Abstract
Background and Aims
Dispersal and establishment ability can influence evolutionary processes such as geographic isolation, adaptive divergence and extinction probability. Through these population-level dynamics, dispersal ability may also influence macro-evolutionary processes such as species distributions and diversification. This study examined patterns of evolution of dispersal-related fruit traits, and how the evolution of these traits is correlated with shifts in geographic range size, habitat and diversification rates in the tribe Brassiceae (Brassicaceae).
Methods
The phylogenetic analysis included 72 taxa sampled from across the Brassiceae and included both nuclear and chloroplast markers. Dispersal-related fruit characters were scored and climate information for each taxon was retrieved from a database. Correlations between fruit traits, seed characters, habitat, range and climate were determined, together with trait-dependent diversification rates.
Key Results
It was found that the evolution of traits associated with limited dispersal evolved only in association with compensatory traits that increase dispersal ability. The evolution of increased dispersal ability occurred in multiple ways through the correlated evolution of different combinations of fruit traits. The evolution of traits that increase dispersal ability was in turn associated with larger seed size, increased geographic range size and higher diversification rates.
Conclusions
This study provides evidence that the evolution of increased dispersal ability and larger seed size, which may increase establishment ability, can also influence macro-evolutionary processes, possibly by increasing the propensity for long-distance dispersal. In particular, it may increase speciation and consequent diversification rates by increasing the likelihood of geographic and thereby reproductive isolation.
Keywords: Brassica, Brassicaceae, Cakile, extinction, fruit traits, heteroarthrocarpy, limited dispersal, long-distance dispersal, range size, speciation, species diversification
INTRODUCTION
Dispersal has been recognized as a primary factor that influences evolutionary rates and outcomes since the earliest days of population genetics (Wright, 1951). Dispersal ability and establishment success are central to several major population processes including geographic isolation, adaptive divergence and extinction probability (Levin and Kerster, 1975; Levin et al., 2003). Theoretically, by influencing population dynamics, dispersal can also affect macro-evolutionary patterns of species' distributions and lineage diversification (Bohrer et al., 2005). Here we investigate how fruit traits associated with dispersal ability and establishment are associated with geographic range size, habitat shifts and diversification rates.
There is a long history in plant biology of interpreting morphological attributes of dispersal propagules in terms of how they influence seed dispersal (Cousens et al., 2008; Rubio de Casas et al., 2012). Traditionally ascribed ‘dispersal syndromes' are based on morphological traits of diaspores that determine dispersal vectors, such as wind, water, gravity or animals (van der Pijl, 1982; Tiffney, 1984; Murray, 1986). Indeed, specific fruit traits that traditionally define dispersal syndromes have been shown to influence dispersal ability directly, suggesting that they may evolve under selection for different dispersal regimes (Murray, 1986). For example, in a classic study of 34 tropical trees, Augspurger (1986) found that species with fruits that have higher wing-loading, the ratio of fruit area to mass, have increased dispersal ability because their fruits fall more slowly and thus can be blown further from the maternal tree.
Fruit traits may also coevolve with seed traits (Tiffney, 1984). Seeds dispersed long distances might favour the evolution of large seeds to increase establishment success (Cain et al., 2000). Conversely, large seeds may favour the evolution of dispersal features that promote dispersal to counter-balance the limitations of their mass (Guo et al., 2000; Leishman et al., 2000; Moles et al., 2005; Bolmgren and Eriksson, 2010). Dispersal attributes may coevolve not only with seed size, but also with the number of seeds contained within a dispersal propagule in response to selection against post-dispersal competition or as a physical constraint of available space within the propagule (Imbert, 2002).
Fruit traits may also evolve in response to selective factors unrelated to dispersal, but nonetheless influence dispersal ability. For example, indehiscence – a trait associated with limited dispersal – may evolve as a means to protect seeds from desiccation in dry environments (Ellner and Shmida, 1981; Gutterman, 1994). Dispersal-related traits may also coevolve, either through indirect selection via selection on correlated traits or through correlational selection acting to maintain or compensate for lost dispersal ability.
Dispersal ability and establishment (via seed size) can have important consequences for population processes such as geographic isolation, adaptive divergence and extinction probability (Levin and Kerster, 1975; Levin et al., 2003). For instance, limited dispersal is predicted to limit gene flow between populations, promoting population fragmentation, genetic divergence, local adaptation and endemism (Levin et al., 2003). By increasing gene flow, increased dispersal ability is therefore predicted to impede population fragmentation and local adaptation (Cain et al., 2000). Alternatively, increased dispersal ability may promote long-distance dispersal beyond current range limits, increase range size (Hubbell, 2001; Gaston, 2003; Holt, 2003; Lester et al., 2007; Kubisch et al., 2014), increase the probability of encountering and adapting to novel habitats (Holt and Gomulkiewicz, 1997; Gomulkiewicz et al., 1999), and promote population isolation and diversification (Cain et al., 2000; Levin et al., 2003). Empirical studies of the effect of dispersal ability on range size are mixed (Edwards and Westoby, 1996; Lester et al., 2007; Gove et al., 2009), so the alternative hypotheses concerning the relationship between dispersal ability, range size, and population isolation and divergence are largely unresolved.
The effect of dispersal ability on the population-level processes of gene flow, range size and ecological divergence are likely to have consequences for macro-evolutionary processes such as lineage diversification (Bohrer et al., 2005). Diversification is the net result of speciation and extinction (Ricklefs, 2007). Both of these processes are likely to be influenced by changes in dispersal ability (Birand et al., 2012). For instance, limited dispersal might increase the rate of speciation by promoting population fragmentation and local adaptation, but it might also increase the likelihood of extinction by reducing range size and increasing endemism. In plants, evidence for the role of dispersal in diversification has been limited to taxa with animal-dispersed fruits (Givnish, 2010). These studies have found that the evolution of long-distance dispersal (e.g. frugivory) is associated with higher diversification rates (Eriksson and Bremer, 1991; Tiffney and Mazer, 1995; Smith, 2001; Moore and Donoghue, 2007; Beaulieu and Donoghue, 2013). Whether this pattern is driven by higher speciation rates in lineages with long-distance dispersal or higher extinction rates in dispersal-limited lineages is unclear. Furthermore, it is unknown whether this association applies to traits that promote dispersal by passive dispersal agents or whether it is specific to dispersal by animals.
The tribe Brassiceae (Brassicaceae) exhibits a diversity of fruit morphology that corresponds to distinct dispersal modes that employ passive dispersal agents such as gravity, wind and water. The tribe is also diverse in geographic range size, habitat type and climate niche (Gomez-Campo, 1980; Al-Shehbaz et al., 2006; Warwick et al., 2009). Thus, the Brassiceae offers an ideal system to study how dispersal-related fruit traits have evolved in association with one another as well as with seed traits, range size and shifts in climatic niche and diversification rates.
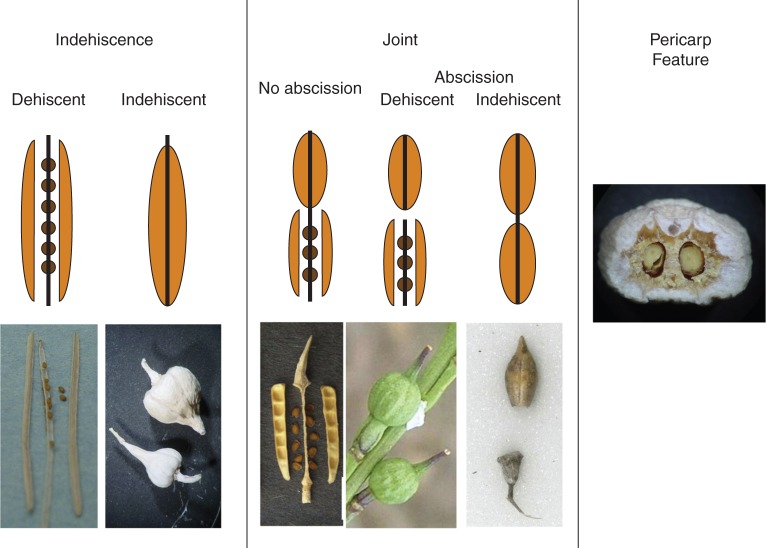
In addition to having members with typical brassicaceous siliques, the tribe Brassiceae has four major morphological fruit traits that correspond to dispersal ability: indehiscence, joints, abscission zones and pericarp features (Fig. 1; Gomez-Campo, 1980; Al-Shehbaz et al., 2006; Hall et al., 2011). Indehiscence is the absence of a dehiscence zone typical of Brassicaceous siliques, which prevents seeds from being released after fruit maturation (Hall et al., 2006). Joints separate the fruit into proximal and distal segments (i.e. heteroarthrocarpy). Joints can be accompanied by the presence of an abscission zone that permits the distal segment to detach, while the proximal segment remains attached to the maternal parent (Rodman, 1974; Payne and Maun, 1981; Donohue, 1998). Pericarp features refer to modifications to the otherwise typical smooth and thin Brassicaceous pericarp, and include corkiness, hooks, barbs and wings. These traits vary widely across the tribe and have evolved several times independently (Hall et al., 2011).
Fig. 1.
Illustration of three major dispersal traits in Brassiceae. Indehiscence is the absence of a dehiscence zone typical of Brassicaceous siliques (fruit on right). Indehiscence restricts seeds from dispersing at maturation. Joints separate the fruit into distinct proximal and distal segments (heteroarthrocarpy). Joints can be accompanied by the presence of an abscission zone that permits the distal segment to disperse. For taxa with a joint, the distal segment is always indehiscent, while the proximal segment can be both indehiscent and dehiscent. Pericarp features are modifications to the pericarp that could potentially influence dispersal ability, such as air pockets that promote dispersal by water.
These four fruit traits – joint, abscission zone, indehiscence and pericarp features – are likely to impact dispersal ability in Brassiceae in several ways. Indehiscence can reduce dispersal by restricting the dispersal of independent seeds (Lu et al., 2010). A joint is also expected to reduce dispersal because it is developmentally associated with indehiscence of the distal segment (Hall et al., 2006). However, the evolution of an abscission zone in conjunction with a joint is likely to increase dispersal as the distal segment can detach and be dispersed as a protected propagule (Imbert, 2002). The evolution of pericarp features is also expected to promote dispersal (Gautier-Hion et al., 1985). In the Brassiceae, the most common pericarp feature, corkiness, is known to promote flotation and dispersal by water (Rodman, 1974; Payne and Maun, 1981), and hooks, barbs and wings have been shown to increase dispersal by animals or wind (Howe and Smallwood, 1982).
Here, we take advantage of the diversity in dispersal-related fruit traits across the Brassiceae to test how the evolution of dispersal ability has influenced the evolution of seed traits, range size, niche and diversification. Specifically, we address the following questions. (1) Do the major dispersal-related fruit characteristics in Brassiceae exhibit correlated evolution? (2) Is the evolution of seed traits correlated with the evolution of fruit traits? (3) Is the evolution of dispersal-related fruit traits and seed traits associated with differences in range size and with shifts in habitat or climatic niche? (4) Are the evolution of dispersal-related fruit traits or transitions in habitat associated with diversification rates?
MATERIALS AND METHODS
Taxon and character sampling
Our phylogenetic analysis included 72 taxa sampled from across the Brassiceae plus four outgroups, representing 64 % of recognized genera and 25 % of recognized species based on Warwick et al. (2009; Supplementary Data Tables S1 and S2). Taxonomic sampling was similar to that in Hall et al. (2011; see Hall et al., 2011 for voucher information), but expanded primarily within the genus Cakile by including 13 additional subspecies or populations (Table S1). Four species were also added using data available through NCBI-GenBank (www.ncbi.nlm.nih.gov/genbank): two additional species of Brassica (B. juncea and B. rapa) and two additional outgroups (Arabidopsis thaliana and Isatis tinctoria). Subsequent trait and climate analysis were performed on a sub-set of 60 taxa within the Brassiceae, primarily by removing sub-specific taxa within Cakile (Table S1).
Leaf material for DNA extractions was obtained from plants grown in the greenhouse. The majority of non-Cakile species were obtained from the Brassicaceae seed bank at la Universidad Politecnica de Madrid, Spain. Additional Cakile specimens were collected along the east coast of the USA, the Great Lakes and the Caribbean from 2004 to 2010. Plants from both the seed stocks and the field were grown in Research Greenhouses at Duke University (Durham, NC, USA).
Low-copy nuclear markers often exhibit higher rates of evolution than chloroplast markers and can be more informative, particularly among recently divergent taxa. However, nuclear markers may also obfuscate resolution because of past hybridization and polyloidization events (Warwick and Hall, 2009). In contrast, chloroplast DNA (cpDNA) is not subject to the complications of hybridization and polyploidy (Wendel and Doyle, 1998), although it typically evolves at a slower rate. Because of their different evolutionary histories, the chloroplast and nuclear genomes may result in different phylogenetic hypotheses for a given clade. In order to capture the potential variation in phylogenetic resolution across genomes, we sampled markers from both genomes.
Six markers were used in our analyses. Four markers, two nuclear (ITS1-ITS4 and Fnr) and two chloroplast (psbA-trnH and Bras4-trnG), were newly sequenced for each taxon (Supplementary Data Table S3). Two markers [nuclear ribosomal DNA (nrDNA), PHYA and cpDNA, matK] were previously sequenced for the same set of taxa (Hall et al., 2011).
DNA extraction, amplification, sequencing and alignment
Total DNA was isolated from fresh leaf material or silica-dried leaves using a Plant DNeasy Mini Kit (Qiagen, Valencia, CA, USA) and cetyltrimethylammonium bromide (CTAB) protocols (Doyle and Doyle, 1987). Amplification and sequencing primers for all regions were from previous studies (Chase et al., 2007; Parisod and Besnard, 2007; Shaw et al., 2007; Song and Mitchell-Olds, 2007; Hall et al., 2011). PCRs for all regions used the mix: template DNA, 1 U of Econo-Taq (Lucigen, Cat. No. 30033-0), 2·5 mm dNTPS, buffer, 10 mm forward primer and 10 mm reverse primer. Reactions were run with an Eppendorf, Master Cycler epigradient S thermal cycler using an initial 5 min denaturation at 80 °C followed by 30 cycles of 95 °C denaturation for 1 min, 1 min annealing at 50 °C, and 4 min extension at 65 °C; followed by 5 min of final extension at 65 °C. PCR products were cleaned using a PCR Purification Kit (Invitrogen K3100-01 Carlsbad, CA, USA).
Nuclear regions were subsequently cloned for a sub-set of taxa to identify multiple copies using a Qiagen Cloning Kit (Qiagen 231122; Venlo, The Netherlands). For Fnr, we cloned 4–7 copies for three different genera within Cakile. Based on Neighbor–Joining analysis, two major copies of Fnr were identified. The copy most similar to sequences for the four additional taxa which we included from NCBI-GenBank, i.e. B. juncea, B. rapa, I. tinctoria and A. thaliana, was identified. We then designed copy-specific primers internal to Fnr to eliminate the need for further cloning (Supplementary Data Table S3). For ITS, all samples were cloned. For each taxon, 3–10 clones were sequenced. The copy used for subsequent analyses was identified by comparison with the four GenBank sequences mentioned above.
Cycle sequencing reactions with the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA) used the thermocycler parameters 94 °C for 5 min, 50 cycles of 94 °C for 1 min and final elongation at 60 °C for 10 min. Samples were electrophoresed on a Beckman Coulter CEQ 8000 sequencer Applied Biosystems 3730 × l automated DNA sequencing instrument, using 96 cm capillary arrays and POP-7 polymer.
Data were analysed using PE-Biosystems v. 3.7 of the program Sequencing Analysis at the DNA Core facility of the University of Missouri, Columbia. DNA sequences were aligned using MUSCLE (Edgar, 2004), then further adjusted by hand in Geneious v. ‘diversitree’ 5·5 (http://www.geneious.com/). All sequences were submitted to NCBI-GenBank (Supplementary Data Table S4) and gene alignments were submitted to TreeBASE (http://treebase.org, Study #15668).
Phylogenetic inference
We inferred phylogenetic relationships for both the nuclear and chloroplast genomes separately, as well as in combination. Each nuclear region was treated as a separate partition in both the nuclear and combined analysis. The three chloroplast regions were treated as a single region, taking into account the uniparental inheritance and lack of recombination in the genome.
We inferred phylogenetic relationships using maximum likelihood (ML) and Bayesian inference (BI). Phylogenies were generated on the Cyberinfrastructure for Phylogenetic Research (CIPRES) v. 3.0 (http://www.phylo.org; Miller et al., 2010). Phylogenetic analyses were rooted using three outgroups based on recent phylogenies published for the family (Beilstein et al., 2008, 2010).
The optimal model of sequence evolution for each region was determined using jModeltest v. 2.0 (Posada, 2008). We used GARLI v. 1.0 (Zwickl, 2006) for ML analyses. We used BEAST v. 1.7.2 (Drummond et al., 2012) for BI analyses, using the default priors (Alfaro and Holder, 2006). For BI analyses, we ran two independent runs of 5 × 107 generations with trees sampled every 5000 generations (see Supplementary Data Table S5 for parameter estimates and effective sample size). The first 10 % of runs were discarded as burn-in. The remaining trees from both runs were combined with LogCombiner v. 1.7.2 (Drummond et al., 2012). A majority-rule consensus tree with a 50 % threshold based on posterior probabilities was constructed with TreeAnnotator v. 1.7.2 (Drummond et al., 2012).
Divergence time estimates
To estimate divergence times, we used BEAST v. 1.7.2 (Drummond et al., 2012). We calibrated the divergence time using two calibration points from Beilstein et al. (2010) with maximum and minimum bounds derived from 95 % confidence intervals of the original estimates: Lineage II [mean = 30·8 million years ago (mya), max = 37·8 mya, min = 23·7 mya] and the Arabidopsis–Brassica split (mean = 43·2 mya, max = 50·7 mya, min = 36·6 mya). We used a normal distribution around the mean with a standard deviation of 1 for the prior (Ho and Phillips, 2009). Dating of the tree was done simultaneously with the phylogenetic estimates described above. We used TreeAnnotator v. 1.7.2 to produce maximum clade credibility trees from posterior probabilities and to determine the 95 % probability density of ages for all nodes in the tree. It should be noted that our branch length estimates are based on previously published branch length estimates (Beilstein et al., 2010), and thus are likely to exhibit greater uncertainty. We have attempted to control for this uncertainty in all subsequent analyses (see below).
Fruit, seed, habitat, range and climatic data
All taxa were scored for the following dispersal-related fruit characters: joint (no joint, joint, abscission joint), dehiscence (fully or partially dehiscent vs. non-dehiscent) and pericarp features (present/absent; encompassing wings, hooks and corkiness) (Fig. 1; Supplementary Data Tables S1 and S2). The majority of data on heteroarthrocarpy (dehiscence, joint and abscission) were taken from Hall et al. (2011) or references therein. At the generic level, taxa used in this study were representative of the overall proportion of fruit trait diversity observed in this tribe for the presence of a joint (approx. 58 % of taxa in the study; approx. 56 % overall), an abscission zone (approx. 38 %; approx. 39 %) and indehiscence (approx. 42 %; approx. 47 %), but lower for the presence of pericarp features (approx. 26 %; approx. 69 %). At the specific level, taxa used in this study were representative of the overall fruit trait diversity in the tribe for the presence of a joint (approx. 65 % of taxa in the study; approx. 59 % overall), but slightly higher for an abscission zone (approx. 42 %; approx. 34 %) and indehiscence (approx. 39 %; approx. 36 %), but lower for pericarp features (approx. 30 %; approx. 50 %). In combination, at the specific level approx. 50 % of the taxa in the study (vs. approx. 52 % overall) exhibited traits associated with increased dispersal ability (abscission zone; pericarp feature), while approx. 20 % (vs. approx. 21 % overall) exhibited only traits associated with limited dispersal (indehiscence; joint, but no abscission zone).
Seed mass and seed number data (Supplementary Data Table S2) were collected from dried seeds collected from a combination of greenhouse-grown plants, the Kew Seed Database and the literature. Data collected from living specimens came from plants grown at either the Harvard University Glasshouse in 2004 or the Duke Greenhouses in 2010. In both cases, 1–6 individual plants were grown per taxon. Flowers were self-pollinated (for highly selfing taxa) and cross-pollinated (for taxa with self-incompatibility) by hand to ensure fruit-set. Two to seven fruits were collected per individual and used to score fruit and seed traits. Seed number was counted per fruit and averaged across fruits and individuals for each taxon. Individual seed mass was measured as the mass (mg) of a group of seeds (2–85) divided by the total number of seeds weighed. For a small number of taxa that we were not able to grow, we obtained additional measures of seed number from a collection of regional floras (see Hall et al., 2011 for references). Additionally, we collected seed number and seed mass data from the Kew SEED Information Database (http://data.kew.org/sid/).
Geo-co-ordinates for each taxon were obtained from the Global Biodiversity Information Facility (GBIF; http://www.gbif.org/), herbarium records and personal observation. Geo-co-ordinates from the GBIF were vetted by hand to remove any errant points, based on broader distribution data from regional floras. Geo-co-ordinates were used to calculate latitudinal range as the difference between the minimum and maximum latitude.
Climatic data were collected for each taxon from the WorldClim Global Climate Database (http://www.worldclim.org/). These data include 19 climatic variables and altitude at a resolution of approx. 1 km2, and were extracted using Maxent 3.3.2 (Phillips et al., 2006; Phillips and Dudík, 2008). The median of these observations was taken per taxon per variable. To reduce the effect of auto-correlation between climate variables, we performed principal component analysis on the median values of all 19 climate variables and altitude across all taxa using the ‘principal’ function with ‘varimax’ rotation in R v. 2.15 (R Development Core Team, 2013; Supplementary Data Tables S5 and S6). Taxa were scored for habitat based on descriptions from regional floras (see Hall et al., 2011). Habitats included: ‘field’ – inland herbaceous community; ‘ruderal’ – disturbed inland herbaceous community; ‘desert’ – xeric inland herbaceous community; and ‘coastal’ – coastal dune herbaceous community (Table S5).
Fruit trait, seed, habitat, range and climate correlations
To test for correlated evolution among dispersal-related fruit characters and between fruit traits and habitat, we used Pagel's method (Pagel; 1994; Pagel and Meade, 2006) for testing for correlated evolution of binary traits. This method compares two evolutionary models: one in which the evolution of the two traits is independent and one in which transitions between states in one trait can depend on transitions between states in the other trait. Maximum likelihood was used to estimate model parameters. A log-likelihood ratio test was used to test for significant differences between the models.
To test for the correlated evolution of dispersal-related fruit characters with seed and environmental traits, we used phylogenetic generalized least squares (PGLS) models implemented in the R package ‘caper’ v. 0.5 (Orme et al., 2012). To correct for multiple comparisons across several geo-spatial and environmental independent variables, we used a standard Bonferonni correction based on the number of comparisons.
Trait-dependent diversification rates
To test if diversification rates were associated with the evolution of dispersal-related fruit traits and habitat shifts, we used either the binary state speciation and extinction (BiSSE) model or the multiple state speciation and extinction (MuSSE) model, depending on the number of character states present (Maddison et al., 2007; FitzJohn et al., 2009), implemented in the R package ‘diversitree’ v. 0.9-3 (FitzJohn, 2012). The BiSSE and MuSSE models use ML to estimate speciation (λ) and extinction (μ) rates among character states. The net diversification (r) rate is estimated as λ minus μ. We tested if rates of λ and μ were significantly different between character states by using a log-likelihood test of models when either λ or μ was constrained, vs. when they were allowed to vary. For diversification analyses, we corrected for incomplete sampling by specifying the fraction of unsampled species that had each character state (FitzJohn, 2012). To compare diversification across habitat types, however, we accounted only for the overall fraction of unsampled species, given the challenges of identifying habitat types for rare or poorly studied taxa. For these analyses, we accounted for phylogenetic uncertainty as explained below.
Phylogenetic uncertainty
To address the influence of branch length and topological uncertainty on tests for correlated evolution and on estimates of diversification rates, we ran analyses across a sub-set of 100 ML and Bayesian divergence-time estimated trees sampled from bootstrap and posterior distributions, respectively. Median values for every estimate were computed across all 100 analyses. Median results did not differ qualitatively in terms of significance or direction between ML and Bayesian tree sets; thus, only median values from the Bayesian tree set are presented.
To address the influence of possible incongruences between the chloroplast and nuclear genomes in the Brassiceae (Franzke et al., 2011), we ran analyses across both sets of phylogenies separately, as well as on the combined phylogeny.
RESULTS
Phylogeny
Combined marker analysis resulted in the greatest resolution across the tribe for both Bayesian and ML analyses (Supplementary Data Figs S1 and S2), and the composition of major clades was consistent across genomes (Figs S3–S6). For Bayesian analysis, we found adequacy of mixing of parameter estimates across the posterior distribution (Table S7).
In contrast to previous studies of the Brassiceae (Hall et al., 2011; Arias and Pires, 2012), Bayesian analysis resolved the genus Cakile as monophyletic, with Cakile arabica appearing as the basal lineage (Supplementary Data Fig. S1). This result, however, was not supported by ML analysis (Fig. S2). Our tree topologies were largely concordant with those reported by Hall et al. (2011) and Arias and Pires (2012) regarding the composition of major groups within the Brassiceae: Zilla, Vella, Cakilinae, Nigra and Oleracea. A notable exception was the placement of Henophyton deserti and Pseuderucaria teretifolia (Henophyton group). Our analysis, similar to that of Hall et al. (2011), placed the Henophyton group within the core Brassiceae (Cakilinae + Nigra + Oleracea) as either unresolved (nrDNA and cpDNA: Figs S3–S6) or affiliated with the Oleracea group (Combined: Figs S1 and S2). In contrast, Arias and Pires (2012) found support for the Henophyton group as sister to the core Brassiceae. We also found discordant resolution between the Cakilinae, Nigra and Oleracea groups across genomes (nrDNA and cpDNA: Figs S3–S6). For the chloroplast markers, we found Cakilinae to be sister to Nigra, albeit with limited support (Figs S3 and S4), similar to the results from the analysis of four chloroplast regions by Arias and Pires (2012). In contrast, based on our analysis of nuclear (Figs S5 and S6) and combined markers (Figs S1 and S2), we found Oleracea to be sister to Nigra. All trees are available on TreeBase (http://treebase.org, Study #15668).
Evolution of fruit and seed traits
Fruit traits were significantly evolutionarily associated with one another (Table 1). The evolution of a joint (creating an indehiscent distal fruit segment in a heteroarthrocarpic fruit) was significantly associated with the evolution of an abscission zone (Table 1), which results in dispersing distal fruit segments. The evolution of indehiscence was also associated with the evolution of pericarp features (Table 1), which is to be expected, since pericarp features seem unlikely to evolve if they do not surround a dispersing propagule. More interestingly, the evolution of indehiscence was significantly associated with the evolution of an abscission zone on the joint, which was marginally significantly associated with pericarp features (Table 1), suggesting that when indehiscence evolves to enclose proximal segments, other features evolve that enhance dispersal. In contrast, there was no association between the evolution of a joint per se (including non-abscising joints) and indehiscence or pericarp features, suggesting that heteroarthrocarpy itself (defined by the joint) does not necessarily alter dispersal via correlated evolution of other traits. Dispersal-related modifications of heteroarthrocarpy, however, such as indehiscence or joint abscission, appear to have promoted the evolution of the tightly linked complex of indehiscence, abscission and pericarp features. Correlations among fruit traits were similar across both nuclear and chloroplast phylogenies, with the exception that the marginal association between abscission and pericarp features was non-significant when tested across the nuclear and chloroplast phylogenies separately (Supplementary Data Table S8).
Table 1.
Test of correlated evolution of fruit traits
| Trait A | Trait B | Log likelihood |
ΔlnLik | P-value | |
|---|---|---|---|---|---|
| Dependent | Independent | ||||
| Abscission zone | Pericarp features | –74.36 | –78·66 | 8.59 | 0.0293 |
| Abscission zone | Indehiscence | –67·11 | –80·85 | 27·48 | 0·0000 |
| Indehiscence | Pericarp features | –69·13 | –77·24 | 16·21 | 0·0012 |
| Joint | Abscission zone | –68·40 | –80·50 | 24·20 | 0·0000 |
| Joint | Pericarp features | –75·80 | –76·39 | 1·19 | 0·1641 |
| Joint | Indehiscence | –77·02 | –79·42 | 4·79 | 0·1093 |
The test consists of comparing likelihoods of two models: one in which transitions between two traits are dependent on one another, and the other in which the transitions are independent of one another. A significant difference between models indicates that the traits are evolutionarily linked (significant results in bold). Relative transition rates between trait pairs are on the left. Significance was adjusted for multiple comparisons (three per trait) using Bonferonni correction (α0·05 = 0·0167). Significant associations are in bold (P < 0·0167, corresponding to an unadjusted significance of P < 0·05), and marginally significant associations are in italics (P < 0·033, corresponding to an unadjusted significance of P < 0·1). Values represent the median estimate from analysis conducted across 100 trees sampled from the Bayesian posterior distribution.
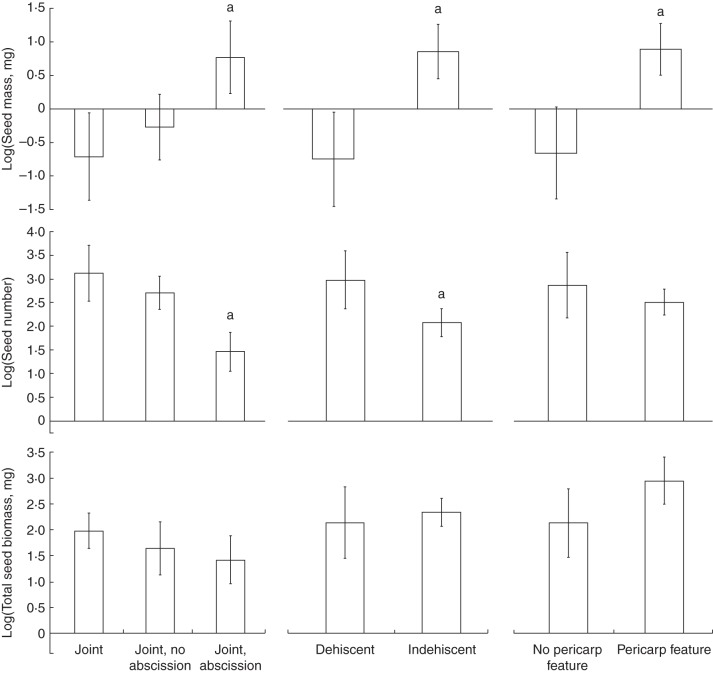
Seed size had a significant negative correlation with seed number per fruit [phylogenetic generalized linear model (PGLM) β = –0·48 ± 0·17, P < 0·01]. Consistent with this association, fruit traits that were positively associated with seed size were negatively associated with seed number per fruit, leading to no significant association with total seed biomass per fruit (Fig. 2; Supplementary Data Table S10). The evolution of a joint per se was not associated with seed size or number (Fig. 2; Table S10). However, the subsequent evolution of an abscission zone on the joint, indehiscence and pericarp features were associated with an increase in seed size and a decrease in seed number, although in the case of pericarp features the decrease in seed number was not significant (Fig. 2; Table S10). Total biomass per fruit was not associated with fruit traits (Fig. 2; Table S10). As above, the evolution of heteroarthrocarpy per se (defined by the presence of a joint) does not appear to have altered the evolution of seed size or number, but seed size and number do seem to have coevolved with traits associated with dispersal that follow the evolution of heteroarthrocarpy (Fig. 2; Table S10). Correlations among fruit and seed traits were similar across both nuclear and chloroplast phylogenies (Tables S9 and S10).
Fig. 2.
Evolution of fruit and seed traits. Associations were tested for using phylogenetic generalized linear models (PGLMs; Supplementary Data Table S10). Bar graphs are provided of phylogenetically corrected mean estimates of seed traits (y-axis) for each fruit trait state (x-axis). Error bars indicate the standard error (s.e.). Significant differences are indicated with a letter (a, different from first state; b, different from second state). Significance was adjusted for multiple comparisons (three seed traits) using Bonferonni correction (α0·05 = 0·0167). Mean and s.e. estimates represent the median values from each analysis run across 100 trees sampled from the Bayesian posterior distribution.
Evolution of fruit and seed traits with habitat, climatic niche and range
The first principle component (PC1) accounted for 90 % of the variation in the climate variables (Supplementary Data Table S6), and was weighted predominantly by temperature variables, including mean annual temperature. While the next three principle components only explained 8 % of the climate variable variation, we included them as they represented important aspects of most environments including: precipitation (PC2), temperature seasonality (PC3) and altitude (PC4) (Table S6).
Habitat was not strongly associated with the evolution of any fruit trait (Table 2). However, seed traits were associated with habitat (Fig. 3; Supplementary Data Table S12). Coastal dune taxa had significantly larger seeds than those found in fields, deserts and ruderal habitats (Fig. 3; Table S12). Desert taxa had significantly more seeds per fruit than taxa that occur in fields, but not other habitats (Fig. 3; Table S12). Total seed biomass per fruit was maintained across all habitats except the coastal done, where there was a significant increase in total seed mass (Fig. 3; Table S12). Correlations between habitat and fruit and seed traits were largely similar across both nuclear and chloroplast phylogenies, with a few exceptions (Tables S11 and S12). First, for both the chloroplast and nuclear phylogenies, there was marginal to fully significant association between the presence of pericarp features and coastal dune habitat (Tables S11 and S12). Secondly, for the chloroplast phylogeny, there was a marginally significant association between the presence of an abscission zone and ruderal habitat and a significant association between the presence of an abscission zone and field habitat (Table S12).
Table 2.
Test of correlated evolution of fruit traits and habitat is conducted across 100 trees sampled from the Bayesian posterior distribution.
| Trait A | Trait B | Log likelihood |
ΔlnLik | P-value | |
|---|---|---|---|---|---|
| Dependent | Independent | ||||
| Abscission zone | Coast | –61·39 | –63·66 | 4·55 | 0·117 |
| Indehiscence | Coast | –60·68 | –61·77 | 2·18 | 0·1833 |
| Joint | Coast | –63·79 | –61·13 | 5·33 | 0·0927 |
| Pericarp features | Coast | –55·04 | –58·65 | 7·22 | 0·0489 |
| Abscission zone | Desert | –75·02 | –74·02 | 2 | 0·1839 |
| Indehiscence | Desert | –73·28 | –72·93 | 0·7 | 0·1234 |
| Joint | Desert | –77·02 | –72·93 | 8·19 | 0·0341 |
| Pericarp features | Desert | –71·49 | –70·32 | 2·34 | 0·1815 |
| Abscission zone | Field | –65·53 | –66·38 | 1·69 | 0·1815 |
| Indehiscence | Field | –64·93 | –65·44 | 1·02 | 0·1528 |
| Joint | Field | –65·11 | –69·35 | 8·48 | 0·0305 |
| Pericarp features | Field | –63·88 | –64·42 | 1·1 | 0·1584 |
| Abscission zone | Ruderal | –76·9 | –79·98 | 6·15 | 0·071 |
| Indehiscence | Ruderal | –77·79 | –78·83 | 2·09 | 0·1838 |
| Joint | Ruderal | –76·95 | –78·5 | 3·09 | 0·1648 |
| Pericarp features | Ruderal | –76·85 | –75·81 | 2·1 | 0·1837 |
The test consists of comparing likelihoods of two models: one in which transitions between two traits are dependent on one another, and the other in which the transitions are independent of one another. A significant difference between models indicates that the traits are evolutionarily linked (significant results in bold). Relative transition rates between trait pairs are on the left. Significance was adjusted for multiple comparisons (three per trait) using Bonferonni correction (α0·05 = 0·0167). Marginally significant associations are in italics (P < 0·033, corresponding to an unadjusted significance of P < 0·1). Values represent the median estimate from analysis conducted across 100 trees sampled from the Bayesian posterior distribution.
Fig. 3.
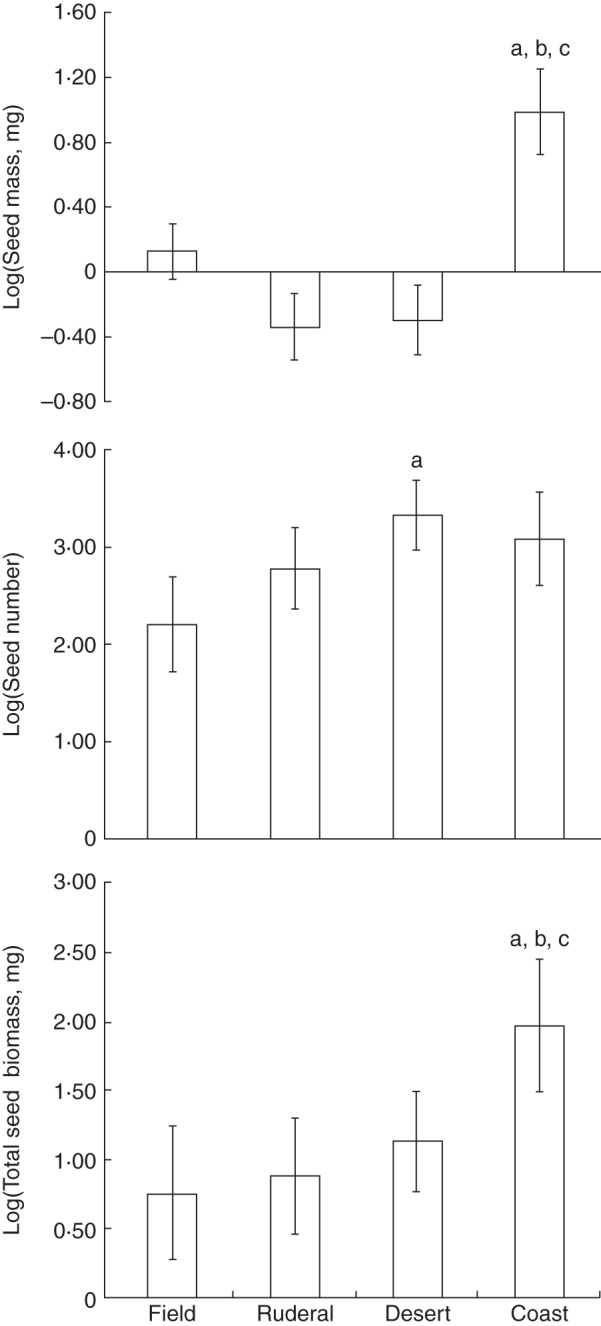
Correlated evolution of habitat and seed traits. Associations were tested for using phylogenetic generalized linear models (PGLMs; Supplementary Data Table S12). Bar graphs are provided of phylogenetically corrected mean estimates of seed traits (y-axis) for each fruit trait state (x-axis). Error bars indicate the standard error (s.e.). Significant differences are indicated with a letter (a, different from first state; b, different from second state; c, different from third state). Significance was adjusted for multiple comparisons (three seed traits) using Bonferonni correction (α0·05 = 0·0167). Mean and s.e. estimates represent the median values from each analysis run across 100 trees sampled from the Bayesian posterior distribution.
There were no significant associations between fruit and seed traits and the measured climate or altitude variables (Supplementary Data Tables S13 and S14). There was, however, a marginally significant positive association between seed size and latitudinal range (Table S14), where taxa with larger seeds tended to have wider ranges. This relationship held when other fruit traits and seed number were included in the model, but it was not significant after correcting for multiple comparisons (Table S14). Correlations among climate and fruit and seed traits were similar, though stronger in the case of seed mass, when controlling for the nuclear and chloroplast phylogenies separately (Tables S13 and S14).
Trait- and habitat-dependent diversification rates
The evolution of fruit traits and the transition to coastal dune habitat were significantly associated with diversification rates (Fig. 4; Supplementary Data Table S15a). The diversification rate of taxa that have a joint was higher than that of taxa without a joint (Fig. 4; Table S15a), but this association appears to be driven by the evolution of an abscission zone. Diversification of taxa with an abscission zone was significantly greater than that of taxa with no joint or taxa with a joint but no abscission zone (Fig. 4; Table S15a). The evolution of indehiscence and dispersal-related pericarp features were also associated with significantly greater diversification rates (Fig. 4; Table S15a). Although extinction rates did differ significantly between taxa with different habitat and joint states, differences in extinction rates were relatively minor or non-significant compared with the magnitude of difference in speciation rates (Table S15a). These patterns were consistent across both nuclear and chloroplast phylogenies.
Fig. 4.
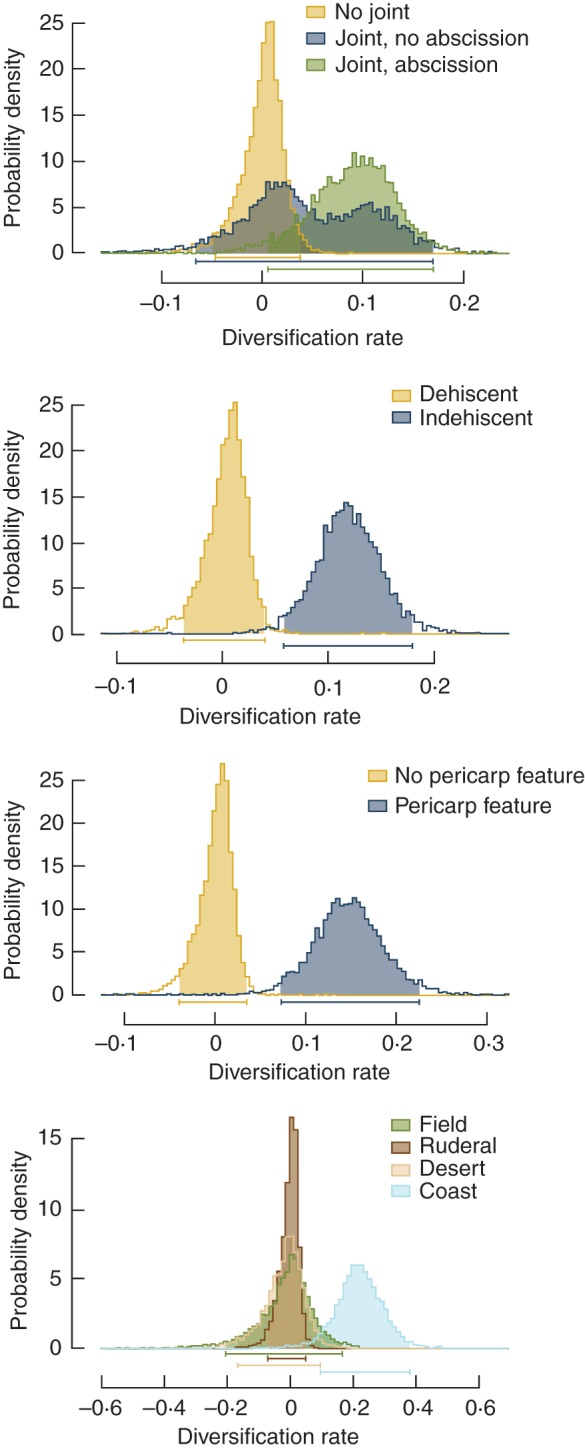
Diversification rates of taxa with different fruit traits (see keys in graphs) and habitat. Diversification rates were calculated as the difference between speciation and extinction rates. These rate estimates are from diversification analysis on the Bayesian consensus tree as an illustration of the patterns found in a more thorough analysis accounting for unsampled taxa and phylogenetic uncertainty (see Supplementary Data Table S15 for estimated speciation and extinction rates separately).
Accounting for taxon sampling biases (i.e. by including the overall proportion of missing taxa) did not alter the overall patterns of fruit trait-related diversification (Supplementary Data Table S15b), with the exception that the association between coastal dune habitat and higher rates of diversification was weaker, due to relatively higher extinction rates. Accounting for ‘unresolved’ clades (i.e. by including unsampled taxa within the phylogeny based on their generic association), however, resulted in no significant difference between diversification rates for joint, indehiscence or pericarp features (Table S15c). The results for the phylogenetic uncertainty analysis also differed across nuclear and chloroplast phylogenies. Given the phylogenetic uncertainty and polyphyletic tendency of multiple genera within the tribe, however, these sensitivity analyses are difficult to interpret. Conservatively, diversification results can be interpreted as pertaining to deeper or ancient events within the tribe (i.e. below the generic level), where we have more complete sampling.
DISCUSSION
This study found evidence that fruit and seed traits coevolved in ways that influence dispersal and possibly establishment. First, while the evolution of heteroarthrocarpy itself did not necessarily correspond to changes in dispersal or seed traits, it preceded the evolution of dispersal-related traits such as abscission, indehiscence, and dispersal-enhancing pericarp features. Secondly, the additional evolution of indehiscence and pericarp features was accompanied by the evolution of larger seeds, but a corresponding trade-off in seed number. Thirdly, there was a trend for coastal dune species and species with larger latitudinal ranges to have larger seeds. Finally, we found that taxa with dispersal-enhancing traits and that inhabit coastal dune habitats have higher diversification rates, driven primarily by increased speciation rates. In summary, the evolution of dispersal-enhancing traits appears to result from a series of correlated morphological changes that have significant effects on species' establishment ability, niche and diversification.
Dispersal-related fruit traits in the Brassiceae appear to have strongly coevolved. Indehiscence, a trait thought to limit dispersal (Ellner and Shmida, 1981), evolved together with dispersal-enhancing features such as joint abscission, pericarp features and a reduction of the number of seeds per propagule (be it the whole fruit or individual segments of it). In combination, these traits functionally represent the evolution of increased dispersal ability. The two traits that have a more obvious influence on dispersal ability (joint abscission and pericarp features) are not themselves correlated. While they can evolve in the same taxa, they appear to be largely independent mechanisms that promote dispersal. Thus, while specific mechanisms may differ among taxa, there appears to be a general trend toward a compensatory evolution of dispersal ability in taxa that evolved indehiscence. Our results are similar to those of a study in the family Melastomataceae that found a significant correlation between the evolution of indehiscence and the evolution of fleshy pericarps associated with frugivory and long-distance dispersal (Clausing et al., 2000). In combination with Clausing et al. (2000), our results hint at a broader trade-off between the evolution of indehiscence and the evolution of long-distance dispersal across angiosperms.
Indehiscence is often found among desert species, an association thought to have evolved either as a mechanism to retain maternal-site advantage (Friedman and Stein, 1980) or as a means to protect seeds from the adverse environment (Ellner and Shmida, 1981). In contrast, we did not observe a significant correlation between indehiscence and desert habitat. One explanation for this incongruity is that desert communities filter taxa from clades with a high proportion of indehiscence. Thus, while the desert community may appear to be disproportionately rich in indehiscence taxa, the taxa themselves may not represent a disproportionate sample of their broader phylogenetic group. In Brassiceae, indehiscence is found, nearly evenly, across all four habitats (comprising 7–12 % of the taxa for a given habitat). Thus, at least in the Brassiceae, any advantages that indehiscence may give desert taxa do not limit indehiscent taxa from occupying other environments. Other potential factors, not considered in this study, are variation in life history traits such as dormancy, germination and life cycle, which are known to coevolve with dispersal and seed size as well as to influence range size and habitat preference (Buoro and Carlson, 2014). For instance, dormancy is expected to coevolve with dispersal and seed size as a bet-hedging strategy in variable environments, such as deserts (Venable and Lawlor, 1980; Gutterman, 1994). While we were unable to consider these important life history traits in our study, their inclusion in future studies of dispersal would probably provide important information on the evolution of propagule traits and their relationship to habitat and range size.
Fruit traits also coevolved with seed size. Traits associated with greater dispersal ability, i.e. indehiscence plus pericarp features, were also associated with the evolution of larger seeds. Larger seeds have been shown to enhance the probability of seedling establishment in both species that are animal dispersed (Aizen and Patterson, 1990; Edwards and Westoby, 1996) and passively dispersed (Moles and Westoby, 2004). Large seeds may be especially important for establishment after long-distance dispersal events into novel habitats or unpredictable environments (Cain et al., 2000; Wang and Smith, 2002; Moles and Westoby, 2004). In this study, for example, larger seeds were marginally associated with a shift to the coastal dune habitat.
The enhanced establishment ability of larger seeds after long-distance dispersal is supported by the marginally significant, positive relationship between seed size and latitudinal range size in the tribe. It has been suggested that larger seeds should have wider geographic ranges because they are better able to establish post-dispersal (Edwards and Westoby, 1996). The evolution of indehiscence or dispersal-related pericarp features had no direct association with range size. Thus, while these traits might facilitate dispersal, it appears that without the appropriate resources to foster establishment after dispersal, range size will remain static. Therefore selection may favour an association, such as that observed here, between large seeds and long-distance dispersal in a manner that enhances colonization after dispersal and increases in range size. However, large seeds have also been argued to be an adaptation to promote establishment in coastal dune habitats in particular, given the limited resources available in most dune systems (Maun, 1994). Thus, it is possible that seed size may have increased initially as an adaptation to the coastal dune environment, rather than in conjunction with fruit traits that enhanced long-distance dispersal.
The evolution of dispersal-enhancing traits was associated with increased rates of diversification within the tribe Brassiceae. The evolution of an abscission zone and pericarp features was associated with elevated rates of diversification, in both cases due to increased speciation rates. This finding is in contrast to some existing theory and empirical evidence. Increased dispersal ability is predicted to limit the process of speciation by increasing gene flow and thereby mitigating adaptive divergence (Coyne and Orr, 2004; Givnish, 2010). In past studies, when diversification rates have been compared between clades with and without increased dispersal ability, diversification rates were shown to be higher in clades with lower dispersal ability (Givnish, 2010). In contrast to these past results, it is possible that increased dispersal ability might promote speciation by increasing the likelihood of long-distance dispersal events and subsequent allopatric isolation (Cain et al., 2000; Bohrer et al., 2005). This argument is often invoked within the context of island biogeography and a role for vicariance vs. dispersal in driving geographic distributions (Roy et al., 2009). In Brassiceae, this dynamic of long-distance colonization and allopatry could be one explanation for the elevated rates of speciation we see associated with fruit traits that increase dispersal ability.
Another explanation for the observed association between increased speciation and traits associated with enhanced dispersal is that long-distance dispersal may facilitate shifts to a novel habitat. For example, in the Brassiceae, dispersal-enhancing traits may be indirectly associated, through their association with seed size, with a shift to the novel coastal dune habitat, which was in turn associated with increased speciation. The shift to the coast might have opened up new niche opportunities that promoted speciation via adaptive radiation and may also have increased geographic range. Given the tight coevolution of these traits, however, it is difficult to disentangle which process has played a dominant role in the diversification of the Brassiceae.
In conclusion, in the Brassiceae, the evolution of enhanced dispersal can occur in multiple ways through the correlated evolution of different fruit traits. Enhanced dispersal was generally associated with larger seed size, which probably facilitates establishment and for which there is some evidence of an associated shift in habitat and increase in range size. The combined effect of these evolutionary transitions in dispersal ability and habitat appears to have accelerated the rate of diversification within the tribe. The evolution of increased dispersal, when accompanied by seed features that increase the ability to establish in isolated or novel habitats, may therefore influence macro-evolutionary processes of diversification.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
This work was supported in part by NSF Doctoral Dissertation Improvement Grant #1011329. The authors thank Elena Kramer, Paul Manos, Cliff Cunningham, Mark Rausher, John Willis and members of the Donohue Lab for comments on the manuscript, and Tracy Tisdale and Jamie Sabo for their participation in this project.
LITERATURE CITED
- Aizen MA, Patterson WA. Acorn size and geographical range in the North American oaks (Quercus L.) Journal of Biogeography. 1990;17:327–332. [Google Scholar]
- Alfaro ME, Holder MT. The posterior and the prior in bayesian phylogenetics. Annual Review of Ecology, Evolution, and Systematics. 2006;37:19–42. [Google Scholar]
- Al-Shehbaz IA, Beilstein MA, Kellogg EA. Systematics and phylogeny of the Brassicaceae (Cruciferae): an overview. Plant Systematics and Evolution. 2006;259:89–120. [Google Scholar]
- Arias T, Pires JC. A fully resolved chloroplast phylogeny of the brassica crops and wild relatives (Brassicaceae: Brassiceae): novel clades and potential taxonomic implications. Taxon. 2012;61:980–988. [Google Scholar]
- Augspurger CK. Morphology and dispersal potential of wind-dispersed diaspores of neotropical trees. American Journal of Botany. 1986;73:353–363. [Google Scholar]
- Beaulieu JM, Donoghue MJ. Fruit evolution and diversification in campanulid angiosperms. Evolution. 2013;67:3132–3144. doi: 10.1111/evo.12180. [DOI] [PubMed] [Google Scholar]
- Beilstein M, Al-Shehbaz I, Mathews S, Kellogg E. Brassicaceae phylogeny inferred from phytochrome A and ndhF sequence data: tribes and trichomes revisted. American Journal of Botany. 2008;95:1307–1327. doi: 10.3732/ajb.0800065. [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proceedings of the National Academy of Sciences; USA. 2010. pp. 18724–18728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birand A, Vose A, Gavrilets S. Patterns of species ranges, speciation, and extinction. American Naturalist. 2012;179:1–21. doi: 10.1086/663202. [DOI] [PubMed] [Google Scholar]
- Bohrer G, Nathan R, Volis S. Effects of long distance dispersal for metapopulation survival and genetic structure at ecological time and spatial scales. Journal of Ecology. 2005;93:1029–1040. [Google Scholar]
- Bolmgren K, Eriksson O. Seed mass and the evolution of fleshy fruits in angiosperms. Oikos. 2010;119:707–718. [Google Scholar]
- Buoro M, Carlson SM. Life-history syndromes: integrating dispersal through space and time. Ecology Letters. 2014;17:756–767. doi: 10.1111/ele.12275. [DOI] [PubMed] [Google Scholar]
- Cain ML, Milligan BG, Strand AE. Long-distance seed dispersal in plant populations. American Journal of Botany. 2000;87:1217–1227. [PubMed] [Google Scholar]
- Chase M, Cowan R, Hollingsworth PM, et al. A proposal for a standardised protocol to barcode all land plants. Taxon. 2007;56:295–299. [Google Scholar]
- Clausing G, Meyer K, Renner SS. Correlations among fruit traits and evolution of different fruits within Melastomataceae. Botanical Journal of the Linnean Society. 2000;133:303–326. [Google Scholar]
- Cousens R, Dytham C, Law R. Dispersal in plants: a population perspective. Oxford: Oxford University Press; 2008. [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer Associates, Inc; 2004. [Google Scholar]
- Donohue K. Maternal determinants of seed dispersal in Cakile edentula: fruit, plant, and site traits. Ecology. 1998;79:2771–2788. [Google Scholar]
- Doyle J, Doyle J. CTAB DNA extraction in plants. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1·7. Molecular Biology and Evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards W, Westoby M. Reserve mass and dispersal investment to geographic range of plant species: phylogenetically independent contrasts. Journal of Biogeography. 1996;23:329–338. [Google Scholar]
- Ellner S, Shmida A. Why are adaptations for long-range seed dispersal rare in desert plants? Oecologia. 1981;51:133–144. doi: 10.1007/BF00344663. [DOI] [PubMed] [Google Scholar]
- Eriksson O, Bremer B. Fruit characteristics, life forms, and species richness in the plant family Rubiaceae. American Naturalist. 1991;138:751–761. [Google Scholar]
- FitzJohn RG. Diversitree: comparative phylogenetic analyses of diversification in R. Methods in Ecology and Evolution. 2012;3:1084–1092. [Google Scholar]
- FitzJohn RG, Maddison W, Otto SP. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Systematic Biology. 2009;58:595–611. doi: 10.1093/sysbio/syp067. [DOI] [PubMed] [Google Scholar]
- Franzke A, Lysak MA, Al-Shehbaz IA, Koch MA, Mummenhoff K. Cabbage family affairs: the evolutionary history of Brassicaceae. Trends in Plant Science. 2011;16:108–16. doi: 10.1016/j.tplants.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Friedman J, Stein Z. The influence of seed dispersal mechanisms on the dispersion of Anastatica hierochuntica (Cruciferae) in the Negev desert of Israel. Journal of Ecology. 1980;68:43–50. [Google Scholar]
- Gaston K. The structure and dynamics of geographic ranges. Oxford: Oxford University Press; 2003. [Google Scholar]
- Gautier-Hion A, Duplantier J-M, Quris R, et al. Fruit characters as a basis of fruit choice and seed dispersal in a tropical forest vertebrate community. Oecologia. 1985;65:324–337. doi: 10.1007/BF00378906. [DOI] [PubMed] [Google Scholar]
- Givnish TJ. Ecology of plant speciation. Taxon. 2010;59:1326–1366. [Google Scholar]
- Gomez-Campo C. Morphology and morpho-taxonomy of the tribe Brassiceae. In: Tsunoda S, Hinata K, Gomez-Campo C, editors. Brassica crops and wild allies. Biology and breeding. Tokyo: Japan Scientific Societies Press; 1980. pp. 3–31. [Google Scholar]
- Gomulkiewicz R, Holt R, Barfield M. The effects of density dependence and immigration on local adaptation and niche evolution in a black-hole sink environment. Theoretical Population Biology. 1999;55:283–296. doi: 10.1006/tpbi.1998.1405. [DOI] [PubMed] [Google Scholar]
- Gove AD, Fitzpatrick MC, Majer JD, Dunn RR. Dispersal traits linked to range size through range location, not dispersal ability, in Western Australian angiosperms. Global Ecology and Biogeography. 2009;18:596–606. [Google Scholar]
- Guo QF, Brown JH, Valone TJ, Kachman SD. Constraints of seed size on plant distribution and abundance. Ecology. 2000;81:2149–2155. [Google Scholar]
- Gutterman Y. Strategies of seed dispersal and germination in plants inhabiting deserts. Botanical Review. 1994;60:373–425. [Google Scholar]
- Hall J, Tisdale T, Donohue K, Kramer E. Developmental basis of an anatomical novelty: heteroarthrocarpy in Cakile lanceolata and Erucaria erucarioides (Brassicaceae) International Journal of Plant Sciences. 2006;167:771–789. [Google Scholar]
- Hall JC, Tisdale TE, Donohue K, Wheeler A, Al-Yahya MA, Kramer EM. Convergent evolution of a complex fruit structure in the tribe Brassiceae (Brassicaceae) American Journal of Botany. 2011;98:1989–2003. doi: 10.3732/ajb.1100203. [DOI] [PubMed] [Google Scholar]
- Ho SY, Phillips MJ. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Systematic Biology. 2009;58:367–380. doi: 10.1093/sysbio/syp035. [DOI] [PubMed] [Google Scholar]
- Holt R. On the evolutionary ecology of species’ ranges. Evolutionary Ecology Research. 2003;5:159–178. [Google Scholar]
- Holt R, Gomulkiewicz R. How does immigration influence local adaptation? A reexamination of a familiar paradigm. American Naturalist. 1997;149:563–572. [Google Scholar]
- Howe HF, Smallwood J. Ecology of seed dispersal. Annual Review of Ecology and Systematics. 1982;13:201–228. [Google Scholar]
- Hubbell SP. The unified neutral theory of biodiversity and biogeography. Princeton, NJ: Princeton University Press; 2001. [Google Scholar]
- Imbert E. Ecological consequences and ontogeny of seed heteromorphism. Perspectives in Plant Ecology Evolution and Systematics. 2002;5:13–36. [Google Scholar]
- Kubisch A, Holt RD, Poethke H-J, Fronhofer EA. Where am I and why? Synthesizing range biology and the eco-evolutionary dynamics of dispersal. Oikos. 2014;123:5–22. [Google Scholar]
- Leishman MR, Wright IJ, Moles AT, Westoby M. The evolutionary ecology of seed size. In: Fenner M, editor. Seeds: the ecology of regeneration in plant communities. New York: CABI Publishing, 31–57; 2000. [Google Scholar]
- Lester SE, Ruttenberg BI, Gaines SD, Kinlan BP. The relationship between dispersal ability and geographic range size. Ecology Letters. 2007;10:745–758. doi: 10.1111/j.1461-0248.2007.01070.x. [DOI] [PubMed] [Google Scholar]
- Levin DA, Kerster HW. Gene flow in seed plants. In: Dobzhansky T, Hecht MK, Steere WC, editors. Evolutionary biology. New York: Springer; 1975. pp. 139–220. [Google Scholar]
- Levin S, Muller-Landau H, Nathan R, Chave J. The ecology and evolution of seed dispersal: a theoretical perspective. Annual Review of Ecology, Evolution, and Systematics. 2003;34:575–604. [Google Scholar]
- Lu J, Tan D, Baskin JM, Baskin CC. Fruit and seed heteromorphism in the cold desert annual ephemeral Diptychocarpus strictus (Brassicaceae) and possible adaptive significance. Annals of Botany. 2010;105:999–1014. doi: 10.1093/aob/mcq041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison W, Midford P, Otto S. Estimating a binary character's effect on speciation and extinction. Systematic Biology. 2007;56:701–710. doi: 10.1080/10635150701607033. [DOI] [PubMed] [Google Scholar]
- Maun MA. Adaptations enhancing survival and establishment of seedlings on coastal dune systems. Vegetatio. 1994;111:59–70. [Google Scholar]
- Miller M, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); New Orleans, LA: IEEE; 2010. pp. 1–8. [Google Scholar]
- Moles AT, Westoby M. Seedling survival and seed size: a synthesis of the literature. Journal of Ecology. 2004;92:372–383. [Google Scholar]
- Moles AT, Ackerly DD, Webb CO, et al. A brief history of seed size. Science. 2005;307:576–580. doi: 10.1126/science.1104863. [DOI] [PubMed] [Google Scholar]
- Moore BR, Donoghue MJ. Correlates of diversification in the plant clade Dipsacales: geographic movement and evolutionary innovations. American Naturalist. 2007;170:S28–S55. doi: 10.1086/519460. [DOI] [PubMed] [Google Scholar]
- Murray D. Seed dispersal. San Diego: Academic Press; 1986. [Google Scholar]
- Orme D, Freckleton RP, Thomas GH, et al. caper: comparative analysis of phylogenetics and evolution in R. Methods in Ecology and Evolution. 2012;3:145–151. [Google Scholar]
- Pagel M. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proceedings of the Royal Society B: Biological Sciences. 1994;255:37–45. [Google Scholar]
- Pagel M, Meade A. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. American Naturalist. 2006;167:808–825. doi: 10.1086/503444. [DOI] [PubMed] [Google Scholar]
- Parisod C, Besnard G. Glacial in situ survival in the Western Alps and polytopic autopolyploidy in Biscutella laevigata L. (Brassicaceae) Molecular Ecology. 2007;16:2755–2767. doi: 10.1111/j.1365-294X.2007.03315.x. [DOI] [PubMed] [Google Scholar]
- Payne AM, Maun MA. Dispersal and floating ability of dimorphic fruit segments of Cakile edenulta var. lacustris. Canadian Journal of Botany-Revue Canadienne De Botanique. 1981;59:2595–2602. [Google Scholar]
- Phillips S, Anderson RP, Schapire R. Maximum entropy modeling of species geographic distributions. Ecological Modelling. 2006;190:231–259. [Google Scholar]
- Phillips SJ, Dudík M. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography. 2008;31:161–175. [Google Scholar]
- van der Pijl L. Principles of dispersal in higher plants. Berlin: Springer-Verlang; 1982. [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- Ricklefs R. History and diversity: explorations at the intersection of ecology and evolution. American Naturalist. 2007;170:S56–S70. doi: 10.1086/519402. [DOI] [PubMed] [Google Scholar]
- Rodman J. Systematics and evolution of the genus Cakile (Cruciferae) Contributions from the Gray Herbarium. 1974;No. 205:3–146. [Google Scholar]
- Roy K, Hunt G, Jablonski D, Krug AZ, Valentine JW. A macroevolutionary perspective on species range limits. Proceedings of the Royal Society B: Biological Sciences. 2009;276:1485–93. doi: 10.1098/rspb.2008.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio de Casas R, Willis CG, Donohue K. Plant dispersal phenotypes: a seed perspective of maternal habitat selection. In: Colbert J, Baguette M, Benton T, Bullock J, editors. Dispersal ecology and evolution. 2012. p. 171. Oxford Scholarship Online. [Google Scholar]
- Shaw J, Lickey E, Schilling E, Small R. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in Angiosperms: the tortoise and the hare III. American Journal of Botany. 2007;94:275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- Smith JF. High species diversity in fleshy-fruited tropical understory plants. American Naturalist. 2001;157:646–653. doi: 10.1086/320625. [DOI] [PubMed] [Google Scholar]
- Song B-H, Mitchell-Olds T. High genetic diversity and population differentiation in Boechera fecunda, a rare relative of Arabidopsis. Molecular Ecology. 2007;16:4079–4088. doi: 10.1111/j.1365-294X.2007.03500.x. [DOI] [PubMed] [Google Scholar]
- Tiffney BH. Seed size, dispersal syndromes, and the rise of the angiosperms: evidence and hypothesis. Annals of the Missouri Botanical Garden. 1984;71:551–576. [Google Scholar]
- Tiffney BH, Mazer SJ. Angiosperm growth habit, dispersal and diversification reconsidered. Evolutionary Ecology. 1995;9:93–117. [Google Scholar]
- Venable DL, Lawlor L. Delayed germination and dispersal in desert annuals – escape in space and time. Oecologia. 1980;46:272–282. doi: 10.1007/BF00540137. [DOI] [PubMed] [Google Scholar]
- Wang BC, Smith TB. Closing the seed dispersal loop. Trends in Ecology and Evolution. 2002;17:379–386. [Google Scholar]
- Warwick SI, Francis A, Gugel RK. Guide to wild germplasm of Brassica and allied crops (tribe Brassiceae, Brassicaceae) I. Taxonomic checklist and life history, ecological, and geographical data. 3rd edn. Ottawa, Canada: Agriculture and Agri-Food Canada, Eastern Cereal and Oilseed Research Centre; 2009. [Google Scholar]
- Warwick SI, Hall JC. Phylogeny of Brassica and wild relatives. In: Gupta SK, editor. Biology and breeding of crucifers. Boca Raton, FL: CRC Press; 2009. pp. 37–68. [Google Scholar]
- Wendel J, Doyle J. Phylogenetic incongruence: window into genome history and molecular evolution. In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular systematics of plants II. Boston, MA: Springer; 1998. pp. 265–296. [Google Scholar]
- Wright S. The genetical structure of populations. Annals of Eugenics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ. 2006. GARLI: genetic algorithm for rapid likelihood inference. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.