Abstract
FBW7 (F-box and WD repeat domain containing 7), also known as FBXW7 or hCDC4, is a tumor suppressor gene mutated in a broad spectrum of cancer cell types. As a component of the SCF E3 ubiquitin ligase, FBW7 is responsible for specifically recognizing phosphorylated substrates, many important for tumor progression, and targeting them for ubiquitin-mediated degradation. Although the role of FBW7 as a tumor suppressor is well established, less well studied is how FBW7-mutated cancer cells might be targeted for selective killing. To explore this further, we undertook a genome-wide RNAi screen using WT and FBW7 knockout colorectal cell lines and identified the spindle assembly checkpoint (SAC) protein BUBR1, as a candidate synthetic lethal target. We show here that asynchronous FBW7 knockout cells have increased levels of mitotic APC/C substrates and are sensitive to knockdown of not just BUBR1 but BUB1 and MPS1, other known SAC components, suggesting a dependence of these cells on the mitotic checkpoint. Consistent with this dependence, knockdown of BUBR1 in cells lacking FBW7 results in significant cell aneuploidy and increases in p53 levels. The FBW7 substrate cyclin E was necessary for the genetic interaction with BUBR1. In contrast, the establishment of this dependence on the SAC requires the deregulation of multiple substrates of FBW7. Our work suggests that FBW7 knockout cells are vulnerable in their dependence on the mitotic checkpoint and that this may be a good potential target to exploit in FBW7-mutated cancer cells.
Keywords: FBW7, BUBR1, spindle assembly checkpoint, synthetic lethality, RNAi
TUMORIGENESIS is a multistep process that relies on the accumulation of multiple mutations and phenotypes to reach the fully metastatic, cancerous state (Luo et al. 2009; Hanahan and Weinberg 2011). Genes that have roles in a multitude of cellular processes or act as “hubs” are often optimal mutational targets for tumors as their disruption or deregulation may affect various aspects of cell growth and survival. One such pleotropic target is FBW7 (F-box and WD40 containing protein 7), a tumor suppressor known to affect a wide network of signaling pathways involved in cancer progression.
The gene that encodes FBW7 has been shown to have an overall mutation frequency of ∼6% across all human tumor types with high mutation rates in specific cancers including T-ALL and endometrial, bladder, and colorectal cancers (Akhoondi et al. 2007; Davis et al. 2014). Analysis of the FBW7 mutations found in cancer has revealed an unexpectedly high number of single missense mutations. These are concentrated mainly to three “hotspot” arginine residues that lie in the WD40 domain of FBW7 that is responsible for substrate binding (Rajagopalan et al. 2004; Akhoondi et al. 2007; Davis et al. 2014). Although these single nucleotide changes usually occur on only one allele, studies have shown that the mutation can act in a dominant-negative manner on several FBW7 substrates and phenotypes (Akhoondi et al. 2007; Davis et al. 2011; King et al. 2013; Welcker et al. 2013). Additionally, recent data have highlighted the roles of upstream signaling, miRNAs, and promoter hypermethylation in the regulation of FBW7 expression, suggesting the existence of multiple potential mechanisms to downregulate FBW7 activity in cancer (Kimura et al. 2003; Akhoondi et al. 2010; Xu et al. 2010; Wang et al. 2014).
FBW7 is a component of the SCF (SKP1, CUL1, F-box protein) E3 ubiquitin ligase complex. It binds one or more phosphorylated sequences in protein substrates, which targets them for degradation via ubiquitin-mediated proteolysis. Many FBW7 substrates, including cyclin E, c-MYC, c-JUN, NOTCH, NF1, and MCL1, have established roles in oncogenesis (Wang et al. 2012). When FBW7 function is lost, these oncogenic substrates can become deregulated and accumulate in cells. Several experiments with conditional FBW7 alleles in mice have confirmed a role for FBW7 in cancer progression through the deregulation of one or more of these substrates (Wang et al. 2012; King et al. 2013; Davis et al. 2014).
Although the mechanism behind the function of FBW7 as a tumor suppressor has been extensively studied, less well known is how we might target loss or mutation of FBW7 therapeutically. Since many of the substrates of FBW7 are not easily druggable, and as a tumor suppressor gene, loss of FBW7 activity cannot be targeted directly, we chose to use a strategy by which we looked for synthetic lethal partners of FBW7 using RNAi screening in wild-type and FBW7 knockout cell lines. Here, we show that cells lacking FBW7 are sensitive to knockdown of the spindle assembly checkpoint (SAC) protein BUBR1. Furthermore, we provide evidence that FBW7 knockout cells are singularly dependent on the SAC such that after downregulation of the mitotic checkpoint, these cells acquire extensive aneuploidy. Finally, to elucidate how we might leverage this synthetic lethal interaction for potential therapy, we determine whether vulnerability to SAC knockdown is linked to the expression of specific FBW7 substrates.
Materials and Methods
Cell culture
HT-29 and HCT116 wild-type cells were obtained from American Type Culture Collection while HCT116 FBW7 −/− cells were generously provided by Bert Vogelstein. HEK293T cells were provided by Brett Finlay. Cells were cultured in McCoy’s 5A (HCT116, HT29) or DMEM (HEK293T) medium (Life Technologies) supplemented with 10% FBS at 37° and 5% CO2. During lentiviral experiments, 1 μg/ml of puromycin, 20 μg/ml of blasticidin (both from Sigma), or 1 mg/ml of hygromycin B (Roche) were used for selection of cells with the appropriate resistance gene.
Plasmids and shRNAs
A list of the main shRNAs used in these studies and their origin is given in Supporting Information, Table S1. For those shRNAs that required cloning, oligonucleotides were annealed and ligated into the AgeI and EcoRI sites of the plko.1-puro vector. Other drug selections besides puromycin were subcloned into the plko.1 vector using the BamHI and KpnI sites. To express cancer-specific mutations of FBW7 or nondegradable forms of cyclin E and MCL1, pCR4-TOPO-FBW7, and pOTB7-cyclin E were obtained from Open Biosystems while pTOPO-MCL1-S159A was a gift from Ulrich Maurer (Addgene plasmid no. 21606). The genes were cloned into the pENTR4 Gateway entry vector with (for FBW7) or without (for cyclin E and MCL1) an N-terminal FLAG tag. Site-directed mutations were made using primers in Table S2 and the QuikChange II site-directed mutagenesis kit (Agilent). Mutants were then Gateway cloned into destination vectors pLenti PGK Hygro DEST or pLenti PGK Blast DEST. Both entry and destination vectors were gifts from Eric Campeau (Addgene plasmid no. 17423, 19065 and 19066). Lentivirus was made in HEK293T cells using MISSION lentiviral packaging mix (Sigma) and Fugene 6 (Promega) optimized according to manufacturer’s instructions.
Genome-wide shRNA screen
Protocols for genome-wide screening of HCT116 cells have been described previously (Blakely et al. 2011; Marcotte et al. 2012; Vizeacoumar et al. 2013). Briefly, FBW7 +/+ and FBW7 −/− cells were infected with a pool of lentivirus containing ∼80,000 shRNAs targeting ∼16,000 genes that was developed by the RNAi Consortium (Moffat et al. 2006; Root et al. 2006). Infected cells were selected using puromycin and multiplicity of infection for both lines was determined to be between 0.3 and 0.4. Dropout of shRNAs from the pool over time was then determined by growing out cells in nonselective media and pelleting aliquots of cells at time points of 0, 8, and 12 days. Genomic DNA was prepared from cell pellets, PCR amplified, digested, and hybridized onto GMAP arrays (Affymetrix Inc.) as was described previously(Blakely et al. 2011; Vizeacoumar et al. 2013). The shRNA Activity Ranking Profile (shARP) score to determine the dropout rate of each shRNA was calculated as determined previously (Ketela et al. 2011; Vizeacoumar et al. 2013) and the Gene Activity Ranking Profile (GARP) score for each gene was averaged from the two lowest shARP scores. To obtain candidate synthetic lethal genes, the differences between the GARP scores of the FBW7 +/+ and FBW7 −/− lines were calculated and the differentials that fell >3 SD away from the mean were considered as “hits.”
A subset of shRNAs were then further tested by infecting FBW7 +/+ and FBW7 −/− cells in 96-well plates, selecting shRNA-positive cells using puromycin and outgrowing in nonselective media for ∼7 days. Cells were fixed and stained with Hoechst 33342 so nuclei could be counted using a Cellomics ArrayScan VTI. After normalization to the shLUC control, cell number differentials were compared over three replicates per cell line for each experiment using a paired Student’s t-test.
Cell counting and clonogenic assays
For cell proliferation experiments, cell lines were infected with control and experimental shRNAs for 1 day and then were grown for 3 days in media containing puromycin and for 1 day in normal media. Each infection was collected and normalized before plating in 96-well plates for 5–7 days. Cells were then fixed in 3.7% paraformaldehyde and stained with Hoechst 33342 before nuclei were counted using a Cellomics ArrayScan. For clonogenic assays, cells were infected with shRNAs at low MOI in six-well plates (three per shRNA) and selected with puromycin for 3 days before being grown in drug-free media for 10–14 days. Colonies were then stained with 0.1% Cystal violet in 95% ethanol for counting. Cell lines were normalized to control and compared using either a two-tailed (cell proliferation assays) or one-tailed (clonogenic assays) unmatched Student’s t-test.
Immunoblotting and flow cytometry
Cell lines were infected with shRNA and selected and released into drug-free medium. Cells were collected after trypsinization by centrifugation. For immunoblotting, pellets were resuspended in 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 10% glycerol, 1% Triton X-100, and protease inhibitors (Roche). Cells were lysed by sonication and centrifuged to remove debris. Lysates were separated by SDS–PAGE, transferred to PVDF, and blotted with antibodies:
From Abcam: GAPD (ab9485), α-tubulin (ab18251), cyclin E ([HE12], ab3927), MCL1 ([Y37], ab32087), BUB1 (ab54893), MPS1 ([N1], ab11108), Securin ([DCS-280], ab3305).
From BD Biosciences: PARP (556494), BUBR1 (612503).
From Santa Cruz Biotechnology: cyclin B ([GNS1], sc-245), p53 ([DO-1], sc-126).
For flow cytometry, cells pelleted as above were fixed in cold 70% ethanol. Where indicated, cells were first stained with pS10 Histone H3 antibody (Abcam, ab5176) followed by anti-rabbit conjugated to Alexa Fluor 488 (Jackson Laboratories), before being incubated with propidium iodide and RNase A. Cell-cycle analysis was performed using Flow Jo and experiments were repeated two more times. Cell lines were compared using a two-tailed, matched Student t-test.
Abnormal anaphases and mitotic profiles
Cells were grown on coverslips for 3–4 days and fixed with 3.7% paraformaldehyde. Coverslips were then co-immunostained with pS10 Histone H3 antibody and α-tubulin ([DM1A], ab7291) followed by DAPI and viewed on a Zeiss Axioplan 2 Fluorescence microscope using MetaMorph (v7.7) software. Stages of mitosis (prophase, prometaphase, metaphase, anaphase, and telophase) were identified manually. Cell lines were compared using a one-tailed unmatched Student t-test. For abnormal anaphase percentages, both abnormal and normal anaphases were counted and cell lines compared using a two-tailed matched Student t-test.
Data availability
All plasmids constructed for this paper are available upon request. All data from the genome-wide shRNA screen is included in Table S3, Table S4 and Table S5.
Results
A genome-wide RNAi screen identifies a genetic interaction between FBW7 and BUBR1
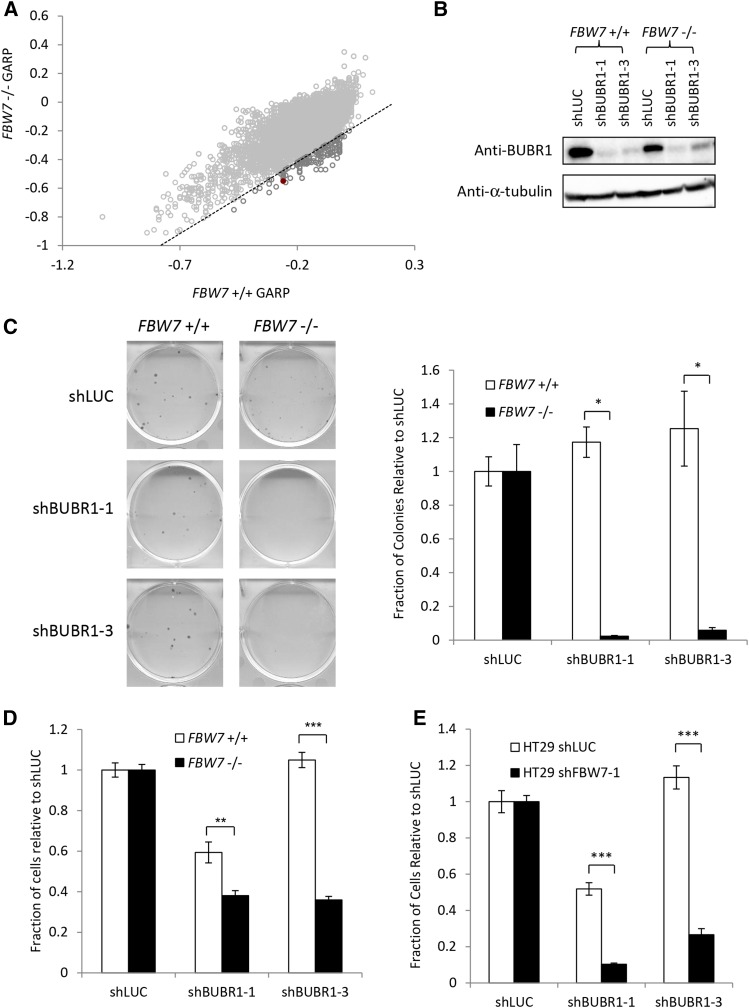
To investigate potential genes to target for selective killing in cells lacking FBW7, we used the HCT116 FBW7 −/− line previously generated by Rajagopalan et al. (2004). This line is homozygous null for FBW7 due to a knockout of exon 8 in both alleles of FBW7 and was originally shown to have increases in cyclin E substrate levels and chromosome instability (CIN) (Rajagopalan et al. 2004). The genome-wide screen was performed using a pooled lentiviral shRNA library representing ∼16,000 human genes (Moffat et al. 2006; Marcotte et al. 2012; Vizeacoumar et al. 2013). This library was then infected into a matched pair of cell lines, HCT116 FBW7 +/+ and HCT116 FBW7 −/− to determine which shRNAs and genes were lost from the pool over time. Comparison of the GARP scores (see Materials and Methods) between these two lines revealed 122 potential negative interactions (Figure 1A, Table S3, Table S4, and Table S5). One of the candidate genes, BUBR1, validated well when a subset of shRNAs were further tested (Figure S1).
Figure 1.
BUBR1 as a synthetic lethal candidate for FBW7 knockout cells. (A) Results from a genome-wide shRNA screen using HCT116 wild-type (FBW7 +/+) and FBW7 knockout (FBW7 −/−) cells. Genes considered as candidate negative genetic interactions are shown in dark gray. The data point representing the BUBR1 gene is shown as a solid red circle. (B) Knockdown of BUBR1 by Western blot after infection with shRNA. (C) Validation of the genetic interaction between FBW7 and BUBR1 using a clonogenic assay. The graph shown on the right shows the average of three replicates. (D) Validation that BUBR1 shRNA is more detrimental to FBW7 −/− cells than FBW7 +/+ cells when compared to a control shRNA (luciferase) in a 96-well nuclei-counting assay. (E) Differential growth after knockdown of BUBR1 in another colorectal cell line (HT29) that was engineered to express either control (luciferase) or FBW7 shRNA. Cells were counted in 96 well after nuclei staining. (*) P < 0.05; (**) P < 0.01; (***) P < 0.0001.
Knockdown of BUBR1 was confirmed as being selectively detrimental to the proliferation of FBW7 −/− cells using independent shRNAs not in the original pooled screen (Figure 1B). Both clonogenic assays and high-content microscopy counting showed a significant decrease in proliferation of FBW7 −/− cells after BUBR1 knockdown when compared with similarly infected FBW7 +/+ cells (Figure 1, C and D). BUBR1 was also shown to be needed to maintain proliferation in another colorectal cell line, HT29, where FBW7 was depleted by shRNA, further establishing a negative genetic interaction between FBW7 and BUBR1 (Figure 1E and Figure S2A).
HCT116 FBW7 −/− cells are dependent on the spindle assembly checkpoint
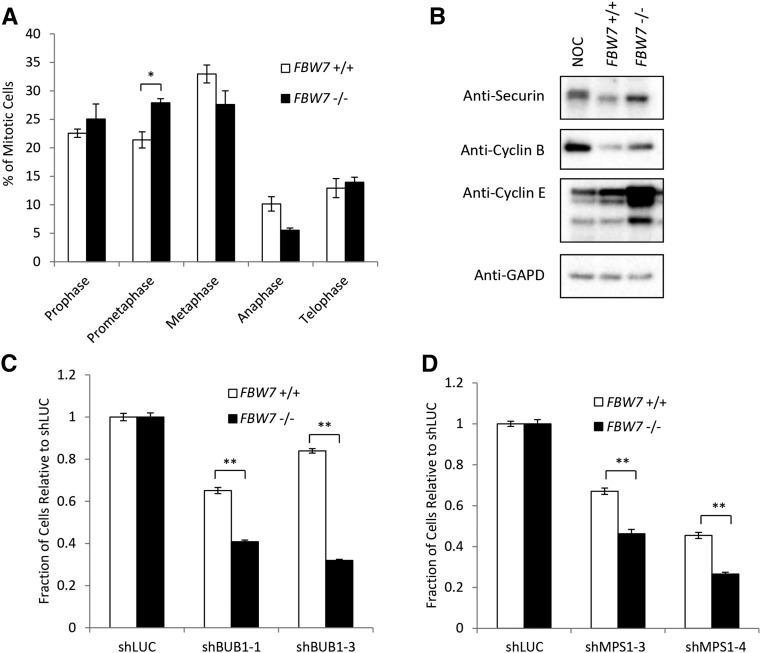
The most-well-known role of BUBR1 is as a component of the mitotic SAC, which, when activated, prevents cells from entering anaphase by blocking the activity of the APC/CCDC20 ubiquitin ligase (Jia et al. 2013). Consistent with this role in maintaining proper chromosome segregation, HCT116 FBW7 −/− mitotic cells have difficulty aligning their chromosomes at the metaphase plate, which leads to increased segregation errors (Rajagopalan et al. 2004). We similarly observed a higher percentage of mitotic cells in prometaphase in FBW7 −/− cells when compared with FBW7 +/+ cells (Figure 2A). FBW7 −/− cells also had an increase in the levels of the APC/C substrates cyclin B and Securin (Figure 2B). This strongly indicates that cells that lack FBW7 depend on an intact SAC.
Figure 2.
FBW7 knockout cells are more sensitive to knockdown of the SAC. (A) Mitotic profiles of FBW7 wild-type and knockout cells expressed as a percentage of the total number of cells in mitosis. (B) Levels of the APC/C targets securin and cyclin B in FBW7 wild-type and knockout cells. (NOC, FBW7 +/+ cells arrested with nocodazole). (C and D) Differential growth between FBW7 +/+ and FBW7 −/− cells using a 96-well nuclei counting assay after infection with shRNA to the SAC components BUB1 (D) and MPS1 (E). (*) P < 0.05; (**) P < 0.001.
To confirm this, we used shRNA to knock down two other proteins with known roles in the SAC, BUB1, and MPS1 (also called TTK). Knockdown of either of these components caused a significant decrease in cell growth in FBW7 −/− cells when compared with FBW7 +/+ cells (Figure 2, C and D, and Figure S2, B and C). We also found that cells depleted of FBW7 showed little or no resistance to low levels of the spindle poisons nocodazole or paclitaxel, which activate the SAC (Figure S2, D and E). Taken into consideration with previous studies that have shown that FBW7 activity is needed for cell sensitivity to high levels of anti-tubulin chemotherapeutics (Finkin et al. 2008; Wertz et al. 2011), we propose knockdown of the SAC as an alternative anti-mitotic strategy to traditional chemotherapeutics targeting the spindle.
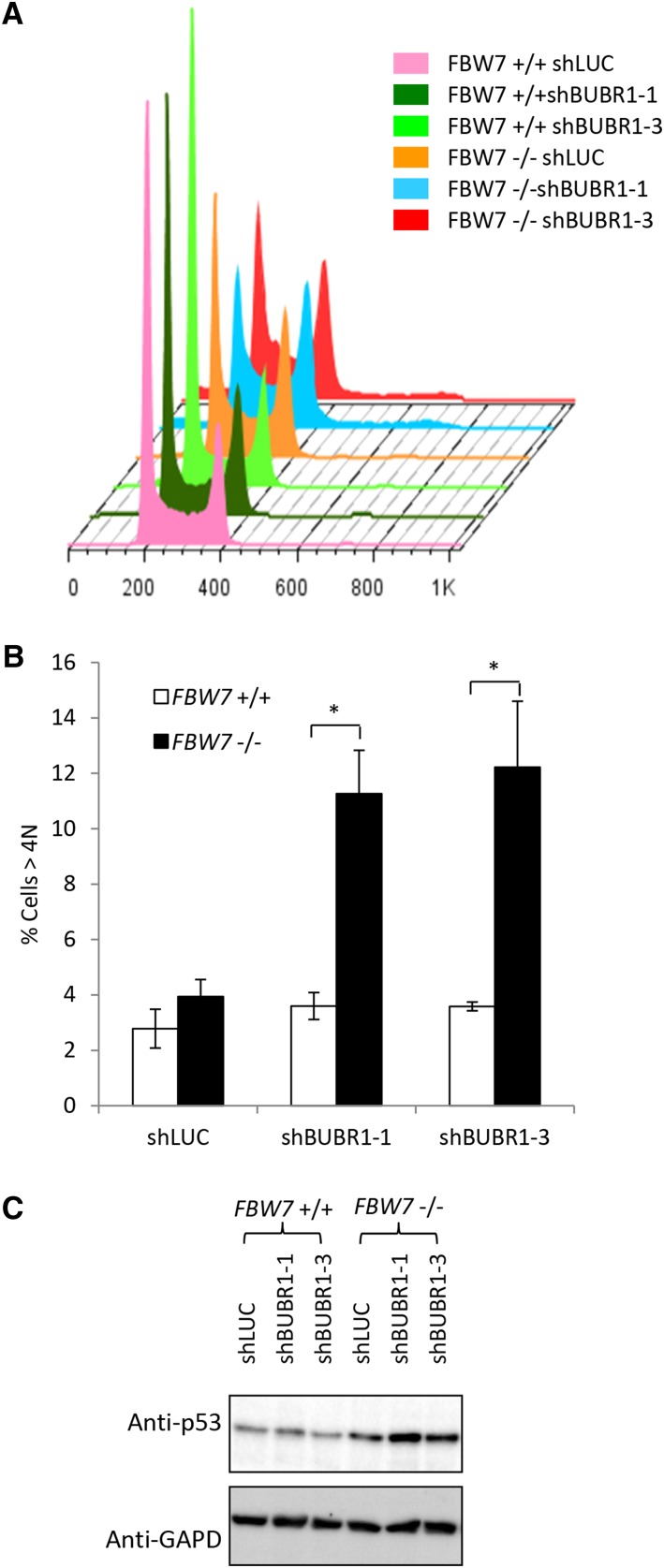
BUBR1 knockdown increases aneuploidy in cells lacking FBW7
To determine the consequences of decreasing SAC function in cells lacking FBW7, we compared the cell-cycle profiles of FBW7 +/+ and FBW7 −/− cells after knockdown of BUBR1 with shRNA (Figure 3A and Figure S3A). FBW7 −/− cells infected with BUBR1 shRNA showed a more than twofold increase in the >4 N DNA over BUBR1 shRNA-infected FBW7 +/+ cells (Figure 3B). Western blotting of lysates after BUBR1 knockdown showed increases in the level of p53 (Figure 3C), but no cleavage of the apoptotic marker PARP (Figure S3B), suggesting that the decrease in cell proliferation is not due to apoptotic cell death. Furthermore, staining of the cells with pH 3 (a marker for mitosis) showed that, as has been shown, knockdown of the SAC with BUBR1 shRNA decreased the percentage of cells in mitosis (Moffat et al. 2006) (Figure S3C). This analysis implies that after knockdown of BUBR1, cells lacking FBW7 do not delay in mitosis at the SAC, but become highly aneuploid and arrest in a p53-dependent manner.
Figure 3.
Knockdown of the SAC increases aneuploidy in FBW7 −/− cells. (A) Flow cytometry profiles of FBW7 +/+ and FBW7 −/− cells stained with propidium iodide and infected with BUBR1 shRNA. (B) Quantification of the >4 N cells as an average over three independent experiments. (C) Western blot of p53 levels after knockdown of BUBR1. (*) P < 0.05.
FBW7 mutations found in cancer display several dominant-negative phenotypes
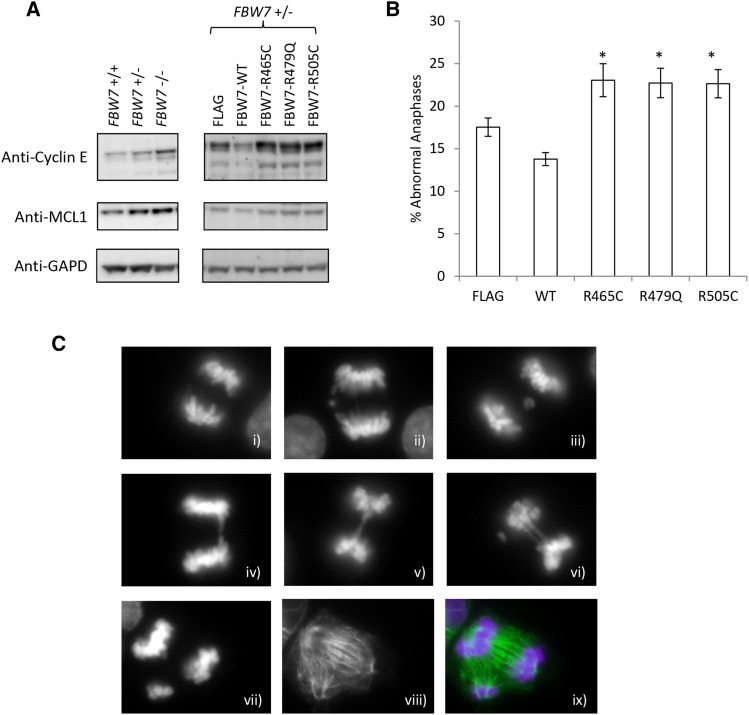
Mutations observed in FBW7 in cancer are often single allele, missense mutations (Davis et al. 2014). To determine how these mutations affect FBW7 substrate levels and phenotypes, we expressed several common mutations (R465C, R479Q, R505C) in HCT116 cells and compared these cells to parental cells expressing vector alone. These three target arginine residues are responsible for the coordination of the substrate phosphate moiety in the binding pocket of FBW7 and, when combined, have a higher frequency of mutation compared with the rest of the FBW7 protein (Rajagopalan et al. 2004; Davis et al. 2014). We determined that R465C, R479Q, and R505C all displayed several dominant-negative phenotypes when expressed in a heterozygous FBW7 background. These phenotypes included changes in cyclin E and MCL1 substrate expression as well as anaphase bridges (Figure 4 and Figure S4A). In contrast, in a homozygous null FBW7 background these mutations showed the same phenotypes as the vector control Figure S4, B–D). We also observed that after BUBR1 knockdown, there was a decrease in colony number when cells were infected with cancer-specific mutations as compared to those infected with wild-type FBW7 (Figure S4E).
Figure 4.
FBW7 cancer-specific arginine mutants have a dominant-negative phenotype. (A) Western blot of cyclin E and MCL1 levels in FBW7 wild-type, FBW7 heterozygous and FBW7 homozygous knockout HCT116 cells and FBW7 heterozygous knockout HCT116 cells with FBW7 wild-type and arginine mutations expressed. (B) Abnormal anaphases (expressed as a percentage of total anaphases) in asynchronous FBW7 +/− cells expressing wild-type FBW7 and FBW7 arginine mutations. (C) Examples of abnormal anaphases found in HCT116 cells include: normal anaphases (i), lagging chromosomes (ii and iii), anaphase bridges (iv and v), and anaphases containing multiple phenotypes (vi). Multipolar anaphases were also occasionally observed as slides were costained with DAPI (vii) and an antibody for α-tubulin (viii). (ix) Merged image. (*) P < 0.05.
The FBW7 substrate cyclin E is necessary for sensitivity to SAC knockdown
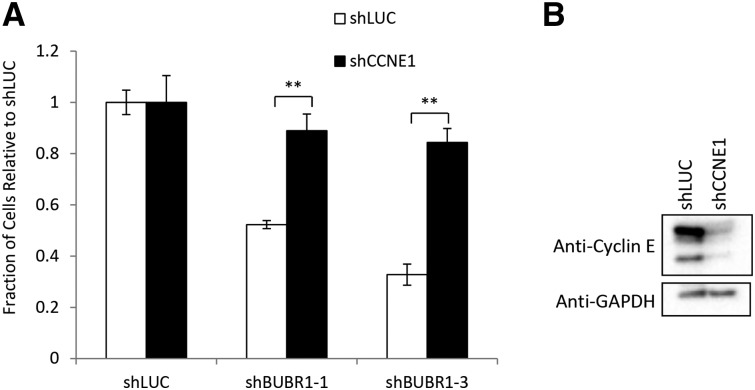
FBW7 has several substrates that are involved in mitosis that could potentially activate the SAC. In particular, we wanted to investigate the function of the cell-cycle regulator cyclin E in the genetic interaction between FBW7 and BUBR1 given that the CIN phenotype in FBW7 knockout HCT116 cells was previously associated with the deregulation of this FBW7-associated substrate (Rajagopalan et al. 2004). We also wanted to look at the role of the survival protein MCL1, which is degraded by FBW7 only after the SAC is activated and, unlike cyclin E, should not be needed for sensitivity to SAC knockdown (Inuzuka et al. 2011; Wertz et al. 2011). In accordance with this, we found that high levels of cyclin E were necessary for the decrease in proliferation of FBW7 −/− cells after BUBR1 knockdown (Figure 5). In contrast, knockdown of MCL1 did not show any rescue of the BUBR1/FBW7 genetic interaction (Figure S5, A and B) suggesting that high levels of cyclin E are needed for the genetic interaction between FBW7 and BUBR1 in HCT116 cells.
Figure 5.
Cyclin E is necessary for sensitivity of FBW7 knockout cells to knockdown of the SAC. (A) FBW7 knockout cells infected with BUBR1 shRNA in a 96-well nuclei-counting assay after knockdown of cyclin E1 (CCNE1). (B) Western blot confirming knockdown of cyclin E after shRNA infection. (**) P < 0.001.
Overexpression of both truncated cyclin E and MCL1 are required for establishing sensitivity to BUBR1 knockdown
Similar to the experiments described above, we also wanted to investigate how overexpression of FBW7 substrates could result in a dependency on BUBR1 and the SAC. High levels of cyclin E have previously been shown to increase genome instability and mitotic defects implicating it as a likely candidate in establishing vulnerability to the mitotic checkpoint (Spruck et al. 1999; Rajagopalan et al. 2004; Minella et al. 2007). Along with a full-length nondegradable cyclin E, we generated an N terminally truncated cyclin E, termed Δ65, for expression in HCT116 cells. Studies have shown that this version of cyclin E is similar to one of the post-translationally processed low-molecular-weight cyclin E isoforms that occur endogenously in cell lines like HCT116 (Porter et al. 2001; Corin et al. 2006). Low-molecular-weight cyclin E isoforms have not been seen in normal cells, but are seen in several types of cancer including colorectal, breast, ovarian, and bladder cancer where they are often correlated with poor survival (Porter et al. 2001; Akli et al. 2004; Bales et al. 2005; Corin et al. 2006; Davidson et al. 2007; Zhou et al. 2011; Akli et al. 2012). Cyclin E isoforms like Δ65 have been shown to cause hyperactive CDK activity and their overexpression in cells can lead to faster cell-cycle progression, increased centrosome amplification, and CIN (Porter et al. 2001; Akli et al. 2004; Bagheri-Yarmand et al. 2010).
In addition to both full-length and truncated cyclin E, we also overexpressed nondegradable MCL1. Although this protein is not known to be involved in activation of the mitotic checkpoint, it is important for cell survival in response to chemically induced mitotic stress and SAC activation (Inuzuka et al. 2011; Wertz et al. 2011). We were therefore interested in how MCL1 could affect the mitotic phenotypes associated with the overexpression of cyclin E, an issue that has remained unexplored.
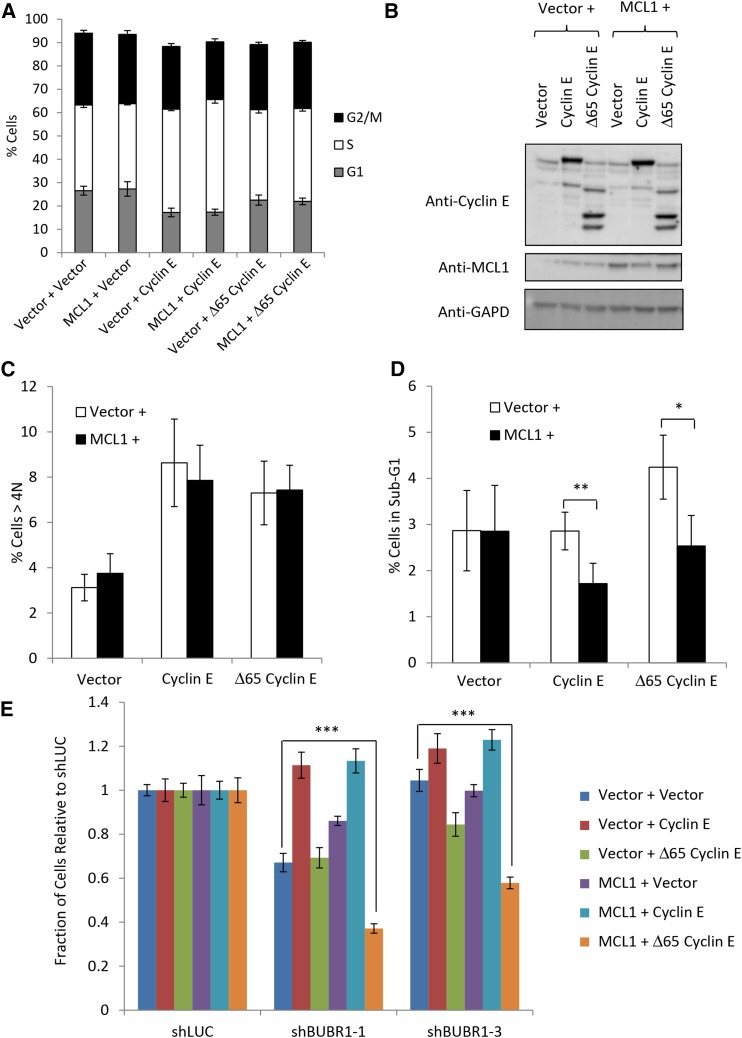
Cells infected to express full-length nondegradable cyclin E produced cell lines that grew much more slowly than wild type and had a higher percentage of cells in S-phase (Figure 6, A and B, and Figure S5C). This defect in cell-cycle progression is consistent with what has been reported previously (Spruck et al. 1999). Interestingly, expression of Δ65 cyclin E resulted in cell-cycle profiles similar to vector alone cell lines (Figure 6A and Figure S5C) suggesting that LMW cyclin E did not perturb the cell cycle to the extent of full-length cyclin E. Both forms of cyclin E increased the percentage of cells with >4 N DNA (Figure 6C) consistent with previous studies that cyclin E overexpression can cause CIN (Spruck et al. 1999; Akli et al. 2004; Bagheri-Yarmand et al. 2010). Unexpectedly, coexpression of MCL1 with cyclin E isoforms did not significantly increase CIN as measured by flow cytometry. It did, however, decrease the fraction of sub-G1 cells (Figure 6D) suggesting that after overexpression of cyclin E, MCL1 has a role in cell survival in this system.
Figure 6.
Establishment of sensitivity to SAC knockdown in FBW7 knockout cells requires multiple substrates. (A) Percentages of G1-, S-, and G2/M-phase cells in HCT116 cells overexpressing nondegradable forms of cyclin E and MCL1. (B) Western blot of HCT116 colorectal cell lines overexpressing nondegradable forms of MCL1 and full-length cyclin E or cyclin E lacking the N-terminal 65 residues (Δ65 cyclin E). (C) Percentages of >4 N cells in three independent experiments. (D) Percentages of cells in the sub-G1 range in three independent experiments. (E) HCT116 cells show sensitivity to BUBR1 knockdown only after expression of MCL1 and the Δ65 form of cyclin E. (*) P < 0.05; (**) P < 0.005; (***) P < 0.0005.
Knockdown of the SAC using BUBR1 shRNA in these cell lines showed that overexpression of cyclin E in either form was not sufficient to establish dependence of HCT116 cells on the SAC (Figure 6E). Instead, overexpression of both Δ65 and MCL1 was required for sensitivity to knockdown of BUBR1. Exactly why the Δ65 isoform, but not full-length cyclin E, was needed to establish sensitivity to BUBR1 knockdown is not known. It is possible that either the slow growth and cell-cycle perturbation of the full-length cyclin E-expressing lines or the consistently lower protein levels of MCL1 in the lines coexpressing full-length cyclin E and MCL1 could be a factor. Together, the results from these cell lines illustrate the complexity that can underlie the mechanisms of CIN in cells and emphasize the need for a better understanding of these mechanisms if we are to target them therapeutically.
Discussion
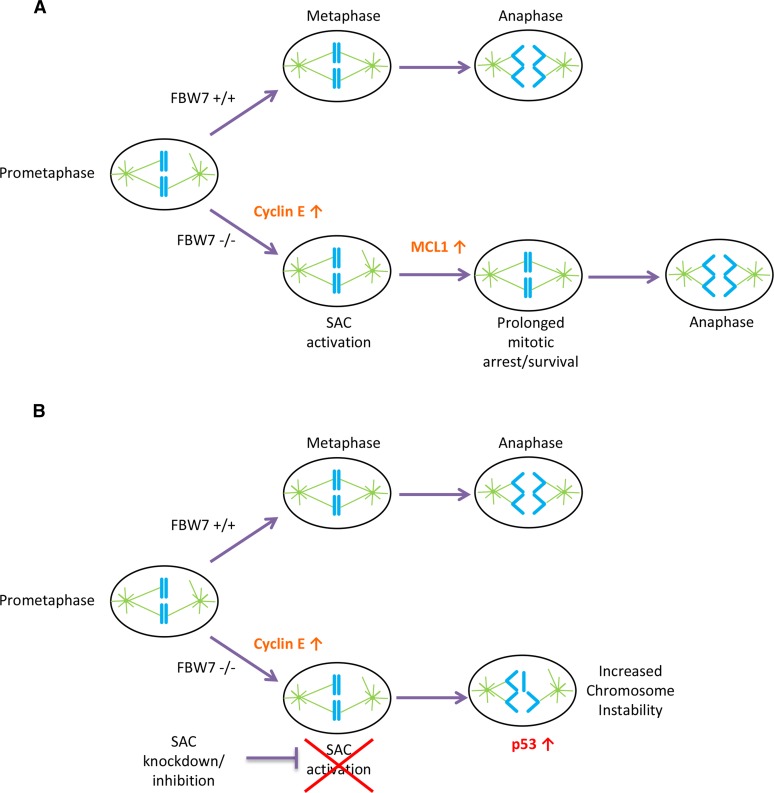
Synthetic lethality has been proposed as a therapeutic strategy for treating cancer. An advantage of this approach is that the synthetic lethal drug target is distinct from the somatically mutated cancer gene product, which itself may be undruggable. In this article, we use this strategy to identify potential synthetic lethal candidates for FBW7, a currently undruggable tumor suppressor gene with several oncogenic substrates that are similarly difficult to target. We identified the gene BUBR1 as a synthetic lethal partner and found that FBW7 knockout cells were vulnerable because of a dependence of these cells on the mitotic checkpoint. Cells that lacked FBW7 and the mitotic checkpoint had not only decreased proliferation but also a significant increase in aneuploidy likely due to premature anaphase entry (Figure 7).
Figure 7.
A model for SAC dependence in cells lacking FBW7. (A) FBW7 +/+ cells in prometaphase efficiently align their chromosomes and correctly segregate properly in anaphase with less reliance on the SAC. FBW7 −/− cells have an increase in cyclin E, which causes problems in mitosis and may lead to SAC activation. FBW7 −/− cells can survive prolonged SAC activation in part because of stabilization of MCL1 allowing more time for chromosome alignment. (B) A decrease in SAC activation by knockdown of BUBR1 may significantly shorten the time available for chromosome alignment in FBW7 −/− cells, resulting in improper chromosome segregation and intolerable levels of chromosome instability.
FBW7 is functionally pleiotropic and its knockout in cells can result in several different observable phenotypes including increased CIN (Rajagopalan et al. 2004). Interestingly, our model presented in Figure 7 demonstrates that targeting the CIN phenotype in FBW7 −/− cells is a viable synthetic lethal strategy. This is consistent with the hypothesis that there is an optimal or permissible level of CIN that can be tolerated in tumor cells but is not found in normal cells (Komarova and Wodarz 2004; Bakhoum and Compton 2012; Janssen and Medema 2013). It follows then that it may be possible to target the cellular stress of CIN therapeutically as it represents a cancer-specific vulnerability (Luo et al. 2009; Bakhoum and Compton 2012). Support for this hypothesis has been seen in genetic studies in mice where there appears to be a threshold of survivable CIN, but levels of instability beyond this threshold are deleterious to tumor formation (Janssen and Medema 2013; Silk et al. 2013). Our experiments suggest that the stress of CIN in HCT116 FBW7 knockout cells can also be exploited using this strategy.
The increase in aneuploidy in FBW7 knockout cells after BUBR1 knockdown was also accompanied by an increase in p53, a result consistent with the role of p53 in cell-cycle arrest after defects in mitosis (Uetake and Sluder 2010; Thompson and Compton 2010; Orth et al. 2012). Studies have shown that cells without p53 can continue cycling even after accumulating aneuploidy, suggesting that the presence of p53 may help to eliminate cells with high aneuploidy or CIN (Thompson and Compton 2010; Silk et al. 2013). The molecular relationship between FBW7 and p53 is complex as FBW7 lies both upstream and downstream of p53 signaling pathways (Kimura et al. 2003; Finkin et al. 2008). Studies in mice, however, show that FBW7 and p53 mutations are cooperative in tumorigenesis and together these proteins help restrain cyclin E genome instability in cells (Mao et al. 2004; Minella et al. 2007; Grim et al. 2012). In contrast, a previous analysis of mutations in gastric cancer found no cooperation between FBW7 and p53 (Akhoondi et al. 2007). Furthermore, using TCGA data available in cBioPortal (Cerami et al. 2012; Gao et al. 2013), we also did not find a significant co-occurrence between FBW7 and p53 in two cancer types highly mutated for FBW7: colorectal adenocarcinoma and uterine endometrial carcinoma, although the two mutations are seen together. Nevertheless, p53 status in FBW7 tumors should be considered before using SAC inhibition as a selective killing strategy in FBW7-mutated cells, since p53 status has been shown to be important in surviving high levels CIN.
FBW7 −/− cells are vulnerable to knockdown not just of BUBR1, but also to two other members of the SAC, BUB1, and MPS1 (Figure 2, C and D). These cells show a higher percentage of cells in prometaphase and higher levels of APC/C substrates when compared with wild-type cells (Figure 2, A and B) suggesting that FBW7 knockout cells may be more dependent on SAC function. In cancer, SAC mutations are rare, but upregulation of SAC proteins and mRNA are seen and may associate with CIN (Carter et al. 2006; Yuan et al. 2006; Janssen and Medema 2013), suggesting that the requirement of an intact mitotic checkpoint is not limited to FBW7 knockout cells.
Traditional antimitotic therapies such as paclitaxel, which interferes with the normal depolymerization of microtubules, focus on stressing or overwhelming the mitotic checkpoint. But targeting the microtubules in cells can have significant side effects even in nonproliferating cells (Luo et al. 2009; Chan et al. 2012). In contrast, inhibition of the SAC would prevent cancer cells from coping with endogenous or induced mitotic stress. This inhibition of the SAC has previously been proposed by Janssen et al. (2009) as a strategy to target cancer cell lines after they found that knockdown of SAC components made cells more sensitive to low doses of paclitaxel. We are proposing a similar strategy here as our data show that the knockdown of SAC components in FBW7 −/− cells, which are intrinsically mitotically stressed, causes a significant decrease in cell proliferation when compared to unstressed FBW7 +/+ cells.
We determined in our experiments that cyclin E was required for cells lacking FBW7 to be sensitized to SAC knockdown (Figure 5). High or dysregulated levels of cyclin E are known to cause mitotic defects such as centrosome amplification and increases in chromosome breaks and translocations (Hinchcliffe et al. 1999; Lacey et al. 1999; Loeb et al. 2005; Bagheri-Yarmand et al. 2010). Notably, cells overexpressing cyclin E can also inhibit APC/CCdh1 activity and increase the APC/C substrates cyclin B1 and Securin (Keck et al. 2007). These cells have increases in the number of cells in prometaphase and unaligned metaphase due to a mitotic delay as chromosomes misalign at the metaphase plate (Keck et al. 2007). Consistent with their ability to dysregulate cyclin E, FBW7 −/− cells show many similar phenotypes to cyclin E-overexpressing cells. They have been shown previously to have increased centrosome number (Rajagopalan et al. 2004). As well, up to 30% of HCT116 FBW7 −/− cells do not align their chromosomes at the metaphase plate during mitosis (Rajagopalan et al. 2004). We also show here that FBW7 −/− cells have an increase in the number of cells in prometaphase and higher levels of APC/C substrates when compared with FBW7 +/+ cells (Figure 2). This suggests that defects in cyclin E levels could directly contribute to a prolonged metaphase arrest and SAC activation in FBW7 −/− cells.
As shown in Figure 6E, the establishment of SAC sensitivity in wild-type cells required the overexpression of both truncated cyclin E and the prosurvival factor MCL1. This kind of analysis is important for multifunctional tumor suppressors like FBW7 as not all functions associated with the protein may be relevant across all tumor types. In fact, work by Ekholm-Reed et al. (2004) has suggested that while FBW7 mutation can deregulate cyclin E in endometrial tumors, this does not necessarily lead to high cyclin E levels; whether this is due to a more recently identified feedback loop or some other mechanism is unknown (Xu et al. 2010). Similarly, studies in colorectal tumors suggest that the CIN phenotype of FBW7 knockout cells is found in some, but not all, FBW7-mutated tumors (Rajagopalan et al. 2004; Kemp et al. 2005). This suggests that further stratification of FBW7-mutated cells may be warranted when considering SAC inhibition as a potential therapy. As well, the effect of other FBW7 substrates, which converge on the SAC including the crucial mitotic kinases Aurora A and Aurora B, and the regulator of chromatid cohesion Jun B, may require further study (Teng et al. 2012; Perez-Benavente et al. 2013).
Using RNAi screening in isogenic cell lines either wild-type or homozygously deleted for FBW7, a highly mutated tumor suppressor, we uncovered a novel genetic interaction between FBW7 and BUBR1. By characterizing this interaction further, we found that cells lacking FBW7 are dependent on the mitotic checkpoint and prone to intolerable levels of aneuploidy after checkpoint downregulation. Our data indicate that, in the future, the threshold of CIN in cancer cells lacking FBW7 should be further investigated as a potential vulnerability for therapeutics.
Supplementary Material
Acknowledgments
We thank Bert Vogelstein for FBW7 HCT116 knockout cell lines and Hong Bing Yu and Brett Finlay for help setting up lentivirus protocols and HEK293T cells. We also thank Kevin Brown and Dahlia Kasimer for technical assistance and the Ontario Research Fund GL2 program. This work was funded by grants from the Canadian Institutes of Health Research (MOP 38096) and the National Institutes of Health (R01CA158162) to P.H. and a Michael Smith Foundation for Health Research post-doctoral fellowship to M.L.B. P.H. and J.M. are Senior Fellows in the Genetics Networks program at the Canadian Institute for Advanced Research. J.M. holds a Canada Research Chair in Functional Genomics of Cancer. The authors declare no conflict of interest.
Footnotes
Communicating editor: S. K. Sharan
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.180653/-/DC1.
Literature Cited
- Akhoondi S., Sun D., von der Lehr N., Apostolidou S., Klotz K., et al. , 2007. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 67: 9006–9012. [DOI] [PubMed] [Google Scholar]
- Akhoondi S., Lindstrom L., Widschwendter M., Corcoran M., Bergh J., et al. , 2010. Inactivation of FBXW7/hCDC4-beta expression by promoter hypermethylation is associated with favorable prognosis in primary breast cancer. Breast Cancer Res. 12: R105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akli S., Zheng P. J., Multani A. S., Wingate H. F., Pathak S., et al. , 2004. Tumor-specific low molecular weight forms of cyclin E induce genomic instability and resistance to p21, p27, and antiestrogens in breast cancer. Cancer Res. 64: 3198–3208. [DOI] [PubMed] [Google Scholar]
- Akli S., Zhang X. Q., Bondaruk J., Tucker S. L., Czerniak P. B., et al. , 2012. Low molecular weight cyclin E is associated with p27-resistant, high-grade, high-stage and invasive bladder cancer. Cell Cycle 11: 1468–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri-Yarmand R., Biernacka A., Hunt K. K., Keyomarsi K., 2010. Low molecular weight cyclin E overexpression shortens mitosis, leading to chromosome missegregation and centrosome amplification. Cancer Res. 70: 5074–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum S. F., Compton D. A., 2012. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J. Clin. Invest. 122: 1138–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bales E., Mills L., Milam N., McGahren-Murray M., Bandyopadhyay D., et al. , 2005. The low molecular weight cyclin E isoforms augment angiogenesis and metastasis of human melanoma cells in vivo. Cancer Res. 65: 692–697. [PubMed] [Google Scholar]
- Blakely K., Ketela T., Moffat J., 2011. Pooled lentiviral shRNA screening for functional genomics in mammalian cells. Methods Mol. Biol. 781: 161–182. [DOI] [PubMed] [Google Scholar]
- Carter S. L., Eklund A. C., Kohane I. S., Harris L. N., Szallasi Z., 2006. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 38: 1043–1048. [DOI] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., et al. , 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. S., Koh C. G., Li H. Y., 2012. Mitosis-targeted anti-cancer therapies: where they stand. Cell Death Dis. 3: e411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corin I., Di Giacomo M. C., Lastella P., Bagnulo R., Guanti G., et al. , 2006. Tumor-specific hyperactive low-molecular-weight cyclin E isoforms detection and characterization in non-metastatic colorectal tumors. Cancer Biol. Ther. 5: 198–203. [DOI] [PubMed] [Google Scholar]
- Davidson B., Skrede M., Silins I., Shih I., Trope C. G., et al. , 2007. Low-molecular weight forms of cyclin E differentiate ovarian carcinoma from cells of mesothelial origin and are associated with poor survival in ovarian carcinoma. Cancer 110: 1264–1271. [DOI] [PubMed] [Google Scholar]
- Davis H., Lewis A., Spencer-Dene B., Tateossian H., Stamp G., et al. , 2011. FBXW7 mutations typically found in human cancers are distinct from null alleles and disrupt lung development. J. Pathol. 224: 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. J., Welcker M., Clurman B. E., 2014. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell 26: 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekholm-Reed S., Spruck C. H., Sangfelt O., van Drogen F., Mueller-Holzner E., et al. , 2004. Mutation of hCDC4 leads to cell cycle deregulation of cyclin E in cancer. Cancer Res. 64: 795–800. [DOI] [PubMed] [Google Scholar]
- Finkin S., Aylon Y., Anzi S., Oren M., Shaulian E., 2008. Fbw7 regulates the activity of endoreduplication mediators and the p53 pathway to prevent drug-induced polyploidy. Oncogene 27: 4411–4421. [DOI] [PubMed] [Google Scholar]
- Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., et al. , 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grim J. E., Knoblaugh S. E., Guthrie K. A., Hagar A., Swanger J., et al. , 2012. Fbw7 and p53 cooperatively suppress advanced and chromosomally unstable intestinal cancer. Mol. Cell. Biol. 32: 2160–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A., 2011. Hallmarks of cancer: the next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E. H., Li C., Thompson E. A., Maller J. L., Sluder G., 1999. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science 283: 851–854. [DOI] [PubMed] [Google Scholar]
- Inuzuka H., Shaik S., Onoyama I., Gao D., Tseng A., et al. , 2011. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature 471: 104–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A., Medema R. H., 2013. Genetic instability: tipping the balance. Oncogene 32: 4459–4470. [DOI] [PubMed] [Google Scholar]
- Janssen A., Kops G. J., Medema R. H., 2009. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc. Natl. Acad. Sci. USA 106: 19108–19113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L., Kim S., Yu H., 2013. Tracking spindle checkpoint signals from kinetochores to APC/C. Trends Biochem. Sci. 38: 302–311. [DOI] [PubMed] [Google Scholar]
- Keck J. M., Summers M. K., Tedesco D., Ekholm-Reed S., Chuang L. C., et al. , 2007. Cyclin E overexpression impairs progression through mitosis by inhibiting APC(Cdh1). J. Cell Biol. 178: 371–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp Z., Rowan A., Chambers W., Wortham N., Halford S., et al. , 2005. CDC4 mutations occur in a subset of colorectal cancers but are not predicted to cause loss of function and are not associated with chromosomal instability. Cancer Res. 65: 11361–11366. [DOI] [PubMed] [Google Scholar]
- Ketela T., Heisler L. E., Brown K. R., Ammar R., Kasimer D., et al. , 2011. A comprehensive platform for highly multiplexed mammalian functional genetic screens. BMC Genomics 12: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Gotoh M., Nakamura Y., Arakawa H., 2003. hCDC4b, a regulator of cyclin E, as a direct transcriptional target of p53. Cancer Sci. 94: 431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King B., Trimarchi T., Reavie L., Xu L., Mullenders J., et al. , 2013. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 153: 1552–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova N. L., Wodarz D., 2004. The optimal rate of chromosome loss for the inactivation of tumor suppressor genes in cancer. Proc. Natl. Acad. Sci. USA 101: 7017–7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey K. R., Jackson P. K., Stearns T., 1999. Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. USA 96: 2817–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb K. R., Kostner H., Firpo E., Norwood T., Tsuchiya K. D., et al. , 2005. A mouse model for cyclin E-dependent genetic instability and tumorigenesis. Cancer Cell 8: 35–47. [DOI] [PubMed] [Google Scholar]
- Luo J., Solimini N. L., Elledge S. J., 2009. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136: 823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J. H., Perez-Losada J., Wu D., Delrosario R., Tsunematsu R., et al. , 2004. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature 432: 775–779. [DOI] [PubMed] [Google Scholar]
- Marcotte R., Brown K. R., Suarez F., Sayad A., Karamboulas K., et al. , 2012. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2: 172–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minella A. C., Grim J. E., Welcker M., Clurman B. E., 2007. p53 and SCFFbw7 cooperatively restrain cyclin E-associated genome instability. Oncogene 26: 6948–6953. [DOI] [PubMed] [Google Scholar]
- Moffat J., Grueneberg D. A., Yang X., Kim S. Y., Kloepfer A. M., et al. , 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124: 1283–1298. [DOI] [PubMed] [Google Scholar]
- Orth J. D., Loewer A., Lahav G., Mitchison T. J., 2012. Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction. Mol. Biol. Cell 23: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Benavente B., Garcia J. L., Rodriguez M. S., Pineda-Lucena A., Piechaczyk M., et al. , 2013. GSK3-SCF(FBXW7) targets JunB for degradation in G2 to preserve chromatid cohesion before anaphase. Oncogene 32: 2189–2199. [DOI] [PubMed] [Google Scholar]
- Porter D. C., Zhang N., Danes C., McGahren M. J., Harwell R. M., et al. , 2001. Tumor-specific proteolytic processing of cyclin E generates hyperactive lower-molecular-weight forms. Mol. Cell. Biol. 21: 6254–6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan H., Jallepalli P. V., Rago C., Velculescu V. E., Kinzler K. W., et al. , 2004. Inactivation of hCDC4 can cause chromosomal instability. Nature 428: 77–81. [DOI] [PubMed] [Google Scholar]
- Root D. E., Hacohen N., Hahn W. C., Lander E. S., Sabatini D. M., 2006. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat. Methods 3: 715–719. [DOI] [PubMed] [Google Scholar]
- Silk A. D., Zasadil L. M., Holland A. J., Vitre B., Cleveland D. W., et al. , 2013. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc. Natl. Acad. Sci. USA 110: E4134–E4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruck C. H., Won K. A., Reed S. I., 1999. Deregulated cyclin E induces chromosome instability. Nature 401: 297–300. [DOI] [PubMed] [Google Scholar]
- Teng C. L., Hsieh Y. C., Phan L., Shin J., Gully C., et al. , 2012. FBXW7 is involved in Aurora B degradation. Cell Cycle 11: 4059–4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S. L., Compton D. A., 2010. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J. Cell Biol. 188: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y., Sluder G., 2010. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr. Biol. 20: 1666–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizeacoumar F. J., Arnold R., Vizeacoumar F. S., Chandrashekhar M., Buzina A., et al. , 2013. A negative genetic interaction map in isogenic cancer cell lines reveals cancer cell vulnerabilities. Mol. Syst. Biol. 9: 696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Ye X., Liu Y., Wei W., Wang Z., 2014. Aberrant regulation of FBW7 in cancer. Oncotarget 5: 2000–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Inuzuka H., Zhong J., Wan L., Fukushima H., et al. , 2012. Tumor suppressor functions of FBW7 in cancer development and progression. FEBS Lett. 586: 1409–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M., Larimore E. A., Swanger J., Bengoechea-Alonso M. T., Grim J. E., et al. , 2013. Fbw7 dimerization determines the specificity and robustness of substrate degradation. Genes Dev. 27: 2531–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz I. E., Kusam S., Lam C., Okamoto T., Sandoval W., et al. , 2011. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature 471: 110–114. [DOI] [PubMed] [Google Scholar]
- Xu Y., Sengupta T., Kukreja L., Minella A. C., 2010. MicroRNA-223 regulates cyclin E activity by modulating expression of F-box and WD-40 domain protein 7. J. Biol. Chem. 285: 34439–34446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B., Xu Y., Woo J. H., Wang Y., Bae Y. K., et al. , 2006. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin. Cancer Res. 12: 405–410. [DOI] [PubMed] [Google Scholar]
- Zhou Y. J., Xie Y. T., Gu J., Yan L., Guan G. X., et al. , 2011. Overexpression of cyclin E isoforms correlates with poor prognosis in rectal cancer. Eur. J. Surg. Oncol. 37: 1078–1084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All plasmids constructed for this paper are available upon request. All data from the genome-wide shRNA screen is included in Table S3, Table S4 and Table S5.