered surveys of molecular genetic variation, first using protein electrophoresis to study allozyme variation (Lewontin and Hubby 1966) and then DNA sequence polymorphism (Kreitman 1983). DNA sequence variation holds information on the frequency, mode, and strength of natural selection acting in the genome. To make inferences from DNA sequence variation data, Drosophila population geneticists spearheaded statistical methods to detect the effects of deleterious mutations (Charlesworth et al. 1993), infer population demographic history [e.g., Glinka et al. 2003; Haddrill et al. 2005) and adaptive evolution (e.g., the McDonald–Kreitman test (McDonald and Kreitman 1991), and the Hudson–Kreitman–Aguade test (Hudson et al. 1987)]. Several software packages exist to run tests using these and other methods on user-supplied data, such as DNAsp (Rozas et al. 2003) and libsequence (Thornton 2003). Studies in a wide variety of species have employed these tests and their derivatives to study adaptive evolution throughout the genome (reviewed in Eyre-Walker 2006).
Population genomics, quantitative genetics, and “evo-devo”
Recent high throughput sequencing technologies (reviewed in van Dijk et al. 2014) have made it feasible to sequence and compare entire genomes of many individuals—the basis of a field now known as population genomics. The Drosophila community boasts powerful resources for population genomics and quantitative genetics, with genome sequences available for hundreds of D. melanogaster individuals from different geographic populations. The Drosophila Genetic Reference Panel (DGRP) is a panel of >200 inbred, mostly homozygous lines of D. melanogaster from Raleigh, NC, whose genome sequences (Mackay et al. 2012; Huang et al. 2014) combined with phenotypes assayed from the same lines are an unprecedented resource for genome-wide association studies (GWAS) (Ober et al. 2012). To map a trait of interest, a researcher can order the DGRP flies from the Bloomington Stock Center (http://flystocks.bio.indiana.edu/), measure phenotypes in these lines, and access the genome data to perform analyses (http://dgrp2.gnets.ncsu.edu/). GWAS analyses using these lines have identified significant genomic associations with sleep traits (Harbison et al. 2013), life span and fecundity (Durham et al. 2014), olfactory behavior (Arya et al. 2015), abdominal pigmentation (Dembeck et al. 2015), starvation resistance, and startle response (Mackay et al. 2012; Ober et al. 2012). In several cases, the genes identified in these studies have orthologs implicated in similar phenotypes in humans (e.g., Harbison et al. 2013).
Complementary to the DGRP is the Drosophila Population Genomics Project (DPGP)—a large-scale resequencing project focused on the genomes of D. melanogaster from populations in Sub-Saharan Africa and France (Pool et al. 2012; Lack et al. 2015). The latest version of DPGP (DPGP3; http://www.dpgp.org/dpgp3/DPGP3.html) involves the sequencing of >300 individuals from a single population in Zambia, in what is presumed to be the ancestral range of the species (Pool et al. 2012; Lack et al. 2015). The Drosophila Genome Nexus is a recent compilation of each of these population genomic sequences assembled against a single common reference genome assembly, which facilitates direct comparisons among datasets (Lack et al. 2015). In total, the Nexus contains 623 genomes with representation from populations that span much of D. melanogaster’s current geographic range. These population data allow for the study of D. melanogaster demographic and migratory history (Pool et al. 2012), natural selection and the genetic basis of local adaptation (Langley et al. 2012), chromosomal inversion polymorphism (Corbett-Detig and Hartl 2012), and copy number variation (Langley et al. 2012). For example, D. melanogaster has recently evolved resistance to some common pesticides. The molecular basis of resistance is well studied. Single nucleotide changes in acetylcholinesterase (Ace) cause resistance to organophosphates and carbamates (Mutero et al. 1994; Menozzi et al. 2004), whereas TE insertions and copy number variants of the cytochrome P450 gene Cyp6g1 causes DDT resistance (Schmidt et al. 2010). Studies using the DGRP identified these and other regions of the genome as targets of recent positive selection (Garud et al. 2015).
Prior to the early work of Dobzhansky (1937) and Muller (1942), little was known of the genetic basis of species differences. What is now known as the Dobzhansky–Muller model of speciation is the basis of many contemporary studies of speciation genetics (e.g., Coyne and Orr 1989, 1997). Owing to the abundance of new genetic and molecular tools, the field of speciation genetics is growing (reviewed in Presgraves 2010). Population genomic resources now exist for several species closely related to D. melanogaster, including D. simulans (Rogers et al. 2014), D. mauritiana (Nolte et al. 2013; Garrigan et al. 2015), and D. yakuba (Rogers et al. 2014) with several additional datasets on the way (http://www.dpgp.org/1K_300genomes.html). Population-level data on transcriptomes (RNA sequence datasets) are also available for studying gene regulatory evolution and de novo genes (Zhao et al. 2014). Because Drosophila species vary widely in many phenotypic aspects (Markow and O’Grady 2007), these resources enable Drosophila researchers to study the function and evolution of species genomes in a comparative framework (Figure 3B).
Comparative population genomic resources also empower the field of evolutionary developmental biology (evo-devo), which arose from an effort to understand species morphological differences by studying the evolution of developmental processes. The pathways involved in building organism body plans are remarkably conserved between distantly related species (Carroll 2008). The homeobox—a conserved motif found in genes involved in patterning during development across metazoans, including the HOX genes described above—was first identified in D. melanogaster (McGinnis et al. 1984; Scott and Weiner 1984). Through the genetic manipulation of cis-regulatory elements (regulatory sequences that govern the spatial and temporal patterns of gene expression) (Arnone and Davidson 1997) Drosophilists led the evo-devo studies of traits like pigmentation (Gompel et al. 2005; Jeong et al. 2008; Williams et al. 2008) and trichome patterns (Sucena et al. 2003; Frankel et al. 2011).
Drosophila Databases and Online Resources
As the fly community expanded in the mid-20th century, the journal Drosophila Information Service catalogued the growing list of known mutants. Eventually the “Red Book” collected descriptions of mutant strains and genes associated with phenotypes (Lindsley and Grell 1968; Lindsley and Zimm 1992). Now, in the internet and postgenomic age, many online resources are available to help researchers find strains, molecular reagents, and data on genomes, genes, proteins, and molecular interactions.
FlyBase, http://flybase.org, (St Pierre et al. 2014) is the typical starting point when a researcher wants to find information on a particular gene or a genomic region. Its user-friendly interface provides access to genome data and annotations thereof from multiple Drosophila species. FlyBase also enables searching for batches of genes based on expression pattern or other criteria. A gene entry includes information on gene structure, genomic neighborhood, protein sequence, homologs, known alleles and phenotypes, and references in the literature; it also provides links to the relevant sections of other online databases. Genome data and annotations available via FlyBase continue to be refined and updated by the Berkeley Drosophila Genome Project (BDGP), http://www.fruitfly.org/ (e.g., Hoskins et al. 2007). Another useful starting point is FlyMine, http://www.flymine.org/, which integrates genome and protein data from multiple sources to allow for complex queries (Lyne et al. 2007). Data from the modENCODE project (described above) can be accessed at http://www.modencode.org/ and through various linked interfaces such as modMine, http://intermine.modencode.org/ (Contrino et al. 2012). For beginning researchers, the Interactive Fly, http://www.sdbonline.org/sites/fly/aimain/1aahome.htm, is a good starting point for exploring the roles of genes in developmental processes.
Beyond those broadly applicable sites, published reviews of Drosophila resources (Matthews et al. 2005; Cook et al. 2010; Mohr et al. 2014) are good starting points to find specialized resources; however, since these reviews go out of date quickly, FlyBase maintains updated lists of links to databases and sources for stocks, reagents, and services (including both nonprofit and for-profit operations) at http://flybase.org/static_pages/allied-data/external_resources5.html. A few examples of online resources with more narrow foci are described below, listed according to the questions they help address.
How can I find flies with mutations in my gene of interest?
The Gene Disruption Project, http://flypush.imgen.bcm.tmc.edu/pscreen/about.html (Bellen et al. 2011), has yielded >12,000 insertion mutants, with the majority of genes represented. This project represents an additional aspect of the many efforts of the BDGP.
Where and when is a gene expressed?
FlyAtlas2, http://flyatlas.gla.ac.uk/flyatlas/index.html, catalogs gene expression at the level of mRNA enrichment across multiple tissues for most genes, based on microarray data (Chintapalli et al. 2007; Robinson et al. 2013). The BDGP embryo in situ database, http://insitu.fruitfly.org/cgi-bin/ex/insitu.pl, documents specific gene expression patterns within embryos (Tomancak et al. 2007).
Where in the genome is a certain transcription factor predicted to bind?
The Drosophila Transcription Factor Database, http://www.flytf.org/, allows exploration of genome and protein data for transcription factors with DNA sequence specificity (Adryan and Teichmann 2006).
With what genes and proteins does a gene/protein interact?
DroID, the Drosophila Interactions Database, http://www.droidb.org/ (Murali et al. 2011), includes protein interaction data from the Drosophila Protein Interaction Map, https://interfly.med.harvard.edu/ (Guruharsha et al. 2011), and other sources in a searchable format.
In the fly brain, what genes are active in what cells, and how do neurons connect?
Virtual FlyBrain, http://www.virtualflybrain.org/site/vfb_site/home.htm (Milyaev et al. 2012), integrates images and other data on brain anatomy at the cellular level.
Where can I find data on genetic variants comprising a population?
The Drosophila Population Genomics Project (DPGP), http://dpgp.org/, has made available sequence data from multiple genomes within several different populations of flies.
Where can I obtain fly stocks?
Tens of thousands of genetically distinct fly stocks of D. melanogaster, from mutants to those with fluorescently tagged genes to those carrying RNAi-inducing transgenes, are available from various sources including the following three large stock centers:
Bloomington Drosophila Stock Center (Indiana University), http://flystocks.bio.indiana.edu/.
Kyoto Drosophila Genetic Resource Center, https://kyotofly.kit.jp/cgi-bin/stocks/index.cgi.
Vienna Drosophila RNAi Center, http://stockcenter.vdrc.at/control/main.
In addition, stocks from related Drosophila species can be obtained from the Drosophila Species Stock Center, https://stockcenter.ucsd.edu/info/welcome.php.
Where can I obtain cell lines, vectors, reporter constructs, cloned fly cDNAs, and other molecular reagents?
The Drosophila Genomics Resource Center (DGRC), https://dgrc.cgb.indiana.edu/Home,
collects and distributes cellular and molecular reagents generated by many individuals and organizations, including the BDGP (e.g., Stapleton et al. 2002).
Glossary
Balancer chromosomes
Chromosomes engineered with multiple inversions, dominant markers, and recessive lethal mutations that, respectively, prevent viable recombination products from being produced during meiosis, allow easy discernment of which progeny inherit which chromosome, and keep mutations on the homologous chromosome from being selected out of the population.
Cellularization
The stage when nuclei in the early syncytial embryo become surrounded by their own individual plasma membranes.
CRISPR/Cas9
A powerful gene editing system, used in many organisms and contexts, that enables the generation of mutations in specific genes through targeting the Cas9 endonuclease to a desired DNA region with a complementary “guide RNA.”
Eclosion
The process at the end of metamorphosis during which a new adult fly emerges from the pupal case.
Enhancer trap
An approach that allows for the identification of promoter/enhancers that express in unique cell types or during specific developmental stages. Typically this approach involves random integration of a reporter gene into the genome, followed by screening for expression of the reporter gene in the desired cell type/temporal pattern.
Enhancer/suppressor screen
A type of genetic screen used to identify additional genes that interact with an already identified gene in a biological process. Additional mutations need only be heterozygous in some cases. The approach starts with an allele of a gene with marginal function such that any additional increase or decrease in function in the pathway results in a change in the phenotypic severity. Heterozygous mutations in the additional genes may be sufficient to have a detectable phenotypic difference in combination with the initial allele. Not requiring homozygosity at loci greatly simplifies the scheme for the screen.
FLP/FRT clonal analysis
Site-directed mitotic recombination approach frequently used to generate patches of cells homozygous for a particular mutant allele in an otherwise heterozygous background. Clonal analysis is especially useful to study the role of a gene in a cell population when animals fully mutant for the gene are lethal, and it involves expression of the flippase recombinase (FLP) to mediate mitotic recombination at the flippase recombination target (FRT) sequences, downstream of which the original cell is heterozygous for a mutation. In a subset of mitotic recombination instances, chromosome segregation to daughter cells results in a homozygous mutant cell that subsequently divides to create a patch (or “clone”) of homozygous cells whose phenotype can be assessed.
Forward genetic screen
A screening approach used to identify genes involved in a particular biological process, typically involving random mutagenesis (by chemicals or radiation) and searching among animals in subsequent generations for mutant animals displaying a desired phenotype.
GAL4/UAS system
A modular experimental method that allows for controlled spatial and temporal expression of a gene or RNAi knockdown construct. Transgenic flies expressing the yeast transcription factor GAL4 under the control of a desired promoter/enhancer are selected and these are crossed to flies with a second transgene carrying the upstream activating sequence (UAS), which responds to GAL4, upstream of the gene of interest. Offspring with both transgenes will express the gene of interest in the pattern dictated by the chosen promoter.
Germline clones
Specific type of clonal analysis used to assess mutant phenotypes in the absence of both maternal contribution and zygotic expression. Typically accomplished using a modified FLP/FRT protocol that incorporates the dominant female sterile ovoD (ensures that only females where recombination occurred in the germline can lay eggs).
Imaginal discs
Tissues within a larva that, during metamorphosis, will undergo dramatic changes to become a particular adult structure like an eye, wing, or leg.
MARCM
Mosaic analysis with a repressible cell marker (MARCM) is a specific type of clonal analysis in which the cells to be analyzed are recognized by expression of a reporter gene. This utilizes the Gal80 repressor, which prevents expression of UAS/Gal4 constructs. The genetics are designed such that the desired recombination also removes the Gal80 repressor, allowing reporter gene expression to be activated only in cells where recombination has occurred.
modENCODE
A large scale collaborative project to define functional elements in the genomes of Drosophila and the nematode C. elegans.
Muller elements
Conserved chromosome arms where, across species, the gene content of an arm is conserved but may differ in order or orientation. Drosophila species have five Muller elements labeled A through F.
Ommatidium
Unit of the Drosophila compound eye consisting of eight photoreceptor cells and surrounded by pigment and support cells.
PhiC31 system
An approach to generate transgenic lines in which the genomic integration site is not random, but rather can be selected from a number of available landing sites. Avoids the difficulties of position affect variation associated with random insertions as integration sites can be selected that are known to express well. Transgenes can also be inserted at the same location, allowing direct phenotypic comparison of different transgenes at the same site.
Polytene chromosomes
Chromosomes that form after multiple rounds of endoreplication (DNA replication without mitosis), which results in multiple sister chromatids that remain synapsed. The banding pattern of polytene chromosomes can be used to visualize genomic reorganizations and deletions.
Puparium
Hard, exoskeleton of the last larval stage. Serves to protect the fly as it undergoes metamorphosis inside the pupal case.
Reverse genetics
An experimental approach to determine the function of a gene in which the phenotypes associated with mutation or knock-down of that gene are studied. It is considered “reverse” genetics only because the forward approach was practiced first, and the reverse approach became much more common only after the D. melanogaster genome was sequenced, with many uncharacterized genes revealed.
Syncytium
Developmental state in which multiple nuclei share a common cytoplasm as a result of mitosis without cytokinesis, as in the early Drosophila embryo.
Drosophila Milestones and Timeline to the Present
The first scientist to adopt D. melanogaster as a research organism was William Castle in 1900, and by a decade later a few others had followed his lead. Thomas Hunt Morgan’s group at Columbia University (and later at the California Institute of Technology), including students Alfred Sturtevant, Calvin Bridges, and Hermann Muller, made groundbreaking discoveries that established D. melanogaster as a valuable genetic model organism. Morgan ultimately became known as the founder of Drosophila genetics—all current Drosophila researchers can trace their intellectual pedigree back to Morgan.
Morgan’s group provided important evidence for the chromosome theory of heredity through correlating the inheritance of red- and white-eyed variants with the segregation of the X chromosome, especially in rare offspring resulting from sex chromosome nondisjunction (Morgan 1910; Bridges 1916). They were the first to deduce linkage of genes in a linear order on a chromosome (Sturtevant 1913) and to dissect chromosomal rearrangements, starting with deficiencies (Bridges 1917). Morgan was later awarded the 1933 Nobel Prize in Physiology or Medicine. Stories of the work in Morgan’s “Fly Room” at Columbia became legendary and are recounted in various books such as Lords of the Fly (Kohler 1994), Fly (Brookes 2001), and Mendel’s Legacy (Carlson 2004). The 2014 film The Fly Room, produced by Alexis Gambis, is told from the perspective of Calvin Bridges’s daughter; the production included construction of a detailed replica of the laboratory space that was temporarily on display in a New York gallery in 2013, and which can still be seen online.
Another Fly Room accomplishment involved polytene chromosomes, which consist of many replicated but not separated DNA molecules adhering in register; these are easily stained and visualized, with each chromosome showing a unique banding pattern. After their discovery in the 1930s in other insects, polytene chromosomes were described in D. melanogaster salivary gland cells (Painter 1933) and established by Bridges (still in Morgan’s group after their move to CalTech) as a primary tool for chromosome mapping (Bridges 1935). His meticulous drawings of polytene chromosomes are still in use 80 years later.
The Soviet-born geneticist Theodosious Dobzhansky spent time in Morgan’s Fly Room and through his studies connected ideas of genetic variability in a population with how populations shift over time to become reproductively isolated new species. His book Genetics and the Origin of Species (Dobzhansky 1937) was crucial in the formulation of the Modern Synthesis, which connected genetics with other areas to create a broad understanding of the mechanism of evolution.
After leaving Morgan’s group in 1915, Herman Muller discovered through examining offspring of irradiated flies that gene mutations are inducible by X-rays in a dose-dependent way (Muller 1927). This seminal work on the nature and origin of mutations earned Muller a Nobel Prize in 1946 and set the stage for scientists to generate (using radiation, and, later, chemicals) vast collections of mutant strains for dissecting the biology of many processes.
Ed Lewis was a driving force in the Drosophila community from the 1940s until his death in 2004; his early work, among many other things, helped develop models for the nature of the gene based on intragenic recombination. His later work using radiation-induced and spontaneous mutants focused on development of the body plan in Drosophila, setting the stage for Drosophila as an important model organism in developmental biology (Lewis 1978). His work on patterning earned him a share of the 1995 Nobel Prize.
Christiane Nusslein-Volhard and Eric Wieschaus were the other corecipients of the 1995 Nobel Prize for their mutagenesis screens to identify and describe mutations affecting the early patterning steps of embryonic development (Nusslein-Volhard and Wieschaus 1980); this later allowed in-depth molecular characterization of conserved genes that govern the basic body plan of most metazoans.
David Hogness, along with Ed Lewis and colleagues performed the first cloning and genomic analysis of a gene, Ubx, within the Bithorax complex (Bender et al. 1983), setting the stage for the broad use of molecular biology in Drosophila genetics.
Mutational analysis also led to important advancements in behavioral genetics, as Seymour Benzer and colleagues identified the first known genes associated with circadian rhythms and learning and memory by analysis of flies with relevant defects (Konopka and Benzer 1971; Dudai et al. 1976; Weiner 1999).
The 1980s saw Allan Spradling and Gerry Rubin’s discovery of methods for making transgenic flies revolutionize the experimental capabilities of the fly community (Rubin and Spradling 1982; Spradling and Rubin 1982). For example, researchers could test whether an added wild-type copy of a gene rescued a mutant phenotype, and they could add tagged versions of genes to the genome to trace protein localization and isolate interactors.
Molecular characterization of D. melanogaster genes exploded in the 1980s and 1990s, making clear how conserved many developmental pathways were between flies and mammals. For example, Walter Gehring’s work on the Pax6 gene as a master regulator of eye development showed that this pathway was shared between flies and humans (reviewed in Gehring 2002).
While gene knock-out technology was more difficult to achieve in flies than in other model systems, Kent Golic’s group developed such techniques around the turn of the 21st century (Rong and Golic 2000, 2001). The sequencing of the fly genome in 2000 by a collaboration between the company Celera and the Berkeley Drosophila Genome Project was a major milestone (Adams et al. 2000) and served as a trial run in preparation for Celera’s contributions to the Human Genome Project. The 21st century thus far has seen development of new targeted gene knock-down and knock-out approaches using RNA interference and the CRISPR/Cas9 system, the latter of which is quickly and dramatically expanding in popularity and use (Gratz et al. 2013; Bassett and Liu 2014).
The most recent Nobel Prize for work using D. melanogaster was awarded to Jules Hoffman in 2011 for his 1996 work on the innate immune response. With a wide range of genetic manipulations available, and with close conservation between a large number of fly and human genes, the fly continues to be used for molecular characterization of basic cell biological and other processes, many of which directly relate to medically relevant human conditions such as neurodegeneration, cancer, and aging.
Conclusion
As we begin the second century of using Drosophila as an experimental tool in the fields of genetics, cell biology, developmental biology, neurobiology, and evolutionary biology, the groundwork laid during the first century will enable the tractable complexity of the fly to continue offering valuable insights into basic science as well as applied, translational research toward human health. Students entering a research career will continue to find exciting and groundbreaking opportunities to contribute to scientific knowledge in the many Drosophila labs around the world.
Acknowledgments
We thank Kevin Cook from the Bloomington Drosophila Stock Center for helpful information, Beth De Stasio, Daven Presgraves and four reviewers for comments on the manuscript, and the many members of the Drosophila community for generously making vast databases and sets of reagents available. Since this primer is meant as an introduction to the field for beginners, we are sorry that we could not cite contributions from so many people who have added to the community’s toolbox and body of knowledge. Work in K.G.H.’s lab is supported by National Institutes of Health AREA grant R15GM080689, National Science Foundation Research in Undergraduate Institutions grant 1158024, and Davidson College. Work in C.A.K.’s lab is supported by the Undergraduate and Creative Activities Office at the College of Charleston and the College of Charleston’s Howard Hughes Medical Institute grant through the Pre-College and Undergraduate Science Education Program. Work in A.M.L.’s lab is supported by the University of Rochester. Work in D.M.R.’s lab is supported by National Institutes of Health AREA grant R15 GM107796.
Footnotes
Communicating editor: E. A. De Stasio
Literature Cited
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., et al. , 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Adoutte A., Balavoine G., Lartillot N., Lespinet O., Prud’homme B., et al. , 2000. The new animal phylogeny: reliability and implications. Proc. Natl. Acad. Sci. USA 97: 4453–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adryan B., Teichmann S. A., 2006. FlyTF: a systematic review of site-specific transcription factors in the fruit fly Drosophila melanogaster. Bioinformatics 22: 1532–1533. [DOI] [PubMed] [Google Scholar]
- Armknecht S., Boutros M., Kiger A., Nybakken K., Mathey-Prevot B., et al. , 2005. High-throughput RNA interference screens in Drosophila tissue culture cells. Methods Enzymol. 392: 55–73. [DOI] [PubMed] [Google Scholar]
- Arnone M. I., Davidson E. H., 1997. The hardwiring of development: organization and function of genomic regulatory systems. Development 124: 1851–1864. [DOI] [PubMed] [Google Scholar]
- Arya G. H., Magwire M. M., Huang W., Serrano-Negron Y. L., Mackay T. F., et al. , 2015. The Genetic Basis for Variation in Olfactory Behavior in Drosophila melanogaster. Chem. Senses 40: 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., 1989. Drosophila: A Laboratory Handbook, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Assis R., Bachtrog D., 2013. Neofunctionalization of young duplicate genes in Drosophila. Proc. Natl. Acad. Sci. USA 110: 17409–17414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B. S., Nagoshi R. N., Burtis K. C., 1987. Molecular genetic aspects of sex determination in Drosophila. Bioessays 6: 66–70. [DOI] [PubMed] [Google Scholar]
- Bargiello T. A., Jackson F. R., Young M. W., 1984. Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature 312: 752–754. [DOI] [PubMed] [Google Scholar]
- Bassett A. R., Liu J. L., 2014. CRISPR/Cas9 and genome editing in Drosophila. J. Genet. Genomics 41: 7–19. [DOI] [PubMed] [Google Scholar]
- Bastock R., St Johnston D., 2008. Drosophila oogenesis. Curr. Biol. 18: R1082–R1087. [DOI] [PubMed] [Google Scholar]
- Baum B., Cherbas L., 2008. Drosophila cell lines as model systems and as an experimental tool. Methods Mol. Biol. 420: 391–424. [DOI] [PubMed] [Google Scholar]
- Bell L. R., Maine E. M., Schedl P., Cline T. W., 1988. Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA binding proteins. Cell 55: 1037–1046. [DOI] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., Liao G., He Y., Carlson J. W., et al. , 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., He Y., Carlson J. W., Evans-Holm M., et al. , 2011. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188: 731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., O’Kane C. J., Wilson C., Grossniklaus U., Pearson R. K., et al. , 1989. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 3: 1288–1300. [DOI] [PubMed] [Google Scholar]
- Bellen H. J., Tong C., Tsuda H., 2010. 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat. Rev. Neurosci. 11: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellmann D., Richardt A., Freyberger R., Nuwal N., Schwarzel M., et al. , 2010. Optogenetically induced olfactory stimulation in Drosophila larvae reveals the neuronal basis of odor-aversion behavior. Front. Behav. Neurosci. 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W., Akam M., Karch F., Beachy P. A., Peifer M., et al. , 1983. Molecular genetics of the bithorax complex in Drosophila melanogaster. Science 221: 23–29. [DOI] [PubMed] [Google Scholar]
- Berleth T., Burri M., Thoma G., Bopp D., Richstein S., et al. , 1988. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 7: 1749–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer K. J., Carroll D., 2014. Targeted genome engineering techniques in Drosophila. Methods 68: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham P. M., Kidwell M. G., Rubin G. M., 1982. The molecular basis of P-M hybrid dysgenesis: the role of the P element, a P-strain-specific transposon family. Cell 29: 995–1004. [DOI] [PubMed] [Google Scholar]
- Boley N., Wan K. H., Bickel P. J., Celniker S. E., 2014. Navigating and mining modENCODE data. Methods 68: 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolshakov V. N., Topalis P., Blass C., Kokoza E., della Torre A., et al. , 2002. A comparative genomic analysis of two distant diptera, the fruit fly, Drosophila melanogaster, and the malaria mosquito, Anopheles gambiae. Genome Res. 12: 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G., Campbell P., Leiva-Neto J. T., Markow T. A., 2007. Analysis of Drosophila species genome size and satellite DNA content reveals significant differences among strains as well as between species. Genetics 177: 1277–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M., Kiger A. A., Armknecht S., Kerr K., Hild M., et al. , 2004. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303: 832–835. [DOI] [PubMed] [Google Scholar]
- Boyes J. W., Vanbrink J. M., 1965. Chromosomes of Calyptrate Diptera. Can. J. Genet. Cytol. 7: 537. [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Aravin A. A., Stark A., Dus M., Kellis M., et al. , 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Bridges C. B., 1916. Non-disjunction as proof of the chromosome theory of heredity. Genetics 1: 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges C. B., 1917. Deficiency. Genetics 2: 445–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges C. B., 1935. Salivary chromosome maps with a key to the banding of the chromosomes of Drosophila melanogaster. J. Hered. 26: 60–64. [Google Scholar]
- Brookes M., 2001. Fly: An Experimental Life. Weidenfeld & Nicolson, London. [Google Scholar]
- Brown J. B., Boley N., Eisman R., May G. E., Stoiber M. H., et al. , 2014. Diversity and dynamics of the Drosophila transcriptome. Nature 512: 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabernard C., 2012. Cytokinesis in Drosophila melanogaster. Cytoskeleton (Hoboken) 69: 791–809. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega J. A., Hartenstein V., 1997. The Embryonic Development of Drosophila melanogaster, Springer-Verlag, Berlin. [Google Scholar]
- Carlson E. A., 2004. Mendel’s Legacy: The Origin of Classical Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Carroll S. B., 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. [DOI] [PubMed] [Google Scholar]
- Carson H. L., 1975. The genetics of speciation at the diploid level. Am. Nat. 109: 83–92. [Google Scholar]
- Carson H. L., 1982. Evolution of Drosophila on the Newer Hawaiian Volcanos. Heredity 48: 3. [DOI] [PubMed] [Google Scholar]
- Carson H. L., 1997. Sexual selection: A driver of genetic change in Hawaiian Drosophila. J. Hered. 88: 343–352. [DOI] [PubMed] [Google Scholar]
- Celniker, S. E., D. A. Wheeler, B. Kronmiller, J. W. Carlson, A. Halpern et al., 2002 Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: RESEARCH0079. [DOI] [PMC free article] [PubMed]
- Celniker S. E., Dillon L. A., Gerstein M. B., Gunsalus K. C., Henikoff S., et al. , 2009. Unlocking the secrets of the genome. Nature 459: 927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Morgan M. T., Charlesworth D., 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. X., Sturgill D., Qu J., Jiang H., Park S., et al. , 2014. Comparative validation of the D. melanogaster modENCODE transcriptome annotation. Genome Res. 24: 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang A. S., Lin C. Y., Chuang C. C., Chang H. M., Hsieh C. H., et al. , 2011. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr. Biol. 21: 1–11. [DOI] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J., Dow J. A., 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39: 715–720. [DOI] [PubMed] [Google Scholar]
- Chiu J. C., Jiang X., Zhao L., Hamm C. A., Cridland J. M., et al. , 2013. Genome of Drosophila suzukii, the spotted wing drosophila. G3 (Bethesda) 3: 2257–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E., Jimenez E., Kim Y. A., Whitworth C., Neville M. C., et al. , 2014. Sex- and tissue-specific functions of Drosophila doublesex transcription factor target genes. Dev. Cell 31: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium, 1998 Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed]
- Contrino S., Smith R. N., Butano D., Carr A., Hu F., et al. , 2012. modMine: flexible access to modENCODE data. Nucleic Acids Res. 40: D1082–D1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook K. R., Parks A. L., Jacobus L. M., Kaufman T. C., Matthews K. A., 2010. New research resources at the Bloomington Drosophila Stock Center. Fly (Austin) 4: 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig R. B., Hartl D. L., 2012. Population genomics of inversion polymorphisms in Drosophila melanogaster. PLoS Genet. 8: e1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 1989. Patterns of speciation in Drosophila. Evolution 43: 362–381. [DOI] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 1997. “Patterns of speciation in Drosophila” revisited. Evolution 51: 295–303. [DOI] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 1998. The evolutionary genetics of speciation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353: 287–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N., Craigie R., Gellert M., Lambowitz A., 2002. Mobile DNA II, ASM Press, Washington, DC. [Google Scholar]
- Czech B., Malone C. D., Zhou R., Stark A., Schlingeheyde C., et al. , 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton R., 2009. A fly by any other name. Nature 457: 368. [DOI] [PubMed] [Google Scholar]
- Daniels S. B., Peterson K. R., Strausbaugh L. D., Kidwell M. G., Chovnick A., 1990. Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics 124: 339–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David J. R., Capy P., 1988. Genetic variation of Drosophila melanogaster natural populations. Trends in genetics. TIG 4: 106–111. [DOI] [PubMed] [Google Scholar]
- Davis T. R., Wickham T. J., McKenna K. A., Granados R. R., Shuler M. L., et al. , 1993. Comparative recombinant protein production of eight insect cell lines. In Vitro Cell. Dev. Biol. Anim. 29A: 388–390. [DOI] [PubMed] [Google Scholar]
- de Graaf C. A., van Steensel B., 2013. Chromatin organization: form to function. Curr. Opin. Genet. Dev. 23: 185–190. [DOI] [PubMed] [Google Scholar]
- Dembeck L. M., Huang W., Magwire M. M., Lawrence F., Lyman R. F., et al. , 2015. Genetic architecture of abdominal pigmentation in Drosophila melanogaster. PLoS Genet. 11: e1005163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir E., Dickson B. J., 2005. fruitless splicing specifies male courtship behavior in Drosophila. Cell 121: 785–794. [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T., 1937. Genetics and the Origin of Species, Columbia University Press, New York. [Google Scholar]
- Dobzhansky T., Queal M. L., 1938. Genetics of natural populations. I. Chromosome variation in populations of Drosophila pseudoobscura inhabiting isolated mountain ranges. Genetics 23: 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W., Nusslein-Volhard C., 1988. A gradient of bicoid protein in Drosophila embryos. Cell 54: 83–93. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium et al., 2007 Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218. [DOI] [PubMed]
- Dubinin N. P., 1946. On lethal mutations in natural populations. Genetics 31: 21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudai Y., Jan Y. N., Byers D., Quinn W. G., Benzer S., 1976. dunce, a mutant of Drosophila deficient in learning. Proc. Natl. Acad. Sci. USA 73: 1684–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy J. B., 2002. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis 34: 1–15. [DOI] [PubMed] [Google Scholar]
- Durham M. F., Magwire M. M., Stone E. A., Leips J., 2014. Genome-wide analysis in Drosophila reveals age-specific effects of SNPs on fitness traits. Nat. Commun. 5: 4338. [DOI] [PubMed] [Google Scholar]
- Echalier G., Ohanessian A., 1969. [Isolation, in tissue culture, of Drosophila melangaster cell lines] C. R. Acad. Sci. Hebd. Seances Acad. Sci. D 268: 1771–1773. [PubMed] [Google Scholar]
- Echeverri C. J., Perrimon N., 2006. High-throughput RNAi screening in cultured cells: a user’s guide. Nat. Rev. Genet. 7: 373–384. [DOI] [PubMed] [Google Scholar]
- Engels W. R., 1983. The P family of transposable elements in Drosophila. Annu. Rev. Genet. 17: 315–344. [DOI] [PubMed] [Google Scholar]
- Erickson J. W., Quintero J. J., 2007. Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 5: e332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S. M., Yan W., Murillo M. P., Ponce J., Papalopulu N., 1995. Tinman, a Drosophila homeobox gene required for heart and visceral mesoderm specification, may be represented by a family of genes in vertebrates - Xnkx-2.3, a 2nd vertebrate homolog of Tinman. Development 121: 3889–3899. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., 2006. The genomic rate of adaptive evolution. Trends Ecol. Evol. 21: 569–575. [DOI] [PubMed] [Google Scholar]
- Fabian L., Brill J. A., 2012. Drosophila spermiogenesis: big things come from little packages. Spermatogenesis 2: 197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno L., Yizhar O., Deisseroth K., 2011. The development and application of optogenetics. Annu. Rev. Neurosci. 34: 389–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection, Clarendon Press, Oxford. [Google Scholar]
- Fonseca N. A., Morales-Hojas R., Reis M., Rocha H., Vieira C. P., et al. , 2013. Drosophila americana as a model species for comparative studies on the molecular basis of phenotypic variation. Genome Biol. Evol. 5: 661–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini M. E., Bonini N. M., 2000. Modeling human neurodegenerative diseases in Drosophila: on a wing and a prayer. Trends Genet. 16: 161–167. [DOI] [PubMed] [Google Scholar]
- Frankel N., Erezyilmaz D. F., McGregor A. P., Wang S., Payre F., et al. , 2011. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature 474: 598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. J., Watabe H. A., Aotsuka T., Pang J. F., Zhang Y. P., 2007. Molecular phylogeny of the Drosophila obscura species group, with emphasis on the Old World species. BMC Evol. Biol. 7: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D., Kingan S. B., Geneva A. J., Andolfatto P., Clark A. G., et al. , 2012. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res. 22: 1499–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D., Kingan S. B., Geneva A. J., Vedanayagam J. P., Presgraves D. C., 2015. Genome diversity and divergence in Drosophila mauritiana: multiple signatures of faster X evolution. Genome Biol. Evol. 7: 1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garud N. R., Messer P. W., Buzbas E. O., Petrov D. A., 2015. Recent selective sweeps in North American Drosophila melanogaster show signatures of soft sweeps. PLoS Genet. 11: e1005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudry Q., Hong E. J., Kain J., de Bivort B. L., Wilson R. I., 2013. Asymmetric neurotransmitter release enables rapid odour lateralization in Drosophila. Nature 493: 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavis E. R., Lehmann R., 1992. Localization of nanos RNA controls embryonic polarity. Cell 71: 301–313. [DOI] [PubMed] [Google Scholar]
- Gehring W. J., 2002. The genetic control of eye development and its implications for the evolution of the various eye-types. Int. J. Dev. Biol. 46: 65–73. [PubMed] [Google Scholar]
- George J. A., DeBaryshe P. G., Traverse K. L., Celniker S. E., Pardue M. L., 2006. Genomic organization of the Drosophila telomere retrotransposable elements. Genome Res. 16: 1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti M. G., Fuller M. T., 2012. What Drosophila spermatocytes tell us about the mechanisms underlying cytokinesis. Cytoskeleton (Hoboken) 69: 869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka S., Ometto L., Mousset S., Stephan W., De Lorenzo D., 2003. Demography and natural selection have shaped genetic variation in Drosophila melanogaster: a multi-locus approach. Genetics 165: 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic K. G., Lindquist S., 1989. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59: 499–509. [DOI] [PubMed] [Google Scholar]
- Gompel N., Prud’homme B., Wittkopp P. J., Kassner V. A., Carroll S. B., 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433: 481–487. [DOI] [PubMed] [Google Scholar]
- Gordon M. D., Scott K., 2009. Motor control in a Drosophila taste circuit. Neuron 61: 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., et al. , 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg R., Crow J. F., 1960. A comparison of the effect of lethal and detrimental chromosomes from Drosophila populations. Genetics 45: 1153–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan R. J., 1997. Fly Pushing: The Theory and Practice of Drosophila genetics, Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- Grossniklaus U., Bellen H. J., Wilson C., Gehring W. J., 1989. P-element-mediated enhancer detection applied to the study of oogenesis in Drosophila. Development 107: 189–200. [DOI] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R., Calos M. P., 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166: 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillen Y., Rius N., Delprat A., Williford A., Muyas F., et al. , 2014. Genomics of ecological adaptation in cactophilic Drosophila. Genome Biol. Evol. 7: 349–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha K. G., Rual J. F., Zhai B., Mintseris J., Vaidya P., et al. , 2011. A protein complex network of Drosophila melanogaster. Cell 147: 690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill P. R., Thornton K. R., Charlesworth B., Andolfatto P., 2005. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 15: 790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. W., Han M. V., Han S. G., 2007. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 3: e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J. B. S., 1932. The Causes of Evolution, Longmans, Green & Co, New York. [Google Scholar]
- Hannon G. J., 2002. RNA interference. Nature 418: 244–251. [DOI] [PubMed] [Google Scholar]
- Harbison S. T., McCoy L. J., Mackay T. F., 2013. Genome-wide association study of sleep in Drosophila melanogaster. BMC Genomics 14: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris W. A., Stark W. S., Walker J. A., 1976. Genetic dissection of the photoreceptor system in the compound eye of Drosophila melanogaster. J. Physiol. 256: 415–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V., 1993. Atlas of Drosophila Development, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Hoskins R. A., Carlson J. W., Kennedy C., Acevedo D., Evans-Holm M., et al. , 2007. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science 316: 1625–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T. T., Eisen M. B., Thornton K. R., Andolfatto P., 2013. A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence. Genome Res. 23: 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Massouras A., Inoue Y., Peiffer J., Ramia M., et al. , 2014. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24: 1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Kreitman M., Aguade M., 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley J., 1942. Evolution, the Modern Synthesis, George Allen & Unwin, London. [Google Scholar]
- Hwang R. Y., Zhong L., Xu Y., Johnson T., Zhang F., et al. , 2007. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr. Biol. 17: 2105–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomou L., Schneider Y. J., Agathos S. N., 2003. Insect cell culture for industrial production of recombinant proteins. Appl. Microbiol. Biotechnol. 62: 1–20. [DOI] [PubMed] [Google Scholar]
- Jeong S., Rebeiz M., Andolfatto P., Werner T., True J., et al. , 2008. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell 132: 783–793. [DOI] [PubMed] [Google Scholar]
- Kaminker, J. S., C. M. Bergman, B. Kronmiller, J. Carlson, R. Svirskas et al., 2002 The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 3: RESEARCH0084. [DOI] [PMC free article] [PubMed]
- Kaneshiro K. Y., 1997. Perkins’ legacy to evolutionary research on Hawaiian Drosophilidae. Pac. Sci. 51: 450–461. [Google Scholar]
- Kazama H., 2015. Systems neuroscience in Drosophila: conceptual and technical advantages. Neuroscience 296: 3–14. [DOI] [PubMed] [Google Scholar]
- Kennerdell J. R., Carthew R. W., 2000. Heritable gene silencing in Drosophila using double-stranded RNA. Nat. Biotechnol. 18: 896–898. [DOI] [PubMed] [Google Scholar]
- Kidwell M. G., 1983. Evolution of hybrid dysgenesis determinants in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 80: 1655–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R. C., 1970. Ovarian Development in Drosophila melanogaster. Academic Press, New York. [Google Scholar]
- Klemenz R., Weber U., Gehring W. J., 1987. The white gene as a marker in a new P-element vector for gene transfer in Drosophila. Nucleic Acids Res. 15: 3947–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler R., 1994. Lords of the Fly: Drosophila Genetics and the Experimental Life, University of Chicago Press, Chicago. [Google Scholar]
- Kondo S., 2014. New horizons in genome engineering of Drosophila melanogaster. Genes Genet. Syst. 89: 3–8. [DOI] [PubMed] [Google Scholar]
- Konopka R. J., Benzer S., 1971. Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 68: 2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M., 1983. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature 304: 412–417. [DOI] [PubMed] [Google Scholar]
- Kulathinal R. J., Stevison L. S., Noor M. A., 2009. The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 5: e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise, D., M.-L. Cariou, J. David, F. Lemeunier, L. Tsacas et al., 1988 Historical biogeography of the Drosophila melanogaster species subgroup, pp. 159–225 in Evolutionary Biology, edited by M. Hecht, B. Wallace, and G. Prance, Springer-Verlag, New York. [Google Scholar]
- Lack J. B., Cardeno C. M., Crepeau M. W., Taylor W., Corbett-Detig R. B., et al. , 2015. The Drosophila genome nexus: a population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics 199: 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley C. H., Stevens K., Cardeno C., Lee Y. C., Schrider D. R., et al. , 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192: 533–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko P., 2012. mRNA localization and translational control in Drosophila oogenesis. Cold Spring Harb. Perspect. Biol. 4: a012294. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Luo L., 2001. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24: 251–254. [DOI] [PubMed] [Google Scholar]
- Lefevre G., Jr, Jonsson U. B., 1962. Sperm transfer, storage, displacement, and utilization in Drosophila melanogaster. Genetics 47: 1719–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leptin M., 1995. Drosophila gastrulation: from pattern formation to morphogenesis. Annu. Rev. Cell Dev. Biol. 11: 189–212. [DOI] [PubMed] [Google Scholar]
- Lewis E. B., 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. [DOI] [PubMed] [Google Scholar]
- Lewontin R. C., Hubby J. L., 1966. A molecular approach to the study of genic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics 54: 595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D. L., Grell E. H., 1968. Genetic Variations of Drosophila melanogaster. Carnegie Institute of Washington, Washington, DC. [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992 The Genome of Drosophila melanogaster. Academic Press, London. [Google Scholar]
- Luo S. D., Baker B. S., 2015. Constraints on the evolution of a doublesex target gene arising from doublesex’s pleiotropic deployment. Proc. Natl. Acad. Sci. USA 112: E852–E861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne R., Smith R., Rutherford K., Wakeling M., Varley A., et al. , 2007. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 8: R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster Genetic Reference Panel. Nature 482: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow T. A., 2015. The secret lives of Drosophila flies. eLife 4: e06793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow T. A., O’Grady P., 2006. Drosophila: A Guide to Species Identification and Use. Academic Press, London. [Google Scholar]
- Markow T. A., O’Grady P. M., 2007. Drosophila biology in the genomic age. Genetics 177: 1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathey-Prevot B., Perrimon N., 2006. Drosophila genome-wide RNAi screens: are they delivering the promise? Cold Spring Harb. Symp. Quant. Biol. 71: 141–148. [DOI] [PubMed] [Google Scholar]
- Matthews K. A., Kaufman T. C., Gelbart W. M., 2005. Research resources for Drosophila: the expanding universe. Nat. Rev. Genet. 6: 179–193. [DOI] [PubMed] [Google Scholar]
- Matunis E. L., Stine R. R., de Cuevas M., 2012. Recent advances in Drosophila male germline stem cell biology. Spermatogenesis 2: 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B., 1950. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 36: 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. H., Kreitman M., 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- McGaugh S. E., Heil C. S., Manzano-Winkler B., Loewe L., Goldstein S., et al. , 2012. Recombination modulates how selection affects linked sites in Drosophila. PLoS Biol. 10: e1001422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W., Garber R. L., Wirz J., Kuroiwa A., Gehring W. J., 1984. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell 37: 403–408. [DOI] [PubMed] [Google Scholar]
- Menozzi P., Shi M. A., Lougarre A., Tang Z. H., Fournier D., 2004. Mutations of acetylcholinesterase which confer insecticide resistance in Drosophila melanogaster populations. BMC Evol. Biol. 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milyaev N., Osumi-Sutherland D., Reeve S., Burton N., Baldock R. A., et al. , 2012. The Virtual Fly Brain browser and query interface. Bioinformatics 28: 411–415. [DOI] [PubMed] [Google Scholar]
- Misra, S., M. A. Crosby, C. J. Mungall, B. B. Matthews, K. S. Campbell et al., 2002 Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol 3: RESEARCH0083. [DOI] [PMC free article] [PubMed]
- modENCODE Consortium et al., 2010 Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797. [DOI] [PMC free article] [PubMed]
- Mohr S. E., Hu Y., Kim K., Housden B. E., Perrimon N., 2014. Resources for functional genomics studies in Drosophila melanogaster. Genetics 197: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. S., DeZazzo J., Luk A. Y., Tully T., Singh C. M., et al. , 1998. Ethanol intoxication in Drosophila: genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell 93: 997–1007. [DOI] [PubMed] [Google Scholar]
- Morgan T. H., 1910. Sex limited inheritance in Drosophila. Science 32: 120–122. [DOI] [PubMed] [Google Scholar]
- Morisato D., Anderson K. V., 1995. Signaling pathways that establish the dorsal-ventral pattern of the Drosophila embryo. Annu. Rev. Genet. 29: 371–399. [DOI] [PubMed] [Google Scholar]
- Muller H. J., 1927. Artificial transmutation of the gene. Science 66: 84–87. [DOI] [PubMed] [Google Scholar]
- Muller H. J., 1940. Bearings of the Drosophila Work on Systematics. Clarendon Press, Oxford. [Google Scholar]
- Muller H. J., 1942. Isolating mechanisms, evolution, and temperature. Biol. Symp. 6: 71–125. [Google Scholar]
- Muqit M. M., Feany M. B., 2002. Modelling neurodegenerative diseases in Drosophila: A fruitful approach? Nat. Rev. Neurosci. 3: 237–243. [DOI] [PubMed] [Google Scholar]
- Murali T., Pacifico S., Yu J., Guest S., Roberts G. G., 3rd, et al. , 2011. DroID 2011: a comprehensive, integrated resource for protein, transcription factor, RNA and gene interactions for Drosophila. Nucleic Acids Res. 39: D736–D743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutero A., Pralavorio M., Bride J. M., Fournier D., 1994. Resistance-associated point mutations in insecticide-insensitive acetylcholinesterase. Proc. Natl. Acad. Sci. USA 91: 5922–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A., Barton N. H., 2003. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution 57: 447–459. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg F. S., Schupbach T., 1993. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell 75: 165–174. [PubMed] [Google Scholar]
- Neville M. C., Nojima T., Ashley E., Parker D. J., Walker J., et al. , 2014. Male-specific fruitless isoforms target neurodevelopmental genes to specify a sexually dimorphic nervous system. Curr. Biol. 24: 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J. Q., Markstein M., Binari R., Pfeiffer B., Liu L. P., et al. , 2008. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat. Methods 5: 49–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T., Neville M. C., Goodwin S. F., 2014. Fruitless isoforms and target genes specify the sexually dimorphic nervous system underlying Drosophila reproductive behavior. Fly (Austin) 8: 95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte V., Pandey R. V., Kofler R., Schlotterer C., 2013. Genome-wide patterns of natural variation reveal strong selective sweeps and ongoing genomic conflict in Drosophila mauritiana. Genome Res. 23: 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor M. A., Grams K. L., Bertucci L. A., Reiland J., 2001. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 98: 12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C., Wieschaus E., 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801. [DOI] [PubMed] [Google Scholar]
- O’Grady P. M., Markow T. A., 2009. Phylogenetic taxonomy in Drosophila. Fly (Austin) 3: 10–14. [DOI] [PubMed] [Google Scholar]
- Ober U., Ayroles J. F., Stone E. A., Richards S., Zhu D., et al. , 2012. Using whole-genome sequence data to predict quantitative trait phenotypes in Drosophila melanogaster. PLoS Genet. 8: e1002685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., Masly J. P., Phadnis N., 2007. Speciation in Drosophila: from phenotypes to molecules. J. Hered. 98: 103–110. [DOI] [PubMed] [Google Scholar]
- Owald D., Lin S., Waddell S., 2015. Light, heat, action: neural control of fruit fly behaviour. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370: 20140211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter T. S., 1933. A new method for the study of chromosome rearrangements and the plotting of chromosome maps. Science 78: 585–586. [DOI] [PubMed] [Google Scholar]
- Palmieri N., Kosiol C., Schlotterer C., 2014. The life cycle of Drosophila orphan genes. eLife 3: e01311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks A. L., Cook K. R., Belvin M., Dompe N. A., Fawcett R., et al. , 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292. [DOI] [PubMed] [Google Scholar]
- Perrimon N., 1984. Clonal analysis of dominant female-sterile, germline-dependent mutations in Drosophila melanogaster. Genetics 108: 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N., 1998. Creating mosaics in Drosophila. Int. J. Dev. Biol. 42: 243–247. [PubMed] [Google Scholar]
- Perrimon N., Mathey-Prevot B., 2007. Applications of high-throughput RNA interference screens to problems in cell and developmental biology. Genetics 175: 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B. D., Jenett A., Hammonds A. S., Ngo T. T., Misra S., et al. , 2008. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105: 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitnick S., Spicer G. S., Markow T. A., 1995. How long is a giant sperm? Nature 375: 109. [DOI] [PubMed] [Google Scholar]
- Pool J. E., Corbett-Detig R. B., Sugino R. P., Stevens K. A., Cardeno C. M., et al. , 2012. Population genomics of sub-saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 8: e1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. R., 1997. Progress and Prospects in Evolutionary Biology. Oxford University Press, New York. [Google Scholar]
- Presgraves D. C., 2010. The molecular evolutionary basis of species formation. Nat. Rev. Genet. 11: 175–180. [DOI] [PubMed] [Google Scholar]
- Richards S., Liu Y., Bettencourt B. R., Hradecky P., Letovsky S., et al. , 2005. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 15: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson B. G., Atkinson N. S., 2013. Is alcoholism learned? Insights from the fruit fly. Curr. Opin. Neurobiol. 23: 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S. W., Herzyk P., Dow J. A., Leader D. P., 2013. FlyAtlas: database of gene expression in the tissues of Drosophila melanogaster. Nucleic Acids Res. 41: D744–D750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers R. L., Cridland J. M., Shao L., Hu T. T., Andolfatto P., et al. , 2014. Landscape of standing variation for tandem duplications in Drosophila yakuba and Drosophila simulans. Mol. Biol. Evol. 31: 1750–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018. [DOI] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2001. A targeted gene knockout in Drosophila. Genetics 157: 1307–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J., Sanchez-DelBarrio J. C., Messeguer X., Rozas R., 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C., 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Kidwell M. G., Bingham P. M., 1982. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell 29: 987–994. [DOI] [PubMed] [Google Scholar]
- Ryder E., Blows F., Ashburner M., Bautista-Llacer R., Coulson D., et al. , 2004. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167: 797–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salz H. K., Erickson J. W., 2010. Sex determination in Drosophila: the view from the top. Fly (Austin) 4: 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapiro A. L., Ihry R. J., Buhr D. L., Konieczko K. M., Ives S. M., et al. , 2013. Rapid recombination mapping for high-throughput genetic screens in Drosophila. G3 (Bethesda) 3: 2313–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J. M., Good R. T., Appleton B., Sherrard J., Raymant G. C., et al. , 2010. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 6: e1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P. S., Matzkin L., Ippolito M., Eanes W. F., 2005. Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution 59: 1721–1732. [PubMed] [Google Scholar]
- Schneider I., 1972. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27: 353–365. [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J., 1984. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc. Natl. Acad. Sci. USA 81: 4115–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharam A. S., Stuart G. W., 2013. Whole genome phylogeny for 21 Drosophila species using predicted 2b-RAD fragments. PeerJ 1: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh C. M., Heberlein U., 2000. Genetic control of acute ethanol-induced behaviors in Drosophila. Alcohol. Clin. Exp. Res. 24: 1127–1136. [PubMed] [Google Scholar]
- Singh N. D., Larracuente A. M., Sackton T. B., Clark A. G., 2009. Comparative genomics on the Drosophila Phylogenetic Tree. Annu. Rev. Ecol. Evol. Syst. 40: 459–480. [Google Scholar]
- Slattery M., Ma L., Spokony R. F., Arthur R. K., Kheradpour P., et al. , 2014. Diverse patterns of genomic targeting by transcriptional regulators in Drosophila melanogaster. Genome Res. 24: 1224–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M., 1982. Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218: 341–347. [DOI] [PubMed] [Google Scholar]
- St Johnston D., 2002. The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet. 3: 176–188. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Nusslein-Volhard C., 1992. The origin of pattern and polarity in the Drosophila embryo. Cell 68: 201–219. [DOI] [PubMed] [Google Scholar]
- St Pierre S. E., Ponting L., Stefancsik R., McQuilton P.; FlyBase Consortium, 2014. FlyBase 102–advanced approaches to interrogating FlyBase. Nucleic Acids Res. 42: D780–D788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton, M., J. Carlson, P. Brokstein, C. Yu, M. Champe et al., 2002 A Drosophila full-length cDNA resource. Genome Biol. 3: RESEARCH0080. [DOI] [PMC free article] [PubMed]
- Stark A., Lin M. F., Kheradpour P., Pedersen J. S., Parts L., et al. , 2007. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450: 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A. H., 1913 The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J. Exp. Zoo. 14: 43–59. [Google Scholar]
- Sturtevant A. H., Dobzhansky T., 1936. Inversions in the third chromosome of wild races of Drosophila pseudoobscura, and their use in the study of the history of the species. Proc. Natl. Acad. Sci. USA 22: 448–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., Novitski E., 1941. The homologies of the chromosome elements in the genus Drosophila. Genetics 26: 517–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucena E., Delon I., Jones I., Payre F., Stern D. L., 2003. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature 424: 935–938. [DOI] [PubMed] [Google Scholar]
- Templeton A. R., 1981. Evolutionary change. Science 214: 900–901. [DOI] [PubMed] [Google Scholar]
- Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A., et al. , 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287. [DOI] [PubMed] [Google Scholar]
- Thornton K., 2003. Libsequence: a C++ class library for evolutionary genetic analysis. Bioinformatics 19: 2325–2327. [DOI] [PubMed] [Google Scholar]
- Toh K. L., Jones C. R., He Y., Eide E. J., Hinz W. A., et al. , 2001. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291: 1040–1043. [DOI] [PubMed] [Google Scholar]
- Tomancak P., Berman B. P., Beaton A., Weiszmann R., Kwan E., et al. , 2007. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 8: R145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J., Karpen G. H., Craig N., Spradling A. C., 1993. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics 133: 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui K., Ueda R., Miyake T., 1987. Cell lines from imaginal discs of Drosophila melanogaster. In Vitro Cell. Dev. Biol. 23: 707–711. [DOI] [PubMed] [Google Scholar]
- van Dijk E. L., Auger H., Jaszczyszyn Y., Thermes C., 2014. Ten years of next-generation sequencing technology. Trends Genet. 30: 418–426. [DOI] [PubMed] [Google Scholar]
- van Eeden F., St Johnston D., 1999. The polarisation of the anterior-posterior and dorsal-ventral axes during Drosophila oogenesis. Curr. Opin. Genet. Dev. 9: 396–404. [DOI] [PubMed] [Google Scholar]
- Venken K. J., Bellen H. J., 2005. Emerging technologies for gene manipulation in Drosophila melanogaster. Nat. Rev. Genet. 6: 167–178. [DOI] [PubMed] [Google Scholar]
- Venken K. J., Bellen H. J., 2007. Transgenesis upgrades for Drosophila melanogaster. Development 134: 3571–3584. [DOI] [PubMed] [Google Scholar]
- Venken K. J., He Y., Hoskins R. A., Bellen H. J., 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751. [DOI] [PubMed] [Google Scholar]
- Venken K. J., Schulze K. L., Haelterman N. A., Pan H., He Y., et al. , 2011. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8: 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst E. C., van de Zande L., Beukeboom L. W., 2010. Insect sex determination: it all evolves around transformer. Curr. Opin. Genet. Dev. 20: 376–383. [DOI] [PubMed] [Google Scholar]
- Vernes S. C., 2014. Genome wide identification of fruitless targets suggests a role in upregulating genes important for neural circuit formation. Sci. Rep. 4: 4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker R. A., Greenleaf A. L., Gyurkovics H., Wisely G. B., Huang S. M., et al. , 1984. Frequent imprecise excision among reversions of a P element-caused lethal mutation in Drosophila. Genetics 107: 279–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall L. B., 2007. Into the mind of a fly. Nature 450: 193–197. [DOI] [PubMed] [Google Scholar]
- Weiner J., 1999. Time, Love, Memory. Vintage, New York. [Google Scholar]
- White J. G., Southgate E., Thomson J. N., Brenner S., 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314: 1–340. [DOI] [PubMed] [Google Scholar]
- Wieschaus E., Nussleinvolhard C., Jurgens G., 1984. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. 3. Zygotic loci on the X–chromosome and 4th chromosome. Wilhelm Rouxs Archives of Developmental Biology 193: 296–307. [DOI] [PubMed] [Google Scholar]
- Williams T. M., Selegue J. E., Werner T., Gompel N., Kopp A., et al. , 2008. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell 134: 610–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C., Pearson R. K., Bellen H. J., O’Kane C. J., Grossniklaus U., et al. , 1989. P-element-mediated enhancer detection: an efficient method for isolating and characterizing developmentally regulated genes in Drosophila. Genes Dev. 3: 1301–1313. [DOI] [PubMed] [Google Scholar]
- Wolfner M. F., 2003. Sex determination: Sex on the brain? Curr. Biol. 13: R101–R103. [DOI] [PubMed] [Google Scholar]
- Yamanaka N., Rewitz K. F., O’Connor M. B., 2013. Ecdysone control of developmental transitions: lessons from Drosophila research. Annu. Rev. Entomol. 58: 497–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehring W. A., Wheeler D. A., Reddy P., Konopka R. J., Kyriacou C. P., et al. , 1984. P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell 39: 369–376. [DOI] [PubMed] [Google Scholar]
- Zhao L., Saelao P., Jones C. D., Begun D. J., 2014. Origin and spread of de novo genes in Drosophila melanogaster populations. Science 343: 769–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Bachtrog D., 2012. Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science 337: 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Bachtrog D., 2015. Ancestral chromatin configuration constrains chromatin evolution on differentiating sex chromosomes in Drosophila. PLoS Genet. 11: e1005331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Zhu H. M., Huang Q. F., Zhao L., Zhang G. J., et al. , 2012. Deciphering neo-sex and B chromosome evolution by the draft genome of Drosophila albomicans. BMC Genomics 13: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann G., Wang L. P., Vaughan A. G., Manoli D. S., Zhang F., et al. , 2009. Manipulation of an innate escape response in Drosophila: photoexcitation of acj6 neurons induces the escape response. PLoS One 4: e5100. [DOI] [PMC free article] [PubMed] [Google Scholar]