Abstract
Background:
Extended spectrum beta lactamases (ESBLs) are an important cause of transferable multidrug resistance (MDR) in gram-negative bacteria. The most described ESBL genes are generally found within integron-like structures as mobile genetic elements. The aim of this study was to identify the accompanying of class 1 integrons and ESBLs in the MDR E. coli isolates.
Methods:
Susceptibility to antimicrobial agents was determined for 33 E. coli strains by the disk diffusion method. Double-disk synergy test was applied for screening ESBL. To identify the strains carrying integrons, the conserved regions of integron-encoded integrase gene intI1 were amplified. For detection of gene cassettes, 5′CS and 3′CS primers were used.
Results:
All E. coli isolates were identified as multi-drug resistant. More than 50% of the isolates were resistant to tetracycline, cephalothin, cefuroxime, amoxicillin-clavulanic acid, and third generation cephalosporines. Nearly all of the isolates displayed sensitivity to piperacillin. There was a significant correlation between production of ESBL and resistance to all antibiotics except for ciprofloxacin and piperacillin (P < 0.01). Thirty two MDR strains (97%) included class 1 integron, and some isolates that included integrons were similar in the size of gene cassettes. The isolates were different in the resistance profiles; however, some others had similar resistance profiles. Of eight ESBL positive isolates, seven (87.5%) carried class 1 integrons.
Conclusion:
Class 1 integrons were frequent in MDR and also ESBL-producing E. coli isolates. High prevalence of class 1 integrons confirms that integron-mediated antimicrobial gene cassettes are important in E. coli resistance profile.
Key Words: Antibiotic, Integrons, Escherichia coli
INTRODUCTION
Despite accessibility to different antibiotics, Urinary tract infections (UTIs) are the second most frequent community-acquired adult infection and the main cause of nosocomial infection [1]. E. coli is one of the major causes of nosocomial infections, especially in patients in intensive care units and the main cause of UTI in developed world, accounting for 70-90% of uncomplicated UTIs [2].
Understanding the molecular mechanism of resistance genes may help to the introduction of new antimicrobial strategies and some preventive procedures to prevent further spreading of resistance determinants among the pathogens [3]. Several resistance genes encode different mechanisms of drug resistance in bacterial genomes and in extra-chromosomal pieces of DNA [4].
Multidrug resistance (MDR) is a major problem worldwide and encoded by resistance genes exists on integrons. Integrons are mobile genetic elements considered to be responsible for the MDR transfer. Most of the integrons reported in clinical isolates are class 1 inegrons. Class 1 integrons carry integrase gene (intI1), which codes for the site-specific recombinase responsible for cassette insertion [5, 6]. Integrase gene also includes the attI1 site, where the cassettes are integrated and a promoter, Pc, is responsible for the transcription of the cassette-encoded genes [5, 7]. Gene cassette contains a single antibiotic resistance gene and a 59-base element (or attC site) downstream of the gene, which is responsible for recombination events [8].
Extended spectrum beta lactamases (ESBLs) are a group of enzymes that are common among Enterobacteriaceae [9]. They are the increasingly important cause of transferable MDR in Gram-negative bacteria throughout the world. ESBLs also have the ability to hydrolyze third and fourth generation cephalosporins and monobactams. ESBL-producing strains are prevented by lactamase inhibitors (clavulanic acid, sulbactam, and tazobactam) [10].
ESBL-encoding genes are generally located on conjugative plasmids (such as blaTEM or blaSHV), although many of the most newly described ESBL genes are usually found within integron-like structures (such as blaCTX-M, blaGES, or blaVEB-1) [11-13]. On the other hand, ESBL-producing isolates usually show resistance to other antibiotics including aminoglyc-sides, tetracyclines, chloramphenicol, trimethoprim, sulfonamides, or quinolones. Resistance to these antibiotics is mostly related to presence of different resistance genes on plasmids, transposons, or integrons as transferable elements or genetic structures generated by combinatorial evolution of different interactive pieces [13-15]. The presence of ESBL genes on integrons can facilitate the distribution of such genetic elements [11, 12].
The aim of this study was to identify whether there are mechanisms of resistance in the E. coli isolates causing UTIs from north of Iran or not.
MATERIALS AND METHODS
Sample collection and identification of bacterial strains. Urine samples were collected from appropriate patients in early morning mid-stream using sterile, wide mouthed glass bottles with screw cap tops between May and July 2012. Samples were maintained in an icebox until laboratory analysis. It did not last more than one hour between sample collection and sample analysis. Urine samples were cultured on nutrient, blood and MacConkey agar plates and incubated at 37°C for 18-24 h. The usual bacteriological methods were applied for cultivation, isolation and identification of isolates from urine samples. The isolates were stored at -70ºC in a tryptic soy broth containing 15% glycerol until processing. The isolates were entitled as E1 to E33.
Antimicrobial susceptibility test. According to Clinical and Laboratory Standards Institute (CLSI) [16], disc diffusion test was applied to identify the susceptibility of the isolates to the following antimicrobials: piperacillin (100 μg), streptomycin (10 μg), tetracycline (30 μg), chloramphenicol (30 μg), cefepime (30 μg), ceftriaxone (30 μg), ceftazidime (30 μg), cephalothin (30 μg), cefotaxime (30 μg), cefuroxime (30 μg), imipenem (10 μg), amoxicillin-clavulanic acid (20/10 μg), and ciprofloxacin (5 μg). E. coli ATCC 25922 and ATCC 35218 were used as the reference strains to control the quality of the applied antimicrobial agents. MDRs were described as resistance to three or more antimicrobials.
Phenotypic detection of ESBLs . Double-disk synergy tests were performed by placing disks of ceftazidime, cefotaxime, and cefepime (30 μg each) at 30 or 20 mm distance (center to center) from a disk containing amoxicillin (20 μg) and clavulanic acid (10 μg). When the cephalosporin zone was expanded by the clavulanate, ESBL production was supposed. It means that the zones produced by the disks with clavulanate were ≥5 mm larger than those without any inhibitor [16].
Detection of class 1 integrons by PCR. The existence of class 1 integrons was identified by PCR using specific primers for the integron integrase genes intI1 (Table 1). A single colony of each isolate was suspended in 25 ml reaction mixture containing 2.5 ml 10× PCR, 1.5 ml 50 mM MgCl2, 2 ml 2.5 mM dNTP, 1 ml primer (forward and reverse) together with 1 unit Taq DNA polymerase (5 U/ml). Volume of the reaction mixture was adjusted by adding filtered deionized water. PCR assays were performed in a DNA thermal cycler (BioRad, USA). The PCR conditions were initial denaturation at 94ºC for 12 min, followed by 30 cycles of 1 min at 94ºC, 30 s at 60ºC for annealing, 2 min at 72ºC for elongation, and final extension was conducted at 72°C for 10 min. A reagent blank was included in every PCR assay containing all components of the reaction mixture except for the bacteria. ATCC E. coli 25922 strain was used as negative control for all PCR assays, and E. coli ur-31 was used as positive control for intI1 gene. PCR products were subjected to horizontal gel electrophoresis on 1% agarose gel (type II, Sigma, USA) in Tris-borate EDTA buffer at 100 volt (50 mA) at room temperature for 1 h. DNA bands were visualized by gel staining with ethidiumbromide (0.5 mg/ml) for 30 min and then photographed [17].
Table 1.
Oligonucleotide primers used in the PCR analysis
| Primer | Oligoneucleotide sequence (5 ´ -3 ´ ) | Amplicon size (bp) |
|---|---|---|
| Int1F | GGTCAAGGATCTGGATTTCG | 491 |
| Int1R | ACATGCGTGTAAATCATCGTC | 491 |
| 5́-CS | GGCATCCAAGCAGCAAG | variable |
| 3́-CS | AAGCAGACTTGACCTGA | variable |
Detection of gene cassettes. To characterize the gene cassettes inserted in the class 1 integrons, the fragments including the cassette regions were amplified by PCR. The primers used to amplify cassettes of class 1 integrons were 5́CS and 3́CS (Table 1) as mentioned previously [18].
RESULTS
Antimicrobial resistance. The resistance patterns of the 33 urinary isolates are shown in the Table 2. All E. coli isolates (33 strains) were identified as multi-drug resistant. More than 50% of the isolates were resistant to tetracycline, cephalothin, cefuroxime, amoxicillin-clavulanic acid, and third generation cephalosporines. Nearly all isolates displayed sensitivity to piperacillin.
Table 2.
Antibiotic resistance profiles for ESBL and non-ESBL E. coli isolates
| Antimicrobial agents (µg) | Diffusion zone (mm) |
ESBL isolates
(n = 8) |
Non-ESBL isolates
(n = 25) |
P value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S* (%) | I* (%) | R* (%) | S (%) | I (%) | R (%) | |||||
| Streptomycin (10 ) | ≤11 | 0 (0) | 5 (62.5) | 3 (37.5) | 4 (16) | 12 (48) | 9 (36) | 0.01 | ||
| Cefepime (30) | ≤14 | 1 (12.5) | 2 (25) | 5 (62.5) | 13 (52) | 3 (12) | 9 (36) | 0.01 | ||
| Ceftriaxone (30) | ≤13 | 1 (12.5) | 0 (0) | 7 (87.5) | 9 (36) | 4 (16) | 12 (48) | 0.01 | ||
| Amoxicillin-clavulanic acid (20/10) | ≤13 | 0 (0) | 0 (0) | 8 (100) | 1 (4) | 3 (12) | 21 (84) | 0.01 | ||
| Chloramphenicol (30) | ≤12 | 3 (37.5) | 0 (0) | 5 (62.5) | 14 (56) | 1 (4) | 10 (40) | 0.01 | ||
| Ceftazidime (30) | ≤14 | 2 (25) | 2 (25) | 4 (50) | 10 (40) | 2 (8) | 13 (52) | 0.01 | ||
| Ciprofloxacin (5) | ≤15 | 3 (37.5) | 2 (25) | 3 (37.5) | 12 (48) | 5 (20) | 8 (32) | Ns* | ||
| Imipenem (10) | ≤15 | 2 (25) | 1 (12.5) | 5 (62.5) | 16 (64) | 3 (12) | 6 (24) | 0.01 | ||
| Cephalothin (30) | ≤14 | 0 (0) | 0 (0) | 8 (100) | 5 (20) | 0 (0) | 20 (80) | 0.01 | ||
| Cefotaxime (30) | ≤14 | 1 (12.5) | 0 (0) | 7 (87.5) | 7 (28) | 5 (20) | 13 (52) | 0.01 | ||
| Tetracyclines (10) | ≤11 | 0 (0) | 0 (0) | 8 (100) | 4 (16) | 2 (8) | 19 (76) | 0.01 | ||
| Cefuroxime (30) | ≤14 | 0 (0) | 1 (12.5) | 7 (87.5) | 6 (24) | 3 (12) | 16 (64) | 0.01 | ||
| Piperacillin(100) | ≤11 | 7 (87.5) | 1 (12.5) | 0 (0) | 22 (88) | 2 (8) | 1 (4) | Ns | ||
ESBL screening. Screening of ESBLs by double-disc synergy test indicated that eight isolates (24%) were ESBL producers. Phenotypic confirmatory disc diffusion test confirmed that 8 of the 33 isolates were ESBL producers. All ESBL isolates were completely resistant to tetracyclines, cephalothin and amoxicillin-clavulanic acid, but rates of resistance to other antibiotics were as bellow: amoxicillin-clavulanic acid 100%, cephalothin 100%, tetracycline 100%, ceftriaxone 87.5%, cefotaxime 87.5%, cefuroxime 87.5%, chloramphenicol 62.5%, cefepime 62.5%, imipenem 62%, ceftazidime 50%, streptomycin 37.5%, ciprofloxacin 37.5%. Antibiotic resistance profiles for ESBL and non-ESBL isolates are shown in Table 2. According to the Table, there was a significant correlation between ESBL production and resistance to all antibiotics except for ciprofloxacin and piperacillin (P < 0.01).
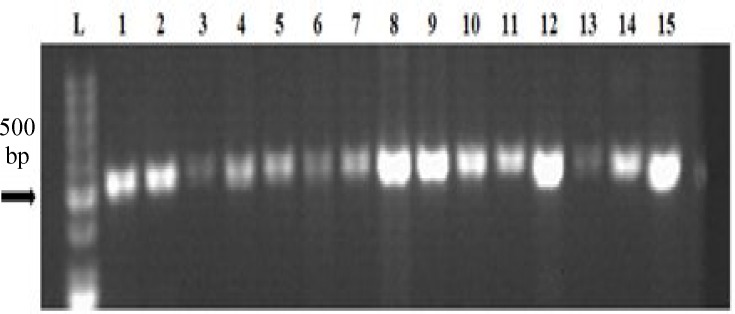
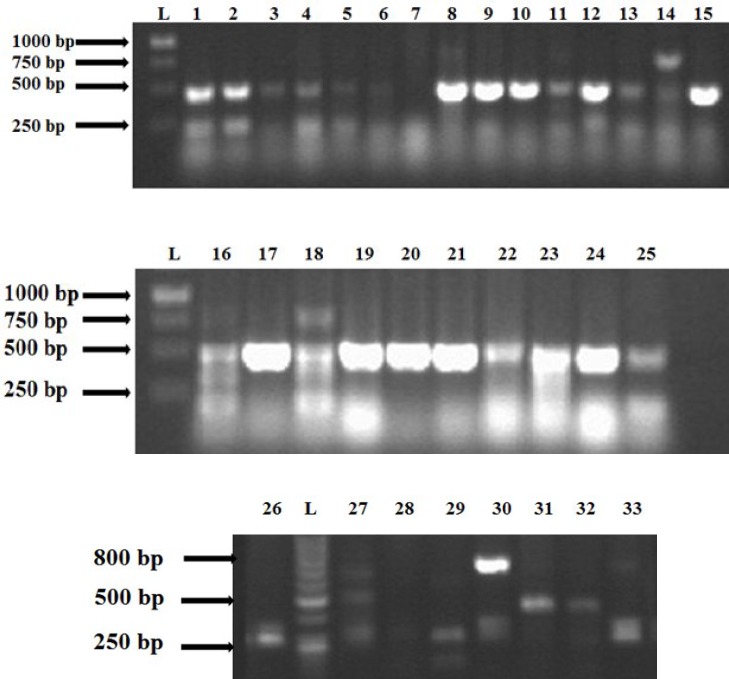
Detection of class 1 integrons and gene cassettes. PCR analysis revealed that 32 strains (97%) contained IntI1 gene (Fig. 1). Among the eight ESBL-producing strains, seven had class 1 integrons. The detected fragment size was 483 bp. The strains including integrons had variable gene cassettes (Fig. 2). One or maximum four gene cassettes was carried by class 1 integrons. Based on PCR amplification with primers 5́CS and 3́CS, the inserted gene cassettes of class 1 integrons were identified as 100, 250, 500, and 700 bp. The isolates similar in the number of gene cassettes were compared based on the results of disk diffusion (Table 3). Some isolates were similar in the size of gene cassettes, which seemed that they were different in the resistance profiles; however, some other had similar resistance profiles.
Fig.1.
PCR amplification of IntI1 gene among some MDR E. coli isolates on 0.7% agarose gel. In all isolates, a fragment of 483 bp was detected. Lane L: DNA ladder. Lane 1, E. coli ATCC 25922 as a positive control for IntI1 gene; Lanes 2-15, Amplified gene of IntI1 in the tested clinical isolates
Fig. 2.
The amplified gene cassettes with different sizes in the tested clinical isolates (1-33). L, DNA ladder
Table 3.
The relation between resistance profiles and the number of gene cassettes
| Isolates | Resistance |
Number of
gene cassettes |
Band(s)
size (bp) |
|---|---|---|---|
| E6, E7 | Amoxicillin-clavulanic acid Tetracyclines |
1 | 250 |
| E1, E2 | Chloramphenicol Tetracyclines |
2 | 250, 500 |
| E17, E19 E22, E23, E24, E25 |
Streptomycin Amoxicillin-clavulanic acid Cephalothin Tetracyclines Ceftriaxone Chloramphenicol Cephalothin Tetracyclines |
2 | 100, 500 |
| E8, E9 | Amoxicillin-clavulanic acid Ceftazidime Cephalothin Tetracyclines Cefuroxime |
2 | 250, 500 |
| E12, E15 | Amoxicillin-clavulanic acid Ceftazidime Cephalothin Cefotaxime |
2 | 250, 500 |
| E16, E18 | Amoxicillin-clavulanic acid Ceftriaxone Ceftazidime Cephalothin Cefotaxime Cefuroxime |
4 | 100, 250,500,700 |
DISSCUSION
Multi-drug resistant bacteria have been recently a problem in hospitalized patients worldwide. The frequency of MDR among clinical isolates is highly variable throughout the world and in different geographic areas and is rapidly changing according to time [19]. MDRs encoded by linked resistance genes exist on integrons, which are potentially movable genetic elements supposed to be involved in transferring MDR. This study was designed to examine the drug resistance pattern, the frequency of the class 1 integrons and ESBLs among the MDR E. coli isolates in UTI patients in north of Iran, Rasht.
Based on the results, E. coli was the most frequent microorganism isolated from urine samples. Other investigators also reported that E. coli was the most commonly isolated aerobic microorganism from UTIs [20, 21]. In our study, similar to Salem et al. survey [17], all E. coli isolates were recognized as MDR. In Ahangarzadeh Rezaee et al. study [22], 84.2% of the isolates were multi-drug resistant.
Nearly all E. coli isolates in this survey were sensitive to piperacillin, while some of them were resistant to other antimicrobials. The isolates displayed variable patterns of resistance to different antibiotics. More than 50% of the isolates were resistant to tetracycline, cephalothin, cefuroxime, amoxicillin-clavulanic acid, and third generation cephalosporines. According to Salem et al. [17], all E. coli isolates displayed resistance to penicillins, amoxicillin, cephalexin, and chloramphenicol. The isolates showed variable patterns of resistance to tetracycline, sulfamethoxazole-trimethoprim, erythromycin, and quinolone group.
In Ahangarzadeh Rezaee et al. [22] study on E. coli isolates from north-west of Iran, antibiotic resistance patterns were as follows: amoxicillin 99.3%, cephalothin 77.8%, co-trimoxazole 75%, tetracycline 72.8%, nalidixic acid 60.7%, norfloxacin 50.7%, ciprofloxacin 47.6%, ceftazidime 46.4%, gentamicin 33.6%, chloramphenicol 20.7%, nitrofurantoin 12.9%, amikacin 12.1%, and imipenem 1.4%.
The antimicrobial resistance genes located on integron-like structures are being dramatically reported worldwide [11]. Most of the newly described ESBL genes are generally found within integron-like structures (such as blaCTX-M, blaGES, or blaVEB-1) [11, 13]. On the other hand, ESBL-producing isolates are usually resistant to other antibiotics, such as aminoglycosides, tetracyclines, chloramphenicol etc. The fact that ESBL genes could be acquired by strains including particular integrons enlarges the possibilities of the selection of these strains by a variety of different antimicrobials. In addition, ESBL genes can be located on integrons, which may simplify the distribution of such genetic elements [11, 12].
For detection of class 1 integrons and ESBLs in this study, PCR amplification of intI1 gene and Double-disk synergy test method were used, respectively. Based on the results, 97% and 24% of the isolates contained class 1 integrons (483 bp in size) and ESBLs, respectively, which is indicative of very high frequency of occurrence of class 1 integrons in E. coli strains. Also, nearly all ESBL isolates carried class 1 integrons.
In 2008, Phongpaichit et al. [23] studied the susceptibility of 175 E. coli isolates from stools against 12 antimicrobial agents and also the presence of class 1 integrons. Their results indicated that 63% of the isolates included class 1 integrons, and the majority of the isolates (85%) were resistant to at least one antimicrobial agent with the following resistance rate: streptomycin 66%, tetracycline 60%, sulfamethoxazole 59%, ampicillin 52%, trimethoprim-sulfamethoxazole 47%, kanamycin 30%, nalidixic acid 27%, ciprofloxacin 23%, norfloxacin 22%, amoxicillin-clavulanic acid 16%, gentamicin 8%, and amikacin 2%.
The presence of class 1 and class 2 integrons were also investigated in Salem et al. study [17]. The results indicated that the class 1 integrons were observed in E. coli isolates (54%), and IntI1 gene yielded a DNA fragment of 1900 bp upon amplification by PCR, but class 2 integrons showed negative results. The absence of IntI2 gene may be attributed that class 2 integrons found in 4 to 20% uropathogenic E. coli strains [24, 25] as well as in other human pathogens [26], other animal pathogens [27], and various commensal bacteria [28, 29]. Essen-Zandbergen et al. [30], reported that among all E. coli isolated from animals, class 1 integrons were found in 76%, and the size of gene cassettes were 600, 1000, 1550, 2000, 2200, and 2500 bp. Martinez-Freijo et al. [31] study represented that 62% of E. coli isolates were integron 1-positive and had different sizes of inserted gene cassette, including 1500, 1600, 1800, 2000, and 3000 bp. In Farshad et al. [32] study performed in Shiraz (Iran), of 96 E. coli isolates from urine samples tested, 6.25% of the strains were the integron 1-positive isolates. In other words, the prevalence of class 1 integrons among the isolates was low.
In Madrid (Spain), Machado et al. [33] investigated the existence of class 1 integrons in ESBL-producing in 133 E. coli isolates and reported that 39% of the isolates were ESBL producing, and class 1 integrons were more common among 67% of the isolates. Thus, based on their study, a significant relationship was not observed between the spread of genetic elements encoding ESBL and the dispersion of integrons in these isolates.
Karimi et al. [34] detected class 1 integrons in ESBL producing in 100 E. coli isolates in Tehran, Iran. Thirty E. coli isolates were positive for ESBLs, and the frequency of class 1 integrons was 52%. Also, 54% of ESBL producers had class 1 integrons. The detected fragment of intI1 was 9000 bp.
In a survey by Al-Assil et al. [35] in Aleppo (Syria), 61.33% of uropathogenic E. coli were ESBL producers, and class 1 integrons were detected in 54.6% of them. Prevalence of class 1 integrons in ESBL isolates was 34%.
In conclusion, this study demonstrates that unlike other researches, the prevalence of class 1 integrons in ESBL producing E. coli strains is very common. The results of the current study also indicated that there are at least two resistance mechanisms in our isolates, and they can be transferred to other clinical strains, then it is very important to identify and control the resistant strains. Increased resistance may be related to the lack of proper research, abuse of chemotherapeutic agents, public misuse of antibiotics, and little or no preventive measure. Thus, piperacillin can be used as a therapeutic agent or the most effective antibiotic in controlling UTI infections in the patients infected by ESB-producing E. coli carrying class 1 integrons in Rasht (Guilan Province, Iran). Nevertheless, more studies are needed to be performed in this area.
ACKNOWLEDGEMENTS
The authors would like to thank the University of Guilan (Iran) for its financial support as well as the staffs from Razi Pathobiology Laboratory for providing samples and for their collaboration in this research.
References
- 1.Huang ES, Stafford RS. National patterns in the treatment of urinary tract infections in women by ambulatory care physicians. Arch Intern Med. 2002 Jun;162(11):41–7. doi: 10.1001/archinte.162.1.41. [DOI] [PubMed] [Google Scholar]
- 2.Gupta K, Sahm DF, Mayfield D. Antimicrobial resistance among uropathogens that cause community-acquired urinary tract infections in women: a nationwide analysis. Clin Infect Dis. 2001 Jun;33(1):89–94. doi: 10.1086/320880. [DOI] [PubMed] [Google Scholar]
- 3.Shapiro JA. Views about evolution are evolving. Am Soc Microbiol News. 1999;65:201–7. [Google Scholar]
- 4.Soulsby L. Resistance to antimicrobials in humans and animals. Br J Med. 2005 Nov;331:1219–20. doi: 10.1136/bmj.331.7527.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy Chowdhury P, Ingold A, Vanegas N, Martínez E, Merlino J, Merkier AK, et al. Dissemination of multiple drug resistance genes by class 1 integrons in Klebsiella pneumoniae isolates from four countries: a comparative study. Antimicrob Agents Chemother. 2011 Apr;55:3140–9. doi: 10.1128/AAC.01529-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betteridge T, Partridge SR, Iredell JR, Stokes HW. Genetic context and structural diversity of class 1 integrons from human commensal bacteria in a hospital intensive care unit. Antimicrob Agents Chemother. 2011 May;55:3939–43. doi: 10.1128/AAC.01831-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stalder T, Barraud O, Casellas M, Dagot C, Ploy M. Integron involvement in environmental spread of antibiotic resistance. Front Microbiol. 2012 Apr;3:119. doi: 10.3389/fmicb.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collis CM, Kim MJ, Stokes HW, Hall RM. Integron-encoded IntI integrases preferentially recognize the adjacent cognate attI site in recombination with a 59-be site. Mol Microbiol. 2002 Dec;46(5):1415–27. doi: 10.1046/j.1365-2958.2002.03260.x. [DOI] [PubMed] [Google Scholar]
- 9.Poole K. Resistance to β-Lactam antibiotics. Cell Mol Life Sci. 2004 Sep;61(17):2200–23. doi: 10.1007/s00018-004-4060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pitout JDD, Hamilton N, Church DL, Nordmann P, Poirel L. Development and clinical validation of a molecular diagnostic assay to detect CTX-M-type β-lactamases in Enterobacteriaceae. Clin Microbiol Infect. 2007 Mar;13(3):291–7. doi: 10.1111/j.1469-0691.2006.01645.x. [DOI] [PubMed] [Google Scholar]
- 11.Bonnet R. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004 Jan;48(1):1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canto´n R, Coque TM, Baquero F. Multi-resistant gram-negative bacilli: from epidemics to endemics. Curr Opin Infect Dis. 2003 Aug;16(4):315–25. doi: 10.1097/00001432-200308000-00003. [DOI] [PubMed] [Google Scholar]
- 13.Jacoby GA, Sutton L. Properties of plasmids responsible for production of extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1991 Jan;35(1):164–9. doi: 10.1128/aac.35.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Preston KE, Graffunder EM, Evans AM, Venezia RA. Survey of plasmid-associated genetic markers in Enterobacteriaceae with reduced susceptibilities to cephalosporins. Antimicrob Agents Chemother. 2003 Jul;47(7):2179–85. doi: 10.1128/AAC.47.7.2179-2185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villa L, Pezzella C, Tosini F, Visca P, Petrucca A, Carattoli A. Multiple-antibiotic resistance mediated by structurally related IncL/M plasmids carrying an extended-spectrum beta-lactamase gene and a class 1 integron. Antimicrob Agents Chemother. 2000 Oct;44(10):2911–4. doi: 10.1128/aac.44.10.2911-2914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing: Twenty-first information supplement M100-S21. Wayne: CLSI; [Google Scholar]
- 17.M. Salem M, Muharram M, Alhosiny IM. Distribution of classes 1 and 2 integrons among multi drug resistant E. coli isolated from hospitalized patients with urinary tract infection in Cairo, Egypt. Aus J Basic Appl Sci. 2010 Jan;4(3):398–407. [Google Scholar]
- 18.Le´ vesque C, Piche´ L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995 Jan;39(1):185–91. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, et al. Multidrug-resistant, extensively drug-resistant and pandrugresistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012 Mar;18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 20.Al-Agamy MH, Ashour MS, Wiegand I. First description of CTX-M β-lactamase-producing clinical Escherichia coli isolates from Egypt. Int J Ant Microb Agents. 2006 Jun;27(6):545–8. doi: 10.1016/j.ijantimicag.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Asour HM, Elsharif A. Species distribution and antimicrobial susceptibility of Gram-negative aerobic bacteria in hospitalized cancer patients. J Trans Med. 2009 Feb;7(2):1–14. doi: 10.1186/1479-5876-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahangarzadeh Rezaee M, Sheikhalizadeh V, Hasani A. Detection of Integrons among multi-drug resistant (MDR) Escherichia Coli strains isolated from clinical specimens in northern wset of Iran. Braz J Microbiol. 2011 Oct;42(4):1308–13. doi: 10.1590/S1517-838220110004000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phongpaichit S, Wuttananupan K, Samasanti W. Class 1 integrons and multidrug resistance among Escherichia coli isolates from human stools. Southeast Asian J Trop M. 2008 Mar;39(2):279–87. [PubMed] [Google Scholar]
- 24.Yu HS, Lee JC, Kang HY, Ro DW, Chung JY, Jeong YS, et al. Changes in gene cassettes of class 1 integrons among Escherichia coli isolates from urine specimens collected in Korea during the last two decades. J Clin Microbiol. 2003 Dec;41(12):5429–33. doi: 10.1128/JCM.41.12.5429-5433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solberg OD, Ajiboye RM, Riley LW. Origin of class 1 and 2 integrons and gene cassettes in a population-based sample of uropathogenic Escherichia coli. J Clin Microbiol. 2006 Apr;44(4):1347–51. doi: 10.1128/JCM.44.4.1347-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranjbar R, Aleo A, Giammanco GM, Dionisi AM, Sadeghifard N, Mammina C. Genetic relatedness among isolates of Shigella sonnei carrying class 2 integrons in Tehran, Iran, 2002–2003. BMC Infect Dis. 2007 Jun;7(6):62–8. doi: 10.1186/1471-2334-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadlec K, Schwarz S. Analysis and distribution of class 1 and class 2 integrons and associated gene cassettes among Escherichia coli isolates from swine, horses, cats and dogs collected in the BfT-GermVet monitoring study. J Antimicrob Chemother. 2008 Sep;62(3):469–73. doi: 10.1093/jac/dkn233. [DOI] [PubMed] [Google Scholar]
- 28.Barlow RS, Pemberton JM, Desmarchelier PM, Gobius KS. Isolation and characterization of integron-containing bacteria without antibiotic selection. Antimicrob Agents Chemother. 2004 Mar;48(3):838–42. doi: 10.1128/AAC.48.3.838-842.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barlow RS, Fegan N, Gobius KS. A comparison of antibiotic resistance integrons in cattle from separate beef meat production systems at slaughter. J Appl Microbiol. 2008 Mar;104(3):651–8. doi: 10.1111/j.1365-2672.2007.03572.x. [DOI] [PubMed] [Google Scholar]
- 30.Essen-Zandbergen AV, Smith H, Veldman K, Mevius D. Occurrence and characteristics of class 1, 2 and 3 integrons in Escherichia coli, Salmonella and Campylobacter spp. in the Netherlands. J Antimicrob Chemother. 2007 Mar;59(4):746–50. doi: 10.1093/jac/dkl549. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Freijo P, Fluit AC, J Schmitz F, Grek VSC, Verhoef J, Jones ME. Class 1 integrons in gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J Antimicrob Chemother. 1998 Dec;42(6):689–96. doi: 10.1093/jac/42.6.689. [DOI] [PubMed] [Google Scholar]
- 32.Frashad S, Japoni A, Hosseini M. Low distribution of integrons among multidrug resistance E. coli strains isolated from children with community-acquired urinary tract infections in Shiraz, Iran. Pol J Microbiol. 2008 Jul;57(3):193–8. [PubMed] [Google Scholar]
- 33.Machado E, Canto´n R, Baquero F, Gala´n GC, Rolla´n A, Peixe L, et al. Integron content of extended-spectrum-β-Lactamase-producing Escherichia coli strains over 12 years in a single hospital in madrid, Spain. J Agents Chemother. 2005 May;49(5):1823–9. doi: 10.1128/AAC.49.5.1823-1829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karimi A, Rahbar M, Fallah F, Navidinia M, Malekan MA. Detection of integron elements and gene groups encoding ESBLs and their prevalence in Escherichia coli and Klebsiella isolated from urine samples by PCR method. Afr J Microbiol Res. 2012 Feb;6(8):1806–9. [Google Scholar]
- 35.Al-Assil B, Mahfoud M, Hamzeh, AR. First report on class 1 integrons and trimethoprim-resistance genes from dfrA group in uropathogenic E. coli (UPEC) from the aleppo area in Syria. Mob Gen Elements. 2013 Jun;3(3):25204–9. doi: 10.4161/mge.25204. [DOI] [PMC free article] [PubMed] [Google Scholar]