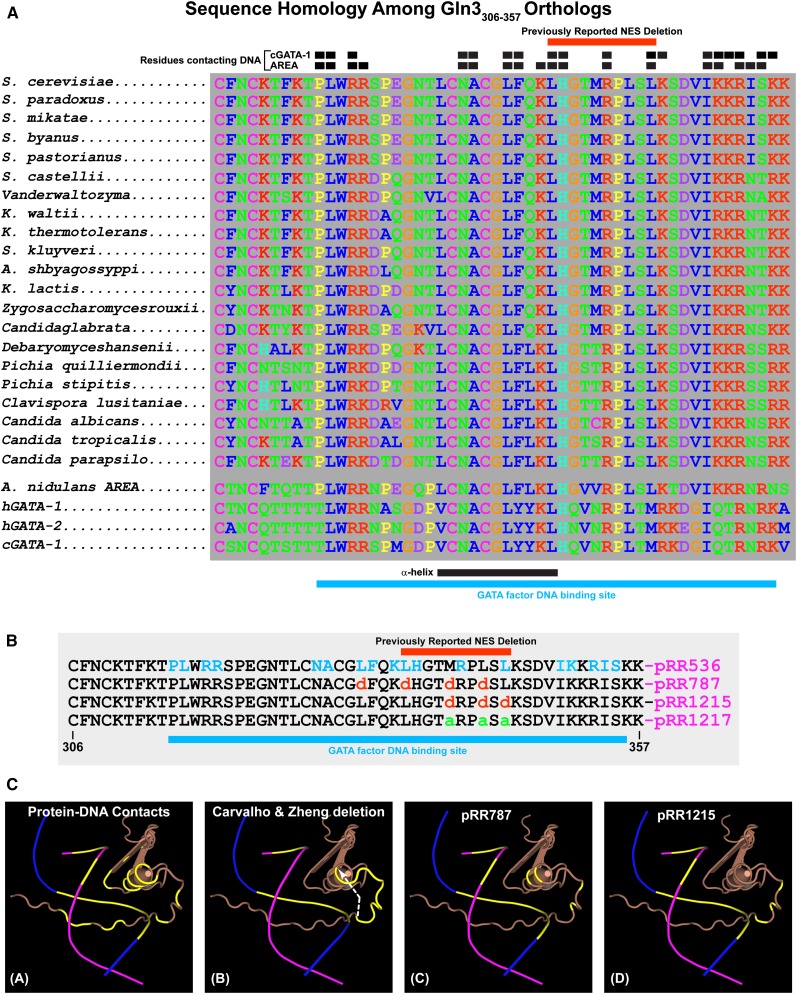
Figure 8.
(A) Sequence homologies among Gln3306–357 orthologs. Black boxes above the homologous sequences indicate residues of the cGATA-1 and AREA GATA factor binding proteins that contact their target DNA sequences. Red bars above the homologous sequences indicate the residues eliminated in the deletion described by Carvalho and Zheng (2003). Black and blue bars beneath the homologous sequences indicate the positions of the α-helix portion of the zinc finger motif and GATA factor DNA binding site, respectively. (B) Amino acid substitutions in the Gln3 wild-type sequence (pRR536) and the plasmids that contain them. Residues that contact the AREA/Gln3 DNA binding site are indicated in blue letters (pRR536). Blue bar beneath the sequences indicates the overall DNA binding site. (C) Three-dimensional renderings of the AREA and, by analogy, Gln3 zinc finger DNA complex. DNA strands appear as blue and maroon lines, with the bases that contact AREA/Gln3 protein indicated in yellow. The AREA/Gln3 protein residues appear in brown. (Image A) Contact residues and bases of AREA/Gln3 protein and DNA are indicated in yellow. (Image B) Bases of the DNA in contact with AREA/Gln3 are indicated in yellow. AREA/Gln3 residues eliminated in the Carvalho and Zheng (2003) deletion are also indicated in yellow. A white arrow indicates the shift in the zinc finger motif’s coiled coil as a result of eliminating the intervening residues, thereby abolishing their interactions with normal DNA contacts. (Images C and D) Residues substituted in pRR787 and pRR1215 are indicated in yellow.