Abstract
Living whole-cell bioreporters serve as environmental biosentinels that survey their ecosystems for harmful pollutants and chemical toxicants, and in the process act as human and other higher animal proxies to pre-alert for unfavorable, damaging, or toxic conditions. Endowed with bioluminescent, fluorescent, or colorimetric signaling elements, bioreporters can provide a fast, easily measured link to chemical contaminant presence, bioavailability, and toxicity relative to a living system. Though well tested in the confines of the laboratory, real-world applications of bioreporters are limited. In this review, we will consider bioreporter technologies that have evolved from the laboratory towards true environmental applications, and discuss their merits as well as crucial advancements that still require adoption for more widespread utilization. Although the vast majority of environmental monitoring strategies rely upon bioreporters constructed from bacteria, we will also examine environmental biosensing through the use of less conventional eukaryotic-based bioreporters, whose chemical signaling capacity facilitates a more human-relevant link to toxicity and health-related consequences.
Keywords: Bioluminescence, Bioremediation, Bioreporter, Ecotoxicology, Fluorescence
1. Introduction
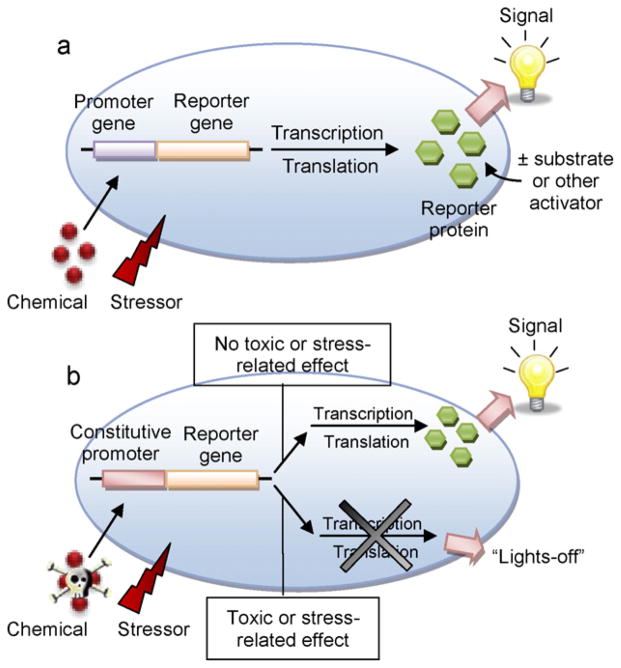
Bioreporters consist of prokaryotic (bacteria) or eukaryotic (fungi, algae, animal) cells that serve as living sensors for priority environmental pollutants and chemicals of toxicological concern. Evolution has provided these cells with unique genetic traits that permit their adaptation to (i.e., for metabolism) or defense against (i.e., a bactericidal toxin) the chemical agents to which they are exposed, thus facilitating their ability to functionally survive and propagate in nearly any ecosystem niche. With an understanding of the genetic mechanisms involved, genetic engineering, synthetic biology, or other nucleic acid-based manipulative techniques can be applied to convert cells into controllable on/off switches tuned to a particular chemical, chemical class, or toxicological effector, the result of which yields what are commonly referred to as ‘lights-on’ or ‘lights-off’ bioreporter constructs (Fig. 1). In a lights-on bioreporter, the cell emits light when exposed to a targeted chemical or toxin. This requires a fundamental understanding of the genetic mechanism(s) involved in that response and the genetic element, or promoter, that controls it. In the native cell, the promoter regulates the genes that respond to the chemical or toxin. In the bioreporter cell, the promoter’s link to these genes is disrupted and replaced with a reporter gene that now, when activated by the promoter, is transcribed and translated to a reporter protein that emits a bioluminescent, fluorescent, or colorimetric signal. Lights-on bioreporters are most often applied in environmental sensing and monitoring schemes, and can be designed to either specifically or nonspecifically report on their interactions with inducer chemicals. If specific, the promoter/reporter gene linkage directly associates with a particular chemical or chemical class, and the generation of light as well as the intensity of light identifies that chemical and indicates its overall concentration. If nonspecific, the promoter/reporter gene construct responds via an increase in light intensity to, for example, general stress or DNA damage related to exposure to a cytotoxic, mutagenic, or genotoxic chemical agent. Although the identity of the particular chemical causing the toxic interaction remains unknown, these bioreporters rapidly and efficiently pre-alert to environmental offenses that can then be more closely examined using chemical-specific bioreporters or conventional analytical techniques such as gas chromatography/mass spectrometry (GC/MS).
Fig. 1.
The fundamentals of whole-cell bioreporter sensing. (a) A ‘lights-on’ bioreporter emits light when its promoter gene is activated upon exposure to a target of interest, and can function either specifically to identify a chemical or chemical class or nonspecifically to identify a toxic or stress-related cellular interaction. (b) A ‘lights-off’ bioreporter emits light continuously and signals the presence of a toxic chemical or stress-related interaction with its constitutive promoter via a reduction in its light emission intensity.
In a lights-off bioreporter, the cell is designed to continuously (constitutively) emit light. A measured decrease in light intensity after chemical exposure indicates that the chemical’s interaction with the bioreporter has caused cellular damage or disrupted general metabolism. Thus, although the chemical itself cannot be identified, it can be classified as displaying toxicity towards a living organism, and pre-warns of potentially analogous toxicity towards higher life forms (i.e., humans). For the most part, these bioreporters are designed around prokaryotic or lower eukaryotic cells that, to date, have served as suitable surrogates for more complex organismal systems, but this is gradually changing as we improve our abilities to manipulate higher eukaryotes towards defined bioreporter sensing strategies.
The emphasis of this review will center on bioreporter sensing technologies that have been applied within real-world environmental settings since this forms the foundation of their practical detection and monitoring capacities. The reader is reminded that there exists a much larger number of bioreporters than those discussed here that still remain laboratory bound, and with capacities to detect a wide array of chemicals and chemical interactions (see recent reviews by Robbens et al. (2010), Diplock et al. (2010), and Hynninen and Virta (2010) for more comprehensive examinations of bioreporter systems).
2. Common reporter elements
Whole-cell bioreporters most commonly incorporate reporter genes that code for signaling elements that emit bioluminescent, fluorescent, or colorimetric endpoints, with bioluminescence being derived from the bacterial (lux) or firefly (luc) genes, fluorescence from the green fluorescent protein (gfp) gene and its other colored variants, and colorimetric endpoints relying upon the β-galactosidase (lacZ) gene.
2.1. Bioluminescence
Bioluminescence–the chemical generation of light within a living organism–is a widely used reporter element in environmental biosensing. The chemical reaction yielding bioluminescence is catalyzed by an enzyme (luciferase) that reacts with a substrate (luciferin) to produce an excited state molecule that generates photons as it relaxes back to its ground state. Biotechnological exploitation of luciferase/luciferin reactions has generally focused on the bacterial bioluminescent system referred to as lux and the firefly bioluminescent system referred to as luc.
2.1.1. Bacterial luciferase (lux)
The luciferin substrate for the bacterial (Vibrio, Photorhabdus, and Photobacterium genera) bioluminescent reaction is a reduced riboflavin phosphate (FMNH2) that is oxidized by the luciferase enzyme in association with a long-chain aldehyde and molecular oxygen. This reaction is controlled genetically by a five gene operon consisting of the luxA, luxB, luxC, luxD, and luxE genes (denoted as luxCDABE to designate the order of the genes in the operon) (Meighen, 1994). The luxA and luxB (luxAB) gene products form a heterodimeric luciferase while the luxC, luxD, and luxE (luxCDE) gene products supply and regenerate the long-chain aldehyde. The remaining required oxygen and FMNH2 reactants are scavenged within the cell through ancillary metabolic processes. The end result is the emission of a blue/green 490 nm light signal. Two classes of lux-based bioreporters are used. The simplest integrates only the luxAB genes, resulting in bioreporters containing only the luciferase enzyme which then requires the exogenous addition of aldehyde, usually in the form of n-decanal, to the reaction. Although this creates a brighter and easier to detect signal due to substrate saturation, it makes signaling a time point dependent occurrence contingent upon the external addition of an activating compound. Nonetheless, numerous bioreporters harboring the luxAB genes have been designed around bacterial, yeast, and mammalian genetic systems and remain well tested within environmental, food, and water-based bioassays.
To permit continuous substrate-independent bioluminescent signaling, bioreporters can alternatively be designed to accommodate the complete luxCDABE gene cassette. These bioreporters contain the full complement of the luciferase/luciferin reaction and can generate bioluminescence spontaneously without any user mandated interventions. This advantageously endows the bioreporter with real-time to near real-time detection capabilities. The luxCDABE genetic operon has been synthetically optimized for efficient gene expression away from its native AT-rich state towards GC-rich microorganisms, thereby allowing for its integration into a wider variety of bacterial hosts (Craney et al., 2007). This is of value because it improves complementary matching of bioreporter hosts to test environments to better accommodate long-term bioreporter maintenance and survival under natural ecological conditions. The luxCDABE operon has also been synthetically restructured for efficient expression in mammalian cells, which is of significant advantage since it allows for the design of autonomously sensing eukaryotic bioreporters whose response kinetics become much more relevant towards human health than that suggested by analogous bacterial or lower eukaryotic bioreporter surrogates (Close et al., 2010).
2.2. Firefly luciferase (luc)
The luc gene, derived most commonly from the firefly Photinus pyralis, is a popular reporter gene due to its high light output and rapid response kinetics (Close et al., 2009). The Luc protein catalyzes the oxidation of a reduced luciferin substrate in the presence of ATP-Mg2+ and oxygen to generate a yellow/green 562 nm light signal, the quantum yield of which is the largest of any of the currently characterized bioluminescent systems. The Luc protein requires no post-translational modifications and is thus immediately available once translated. luc reporter systems require the exogenous addition of the luciferin substrate, so luc-based bioreporters are not able to react autonomously or monitor user defined targets in a continuous fashion. Nonetheless, its maximal light output translates into very sensitive assays that have been applied over a wide range of organic compounds, heavy metals, and environmentally significant estrogenic and endocrine disruptor agents.
2.3. Green fluorescent protein (GFP)
GFP is a photoprotein cloned from the jellyfish Aequorea victoria and is representative of a large family of natural and recombinant photoproteins capable of producing light in a palette of colors (Shaner et al., 2005). It requires no substrate to do so, but is dependent on an external light source to activate its fluorescent output and is thus always tied to instrumentation capable of performing this task. GFP and its variants are capable of functioning semi-continuously and perform well as near real-time sensors in environmental monitoring assessments. Due to their differently colored light emission outputs, they can additionally be implemented in dual-color formats where each color indicates a separate event (Hever and Belkin, 2006).
2.4. β-Galactosidase (lacZ)
The lacZ gene cloned from E. coli encodes a β-galactosidase (β-gal) enzyme that catalyzes the hydrolysis of substrate β-galactoside disaccharides into monosaccharides. lacZ-based bioreporters traditionally yielded a colorimetric signal when supplied with the substrate o-nitrophenyl-β-D-galactoside (ONPG). Commercially available kits such as the SOS Chromotest implement lacZ-based fusions to DNA responsive genes to monitor environmental samples for potential mutagenic or carcinogenic genotoxic compounds (Quillardet et al., 1982). Besides colorimetric endpoints, lacZ bioreporters can be used in conjunction with a variety of β-galactoside substrates that permit luminescent (Nazarenko et al., 2001), chemiluminescent (Jain and Magrath, 1991), or fluorescent (Rowland et al., 1999) endpoints. A disadvantage of lacZ-based biosensing strategies is that β-gal is endogenously present in natural environmental matrices and contributes to elevated background activity. Also, bioreporter cells must be permeabilized upon substrate addition to quantify β-galactosidase activity, which results in discontinuous and oftentimes delayed data accumulation, but electrochemical and amperometric interfaces can be implemented to bypass permeabilization steps (Ron and Rishpon, 2010).
3. Bacterial bioreporter applications in environmental assessment
Bacterial cells, being genetically easy to manipulate and robust enough to survive under various environmental conditions, have been employed as host cells in constructions of the majority of whole-cell bioreporters. By exploiting intrinsic cellular processes such as the stress response, defense against toxins, and catabolism of various compounds, bacterial bioreporters can be designed to detect a variety of chemicals and physical conditions. A substantial number of bacterial bioreporters have been constructed and characterized (recently reviewed in Diplock et al., 2010; Eltzov and Marks, 2011; Ripp et al., 2011; van der Meer and Belkin, 2010), however, their applications in the field are relatively limited, primarily due to regulations limiting or excluding the environmental release of recombinant DNA. To explore what bacterial bioreporters can offer as environmental monitors, this section highlights recent applications of bacterial bioreporters in environmental assessment. Note that one of the primary advantages of bioreporters is their ability to report on bioavailability–that portion of the chemical that is freely available to cross the cellular membrane and therefore having a biological effect. The relationship between bioreporter sensing and bioavailability under environmental influences is a complex process that depends on the bioreporter’s physiology, growth rate, membrane composition, transport mechanisms, and a host of other factors (Harms et al., 2006). Bioavailability is also specific in terms of the receptor, where, for example, a sample that is not bioavailable to bacteria may be bioavailable to plants or animals, or vice versa. Therefore, results obtained using bacterial bioreporters still need to be carefully interpreted as to what they actually measure, and a suite of different bioassays might be more appropriate to obtain comprehensive analyses to satisfy the goals of environmental assessments.
3.1. Bioreporters for the detection and monitoring of heavy metals
Heavy metals are important inorganic contaminants with regard to risk assessment due to their environmental prevalence and biological toxicity toward humans and wildlife. A long history of characterization of microbial metal resistance has accumulated significant knowledge of genetic elements and regulatory mechanisms that specifically respond to metals. Typically, bacteria counteract metal toxicity by means of efflux transporters that actively export the toxic metal ions to the outside of the cell and/or modification enzymes that transform the metals to less toxic forms (Nies, 1999). Because expression of the proteins involved in these defense machineries use energy, thus imposing extra metabolic burden to the host cells, they are only induced when metal ions are present at toxic concentrations. The induction of particular defense machinery often involves a specific transcriptional regulator, which upon metal-binding activates the transcription of downstream defense related genes. For example, defenses against mercury (Hg) and arsenic (As) are regulated by the transcriptional regulators MerR and ArsR, respectively. A generic metal bioreporter can be constructed by fusing the genes encoding the metal-specific transcriptional regulator as well as the promoter/operator of the defense related genes to a promoterless reporter gene (luc, gfp, etc.) or reporter genes (in the case of luxAB and luxCDABE). Upon exposure, the transcriptional regulator is activated by metal binding and subsequently turns on the expression of the reporter gene(s), finally leading to production of a measurable signal.
Because the toxicity of a particular metal is greatly dependent on the form in which it exists, analysis using bioreporters yields more biologically relevant information than conventional chemical methods. Many reports using artificially amended samples have demonstrated in proof-of-concept that whole-cell bacterial bioreporters can be used for assessment of bioavailability and toxicity of metals in different sample matrices (Bondarenko et al., 2008; Brandt et al., 2006; Ivask et al., 2009, 2002) (a more detailed review is available in Hynninen and Virta, 2010). Rather than discussing the entire inventory of bacterial metal bioreporters, here we present a brief overview of their applications in field samples and refer the reader to Table 1 for a more comprehensive list.
Table 1.
Field applications of bacterial bioreporters for heavy metal bioavailability assessment.
| Bioreporter
|
Field application
|
||||||
|---|---|---|---|---|---|---|---|
| Host strain | Reporter construct | Analyte | Response time | Detection limit | Matrix | Response time | Reference |
| Bacillus subtilis BR151 (pTOO24) (Tauriainen et al., 1998) | cadC-luc | Cd | 3 h | 3.3 nM (Cd) (Tauriainen et al., 1998) 3.7 μg/L (Cd) (Hakkila et al., 2004) |
Sediment (water suspension and water extract) Soil (water suspension and water extract) |
2 h 2 h |
Hakkila et al. (2004) Ivask et al. (2011) Ivask et al. (2004) |
| E. coli CM1166 (pC200) (Corbisier et al., 1993) | ars-luxAB | As, Sb | 1 h | <0.1 mg/L (Flynn et al., 2002) | Soil and sediment (water extract) | 1 h | Flynn et al. (2002, 2003) |
| E. coli DH5α (pVLCD1) (Liao et al., 2006) | cadC-gfp | Cd, Pb, Sb | 2 h | 0.1 nM (Cd(II)), 10 nM (Pb(II)), 0.1 nM (Sb(III)) (Liao et al., 2006) | Sediment and soil (water extract) | 2 h | Liao et al. (2006) |
| E. coli DH5α (pJAMA-arsR) (Stocker et al., 2003) | arsR-luxAB | As | 0.5 h | 4–7 μg/L (Stocker et al., 2003; Trang et al., 2005) | Groundwater Tap water |
1.5 h 1 h |
Trang et al. (2005) Stocker et al. (2003) |
| E. coli JM109 (pNM2) (Larose et al., 2011) | merRT-luxCDABE | Hg | 2 h | 0.5 ng/L (Larose et al., 2011) | Snowpack | 2 h | Larose et al. (2011) |
| E. coli MC1061 (parsRluxCDABE) (Hakkila et al., 2002) | arsR-luxCDABE | As | 3–4 h | 141 μg/L (As(V)), 18 μg/L (As(III)) (fiber-optic immobilized); 80 μg/L (As(V)), 8 μg/L (As(III)) (free bacteria) (Ivask et al., 2007) | Sediment (water suspension) | 3–4 h | Ivask et al. (2007) |
| E. coli MC1061 (pmerBRBS luc) (Ivask et al., 2001) | merRB-luc | Organic Hg | 2 h | 0.2 nM (methyl Hg), 1 nM (phenyl Hg), 10 μM (dimethyl Hg) (Ivask et al., 2001) | Sediment (water suspension) | 2 h | Hakkila et al. (2004) |
| E. coli MC1061 (pmerRluxCDABE) (Hakkila et al., 2002) | merR-luxCDABE | Hg | 2–3 h | 2.6–6.3 μg/L (immobilized) (Hakkila et al., 2004; Ivask et al., 2007); 0.03 μg/L (free) (Ivask et al., 2007) | Sediment (water suspension) | 2–3 h | Hakkila et al. (2004), Ivask et al. (2007) |
| E. coli MC1061 (pSLcueR/pDNPcopAluc) (Hakkila et al., 2004) | cueR, copA-luc | Cu | 2 h | NR | Sediment (water suspension) | 2 h | Hakkila et al. (2004) |
| E. coli MC1061 (pTOO11) (Virta et al., 1995) | merR-luc | Hg | 1–2 h | 0.1 fM (Virta et al., 1995); 6.7 mg/L (Hakkila et al., 2004) | Sediment (water suspension) | 2 h | Hakkila et al. (2004), Lappalainen et al. (2000) |
| E. coli MC1061 (pTOO31) (Tauriainen et al., 1999) | arsR-luc | As | 1 h | 33 nM (NaAsO2), 33 μM (Na2 HAsO4) (Tauriainen et al., 1999); 6.1 μg/L (Hakkila et al., 2004) | Sediment (water suspension) Soil (water suspension and water extract) |
2 h 2 h |
Hakkila et al. (2004) Turpeinen et al. (2003) |
| P. fluorescens DF57-Cu15 (Tom-Petersen et al., 2001) | Cu-inducible promoter-luxAB | Cu | 1.5 h | 0.005 mg/L (Cu2+) (Maderova et al., 2011) | Soil (water suspension and water extract) | 1.5 h | Brandt et al. (2006, 2008), Maderova et al. (2011) |
| Ralstonia eutropha AE 1239 (Corbisier et al., 1996) | cop-luxCDABE | Cu | NR | NR | Incinerator fly-ashes (acetate extract and medium suspension) | 4–5 h | Corbisier et al. (1996) |
| R. metallidurans AE1433 (Corbisier et al., 1996) | cupS-luxCDABE | Pb, Zn, Cd, Co | 5 h | 0.4 ppm (Pb) (Magrisso et al., 2009) | Soil (water suspension) | 3 h | Magrisso et al. (2009) |
| R. metallidurans AE2515 (Tibazarwa et al., 2001) | cnrYXH-luxCDABE | Ni, Co | 4–6 h | 0.1 μM (Ni), 9 μM (Co) (Tibazarwa et al., 2001) | Soil (medium suspension) | Continuous monitoring | Everhart et al. (2006) |
| Staphylococcus aureus RN4220 (pTOO24) (Tauriainen et al., 1998) | cadC-luc | Cd, Pb | 2 h | 10 nM (Cd), 33 nM (Pb) (Tauriainen et al., 1998) 1 nM (Sn) |
Sediment (water suspension) Soil (water suspension and water extract) Soil (water extract) |
2 h 2 h 2 h |
Hakkila et al. (2004) Ivask et al. (2004) Turpeinen et al. (2000) |
| Synechococcus sp. CCMP 2669 (Boyanapalli et al., 2007) | PisiAB-luxAB | Fe3+ | 12 h | 1 nM (Boyanapalli et al., 2007) | Seawater | 12 h | Boyanapalli et al. (2007), Breitbarth et al. (2009) |
| Synechococcus sp. KAS101 (Durham et al., 2002) | PisiAB-luxAB | Fe3+ | 12 h | 0.45 nM (Hassler et al., 2006) | Lake water | 12 h | Breitbarth et al. (2009), Hassler et al. (2009), McKay et al. (2005), Porta et al. (2005) |
| Synechocystis sp. PCC 6803 coaLux (Peca et al., 2008) | coaR-PcoaT-luxAB | Co | 3 h | 0.3 μM (Co2+) (Peca et al., 2008) | Soil (acetate extract) | 3 h | Peca et al. (2008) |
| Synechocystis sp. PCC 6803 nrsLux (Peca et al., 2008) | nrsR-PnrsBACD-luxAB | Ni | 3 h | 0.2 μM (Ni2+) (Peca et al., 2008) | Soil (acetate extract) | 3 h | Peca et al. (2008) |
NR: not reported.
3.1.1. Heavy metals in soil and sediment
Industry and mining contribute substantially to heavy metal soil contamination, but sources such as road runoff from automobiles and the spread of metal-containing livestock manure also function as environmental inputs (Bolan et al., 2004). Metal contamination in soils as well as sediments is traditionally evaluated by the total amount of metals determined using analytic chemical methods after acid extraction of solid samples. However, these types of methods cannot distinguish between bioavailable and non-bioavailable forms, and therefore do not indicate actual toxicity towards biological systems. Determination of the bioavailable fraction of metals in soil and sediment samples is of particular interest since a variety of sample attributes, such as soil type, organic matter content, pH, redox potential, and humidity can affect metal bioavailability (Hynninen and Virta, 2010). Therefore, chemically defined metal contents need more biologically relevant interpretations, which can be achieved using whole-cell bioreporters as living biosentinels tuned to heavy metal bioavailability.
Metals are generally present in soil and sediments in two forms, a soluble phase that can be readily extracted by water and a particle-absorbed solid phase that is not water extractable (Degryse et al., 2009). While it is widely accepted that the water extractable fraction is directly bioavailable (Giller et al., 2009), currently there is no agreement on the bioavailability of particle-bound metals (Magrisso et al., 2009). Soil–water suspensions obtained by mixing air-dried soil with water, and soil–water extract, which is the particle-free supernatant formed after centrifugation of the suspension, are the two commonly employed sample preparation methods (Hynninen and Virta, 2010). Bioavailable metal in the soluble fraction can be analyzed using water extracts, whereas bioreporter analysis of soil suspensions wherein bioreporter cells are in direct contact with soil particles allows for measurement of bioavailable metal including the absorbed fraction present in the sample, keeping in mind the imprecision of bioavailability (Harms et al., 2006).
Comparisons between the amount of bioavailable metal measured by bioreporter analysis using either water extracts or soil suspensions to the amount of total metal determined by chemical methods indicate that bioavailable metal only accounts for a small fraction of total metal and that the bioavailable fraction is not always correlated with total metal concentration. Ivask et al. (2007) analyzed 10 soil and sediment samples using an E. coli merR-luxCDABE strain and an E. coli ars-luxCDABE strain to examine Hg and As bioavailability in suspensions. The results showed that only 1.2–6.7% of total Hg and 0.9–4.9% of total As was available for detection by bioreporter cells. In a larger scale study, bioavailable Cd and lead (Pb) were found to be 0.5–56% of total Cd and 0.2–8.6% of total Pb across 50 different soil samples (Ivask et al., 2004). These and other similar findings suggest that chemically determined total metal concentration alone is not an adequate indicator for the purpose of ecological evaluation.
Bioreporter analyses of water extracts of field soil and sediment samples have also revealed that water extractable metal is not always bioavailable. Turpeinen et al. (2000) reported that only 4–6% and 13–43% of total water soluble Pb in humic surface soil and mineral soil sampled from the same site, respectively, were bioavailable as determined using a luc-based Staphylococcus aureus RN4220 (pTOO24) bioreporter. Similar observations were made by Liao and colleagues using a cadCgfp-based E. coli DH5α bioreporter in contaminated sediment and soil samples (Liao et al., 2006). The percentage varies by metal species and soil samples as well (Brandt et al., 2008; Maderova et al., 2011; Turpeinen et al., 2003). As a result, bioavailability (hence, effects on biological systems) cannot be well predicted by chemically measured water soluble metal concentration.
Bioreporter analysis of soil suspensions is of particular interest for another reason; it allows for the assessment of bioavailability of the particle-absorbed metal fraction. Whether absorbed metal is bioavailable is still uncertain, especially in contaminated field samples. A study in which fifty contaminated agricultural soils were tested using two luc-based bioreporters, Bacillus subtilis BR151 (pTOO24) and S. aureus RN4220 (pTOO24) specific for Cd and Pb, respectively, showed that 115-fold more Cd and 40-fold more Pb were bioavailable in soil suspensions than in soil/water extracts (Ivask et al., 2004). Contradictorily, another study by Magrisso et al. (2009) reported that absorbed Pb was unavailable to a luxCDABE-based bioreporter Cupriavidus metallidurans AE1433, as indicated by no bioluminescence induction with soil suspensions despite a chemical analysis acid extraction procedure that suggested significant amounts of total Pb in the soil sample. The uncertainty in bioavailability of bound metals is then translated to the lack of confidence in the evaluation of contamination scales, remediation strategies and efficacies if only chemical methods are used. Therefore, bacterial bioreporters can be included as a complementary analytic tool to conventional chemical methods for more comprehensive assessments of metal contamination in soils and sediments.
3.2. Heavy metals in water
Heavy metal contamination in water systems impacts the growth of waterborne organisms, and with respect to human health, drinking water quality. Therefore, there is an increasing need for rapid, sensitive and cost-effective screening protocols to monitor natural and drinking water systems. Bacterial bioreporters are well suited for this task for several reasons. Since bacteria are very sensitive to toxic heavy metals and have evolved to rapidly eliminate the toxic effects, very low detection limits can be accomplished using bacteria as host cells for bioreporter construction (Hynninen and Virta, 2010). In fact, most heavy metal bioreporters can detect concentrations below the drinking water safety limit (Ripp et al., 2011). Bacterial bioreporter assays are less time-consuming and less expensive than chemical analysis, making them a suitable method for Tier I screening in large scale surveys.
Arsenic serves as one of the most serious water contaminants (Nordstrom, 2002). Several bacterial bioreporters have been developed for arsenic by taking advantage of the arsenic resistance mechanism found in most bacteria (reviewed in Diesel et al., 2009). The arsenic bioreporter E. coli DH5α (pJAMA-arsR) containing the luxAB genes as reporter elements has been applied in a large scale field testing wherein 194 different groundwater samples from Vietnam were screened for arsenic (Stocker et al., 2003; Trang et al., 2005). After a 1.5 h incubation, this strain exhibited a lower detection limit of 7 μg As/L groundwater with a linear response between 10 and 100 μg As/L. When validated against chemical analysis, the bioreporter assay was more reliable than chemical field test kits, yielding only 2.4% false positive and 8.0% false negative results if using the World Health Organization guideline of 10 μg/L as the threshold value for safety. This advocates the potential monitoring capabilities of arsenic bioreporters, especially in disadvantaged areas where access to expensive chemical analysis equipment is limited.
Another commonly examined metal in aqueous environments is iron (Fe), whose bioavailability is typically monitored in relation to primary production in aqueous ecosystems because it is an essential nutrient for phytoplankton. Fe-responsive bioreporters are normally constructed using ecologically relevant cyanobacteria species belonging to the Synechococcus and Synechocystis genus (Bullerjahn et al., 2010). These bioreporters usually carry a transcriptional fusion of the Vibrio harveyi luxAB genes and the isiAB promoter responsive to iron deficiency. Synechococcus sp. strains PCC 7942 and PCC 7002 have been engineered to carry such reporter constructs, and the resulting bioreporters, designated as KAS101 (Durham et al., 2002) and CCMP2669 (Boyanapalli et al., 2007; Bullerjahn et al., 2010), respectively, have been used to assess Fe bioavailability in the Great Lakes (Hassler et al., 2009; McKay et al., 2005; Porta et al., 2005) and marine environments (Boyanapalli et al., 2007). These bioreporters function such that the bioluminescent response increases with reducing concentration of intracellular Fe3+, making them suitable for low concentration detection. In addition, the sensitivity of the isiAB promoter to Fe3+ repletion permits very low detection limits (<1 nM).
With the aim of understanding the fate of gaseous elemental Hg during atmospheric mercury depletion events (AMDEs), Larose et al. (2011) employed an E. coli mer-luxCDABE bioreporter strain to characterize bioavailable Hg in arctic snowpacks. Hg(0) in gas phase is transported from lower latitudes to polar regions, causing local mercury contamination even without significant anthropogenic sources. By evaluating the bioavailable fraction of Hg in surface and basal snow, it was possible to determine the contribution of AMDEs and other deposition pathways to the bioavailable Hg pool, as well as to obtain a better understanding of Hg cycling within the snowpacks. This could not be achieved using traditional analytical methods because the total amount of Hg, both in bioavailable or non-bioavailable forms, is measured in these methods.
3.3. Bioreporters for the detection and monitoring of organic pollutants
Anthropogenic activities have released various types of organic compounds into the environment. These compounds, despite their industrial value, represent another major group of environmental pollutants as they can evoke adverse effects on human health. Microorganisms have evolved transcriptionally regulated catabolic pathways for the degradation of organic compounds (Tropel and van der Meer, 2004). Similar to the case of metal resistance, the regulation of catabolic pathways at the level of transcription is accomplished by an effector-activated regulatory protein stimulating expression of the genes encoding degradation enzymes. Bacterial bioreporters for organic chemicals are therefore designed around these regulatory proteins and corresponding promoters. For example, a plasmid containing a luc gene fused to the xylR gene encoding the toluene-binding regulatory protein XylR and the XylR-responsive promoter Pu was constructed and introduced into E. coli DH5α, yielding a bioluminescent bioreporter that responded to toluene and related compounds (Willardson et al., 1998). Bioreporters can also be constructed using bacterial strains that mineralize target compounds, allowing for coupled reporting and degradation. Such bioreporters are of particular interest in the aspect that in addition to detecting the presence of target compounds, they can provide insight towards the catabolic potential of bioremediation. Bioreporters using degrading and non-degrading bacteria have been developed for middle-chain alkanes (Sticher et al., 1997), simple aromatic hydrocarbons (such as BTEX (benzene, toluene, ethylbenzene and xylene)) (Applegate et al., 1997, 1998; Selifonova and Eaton, 1996; Stiner and Halverson, 2002; Willardson et al., 1998), two to three ring polycyclic aromatic hydrocarbons (PAHs) (such as naphthalene and phenanthrene) (King et al., 1990; Tecon et al., 2009, 2006), phenolic compounds (Abd-El-Haleem et al., 2002; Hay et al., 2000; Leedjarv et al., 2006; Shingler and Moore, 1994), and polychlorinated biphenyls (PCBs) and their metabolites (Feliciano et al., 2006; Layton et al., 1998). A detailed discussion of these bioreporters is addressed by Tecon and van der Meer (2008) and their applications in field experiments are summarized in Table 2.
Table 2.
Applications of bacterial bioreporters for monitoring organic pollutants in environmental samples.
| Bioreporter
|
Field application
|
||||||
|---|---|---|---|---|---|---|---|
| Host strain | Reporter construct | Analyte | Response time | Detection limit | Matrix | Response time | Reference |
| Burkholderia sartisoli RP007 (pPROBE-phn-luxAB) (Tecon et al., 2010) | phn-luxAB | Naphthalene, phenanthrene | 3 h | 0.17 μM (naphthalene) (Tecon et al., 2010) | Seawater | 3 h | Tecon et al. (2010) |
| E. coli DH5α (pGEc74, pJAMA7) (Sticher et al., 1997) | alkS, PalkB-luxAB | C6-C10 alkane | 15–75 min | 24.5 nM (octane) (Sticher et al., 1997) | Soil (DMSO or water extract) Groundwater Seawater |
1 h 2 h 2 h |
Bundy et al. (2001) Bhattacharyya et al. (2005) Tecon et al. (2010) |
| E. coli DH5α (pHYBP103M3) (Beggah et al., 2008) | hp | 2-hydroxylbiphenyl and biphenyl | 2 h | 0.30 μM (2-hydroxylbiphenyl) (Tecon et al., 2010) | Seawater | 2 h | Tecon et al. (2010) |
| E. coli DH5α (pHYBP109) (Jaspers et al., 2000) | hbpR-PhbpC-luxAB | 2-hydroxylbiphenyl | 2 h | 0.30 μM (2-hydroxylbiphenyl) (Tecon et al., 2010) | Seawater | 2 h | Tecon et al. (2010) |
| E. coli DH5α (pLZCapR) (Shin et al., 2005) | capR-lacZ | Phenolic compounds | 5–7 h | 0.1 μM (phenol) (Shin et al., 2005) | Hospital water Soil (water extract) |
5–7 h 5–7 h |
Shin et al. (2005) Shin et al. (2005) |
| E. coli DH5α (pPROBE-LuxAB-TbuT) (Tecon et al., 2010) | tbuT-luxAB | BTEX | 2 h | 0.24 μM (toluene) (Tecon et al., 2010) | Seawater | 2 h | Tecon et al. (2010) |
| E. coli HMS174 (pOS25) (Selifonova and Eaton, 1996) | ipbR-luxCDABE | Hydrophobic compounds | 100–250 min | Varies among compounds (Selifonova and Eaton, 1996) | Sediment (ethanol extract) Soil (HPCD extract) |
100–250 min 2 h |
Selifonova and Eaton (1996) Diplock et al. (2009) |
| E. coli pGLTUR (Willardson et al., 1998) | xylR-Pu-luc | Toluene | 1 h | 10–20 μM (Willardson et al., 1998) | Soil (ethanol extract) and deep aquifer water Soil (DMSO or water extract) |
1 h 1 h |
Willardson et al. (1998) Bundy et al. (2001) |
| P. fluorescens HK44 (pUTK21) (King et al., 1990) | nahR-PnahG-luxCDABE | Naphthalene | 8–24 min | 12–120 μM (Heitzer et al., 1992) | Soil (DMSO and water extract) Soil Soil (methanol and HPCD extract) |
1 h in situ monitoring 1 h |
Bundy et al. (2001) Ripp et al. (2000) Paton et al. (2009) |
| P. fluorescens OS8 (pDNdmpRlux) (Leedjarv et al., 2006) | dmpR-Po-luxCDABE | Phenolic compounds | 4 h | 0.08 mg/L (phenol), 0.03 mg/L (2-methylphenol) (Leedjarv et al., 2006) | Groundwater and dump leachate | 4 h | Leedjarv et al. (2006) |
| P. putida TVA8 (Applegate et al., 1998) | tod-luxCDABE (Ch) | BTEX | 1–4 h | 0.03–50 mg/L (Applegate et al., 1998) | Soil (HPCD extract) Soil (water extract) Groundwater Wastewater and groundwater |
30 min 2 h 2 h Continuous monitoring |
Diplock et al. (2009) Dawson et al. (2008) Bhattacharyya et al. (2005) Kuncova et al. (2011) |
| Ralstonia eutropha JMP 134-32 (Hay et al., 2000) | tfdRPDII-luxCDABE | 2,4-Dichlorophen-oxyacetic acid | 20– 60 min | 2.0 μM | Soil | 1 h | Toba and Hay (2005) |
Ch: integrated into the chromosome.
3.3.1. Organic pollutants in soil
The fate of organic compounds released into soils is a complicated process that encompasses biodegradation, leaching, volatilization, sequestration, and/or bioaccumulation (Semple et al., 2003). In terms of risk assessment, the fraction undergoing biodegradation is of the most interest, and serves as a suitable monitoring endpoint for bacterial bioreporters. King et al. (1990) constructed the first organic chemical bioreporter Pseudomonas fluorescens HK44 for the detection of naphthalene. Containing a plasmid-borne nahG-luxCDABE fusion, HK44 emits light upon exposure to naphthalene, salicylate, and substituted analogs. A controlled field release of strain HK44 into soils in contained lysimeters was initiated in 1996 to demonstrate bioreporter monitoring of a bioremediation event (Ripp et al., 2000). HK44 cells were sprayed within a PAH contaminated soil matrix and naphthalene-induced bioluminescence was detected over a 2 year monitoring period, with signal strength diminishing in parallel with the bioremediative loss of naphthalene.
Bioreporter assays of soil samples are more conventionally performed on organic solvent extractions of contaminated soils rather than directly within the soil itself. As an example, Dawson et al. (2008) applied the Pseudomonas putida TVA8 luxCDABE BTEX bioreporter to methanol extracts of BTEX-impacted soils to estimate degradation and toxicity. Bioluminescent response was achieved within a 2 h incubation of bioreporter cells and diluted methanol soil extracts. It was also demonstrated that changes in BTEX concentration and toxicity over time could be correlated with the bioluminescent response to methanol soil extracts during the process of degradation. However, additional concerns associated with solvent extraction are raised. First, the extractable amount of organic contaminants from soil samples may vary by different solvents (Kelsey et al., 1997). Second, the alcohol solvent (such as methanol and butanol) used for extraction can be toxic to the bioreporter cells, resulting in loss in viability and a corresponding loss in signal. Third, solvent toxicity can damage the cell membrane, thus increasing the supply of fatty acids to the cell that then serve as a substrate for bacterial luciferase to erroneously increase the bioluminescent response. Although dilution is commonly performed to minimize solvent effects, dilution steps can reduce the test concentration below the bioreporter detection limit, leading to an underestimation of the amount of contaminant present in the original sample. Another problem of solvent extraction is, once again, associated with bioavailability. Many physico-chemical properties of soils can impact the bioavailability of organic compounds (Semple et al., 2003), whereas solvent extraction cannot distinguish between biologically available and unavailable fractions. The bioavailability issue is critical when determining biodegradation potential (i.e., the extent of contaminants that can be degraded by microbes). As demonstrated by Paton et al. (2009), bacterial bioreporters can be used to address these concerns. In their experimental design, soils historically contaminated with naphthalene were subjected to bioremediation by adding a natural naphthalene-degrading microbial species, followed by chemical analysis to determine the amount of naphthalene that had been degraded. A range of non-exhaustive extraction techniques was used to extract naphthalene from the soils prior to biodegradation. By comparing the bioluminescent response of the naphthalene bioluminescent bioreporter HK44 to each extract with the amount of biodegradable naphthalene that was determined chemically, the authors demonstrated a correlation between extractable and bioavailable naphthalene. In addition, the extract was immediately useable in the bioreporter assay without a dilution step, suggesting a more straightforward assay format.
3.3.2. Organic pollutants in water
Compared to soil samples, organic compound detection in water using bacterial bioreporters is more straightforward since the aqueous sample can be directly used in bioreporter assays without prior solvent extraction. Willardson et al. (1998) applied the previously mentioned xylR-Pu-luc bioreporter E. coli DH5α (pGLTUR) for detection of toluene and related compounds in a deep water aquifer. The bioreporter accurately measured contaminant concentrations within 3% of those measured by conventional chemical methods after 1 h of incubation with the water sample.
Detection of hydrocarbons in groundwater and wastewater has also been demonstrated using the BTEX bioluminescent bioreporter P. putida TVA8. Because strain TVA8 also responds to some chlorinated aliphatic hydrocarbons (CAHs) (Shingleton et al., 1998), Bhattacharyya and colleagues employed a set of different constitutive and inducible bioluminescent bioreporters including TVA8 to assess several groundwater samples that were contaminated with CAHs including a known TVA8 inducer trichloroethylene (TCE) (Bhattacharyya et al., 2005). Strain TVA8 linearly responded to TCE within the range from 0 to approximately 2000 μmol/L, covering the concentrations of TCE in the groundwater samples. It was demonstrated that the samples inducing the highest bioluminescence were also the ones that contained the highest chemically-determined TCE concentrations (Bhattacharyya et al., 2005). Kuncova et al. (2011) compared the bioluminescent response of TVA8 to contaminated wastewater influent and effluent after treatment. The removal of the majority of BTEX contamination was indicated by an approximate 100-fold decrease in the maximum bioluminescence induced by the effluent than that induced by the influent. The bioreporter results also agreed with side-by-side chemical analysis, which measured approximately 1000-times less BTEX compounds in the effluent sample. However, the bioreporter assay failed to respond to a groundwater sample with known high concentrations of BTEX, probably due to the presence of uncharacterized toxicants within the sample. Dilution schemes can alleviate such toxic interactions, and the inclusion of constitutive ‘always on’ bioreporters as controls can indicate the presence of interfering toxicants that reduce cell viability.
A phenol bioreporter P. fluorescens OS8 (pDNdmpRlux) containing a plasmid-borne dmpR-Po-luxCDABE fusion was constructed and used to detect phenolic compounds in groundwater and dump leachates (Leedjarv et al., 2006). With a laboratory-determined detection limit of 0.08 mg phenol/L, this bioreporter was able to elicit a detectable bioluminescent response to nine out of ten samples tested. Substantial variation of the bioavailable fractions across samples was also observed, ranging from 6% to 95% of the total amount of phenol determined by chemical methods. One sample, although chemically determined to contain phenol at a total concentration approximately 140-times the bioreporter detection limit, was unsuccessful in inducing bioluminescence in the bioreporter, suggesting that phenol present in the sample was probably not bioavailable. Such results thus support the importance of taking bioavailability into account in environmental risk assessment.
In a contaminated site, for instance an oil spill site, it is not uncommon that different types of pollutants coexist. Tecon et al. (2010) recently demonstrated the application of a suite of multiple bacterial bioreporters for monitoring hydrocarbon mixtures in marine environments. Five bioreporters for the detection of short chain linear alkanes, monoaromatic and polyaromatic compounds, biphenyl, 2-hydroxybiphenyl, and DNA damage were used to evaluate artificial crude oil spills in seawater over a period of 7–10 days. Three strains including the short chain alkane bioreporter E. coli DH5α (pGEc74, pJAMA7), the BTEX bioreporter E. coli DH5α (pPROBE-luxAB-TbuT), and the naphthalene and phenanthrene bioreporter Burkholderia sartisoli RP007 (pPROBE-phn-luxAB) were able to detect significant amounts of inducing compounds in oil-contaminated seawater samples. The short chain alkane bioreporter detected a maximum equivalent octane concentration between 200 and 600 nM 6 h after the spill in individual replicates, whereas chemical analysis also measured maximum concentrations of C11 and C12 alkanes at approximately 200 nM 2 to 6 h after the spill. Similarly, the equivalent naphthalene concentration detected by the bioreporter assay peaked at approximately 1 μM (equivalent to 0.13 μg/mL) 2–3 days after the spill, which was comparable to total naphthalene and phenanthrene concentrations of 0.18–0.80 μg/mL as measured by chemical methods in three other replicates. Although chemical analysis of monoaromatic compounds was not performed due to the discrepancy in the types of compounds that can be measured between chemical methods and bioreporter assays, the concentration of toluene equivalents detected by the BTEX bioreporter evolved in a manner similar to that of octane, with a peak 6 h after the spill followed by a gradual decrease. The authors therefore stated that the trend of concentrations of each type of organic compound determined by bioreporter assays was generally in agreement with chemical analysis as well as data from other studies. Light and volatile species such as alkane and BTEX peaked a few hours after the spill, followed by a gradual decrease. On the other hand, heavier PAH constituents (i.e., naphthalene) were bioavailable to the bioreporters in aqueous phase for several days. They also concluded that this multi-strain bioreporter platform could be used as a simple and rapid tool for monitoring hydrocarbon mixture contamination and potentially as an indicator to predict the time scale of oil spills.
3.4. Bioreporters for the detection and monitoring of environmental toxicity
In addition to specifically sensing single compounds or classes of compounds, whole-cell bioreporters can be designed to detect effects without necessarily identifying the chemical nature of the analytes. These effects can be general, such as cytotoxicity, or somewhat specific, such as genotoxicity, protein damage, or oxidative stress. Bioreporters for cytotoxicity detection are usually constructed by expressing the reporter gene under the control of a strong constitutive promoter and function in a lights-off mode. Exposure to toxicants is reflected by a decrease in reporter signal due to inhibited cellular metabolism. Alternatively, reporter genes can be coupled with a promoter responsive to certain types of stress, allowing for semi-specific detection in which exposure to samples capable of causing the given stress results in an elevated level of signal. With regard to the choice of stress-responsive promoters, the promoter of the recA gene involved in DNA repair is commonly used for the construction of genotoxicity responsive bioreporters. Promoters of genes involved in the heat shock response such as grpE and dnaK are usually employed to detect protein damage. For monitoring environmental samples where the coexistence of various toxic chemicals is routine, using toxicity sensitive whole-cell bioreporters is advantageous in that the collective toxicity from all contaminants present in the sample is reported. Applications of genetically modified whole-cell bioreporters for toxicity assessment in environmental samples are summarized in Table 3.
Table 3.
Environmental toxicity monitoring using genetically modified bacterial bioreporters.
| Bioreporter
|
Field application
|
||||
|---|---|---|---|---|---|
| Host strain | Reporter construct | Effector | Matrix | Pollutants | Reference |
| Acinetobacter baylyi | recA-luxCDABE | DNA damage | Groundwater | Phenolic compounds | Song et al. (2009) |
| ADP1 recA-lux (Song et al., 2009) | |||||
| Acinetobacter DF4/pUTK2 (Abd-El-Haleem et al., 2006) | luxCDABE | General | Wastewater | Metals | Abd-El-Haleem et al. (2006) |
| B. subtilis BR151 (pCSS962/pBL1) (Lampinen et al., 1992) | luxCDABE | General | Soil (water suspension and water extract) | Cd | Ivask et al. (2011, 2004) |
| E. coli DPD2511 (Belkin et al., 1996) | katG-luxCDABE | Oxidative stress | Water source from treatment plant and stream water | Not specified | Gu et al. (2001) |
| E. coli DPD2540 (Bechor et al., 2002) | fabA-luxCDABE | Membrane damage | Water source from treatment plant and stream water | Not specified | Gu et al. (2001) |
| E. coli DPD2794 (Vollmer et al., 1997) | recA-luxCDABE | DNA damage | Tap water and river water Water source from treatment plant and stream water |
Not specified Not specified |
Eltzov et al. (2009) Gu et al. (2001) |
| E. coli HB101 (pUCD607) (Rattray et al., 1990) | luxCDABE | General | Groundwater Water source from treatment plant and stream water Sediment and soil Soil (water or solvent extract) Soil (water extract) Soil (water extract) |
CAH Not specified As and Sb Hydrocarbon Cu and Ni Phenyltin compounds |
Bhattacharyya et al. (2005) Gu et al. (2001) Flynn et al. (2002) Flynn et al. (2003) Bundy et al. (2001), Diplock et al. (2009) Paton et al. (2006a) Paton et al. (2006b) |
| E. coli MC1061 (pTOO02) (Hakkila et al., 2004) | luc | General | Sediment (water suspension) | Metals | Hakkila et al. (2004) |
| E. coli MG1655 (pJAMA8-cda) (Tecon et al., 2010) | cda-luxAB | DNA damage | Seawater | Hydrocarbon | Tecon et al. (2010) |
| E. coli TV1061 (Van Dyk et al., 1994) | grpE-luxCDABE | Protein damage | Tap water and river water Water source from treatment plant and stream water |
Not specified Not specified |
Eltzov et al. (2009) Gu et al. (2001) |
| P. fluorescens 10586 (pUCD607) (Aminhanjani et al., 1993) | luxCDABE | General | Groundwater Sediment and soil Soil (water extract) Soil (water extract) Sediment and groundwater |
CAH As and Sb Cu and Ni Phenyltin compounds BTEX |
Bhattacharyya et al. (2005) Flynn et al. (2002) Flynn et al. (2003) Paton et al. (2006a) Paton et al. (2006b) Sousa et al. (1998) |
| P. putida F1 (pUCD607) (Weitz et al., 2001) | luxCDABE | General | Soil (water or solvent extract) | Hydrocarbon | Bundy et al. (2001), Dawson et al. (2008), Diplock et al. (2009) |
| P. fluorescens DF57-40E7 (Tom-Petersen et al., 2001) | luxAB | General | Soil (water suspension and extract) | Cu | Brandt et al. (2006, 2008), Maderova et al. (2011) |
| P. fluorescens OS8 (pDNLux) (Leedjarv et al., 2006) | luxCDABE | General | Groundwater and dump leachate | Phenolic compounds | Leedjarv et al. (2006) |
| P. fluorescens Shk1 (Kelly et al., 1999) | luxCDABE | General | Wastewater | Metals | Kelly et al. (2004), Lajoie et al. (2003) |
| S. aureus RN4220 (pTOO02) (Ivask et al., 2004) | luc | General | Soil | Pb and Cd | Ivask et al. (2004) |
| Salmonella typhimurium TA1535 (pSWITCH) (Baumstark-Khan et al., 2005) | PcolD-luxCDABE/Plac-gfpuv (bicistronic) | General and DNA damage | Groundwater | Not specified | Baumstark-Khan et al. (2005) |
Because of its autonomous nature and easily measurable light signal, the luxCDABE gene cluster is the most commonly used reporter element for the construction of toxicity responsive bioreporters. In fact, naturally bioluminescent marine bacteria such as Vibrio fischeri were originally exploited for toxicity monitoring. The widely used and commercially available Microtox test was developed on the basic principle that exposure to toxicants can be reflected by the reduction of bioluminescence in V. fischeri (Chang et al., 1981). One limitation, however, is that because V. fischeri is a marine species, the assay needs to be performed in saline, limiting its application in more complex environmental samples. Using recombinant DNA techniques, other environmentally relevant yet naturally non-luminescent bacterial species can be engineered to express the bioluminescence genes and function as toxicity bioreporters. An example is P. fluorescence Shk1, which was constructed by introducing the luxCDABE genes into a P. fluorescence strain isolated from the activated sludge of a wastewater treatment plant (Kelly et al., 1999). Strain Shk1 was shown to respond to a range of toxicants including cadmium, dinitrophenol, and hydroquinone. It also has been successfully used to monitor toxicant loads in wastewater influents (Kelly et al., 2004; Lajoie et al., 2003; Ren and Frymier, 2003). Additionally, plasmid pUCD607 containing the luxCDABE genes isolated from V. fischeri (Shaw and Kado, 1986) has been introduced to non-luminescent bacteria for bioreporter development. Two strains constructed using pUCD607, E. coli HB101 (pUCD607) (Rattray et al., 1990) and P. fluorescens 10586 (pUCD607) (Aminhanjani et al., 1993), have been utilized to assess toxicity in a variety of soil and water samples (Bhattacharyya et al., 2005; Bundy et al., 2001; Dawson et al., 2007; Flynn et al., 2002, 2003; Nissen et al., 2009; Paton et al., 2006a,b; Tiensing et al., 2002). A bioluminescently tagged Acinetobacter strain DF4/pUTK2 was also developed and successfully applied to monitor for heavy metal toxicity in industrial and municipal wastewater samples (Abd-El-Haleem et al., 2006). This group has also designed a lights-on bioreporter that monitors for nitrate and nitrite toxicity in wastewater. Just like heavy metals, these inorganic compounds can have significantly deleterious effects on aquatic life when released in effluent waters (Abd-El-Haleem et al., 2007). By fusing the nitrate/nitrite responsive nasR-like promoter from Klebsiella to the luxCDABE gene cassette, they demonstrated real-time biosensing of these compounds in the influent and effluent waters of Egyptian sewage and industrial wastewater treatment plants, with laboratory demonstrated detection limits of <10 ppm.
In addition to simply detecting the presence of toxicants, lights-off bioreporters can be used to determine bioremediation constraints. Sousa et al. (1998) used the environmentally relevant, constitutively bioluminescent strain P. fluorescens 10586s (pUCD607) as a proxy to evaluate the metabolic burden on bioremediation under various BTEX contaminated sediment and water samples. By assessing the bioluminescent response under different sample manipulations, it was possible to determine if the environmental condition was favorable for microbial remediation to occur, as well as to identify and alleviate potential constraints. When assessing complex samples for a specific target, the compound-specific lights-on bioreporter might be interfered with by the sample matrix, causing inhibition or signal quenching (Brandt et al., 2006). A constitutive toxicity bioreporter therefore can be used in parallel to correct for this potential interference.
4. Emerging whole-cell eukaryotic bioreporters
While bacterial-based bioreporters are well established for the detection and monitoring of a wide variety of environmental conditions, there is an increasing trend towards the use of eukaryotic systems in this role. This shift owes to the increasing desire for determination of the effects and bioavailability of environmental contaminants as they relate to humans and other animals. Bacterial bioreporters are handicapped in this regard for several reasons. One chief concern is that the different ploidities of bacterial organisms can affect the mutagenic or carcinogenic actions of environmental contaminants that trigger the reporter (Cavenee et al., 1983), or can lead to situations where genetic alteration cannot be detected. In addition, these bacterial reporters may simply lack the required components for interacting with the target analyte, as is the case with estrogenic monitoring (Gu et al., 2002). Whole-cell eukaryotic bioreporters overcome these problems because their detection of specific compounds provides bioavailability data that is directly relatable to humans and other animals (Struss et al., 2010).
4.1. Available classes of eukaryotic bioreporter proteins
Eukaryotic whole-cell bioreporters are designed using the same fluorescent and bioluminescent systems that have been incorporated into bacterial bioreporters–fluorescent GFP, bioluminescent lux and luc, and colorimetric lacZ–with each presenting more or less the same advantages and disadvantages as that established in their bacterial counterparts. Also, analogous to bacterial bioreporters, their incorporation into eukaryotic hosts can be implemented constitutively wherein the loss of signal indicates a toxic effect or inducibly wherein an increase in signal indicates the presence of a targeted agent. GFP and its variants are commonly used in eukaryotes to detect environmental contaminants, and can allow for parallel detection of multiple contaminants when several fluorescent proteins with non-overlapping emission wavelengths are used (Shibasaki et al., 2001). However, unlike when expressed in bacteria, their application in eukaryotic cells is hindered by the presence of additional naturally fluorescent compounds within the host. This can lead to high levels of background fluorescence under standard imaging conditions, and therefore reduces the detectability of the reporter signal, especially at lower cell population sizes or under conditions of weak induction. Bioluminescent reporter systems, however, are not subject to these high background levels in eukaryotic hosts and are therefore often preferred over their fluorescent counterparts for imaging of smaller cell population sizes or detection of weakly induced signals. The luc gene is the most commonly used of the bioluminescent reporters, however, because it requires the addition of a chemical substrate prior to bioluminescent emission, it is not well suited for remote or online monitoring, and can itself be toxic to the eukaryotic host (Hollis et al., 2001). The bacterial lux genes were recently optimized for expression under eukaryotic regulatory controls (Close et al., 2010; Gupta et al., 2003). With no substrate addition required, these are the only reporter systems capable of repetitive, real-time signaling, although their resulting bioluminescence emission is not as bright as luc.
4.2. Environmental monitoring using lower eukaryotic hosts
Lower eukaryotes, such as yeast, have proven invaluable as biomonitoring tools due to their ease of use, plentiful genetic manipulation techniques, and single celled nature (Walmsley and Keenan, 2000). Combined with their eukaryotic genetic architecture, this makes them logical replacements for bacterial bioreporters when accessing bioavailability or investigating eukaryotic specific metabolic pathways. The most common use of yeast-based environmental bioreporters has been for the detection and bioavailable assessment of estrogenic or androgenic compounds (Table 4). The similarities between yeast and bacterial growth and maintenance, combined with the ability of yeast cells to express the human estrogen receptor (Metzger et al., 1988), has made them a preferred model organism for the detection and measurement of estrogenic/androgenic compounds from environmental samples.
Table 4.
Selected environmental applications of eukaryotic bioreporters.
| Bioreporter
|
Field application
|
||||||
|---|---|---|---|---|---|---|---|
| Host strain or cell line | Reporter construct | Analyte | Response time | EC50/Detection limit | Matrix | Response time | Reference |
| S. cerevisiae/AhR (Leskinen et al., 2008) | DRE-luc | AhR ligands | 3.5 h | 5.5 nM DHT/0.5 nM DHT | Sediment | 3.5 h | Leskinen et al. (2008) |
| Rat hepatoma cell line H4IIE DR-CALUX (Murk et al., 1996) | DRE-luc | AhR ligands | 24 h | 0.01 nM TCDD/0.5 fM TCDD | Sediment Soil Indoor dust |
24 h 24 h 24 h |
Dindal et al. (2007), Hilscherova et al. (2010), Houtman et al. (2006), Keiter et al. (2008), Murk et al. (1996), Wolz et al. (2008) Andersson et al. (2009), Sidlova et al. (2009) Suzuki et al. (2010) |
| Human breast carcinoma cell line MDA-kb2 (Wilson et al., 2002) | ARE-luc | Androgenic activity | Overnight | NR/0.1 nM DHT | Water | 24 h | He et al. (2011) |
| S. cerevisiae hERα (Gaido et al., 1997) | ERE-lacZ | Estrogenic activity | 20 h | 3.5 nM DHT/2.19 nM DHT | Wastewater | 20 h | Nakada et al. (2004), Nelson et al. (2007) |
| S. cerevisiae BLYES (Sanseverino et al., 2005) | ERE-lux | Estrogenic activity | 4–6 h | 0.24 nM E2/0.045 nM E2 | Surface water and drinking water | 4–6 h | Bergamasco et al. (2011) |
| S. cerevisiae hERα (Leskinen et al., 2005) | ERE-luc | Estrogenic activity | 2.5 h | 0.5 nM E2/0.03 nM E2 | Wastewater | 2.5 h | Salste et al. (2007) |
| S. cerevisiae hER YES (yeast estrogen screen) (Routledge and Sumpter, 1996) | ERE-lacZ | Estrogenic activity | 72 h | 0.44 nM E2/0.045 nM E2 (Sanseverino et al., 2005) | Wastewater Sediment Surface and ground water Sewage |
72 h 24 h 72–96 h 72 h |
Fernandez et al. (2007), Nelson et al. (2007), Thorpe et al. (2006) Grund et al. (2011) Aneck-Hahn et al. (2008), Beek et al. (2006), Leusch et al. (2010) Leusch et al. (2010) |
| S. cerevisiae hERα (Balsiger et al., 2010) | ERE-lacZ | Estrogenic activity | 2 h | 0.145 nM E2/100–200 pM | Wastewater Sediment |
2 h 2 h |
Balsiger et al. (2010) Jeffries et al. (2011) |
| Human breast carcinoma cell line MCF-7 MELN (Balaguer et al., 1999) | ERE-luc | Estrogenic activity | 16 h | 5 pM E2/1 pM E2 | Sediment Sewage Wastewater Surface and ground water |
16 h 16 h 16 h 16 h |
Louiz et al. (2008) Hernandez-Raquet et al. (2007), Leusch et al. (2010) Cargouet et al. (2004), Dagnino et al. (2010) Cargouet et al. (2007) Cargouet et al. (2004), Leusch et al. (2010) |
| Human breast carcinoma cell line T47D-KBluc (Wilson et al., 2004) | ERE-luc | Estrogenic activity | 24 h | 0.01 nM E2/1 pM E2 | Wastewater Water Sewage |
24 h 24 h 24 h |
Wehmas et al. (2011) He et al. (2011), Leusch et al. (2010) Leusch et al. (2010) |
| S. cerevisiae BMA64/luc (Leskinen et al., 2005) | luc | Toxicity | 0.5 h | NR | Soil | 0.5 h | Lankinen et al. (2011) |
hER: human estrogen receptor; hAR: human androgen receptor; AhR: acyl hydrocarbon receptor; ERE: estrogen response element; ARE: androgen response element; DRE: dioxin response element; E2: 17β-estradiol; T: 17β-testosterone; DHT: dihydrotestosterone; TCDD: 2,3,7,8-tetrachlorodibenzo-p-dioxin; NR: not reported.
With the optimization and re-engineering of the lux cassette to function in S. cerevisiae (Gupta et al., 2003), it became possible to use bioluminescent production as a measure of both the presence and bioavailability of estrogenic/androgenic chemicals in a host with relevance to humans. Upon exposure to estrogen, the bioluminescent yeast strains are capable of initiating expression of the lux genes, ultimately leading to the production of bioluminescence. When compared with the traditional lacZ colorimetric-based assays, the lux-based system is able to demonstrate similar dynamic ranges (4.5 × 10−11 to 2.8 × 10−9 M for each) and EC50 values (2.4 (±1.0) × 10−10 for the lux system and 4.4 (±1.1) × 10−10 for the colorimetric system), however, the lux-based system can do so much faster, producing results in as little as 1 h compared to the minimum of 3 d for the colorimetric system (Eldridge et al., 2007; Sanseverino et al., 2005). In a similarly human-relevant application, Bakhrat et al. (2011) proposed that a modified version of the yeast-based lux reporter system be used to evaluate the effectiveness of sunscreens. By replacing the promoter governing expression of the lux genes with the DNA damage-responsive UFO1 promoter, they were able to detect changes in the dose of UV radiation received by yeast expressing the full lux cassette under different sunscreen protection regimens.
More traditional luc-based bioluminescent bioreporters have been incorporated into yeast for the evaluation of heavy metal and other environmental contaminants (Table 4). Lankinen et al. (2011) demonstrated the value of yeast-based biosensing in their comparative analysis of nickel (Ni) responsive fungal, bacterial, and yeast bioreporters. Using fungal bioreporters, it was determined that Ni became cytotoxic at a concentration of 20 mg/L. This was in contrast to the results obtained when using bacterial bioreporters (85 mg Ni/L) and especially that of luc-based yeast bioreporters, which did not display a cytotoxic response until Ni concentrations reached 294 mg/L. Had these tests been performed using only one of the bioreporter organisms, it would not have been possible to draw relevant conclusions concerning the bioavailability of Ni across the full array of community members. The inclusion of the yeast bioreporter is specifically relevant, as it provides the best indication of Ni toxicity to humans and other higher animals.
Similar results were demonstrated in a second study that evaluated the detection of polyhalogenated organic pollutants in river sediments (Leskinen et al., 2008). In this study, detection of these chemicals was compared between traditional chemical analysis and luc-based biodetection using either yeast or rat cells as bioreporter hosts. The various detection strategies all had varying levels of sensitivity, as well as disparate strengths and weaknesses. The mammalian cell-based bioreporters were determined to have EC50 values 15–500 times lower than their yeast-based counterparts, but were not as robust, nor as inexpensive to maintain. As such, the authors suggested that the yeast-based detection system could function as a valuable pre-screening tool for the early detection of polyhalogenated pollutants in environmental samples. Those samples that tested positive for the presence of contaminants with yeast could then be subjected to the more sensitive mammalian or analytical-based detection strategies.
Novel luc-based bioreporters for heavy metal sensing were recently constructed in the ciliate Tetrahymena thermophila (Amaro et al., 2011). The luc reporter gene was fused to heavy metal responsive metallothionein promoters, and these bioreporters emitted increased bioluminescence in proportion to increased heavy metal concentrations. Since ciliates do not have a cell wall in their vegetative state, they more readily take up the chemicals to which they are exposed, and as a consequence display greater sensitivity than their yeast bioreporter counterparts (down to low nanomolar concentrations) and within rapid timeframes (~2 h).
The incorporation of GFP into yeast reporters was uniquely demonstrated by Radhika et al. (2007) in a construct capable of “smelling” the presence of 2,4-dinitrotoluene (DNT), a mimic for the explosive compound trinitrotoluene (TNT). This was accomplished by introducing both the mammalian olfactory receptor Olfr226 and a gfp gene under the control of the G-coupled protein receptor responsive CREBP promoter. Upon detection of DNT by the olfactory receptor, a traditional G-coupled protein receptor cascade was initiated that culminated with the activation of the CREBP promoter and expression of GFP. By exciting the reporter cells at set intervals, it was then possible to continually monitor for the presence of DNT in the environment. By altering the specificity of the olfactory receptor, it should also be possible to screen for alternative chemicals, opening up the possibility of a remote sensor array with direct relevance to human cytotoxicity.
4.3. Environmental monitoring using human whole-cell bioreporters
Driven by the same concerns that lead to the evolution from bacterial to yeast and other lower eukaryotic bioreporters, many research groups are choosing to forego the use of yeast as proxies for human relevance and instead opting to survey for environmental contamination directly in a human cellular host. There are several advantages to this strategy, namely, that the bioavailability of the compound being tested should correlate directly to humans as a whole, and that, through the use of multiple human cell types, the effects of the compound in question can be evaluated in light of individual organ systems. This level of specificity and relevance is impractical in bacterial reporters and has allowed human cell-based reporter systems to gain widespread popularity in the biomedical and biotechnology communities.
Just as with yeast-based reporters, the most popular sensing targets for human cell-based bioreporters have been estrogenic and androgenic compounds. Liu and Lu (2011), for example, modulated the expression of GFP through activation of the human estrogen response element in human lung carcinoma cells. While the main target of this investigation was the estrogenic surfactant nonylphenol, this could be considered a proof-in-principle demonstration of their system since the estrogen response elements used should display broad reactivity to many other estrogenic compounds.
The toxic effects of PAHs were evaluated in a high-throughput assay using luc-incorporated mammalian cell lines stably transfected with the luc gene under the control of a dioxin responsive promoter (Machala et al., 2001). Bioluminescence emission from reporter cells exposed to increasing concentrations of PAHs signaled concentrations at which the toxicants became available to the cell. This methodology allows for rapid detection and screening of multiple conditions simultaneously, and the bright nature and low background of the luc system allows for detection of even weakly induced cells.
The incorporation of lux within mammalian cells was demonstrated by Close et al. (2012) using human kidney cells constitutively expressing bioluminescence. These investigators were able to demonstrate the bioavailability of a toxic aldehyde compound while simultaneously demonstrating during which time periods of the exposure the target cells were processing the aldehyde. This ability is unique to the autonomous bioluminescent expression offered by the lux cassette and, if proven to be a successful technique over time, should be able to offer researchers increased levels of data collection with minimal changes to their existing experimental protocols.
5. A needs assessment of whole-cell bioreporters for environmental applications
Whole-cell bioreporters have some noteworthy advantages. As self-propagating entities, a substantial number of bioreporters can be easily obtained at low cost, bioreporter assays are fast and simple to perform, they maintain sensitivity that typically meets or exceeds necessary standards, they report on chemical bioavailability rather than mere total concentrations, evolution and nature provide a substantial selection of bioreporters for biosensing needs, and fairly straightforward genetic manipulation techniques allow for desired modifications. Despite these advantages, bioreporters rarely if ever reach commercialization stages or conventional field application status due to several persistent obstacles, foremost of which, from a marketing standpoint, is the inability to patent most bioreporters since the reporter gene technology they incorporate is well substantiated and lacks novelty. Without patent protection, there is little profit to be made and consequent little interest in the pursuit.
The classification of bioreporters as recombinant organisms significantly affects their application capacity in real-world environments. The potential for recombinant DNA to relocate from its original host to other members of the microbial community is a complex and poorly understood event whose consequences may impact environmental and public health safety. Government-mandated restrictions and guidelines strictly limit the introduction of recombinant organisms into the environment, and make the process of moving a bioreporter from the lab to the field a challenging, lengthy, and costly undertaking that typically exceeds the resources of an academic lab and proves unprofitable for a commercial enterprise. As a consequence, real-world bioreporter applications are more focused on biosensors where the bioreporter remains entrapped within a monitoring device and is thus not freely released to the environment (Fleming, 2010). However, the challenge then becomes finding an immobilization or encapsulation matrix that sustains long-term bioreporter cell viability and maintains the bioreporters in an immediately responsive state. Many different types of encapsulation matrices are available, with none yet providing the model long-term, instantly responsive characteristics that are needed (Date et al., 2010). There are clever alternatives, such as using sporulating bacteria as the bioreporter scaffold, which allows the bioreporter to be stored as a spore for exceedingly long durations with germination being triggered upon interaction with key environmental intermediates (Knecht et al., 2011). There are pressing needs for other such alternatives.
The definitive metric of a bioreporter is its sensitivity, and with bioreporters as of yet not approaching the sensitivity (nor specificity) of analytical chemical methods, many end-user applications remain off limits. To improve sensitivity, researchers have focused on finding promoter elements that have evolved greater sensitivity or genetically modifying promoter elements to be more sensitive (Behzadian et al., 2011; Norman et al., 2005; Peng et al., 2010). Innovative synthetic biology techniques will certainly assist in these latter efforts and ultimately propel molecular biological activities towards much needed optimizations and restructurings of bioreporter elements (see Ghim et al., 2010; Close et al., 2010; Yagur-Kroll and Belkin, 2011 for examples). Many bioreporters also respond to more than a single target, making it difficult to determine the occurrence and quantity of a specifically desired chemical within a complex mixture. Modifications of the transcriptional regulator involved in the sensing pathway can provide a potential method to improve specificity. Ambiguousness in chemical identification caused by low specificity may even be resolvable without actually improving the bioreporter itself. As demonstrated by several groups, testing with an array of different bioreporters in combination with pattern learning algorithms or decision tree models can potentially identify a chemical by the unique pattern or ‘fingerprint’ of bioreporter signals (Elad et al., 2008; Jouanneau et al., 2011). It is still important to point out that bioreporters are not intended to fully replace chemical analytical methods. On the contrary, bioreporters serve as an ideal complementary chemical analysis approach. Environmental monitoring ordinarily occurs over large geographic areas, and bioreporter assays very effectively provide rapid ‘snapshots’ of contaminant presence/absence, thus delineating where further, more precise analytical sampling should and, more importantly, should not occur. It is this movement away from blind sampling that reduces the exorbitant costs associated with analytical methods that often yield large numbers of unproductive samples simply labeled as ‘below detection limits’.
6. Conclusion
Whole-cell bioreporters, whether existing within the walls of the laboratory or applied as bona fide environmental sensors, have proven to be popular and practical tools for the detection and monitoring of contaminants of ecological concern. Considering the advantages of small size, massive population numbers, robustness, adaptability, and information processing power that even on a per-cell basis exceed current silicon-based technologies, it is clear that living cells are particularly well suited for biosensing applications. Additionally, with researchers endlessly fine-tuning the genetics and reporter gene scaffolding within bioreporter organisms, the biosensing attributes of speed, sensitivity, and specificity will continue to improve. More of a concern at this juncture is how one deals with the massive data downloads that will emanate from collections of bioreporter organisms each continuously and unremittingly monitoring a specific chemical or chemical interaction over hours, days, months, or perhaps even years. The scale of these resulting databases will be unmanageable using the current toolbox of data management software, and we will begin entering the deluge of ‘data-intensive’ science that has been pre-warned by Bell et al. (2009) and Hey et al. (2009). The challenge may therefore not lie within the bioreporter’s ability to successfully gather and communicate data, but in our ability to process and meaningfully interpret what the bioreporter is trying to tell us.
Acknowledgments
Portions of this review reflecting work by the authors were supported by the National Science Foundation Division of Chemical, Bioengineering, Environmental, and Transport Systems under award number CBET-0853780, the Division of Biological Infrastructure under award number DBI-0963854, the National Institutes of Health, National Cancer Institute, Cancer Imaging Program under award number CA127745-01, the USDA National Institute of Food and Agriculture Biotechnology Risk Assessment Program under grant number 2009-39210-20230, and the Army Defense University Research Instrumentation Program.
References
- Abd-El-Haleem D, Ripp S, Scott C, Sayler GS. A luxCDABE-based bioluminescent bioreporter for the detection of phenol. J Ind Microbiol Biotechnol. 2002;29:233–237. doi: 10.1038/sj.jim.7000309. [DOI] [PubMed] [Google Scholar]
- Abd-El-Haleem D, Ripp S, Zaki S, Sayler GS. Detection of nitrate/nitrite bioavailability in wastewater using a luxCDABE-based Klebsiella oxytoca bioluminescent bioreporter. J Microbiol Biotechnol. 2007;17:1254–1261. [PubMed] [Google Scholar]
- Abd-El-Haleem D, Zaki S, Abulhamd A, Elbery H, Abu-Elreesh G. Acine-tobacter bioreporter assessing heavy metals toxicity. J Basic Microbiol. 2006;46:339–347. doi: 10.1002/jobm.200510122. [DOI] [PubMed] [Google Scholar]
- Amaro F, Turkewitz AP, Martin-Gonzalez A, Gutierrez JC. Whole-cell biosensors for detection of heavy metal ions in environmental samples based on metallothionein promoters from Tetrahymena thermophila. Microb Biotechnol. 2011;4:513–522. doi: 10.1111/j.1751-7915.2011.00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminhanjani S, Meikle A, Glover LA, Prosser JI, Killham K. Plasmid and chromosomally encoded luminescence marker systems for detection of Pseudomonas fluorescens in soil. Mol Ecol. 1993;2:47–54. [Google Scholar]
- Andersson E, Rotander A, von Kronhelm T, Berggren A, Ivarsson P, Hollert H, Engwall M. AhR agonist and genotoxicant bioavailability in a PAH-contaminated soil undergoing biological treatment. Environ Sci Pollut Res. 2009;16:521–530. doi: 10.1007/s11356-009-0121-9. [DOI] [PubMed] [Google Scholar]
- Aneck-Hahn NH, Bornman MS, de Jager C. Preliminary assessment of oestrogenic activity in water sources in Rietvlei Nature Reserve, Gauteng, South Africa. Afr J Aquat Sci. 2008;33:249–254. [Google Scholar]
- Applegate B, Kelly C, Lackey L, McPherson J, Kehrmeyer S, Menn FM, Bienkowski P, Sayler G. Pseudomonas putida B2: a tod-lux bioluminescent reporter for toluene and trichloroethylene co-metabolism. J Ind Microbiol Biotechnol. 1997;18:4–9. doi: 10.1038/sj.jim.2900334. [DOI] [PubMed] [Google Scholar]
- Applegate BM, Kehrmeyer SR, Sayler GS. A chromosomally based tod-luxCDABE whole-cell reporter for benzene, toluene, ethybenzene, and xylene (BTEX) sensing. Appl Environ Microbiol. 1998;64:2730–2735. doi: 10.1128/aem.64.7.2730-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhrat A, Eltzov E, Finkelstein Y, Marks RS, Raveh D. UV and arsenate toxicity: a specific and sensitive yeast bioluminescence assay. Cell Biol Toxicol. 2011;27:227–236. doi: 10.1007/s10565-011-9184-8. [DOI] [PubMed] [Google Scholar]
- Balaguer P, Francois F, Comunale F, Fenet H, Boussioux AM, Pons M, Nicolas JC, Casellas C. Reporter cell lines to study the estrogenic effects of xenoestrogens. Sci Total Environ. 1999;233:47–56. doi: 10.1016/s0048-9697(99)00178-3. [DOI] [PubMed] [Google Scholar]
- Balsiger HA, de la Torre R, Lee WY, Cox MB. A four-hour yeast bioassay for the direct measure of estrogenic activity in wastewater without sample extraction, concentration, or sterilization. Sci Total Environ. 2010;408:1422–1429. doi: 10.1016/j.scitotenv.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumstark-Khan C, Cioara K, Rettberg P, Horneck G. Determination of geno- and cytotoxicity of groundwater and sediments using the recombinant SWITCH test. J Environ Sci Health Part A-Toxic/Hazard Subst Environ Eng. 2005;40:245–263. doi: 10.1081/ese-200045529. [DOI] [PubMed] [Google Scholar]
- Bechor O, Smulski DR, Van Dyk TK, LaRossa RA, Belkin S. Recombinant microorganisms as environmental biosensors: pollutants detection by Escherichia coli bearing fabA’:: lux fusions. J Biotechnol. 2002;94:125–132. doi: 10.1016/s0168-1656(01)00423-0. [DOI] [PubMed] [Google Scholar]
- Beek IC, Bruhn R, Gandrass J. Analysis of estrogenic activity in coastal surface waters of the Baltic Sea using the yeast estrogen screen. Chemosphere. 2006;63:1870–1878. doi: 10.1016/j.chemosphere.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Beggah S, Vogne C, Zenaro E, van der Meer JR. Mutant HbpR transcription activator isolation for 2-chlorobiphenyl via green fluorescent protein-based flow cytometry and cell sorting. Microb Biotechnol. 2008;1:68–78. doi: 10.1111/j.1751-7915.2007.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadian F, Barjeste H, Hosseinkhani S, Zarei AR. Construction and characterization of Escherichia coli whole-cell biosensors for toluene and related compounds. Curr Microbiol. 2011;62:690–696. doi: 10.1007/s00284-010-9764-5. [DOI] [PubMed] [Google Scholar]
- Belkin S, Smulski DR, Vollmer AC, Van Dyk TK, LaRossa RA. Oxidative stress detection with Escherichia coli harboring a katG’::lux fusion. Appl Environ Microbiol. 1996;62:2252–2256. doi: 10.1128/aem.62.7.2252-2256.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G, Hey T, Szalay A. Beyond the data deluge. Science. 2009;323:1297–1298. doi: 10.1126/science.1170411. [DOI] [PubMed] [Google Scholar]
- Bergamasco AMD, Eldridge M, Sanseverino J, Sodre FF, Montagner CC, Pescara IC, Jardim WF, Umbuzeiro GD. Bioluminescent yeast estrogen assay (BLYES) as a sensitive tool to monitor surface and drinking water for estrogenicity. J Environ Monit. 2011;13:3288–3293. doi: 10.1039/c1em10464k. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya J, Read D, Amos S, Dooley S, Killham K, Paton GI. Biosensor-based diagnostics of contaminated groundwater: assessment and remediation strategy. Environ Pollut. 2005;134:485–492. doi: 10.1016/j.envpol.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Bolan NS, Adriano DC, Mahimairaja S. Distribution and bioavailability of trace elements in livestock and poultry manure by-products. Crit Rev Environ Sci Technol. 2004;34:291–338. [Google Scholar]
- Bondarenko O, Rolova T, Kahru A, Ivask A. Bioavailability of Cd, Zn and Hg in soil to nine recombinant luminescent metal sensor bacteria. Sensors. 2008;8:6899–6923. doi: 10.3390/s8116899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyanapalli R, Bullerjahn GS, Pohl C, Croot PL, Boyd PW, McKay RML. Luminescent whole-cell cyanobacterial bioreporter for measuring Fe availability in diverse marine environments. Appl Environ Microbiol. 2007;73:1019–1024. doi: 10.1128/AEM.01670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt KK, Holm PE, Nybroe O. Bioavailability and toxicity of soil particle-associated copper as determined by two bioluminescent Pseudomonas fluorescens biosensor strains. Environ Toxicol Chem. 2006;25:1738–1741. doi: 10.1897/05-558r.1. [DOI] [PubMed] [Google Scholar]
- Brandt KK, Holm PE, Nybroe O. Evidence for bioavailable copper-dissolved organic matter complexes and transiently increased copper bioavailability in manure-amended soils as determined by bioluminescent bacterial biosensors. Environ Sci Technol. 2008;42:3102–3108. doi: 10.1021/es071916+. [DOI] [PubMed] [Google Scholar]
- Breitbarth E, Gelting J, Walve J, Hoffmann LJ, Turner DR, Hassellov M, Ingri J. Dissolved iron (II) in the Baltic Sea surface water and implications for cyanobacterial bloom development. Biogeosciences. 2009;6:2397–2420. [Google Scholar]
- Bullerjahn GS, Boyanapalli R, Rozmarynowycz MJ, McKay RML. Cyanobacterial bioreporters as sensors of nutrient availability. In: Belkin S, Gu MB, editors. Whole Cell Sensing Systems II: Applications. Springer-Verlag; Berlin, Germany: 2010. pp. 165–188. [DOI] [PubMed] [Google Scholar]
- Bundy JG, Campbell CD, Paton GI. Comparison of response of six different luminescent bacterial bioassays to bioremediation of five contrasting oils. J Environ Monit. 2001;3:404–410. doi: 10.1039/b103104j. [DOI] [PubMed] [Google Scholar]
- Cargouet M, Perdiz D, Levi Y. Evaluation of the estrogenic potential of river and treated waters in the Paris area (France) using in vivo and in vitro assays. Ecotox Environ Safe. 2007;67:149–156. doi: 10.1016/j.ecoenv.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Cargouet M, Perdiz D, Mouatassim-Souali A, Tamisier-Karolak S, Levi Y. Assessment of river contamination by estrogenic compounds in Paris area (France) Sci Total Environ. 2004;324:55–66. doi: 10.1016/j.scitotenv.2003.10.035. [DOI] [PubMed] [Google Scholar]
- Cavenee WK, Dryja TP, Phillips RA, Benedict WF, Godbout R, Gallie BL, Murphie AL, Strong AL, White RL. Expression of recessive allele by chromosomal mechanism in retinoblastoma. Nature. 1983;305:779–784. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- Chang JC, Taylor PB, Leach FR. Use of the Microtox assay system for environmental samples. Bull Environ Contam Toxicol. 1981;26:150–156. doi: 10.1007/BF01622069. [DOI] [PubMed] [Google Scholar]
- Close DM, Patterson SS, Ripp S, Baek SJ, Sanseverino J, Sayler GS. Autonomous bioluminescent expression of the bacterial luciferase gene cassette (lux) in a mammalian cell line. PLoS ONE. 2010;5:e12441. doi: 10.1371/journal.pone.0012441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close DM, Ripp S, Sayler GS. Reporter proteins in whole-cell optical bioreporter detection systems, biosensor integrations, and biosensing applications. Sensors. 2009;9:9147–9174. doi: 10.3390/s91109147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close DM, Xu T, Smartt AE, Rogers A, Crossley R, Price S, Ripp S, Sayler GS. The evolution of the bacterial luciferase gene cassette (lux) as a real-time bioreporter. Sensors. 2012;12:732–752. doi: 10.3390/s120100732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbisier P, Ji G, Nuyts G, Mergeay M, Silver S. luxAB gene fusions with the arsenic and cadmium resistance operons of Staphylococcus aureus plasmid pI258. FEMS Microbiol Lett. 1993;110:231–238. doi: 10.1111/j.1574-6968.1993.tb06325.x. [DOI] [PubMed] [Google Scholar]
- Corbisier P, Thiry E, Diels L. Bacterial biosensors for the toxicity assessment of solid waste. Environ Toxicol Water Quality. 1996;11:171–177. [Google Scholar]
- Craney A, Hohenauer T, Xu Y, Navani NK, Li YF, Nodwell J. A synthetic luxCDABE gene cluster optimized for expression in high-GC bacteria. Nucleic Acids Res. 2007;35:e46. doi: 10.1093/nar/gkm086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagnino S, Gomez E, Picot B, Cavailles V, Casellas C, Balaguer P, Fenet H. Estrogenic and AhR activities in dissolved phase and suspended solids from wastewater treatment plants. Sci Total Environ. 2010;408:2608–2615. doi: 10.1016/j.scitotenv.2010.02.034. [DOI] [PubMed] [Google Scholar]
- Date A, Pasini P, Daunert S. Fluorescent and bioluminescent cell-based sensors: strategies for their preservation. In: Belkin S, Gu MB, editors. Whole Cell Sensing Systems I: Reporter Cells and Devices. Springer-Verlag; Berlin, Germany: 2010. pp. 57–75. [DOI] [PubMed] [Google Scholar]
- Dawson JJC, Godsiffe EJ, Thompson IP, Ralebitso-Senior TK, Killham KS, Paton GI. Application of biological indicators to assess recovery of hydrocarbon impacted soils. Soil Biol Biochem. 2007;39:164–177. [Google Scholar]
- Dawson JJC, Iroegbu CO, Maciel H, Paton GI. Application of luminescent biosensors for monitoring the degradation and toxicity of BTEX compounds in soils. J Appl Microbiol. 2008;104:141–151. doi: 10.1111/j.1365-2672.2007.03552.x. [DOI] [PubMed] [Google Scholar]
- Degryse F, Smolders E, Parker DR. Partitioning of metals (Cd, Co, Cu, Ni, Pb, Zn) in soils: concepts, methodologies, prediction and applications—a review. Eur J Soil Sci. 2009;60:590–612. [Google Scholar]
- Diesel E, Schreiber M, van der Meer JR. Development of bacteria-based bioassays for arsenic detection in natural waters. Anal Bioanal Chem. 2009;394:687–693. doi: 10.1007/s00216-009-2785-x. [DOI] [PubMed] [Google Scholar]
- Dindal A, Thompson E, Aume L, Billets S. Application of site-specific calibration data using the CALUX by XDS bioassay for dioxin-like chemicals in soil and sediment samples. Environ Sci Technol. 2007;41:8376–8382. doi: 10.1021/es071303x. [DOI] [PubMed] [Google Scholar]
- Diplock EE, Alhadrami HA, Paton GI. Application of microbial bioreporters in environmental microbiology and bioremediation. In: Belkin S, Gu MB, editors. Whole Cell Sensing Systems II. Springer-Verlag; Berlin, Germany: 2010. pp. 189–209. [DOI] [PubMed] [Google Scholar]
- Diplock EE, Mardlin DP, Killham KS, Paton GI. Predicting bioremediation of hydrocarbons: laboratory to field scale. Environ Pollut. 2009;157:1831–1840. doi: 10.1016/j.envpol.2009.01.022. [DOI] [PubMed] [Google Scholar]
- Durham KA, Porta D, Twiss MR, McKay RML, Bullerjahn GS. Construction and initial characterization of a luminescent Synechococcus sp PCC 7942 Fe-dependent bioreporter. FEMS Microbiol Lett. 2002;209:215–221. doi: 10.1111/j.1574-6968.2002.tb11134.x. [DOI] [PubMed] [Google Scholar]
- Elad T, Benovich E, Magrisso S, Belkin S. Toxicant identification by a luminescent bacterial bioreporter panel: application of pattern classification algorithms. Environ Sci Technol. 2008;42:8486–8491. doi: 10.1021/es801489a. [DOI] [PubMed] [Google Scholar]
- Eldridge ML, Sanseverino J, Layton AC, Easter JP, Schultz TW, Sayler GS. Saccharomyces cerevisiae BLYAS, a new bioluminescent bioreporter for detection of androgenic compounds. Appl Environ Microbiol. 2007;73:6012–6018. doi: 10.1128/AEM.00589-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltzov E, Marks RS. Whole-cell aquatic biosensors. Anal Bioanal Chem. 2011;400:895–913. doi: 10.1007/s00216-010-4084-y. [DOI] [PubMed] [Google Scholar]
- Eltzov E, Marks RS, Voost S, Wullings BA, Heringa MB. Flow-through real time bacterial biosensor for toxic compounds in water. Sens Actuator B-Chem. 2009;142:11–18. [Google Scholar]
- Everhart JL, McNear D, Peltier E, van der Lelie D, Chaney RL, Sparks DL. Assessing nickel bioavailability in smelter-contaminated soils. Sci Total Environ. 2006;367:732–744. doi: 10.1016/j.scitotenv.2005.12.029. [DOI] [PubMed] [Google Scholar]
- Feliciano J, Xu SF, Guan XY, Lehmler HJ, Bachas LG, Daunert S. ClcR-based biosensing system in the detection of cis-dihydroxylated (chloro-)biphenyls. Anal Bioanal Chem. 2006;385:807–813. doi: 10.1007/s00216-006-0505-3. [DOI] [PubMed] [Google Scholar]
- Fernandez MP, Campbell PM, Ikonomou MG, Devlin RH. Assessment of environmental estrogens and the intersex/sex reversal capacity for chinook salmon (Oncorhynchus tshawytscha) in primary and final municipal wastewater effluents. Environ Int. 2007;33:391–396. doi: 10.1016/j.envint.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Fleming JT. Electronic interfacing with living cells. In: Belkin S, Gu MB, editors. Whole Cell Sensing Systems I: Reporter Cells and Devices. Springer-Verlag; Berlin, Germany: 2010. pp. 155–178. [Google Scholar]
- Flynn HC, McMahon V, Diaz GC, Demergasso CS, Corbisier P, Meharg AA, Paton GI. Assessment of bioavailable arsenic and copper in soils and sediments from the Antofagasta region of northern Chile. Sci Total Environ. 2002;286:51–59. doi: 10.1016/s0048-9697(01)00962-7. [DOI] [PubMed] [Google Scholar]
- Flynn HC, Meharg AA, Bowyer PK, Paton GI. Antimony bioavailability in mine soils. Environ Pollut. 2003;124:93–100. doi: 10.1016/s0269-7491(02)00411-6. [DOI] [PubMed] [Google Scholar]
- Gaido KW, Leonard LS, Lovell S, Gould JC, Babai D, Portier CJ, McDonnell DP. Evaluation of chemicals with endocrine modulating activity in a yeast-based steroid hormone receptor gene transcription assay. Toxicol Appl Pharmacol. 1997;143:205–212. doi: 10.1006/taap.1996.8069. [DOI] [PubMed] [Google Scholar]
- Ghim CM, Lee SK, Takayama S, Mitchell RJ. The art of reporter proteins in science: past, present and future applications. BMB Reports. 2010;43:451–460. doi: 10.5483/bmbrep.2010.43.7.451. [DOI] [PubMed] [Google Scholar]
- Giller KE, Witter E, McGrath SP. Heavy metals and soil microbes. Soil Biol Biochem. 2009;41:2031–2037. [Google Scholar]
- Grund S, Higley E, Schonenberger R, Suter MJF, Giesy JP, Braunbeck T, Hecker M, Hollert H. The endocrine disrupting potential of sediments from the Upper Danube River (Germany) as revealed by in vitro bioassays and chemical analysis. Environ Sci Pollut Res. 2011;18:446–460. doi: 10.1007/s11356-010-0390-3. [DOI] [PubMed] [Google Scholar]
- Gu MB, Kim BC, Cho J, Hansen PD. The continuous monitoring of field water samples with a novel multi-channel two-stage mini-bioreactor system. Environmental Monitoring and Assessment. 2001;70:71–81. doi: 10.1023/a:1010612727587. [DOI] [PubMed] [Google Scholar]
- Gu MB, Min J, Kim EJ. Toxicity monitoring and classification of endocrine disrupting chemicals (EDCs) using recombinant bioluminescent bacteria. Chemosphere. 2002;46:289–294. doi: 10.1016/s0045-6535(01)00081-9. [DOI] [PubMed] [Google Scholar]
- Gupta RK, Patterson SS, Ripp S, Sayler GS. Expression of the Photorhabdus luminescens lux genes (luxA, BCD, and E) in Saccharomyces cerevisiae. FEMS Yeast Res. 2003;4:305–313. doi: 10.1016/S1567-1356(03)00174-0. [DOI] [PubMed] [Google Scholar]
- Hakkila K, Green T, Leskinen P, Ivask A, Marks R, Virta M. Detection of bioavailable heavy metals in EILATox-Oregon samples using whole-cell luminescent bacterial sensors in suspension or immobilized onto fibre-optic tips. J Appl Toxicol. 2004;24:333–342. doi: 10.1002/jat.1020. [DOI] [PubMed] [Google Scholar]
- Hakkila K, Maksimow M, Karp M, Virta M. Reporter genes lucFF, luxCDABE, gfp, and dsred have different characteristics in whole-cell bacterial sensors. Anal Biochem. 2002;301:235–242. doi: 10.1006/abio.2001.5517. [DOI] [PubMed] [Google Scholar]
- Harms H, Wells MC, van der Meer JR. Whole-cell living biosensors—are they ready for environmental application? Appl Microbiol Biotechnol. 2006;70:273–280. doi: 10.1007/s00253-006-0319-4. [DOI] [PubMed] [Google Scholar]
- Hassler CS, Havens SM, Bullerjahn GS, McKay RML, Twiss MR. An evaluation of iron bioavailability and speciation in western Lake Superior with the use of combined physical, chemical, and biological assessment. Limnol Oceanogr. 2009;54:987–1001. [Google Scholar]
- Hassler CS, Twiss MR, McKay RML, Bullerjahn GS. Optimization of iron-dependent cyanobacterial (Synechococcus Cyanophyceae) bioreporters to measure iron bioavailability. J Phycol. 2006;42:324–335. [Google Scholar]
- Hay AG, Rice JF, Applegate BM, Bright NG, Sayler GS. A bioluminescent whole-cell reporter for detection of 2,4-dichlorophenoxyacetic acid and 2,4-dichlorophenol in soil. Appl Environ Microbiol. 2000;66:4589–4594. doi: 10.1128/aem.66.10.4589-4594.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YH, Wiseman SB, Hecker M, Zhang XW, Wang N, Perez LA, Jones PD, El-Din MG, Martin JW, Giesy JP. Effect of ozonation on the estrogenicity and androgenicity of oil sands process-affected water. Environ Sci Technol. 2011;45:6268–6274. doi: 10.1021/es2008215. [DOI] [PubMed] [Google Scholar]
- Heitzer A, Webb OF, Thonnard JE, Sayler GS. Specific and quantitative assessment of naphthalene and salicylate bioavailability by using a bioluminescent catabolic reporter bacterium. Appl Environ Microbiol. 1992;58:1839–1846. doi: 10.1128/aem.58.6.1839-1846.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Raquet G, Soef A, Delgenes N, Balaguer P. Removal of the endocrine disrupter nonylphenol and its estrogenic activity in sludge treatment processes. Water Res. 2007;41:2643–2651. doi: 10.1016/j.watres.2007.02.039. [DOI] [PubMed] [Google Scholar]
- Hever N, Belkin S. A dual-color bacterial reporter strain for the detection of toxic and genotoxic effects. Eng Life Sci. 2006;6:319–323. [Google Scholar]
- Hey T, Tansley S, Tolle K. The Fourth Paradigm: Data-intensive Scientific Discovery. Microsoft Research; Redmond, Washington: 2009. [Google Scholar]
- Hilscherova K, Dusek L, Sidlova T, Jalova V, Cupr P, Giesy JP, Nehyba S, Jarkovsky J, Klanova J, Holoubek I. Seasonally and regionally determined indication potential of bioassays in contaminated river sediments. Environ Toxicol Chem. 2010;29:522–534. doi: 10.1002/etc.83. [DOI] [PubMed] [Google Scholar]
- Hollis RP, Lagido C, Pettitt J, Porter AJR, Killham K, Paton GI, Glover LA. Toxicity of the bacterial luciferase substrate, n-decyl aldehyde, to Saccharomyces cerevisiae and Caenorhabditis elegans. FEBS Lett. 2001;506:140–142. doi: 10.1016/s0014-5793(01)02905-2. [DOI] [PubMed] [Google Scholar]
- Houtman CJ, Booij P, Jover E, del Rio DP, Swart K, van Velzen M, Vreuls R, Legler J, Brouwer A, Lamoree MH. Estrogenic and dioxin-like compounds in sediment from Zierikzee harbour identified with CALUX assay-directed fractionation combined with one and two dimensional gas chromatography analyses. Chemosphere. 2006;65:2244–2252. doi: 10.1016/j.chemosphere.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Hynninen A, Virta M. Whole-cell bioreporters for the detection of bioavailable metals. In: Belkin S, Gu MB, editors. Whole Cell Sensing Systems II: Applications. Springer-Verlag; Berlin, Germany: 2010. pp. 31–63. [DOI] [PubMed] [Google Scholar]
- Ivask A, Dubourguier HC, Pollumaa L, Kahru A. Bioavailability of Cd in 110 polluted topsoils to recombinant bioluminescent sensor bacteria: effect of soil particulate matter. J Soils Sediments. 2011;11:231–237. [Google Scholar]
- Ivask A, Francois M, Kahru A, Dubourguier HC, Virta M, Douay F. Recombinant luminescent bacterial sensors for the measurement of bioavailability of cadmium and lead in soils polluted by metal smelters. Chemosphere. 2004;55:147–156. doi: 10.1016/j.chemosphere.2003.10.064. [DOI] [PubMed] [Google Scholar]
- Ivask A, Green T, Polyak B, Mor A, Kahru A, Virta M, Marks R. Fibre-optic bacterial biosensors and their application for the analysis of bioavailable Hg and As in soils and sediments from Aznalcollar mining area in Spain. Biosens Bioelectron. 2007;22:1396–1402. doi: 10.1016/j.bios.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Ivask A, Hakkila K, Virta M. Detection of organomercurials with sensor bacteria. Anal Chem. 2001;73:5168–5171. doi: 10.1021/ac010550v. [DOI] [PubMed] [Google Scholar]
- Ivask A, Rolova T, Kahru A. A suite of recombinant luminescent bacterial strains for the quantification of bioavailable heavy metals and toxicity testing. BMC Biotechnol. 2009;9:41. doi: 10.1186/1472-6750-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivask A, Virta M, Kahru A. Construction and use of specific luminescent recombinant bacterial sensors for the assessment of bioavailable fraction of cadmium, zinc, mercury and chromium in the soil. Soil Biol Biochem. 2002;34:1439–1447. [Google Scholar]
- Jain VK, Magrath IT. A chemiluminescent assay for quantitation of beta-galactosidase in the femtogram range—application to quantitation of beta-galactosidase in lacZ-transfected cells. Anal Biochem. 1991;199:119–124. doi: 10.1016/0003-2697(91)90278-2. [DOI] [PubMed] [Google Scholar]
- Jaspers MCM, Suske WA, Schmid A, Goslings DAM, Kohler HPE, van der Meer JR. HbpR, a new member of the XylR/DmpR subclass within the NtrC family of bacterial transcriptional activators, regulates expression of 2-hydroxybiphenyl metabolism in Pseudomonas azelaica HBP1. J Bacteriol. 2000;182:405–417. doi: 10.1128/jb.182.2.405-417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries MKS, Conoan NH, Cox MB, Sangster JL, Balsiger HA, Bridges AA, Cowman T, Knight LA, Bartelt-Hunt SL, Kolok AS. The anti-estrogenic activity of sediments from agriculturally intense watersheds: assessment using in vivo and in vitro assays. Aquat Toxicol. 2011;105:189–198. doi: 10.1016/j.aquatox.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouanneau S, Durand MJ, Courcoux P, Blusseau T, Thouand G. Improvement of the identification of four heavy metals in environmental samples by using predictive decision tree models coupled with a set of five bioluminescent bacteria. Environ Sci Technol. 2011;45:2925–2931. doi: 10.1021/es1031757. [DOI] [PubMed] [Google Scholar]
- Keiter S, Grund S, van Bavel B, Hagberg J, Engwall M, Kammann U, Klempt M, Manz W, Olsman H, Braunbeck T, Hollert H. Activities and identification of aryl hydrocarbon receptor agonists in sediments from the Danube river. Anal Bioanal Chem. 2008;390:2009–2019. doi: 10.1007/s00216-007-1652-x. [DOI] [PubMed] [Google Scholar]
- Kelly CJ, Lajoie CA, Layton AC, Sayler GS. Bioluminescent reporter bacterium for toxicity monitoring in biological wastewater treatment systems. Water Environ Res. 1999;71:31–35. [Google Scholar]
- Kelly CJ, Tumsaroj N, Lajoie CA. Assessing wastewater metal toxicity with bacterial bioluminescence in a bench-scale wastewater treatment system. Water Res. 2004;38:423–431. doi: 10.1016/S0043-1354(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Kelsey JW, Kottler BD, Alexander M. Selective chemical extractants to predict bioavailability of soil-aged organic chemicals. Environ Sci Technol. 1997;31:214–217. [Google Scholar]
- King JMH, Digrazia PM, Applegate B, Burlage R, Sanseverino J, Dunbar P, Larimer F, Sayler GS. Rapid, sensitive bioluminescent reporter technology for naphthalene exposure and biodegradation. Science. 1990;249:778–781. doi: 10.1126/science.249.4970.778. [DOI] [PubMed] [Google Scholar]
- Knecht LD, Pasini P, Daunert S. Bacterial spores as platforms for bioanalytical and biomedical applications. Anal Bioanal Chem. 2011;400:977–989. doi: 10.1007/s00216-011-4835-4. [DOI] [PubMed] [Google Scholar]
- Kuncova G, Pazlarova J, Hlavata A, Ripp S, Sayler GS. Bioluminescent bioreporter Pseudomonas putida TVA8 as a detector of water pollution, Operational conditions and selectivity of free cells sensor. Ecol Indic. 2011;11:882–887. [Google Scholar]
- Lajoie CA, Lin SC, Kelly CJ. Comparison of bacterial bioluminescence with activated sludge oxygen uptake rates during zinc toxic shock loads in a wastewater treatment system. J Environ Eng-ASCE. 2003;129:879–883. [Google Scholar]
- Lampinen J, Koivisto L, Wahlsten M, Mantsala P, Karp M. Expression of luciferase genes from different origins in Bacillus subtilis. Mol Gen Genet. 1992;232:498–504. doi: 10.1007/BF00266255. [DOI] [PubMed] [Google Scholar]
- Lankinen P, Kähkönen M, Rajasärkkä J, Virta M, Hatakka A. The effect of nickel contamination on the growth of litter-decomposing fungi, extracellular enzyme activities and toxicity in soil. Boreal Environ Res. 2011;16:229–239. [Google Scholar]
- Lappalainen JO, Karp MT, Nurmi J, Juvonen R, Virta MPJ. Comparison of the total mercury content in sediment samples with a mercury sensor bacteria test and Vibrio fischeri toxicity test. Environ Toxicol. 2000;15:443–448. [Google Scholar]
- Larose C, Dommergue A, Marusczak N, Coves J, Ferrari CP, Schneider D. Bioavailable mercury cycling in polar snowpacks. Environ Sci Technol. 2011;45:2150–2156. doi: 10.1021/es103016x. [DOI] [PubMed] [Google Scholar]
- Layton AC, Muccini M, Ghosh MM, Sayler GS. Construction of a bioluminescent reporter strain to detect polychlorinated biphenyls. Appl Environ Microbiol. 1998;64:5023–5026. doi: 10.1128/aem.64.12.5023-5026.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leedjarv A, Ivask A, Virta M, Kahru A. Analysis of bioavailable phenols from natural samples by recombinant luminescent bacterial sensors. Chemosphere. 2006;64:1910–1919. doi: 10.1016/j.chemosphere.2006.01.026. [DOI] [PubMed] [Google Scholar]
- Leskinen P, Hilscherova K, Sidlova T, Kiviranta H, Pessala P, Salo S, Verta M, Virta M. Detecting AhR ligands in sediments using bioluminescent reporter yeast. Biosens Bioelectron. 2008;23:1850–1855. doi: 10.1016/j.bios.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Leskinen P, Michelini E, Picard D, Karp M, Virta M. Bioluminescent yeast assays for detecting estrogenic and androgenic activity in different matrices. Chemosphere. 2005;61:259–266. doi: 10.1016/j.chemosphere.2005.01.080. [DOI] [PubMed] [Google Scholar]
- Leusch FDL, De Jager C, Levi Y, Lim R, Puijker L, Sacher F, Tremblay LA, Wilson VS, Chapman HF. Comparison of five in vitro bioassays to measure estrogenic activity in environmental waters. Environ Sci Technol. 2010;44:3853–3860. doi: 10.1021/es903899d. [DOI] [PubMed] [Google Scholar]
- Liao VHC, Chien MT, Tseng YY, Ou KL. Assessment of heavy metal bioavailability in contaminated sediments and soils using green fluorescent protein-based bacterial biosensors. Environ Pollut. 2006;142:17–23. doi: 10.1016/j.envpol.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Liu YQ, Lu XB. Establishing a assay for detection of nonylphenol estrogenic effects. Appl Mech Mater. 2011;71:3003–3006. [Google Scholar]
- Louiz I, Kinani S, Gouze ME, Ben-Attia M, Menif D, Bouchonnet S, Porcher JM, Ben-Hassine OK, Ait-Aissa S. Monitoring of dioxin-like, estrogenic and anti-androgenic activities in sediments of the Bizerta lagoon (Tunisia) by means of in vitro cell-based bioassays: Contribution of low concentrations of polynuclear aromatic hydrocarbons (PAHs) Sci Total Environ. 2008;402:318–329. doi: 10.1016/j.scitotenv.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Machala M, Vondracek J, Blaha L, Ciganek M, Nea J. Aryl hydrocarbon receptor-mediated activity of mutagenic polycyclic aromatic hydrocarbons determined using in vitro reporter gene assay. Mutat Res Genet Toxicol Environ Mutagen. 2001;497:49–62. doi: 10.1016/s1383-5718(01)00240-6. [DOI] [PubMed] [Google Scholar]
- Maderova L, Watson M, Paton GI. Bioavailability and toxicity of copper in soils: Integrating chemical approaches with responses of microbial biosensors. Soil Biol Biochem. 2011;43:1162–1168. [Google Scholar]
- Magrisso S, Belkin S, Erel Y. Lead bioavailability in soil and soil components. Water Air Soil Pollut. 2009;202:315–323. [Google Scholar]
- McKay RML, Porta D, Bullerjahn GS, Al-Rshaidat MMD, Klimowicz JA, Sterner RW, Smutka TM, Brown ET, Sherrell RM. Bioavailable iron in oligotrophic Lake Superior assessed using biological reporters. J Plankton Res. 2005;27:1033–1044. [Google Scholar]
- Meighen EA. Genetics of bacterial bioluminescence. Annu Rev Genet. 1994;28:117–139. doi: 10.1146/annurev.ge.28.120194.001001. [DOI] [PubMed] [Google Scholar]
- Metzger D, White J, Chambon P. The human oestrogen receptor functions in yeast. Nature. 1988;334:31–36. doi: 10.1038/334031a0. [DOI] [PubMed] [Google Scholar]
- Murk AJ, Legler J, Denison MS, Giesy JP, vandeGuchte C, Brouwer A. Chemical-activated luciferase gene expression (CALUX): a novel in vitro bioassay for Ah receptor active compounds in sediments and pore water. Fundam Appl Toxicol. 1996;33:149–160. doi: 10.1006/faat.1996.0152. [DOI] [PubMed] [Google Scholar]
- Nakada N, Nyunoya H, Nakamura M, Hara A, Iguchi T, Takada H. Identification of estrogenic compounds in wastewater effluent. Environ Toxicol Chem. 2004;23:2807–2815. doi: 10.1897/03-699.1. [DOI] [PubMed] [Google Scholar]
- Nazarenko DA, Dertinger SD, Gasiewicz TA. Enhanced detection of beta-galactosidase reporter activation is achieved by a reduction of hemoglobin content in tissue lysates. Biotechniques. 2001;30:776–781. doi: 10.2144/01304st03. [DOI] [PubMed] [Google Scholar]
- Nelson J, Bishay F, van Roodselaar A, Ikonomou M, Law FCP. The use of in vitro bioassays to quantify endocrine disrupting chemicals in municipal wastewater treatment plant effluents. Sci Total Environ. 2007;374:80–90. doi: 10.1016/j.scitotenv.2006.11.031. [DOI] [PubMed] [Google Scholar]
- Nies DH. Microbial heavy-metal resistance. Appl Microbiol Biotechnol. 1999;51:730–750. doi: 10.1007/s002530051457. [DOI] [PubMed] [Google Scholar]
- Nissen S, Alexander BD, Dawood I, Tillotson M, Wells RPK, Macphee DE, Killham K. Remediation of a chlorinated aromatic hydrocarbon in water by photoelectrocatalysis. Environ Pollut. 2009;157:72–76. doi: 10.1016/j.envpol.2008.07.024. [DOI] [PubMed] [Google Scholar]
- Nordstrom DK. Public health—Worldwide occurrences of arsenic in ground water. Science. 2002;296:2143–2145. doi: 10.1126/science.1072375. [DOI] [PubMed] [Google Scholar]
- Norman A, Hansen LH, Sorensen SJ. Construction of a ColD cda promoter-based SOS-green fluorescent protein whole-cell biosensor with higher sensitivity toward genotoxic compounds than constructs based on recA, umuDC, or sul4 promoters. Appl Environ Microbiol. 2005;71:2338–2346. doi: 10.1128/AEM.71.5.2338-2346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton GI, Reid BJ, Sempled KT. Application of a luminescence-based biosensor for assessing naphthalene biodegradation in soils from a manufactured gas plant. Environ Pollut. 2009;157:1643–1648. doi: 10.1016/j.envpol.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Paton GI, Viventsova Ruth E, Kumpene J, Wilson MJ, Weitz HJ, Dawson JJC. An ecotoxicity assessment of contaminated forest soils from the Kola Peninsula. Sci Total Environ. 2006a;355:106–117. doi: 10.1016/j.scitotenv.2005.04.036. [DOI] [PubMed] [Google Scholar]
- Paton GT, Cheewasedtham W, Marr IL, Dawson JJC. Degradation and toxicity of phenyltin compounds in soil. Environ Pollut. 2006b;144:746–751. doi: 10.1016/j.envpol.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Peca L, Kos PB, Mate Z, Farsang A, Vass I. Construction of bioluminescent cyanobacterial reporter strains for detection of nickel, cobalt and zinc. FEMS Microbiol Lett. 2008;289:258–264. doi: 10.1111/j.1574-6968.2008.01393.x. [DOI] [PubMed] [Google Scholar]
- Peng ZX, Yan YL, Xu YQ, Takeo M, Yu HY, Zhao ZL, Zhan YH, Zhang W, Lin M, Chen M. Improvement of an E. coli bioreporter for monitoring trace amounts of phenol by deletion of the inducible sigma(54)-dependent promoter. Biotechnol Lett. 2010;32:1265–1270. doi: 10.1007/s10529-010-0317-6. [DOI] [PubMed] [Google Scholar]
- Porta D, Bullerjahn GS, Twiss MR, Wilhelm SW, Poorvin L, McKay RML. Determination of bioavailable Fe in Lake Erie using a luminescent cyanobacterial bioreporter. J Gt Lakes Res. 2005;31:180–194. [Google Scholar]
- Quillardet P, Huisman O, Dari R, Hofnung M. SOS Chromotest, a direct assay of induction of an SOS function in Escherichia coli K-12 to measure genotoxicity. Proc Natl Acad Sci U S A. 1982;79:5971–5975. doi: 10.1073/pnas.79.19.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhika V, Proikas-Cezanne T, Jayaraman M, Onesime D, Ha JH, Dhanasekaran DN. Chemical sensing of DNT by engineered olfactory yeast strain. Nat Chem Biol. 2007;3:325–330. doi: 10.1038/nchembio882. [DOI] [PubMed] [Google Scholar]
- Rattray EAS, Prosser JI, Killham K, Glover LA. Luminescence-based nonextractive technique for in situ detection of Escherichia coli in soil. Appl Environ Microbiol. 1990;56:3368–3374. doi: 10.1128/aem.56.11.3368-3374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Frymier PD. The use of a genetically engineered Pseudomonas species (Shk1) as a bioluminescent reporter for heavy metal toxicity screening in wastewater treatment plant influent. Water Environ Res. 2003;75:21–29. doi: 10.2175/106143003x140791. [DOI] [PubMed] [Google Scholar]
- Ripp S, Layton AC, Sayler GS. The microbe as a reporter: microbial bioreporter sensing technologies for chemical and biological detection. In: Sen K, Ashbolt NJ, editors. Environmental Microbiology: Current Technology and Water Applications. Caister Academic Press; Norfolk, UK: 2011. pp. 281–308. [Google Scholar]
- Ripp S, Nivens DE, Ahn Y, Werner C, Jarrell J, Easter JP, Cox CD, Burlage RS, Sayler GS. Controlled field release of a bioluminescent genetically engineered microorganism for bioremediation process monitoring and control. Environ Sci Technol. 2000;34:846–853. [Google Scholar]
- Robbens J, Dardenne F, Devriese L, De Coen W, Blust R. Escherichia coli as a bioreporter in ecotoxicology. Appl Microbiol Biotechnol. 2010;88:1007–1025. doi: 10.1007/s00253-010-2826-6. [DOI] [PubMed] [Google Scholar]
- Ron EZ, Rishpon J. Electrochemical cell-based sensors. In: Belkin S, Gu MB, editors. Whole Cell Sensing Systems I: Reporter Cells and Devices. Spring-Verlag; Berlin, Germany: 2010. pp. 77–84. [Google Scholar]
- Routledge EJ, Sumpter JP. Estrogenic activity of surfactants and some of their degradation products assessed using a recombinant yeast screen. Environ Toxicol Chem. 1996;15:241–248. [Google Scholar]
- Rowland B, Purkayastha A, Monserrat C, Casart Y, Takiff H, McDonough KA. Fluorescence-based detection of lacZ reporter gene expression in intact and viable bacteria including Mycobacterium species. FEMS Microbiol Lett. 1999;179:317–325. doi: 10.1111/j.1574-6968.1999.tb08744.x. [DOI] [PubMed] [Google Scholar]
- Salste L, Leskinen P, Virta M, Kronberg L. Determination of estrogens and estrogenic activity in wastewater effluent by chemical analysis and the bioluminescent yeast assay. Sci Total Environ. 2007;378:343–351. doi: 10.1016/j.scitotenv.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Sanseverino J, Gupta RK, Layton AC, Patterson SS, Ripp SA, Saidak L, Simpson ML, Schultz TW, Sayler GS. Use of Saccharomyces cerevisiae BLYES expressing bacterial bioluminescence for rapid, sensitive detection of estrogenic compounds. Appl Environ Microbiol. 2005;71:4455–4460. doi: 10.1128/AEM.71.8.4455-4460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selifonova OV, Eaton RW. Use of an ipb-lux fusion to study regulation of the isopropylbenzene catabolism operon of Pseudomonas putida RE204 and to detect hydrophobic pollutants in the environment. Appl Environ Microbiol. 1996;62:778–783. doi: 10.1128/aem.62.3.778-783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple KT, Morriss AWJ, Paton GI. Bioavailability of hydrophobic organic contaminants in soils: fundamental concepts and techniques for analysis. Eur J Soil Sci. 2003;54:809–818. [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Shaw JJ, Kado CI. Development of a Vibrio bioluminescence gene set to monitor phytopathogenic bacteria during the ongoing disease process in a nondisruptive manner. Nat Biotechnol. 1986;4:560–564. [Google Scholar]
- Shibasaki S, Ueda M, Ye K, Shimizu K, Kamasawa N, Osumi M, Tanaka A. Creation of cell surface-engineered yeast that display different fluorescent proteins in response to the glucose concentration. Appl Microbiol Biotechnol. 2001;57:528–533. doi: 10.1007/s002530100767. [DOI] [PubMed] [Google Scholar]
- Shin HJ, Park HH, Lim WK. Freeze-dried recombinant bacteria for on-site detection of phenolic compounds by color change. J Biotechnol. 2005;119:36–43. doi: 10.1016/j.jbiotec.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Shingler V, Moore T. Sensing of aromatic compounds by the DmpR transcriptional activator of phenol-catabolizing Pseudomonas sp strain CF600. J Bacteriol. 1994;176:1555–1560. doi: 10.1128/jb.176.6.1555-1560.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingleton JT, Applegate BM, Nagel AC, Bienkowski PR, Sayler GS. Induction of the tod operon by trichloroethylene in Pseudomonas putida TVA8. Appl Environ Microbiol. 1998;64:5049–5052. doi: 10.1128/aem.64.12.5049-5052.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidlova T, Novak J, Janosek J, Andel P, Giesy JP, Hilscherova K. Dioxin-like and endocrine disruptive activity of traffic-contaminated soil samples. Arch Environ Contam Toxicol. 2009;57:639–650. doi: 10.1007/s00244-009-9345-4. [DOI] [PubMed] [Google Scholar]
- Song YZ, Li GH, Thornton SF, Thompson IP, Banwart SA, Lerner DN, Huang WE. Optimization of bacterial whole cell bioreporters for toxicity assay of environmental samples. Environ Sci Technol. 2009;43:7931–7938. doi: 10.1021/es901349r. [DOI] [PubMed] [Google Scholar]
- Sousa S, Duffy C, Weitz H, Glover LA, Bar E. Use of a lux-modified bacterial, biosensor to identify constraints to bioremediation of BTEX-contaminated sites. Environ Toxicol Chem. 1998;17:1039–1045. [Google Scholar]
- Sticher P, Jaspers MCM, Stemmler K, Harms H, Zehnder AJB, van der Meer JR. Development and characterization of a whole-cell bioluminescent sensor for bioavailable middle-chain alkanes in contaminated groundwater samples. Appl Environ Microbiol. 1997;63:4053–4060. doi: 10.1128/aem.63.10.4053-4060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiner L, Halverson LJ. Development and characterization of a green fluo-rescent protein-based bacterial biosensor for bioavailable toluene and related compounds. Appl Environ Microbiol. 2002;68:1962–1971. doi: 10.1128/AEM.68.4.1962-1971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker J, Balluch D, Gsell M, Harms H, Feliciano J, Daunert S, Malik KA, van der Meer JR. Development of a set of simple bacterial biosensors for quantitative and rapid measurements of arsenite and arsenate in potable water. Environ Sci Technol. 2003;37:4743–4750. doi: 10.1021/es034258b. [DOI] [PubMed] [Google Scholar]
- Struss AK, Pasini P, Daunert S. Biosensing systems based on genetically engineered whole cells. In: Zourob M, editor. Recognition Receptors in Biosensors. Springer; New York: 2010. pp. 565–598. [Google Scholar]
- Suzuki G, Someya M, Takahashi S, Tanabe S, Sakai S, Takigami H. Dioxin-like activity in Japanese indoor dusts evaluated by means of in vitro bioassay and instrumental analysis: brominated dibenzofurans are an important contributor. Environ Sci Technol. 2010;44:8330–8336. doi: 10.1021/es102021c. [DOI] [PubMed] [Google Scholar]
- Tauriainen S, Karp M, Chang W, Virta M. Luminescent bacterial sensor for cadmium and lead. Biosens Bioelectron. 1998;13:931–938. doi: 10.1016/s0956-5663(98)00027-x. [DOI] [PubMed] [Google Scholar]
- Tauriainen S, Virta M, Chang W, Karp M. Measurement of firefly luciferase reporter gene activity from cells and lysates using Escherichia coli arsenite and mercury sensors. Anal Biochem. 1999;272:191–198. doi: 10.1006/abio.1999.4193. [DOI] [PubMed] [Google Scholar]
- Tecon R, Beggah S, Czechowska K, Sentchilo V, Chronopoulou PM, McGenity TJ, van der Meer JR. Development of a multistrain bacterial bioreporter platform for the monitoring of hydrocarbon contaminants in marine environments. Environ Sci Technol. 2010;44:1049–1055. doi: 10.1021/es902849w. [DOI] [PubMed] [Google Scholar]
- Tecon R, Binggeli O, van der Meer JR. Double-tagged fluorescent bacterial bioreporter for the study of polycyclic aromatic hydrocarbon diffusion and bioavailability. Environ Microbiol. 2009;11:2271–2283. doi: 10.1111/j.1462-2920.2009.01952.x. [DOI] [PubMed] [Google Scholar]
- Tecon R, van der Meer JR. Bacterial biosensors for measuring availability of environmental pollutants. Sensors. 2008;8:4062–4080. doi: 10.3390/s8074062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecon R, Wells M, van der Meer JR. A new green fluorescent protein-based bacterial biosensor for analysing phenanthrene fluxes. Environ Microbiol. 2006;8:697–708. doi: 10.1111/j.1462-2920.2005.00948.x. [DOI] [PubMed] [Google Scholar]
- Thorpe KL, Gross-Sorokin M, Johnson I, Brighty G, Tyler CR. An assessment of the model of concentration addition for predicting the estrogenic activity of chemical mixtures in wastewater treatment works effluents. Environ Health Perspect. 2006;114:90–97. doi: 10.1289/ehp.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibazarwa C, Corbisier P, Mench M, Bossus A, Solda P, Mergeay M, Wyns L, van der Lelie D. A microbial biosensor to predict bioavailable nickel in soil and its transfer to plants. Environ Pollut. 2001;113:19–26. doi: 10.1016/s0269-7491(00)00177-9. [DOI] [PubMed] [Google Scholar]
- Tiensing T, Strachan N, Paton GI. Evaluation of interactive toxicity of chlorophenols in water and soil using lux-marked biosensors. J Environ Monit. 2002;4:482–489. doi: 10.1039/b202070j. [DOI] [PubMed] [Google Scholar]
- Toba FA, Hay AG. A simple solid phase assay for the detection of 2,4-D in soil. J Microbiol Methods. 2005;62:135–143. doi: 10.1016/j.mimet.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Tom-Petersen A, Hosbond C, Nybroe O. Identification of copper-induced genes in Pseudomonas fluorescens and use of a reporter strain to monitor bioavailable copper in soil. FEMS Microbiol Ecol. 2001;38:59–67. [Google Scholar]
- Trang PTK, Berg M, Viet PH, Van Mui N, van der Meer JR. Bacterial bioassay for rapid and accurate analysis of arsenic in highly variable groundwater samples. Environ Sci Technol. 2005;39:7625–7630. doi: 10.1021/es050992e. [DOI] [PubMed] [Google Scholar]
- Tropel D, van der Meer JR. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol Mol Biol Rev. 2004;68:474–500. doi: 10.1128/MMBR.68.3.474-500.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpeinen R, Salminen J, Kairesalo T. Mobility and bioavailability of lead in contaminated boreal forest soil. Environ Sci Technol. 2000;34:5152–5156. [Google Scholar]
- Turpeinen R, Virta M, Haggblom MM. Analysis of arsenic bioavailability in contaminated soils. Environ Toxicol Chem. 2003;22:1–6. [PubMed] [Google Scholar]
- van der Meer JR, Belkin S. Where microbiology meets microengineering: design and applications of reporter bacteria. Nat Rev Microbiol. 2010;8:511–522. doi: 10.1038/nrmicro2392. [DOI] [PubMed] [Google Scholar]
- Van Dyk TK, Majarian WR, Konstantinov KB, Young RM, Dhurjati PS, Larossa RA. Rapid and sensitive pollutant detection by induction of heat shock gene-bioluminescence gene fusions. Appl Environ Microbiol. 1994;60:1414–1420. doi: 10.1128/aem.60.5.1414-1420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virta M, Lampinen J, Karp M. A luminescence-based mercury biosensor. Anal Chem. 1995;67:667–669. [Google Scholar]
- Vollmer AC, Belkin S, Smulski DR, VanDyk TK, LaRossa RA. Detection of DNA damage by use of Escherichia coli carrying recA’-lux, uvrA’-lux, or alkA’-lux reporter plasmids. Appl Environ Microbiol. 1997;63:2566–2571. doi: 10.1128/aem.63.7.2566-2571.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walmsley RM, Keenan P. The eukaryote alternative: advantages of using yeasts in place of bacteria in microbial biosensor development. Biotechnol Bio-process Eng. 2000;5:387–394. [Google Scholar]
- Wehmas LC, Cavallin JE, Durhan EJ, Kahl MD, Martinovic D, Mayasich J, Tuominen T, Villeneuve DL, Ankley GT. Screening complex effluents for estrogenic activity with the T47D-KBluc cell bioassay: assay optimization and comparison with in vivo responses in fish. Environ Toxicol Chem. 2011;30:439–445. doi: 10.1002/etc.388. [DOI] [PubMed] [Google Scholar]
- Weitz HJ, Ritchie JM, Bailey DA, Horsburgh AM, Killham K, Glover LA. Construction of a modified mini-Tn5 luxCDABE transposon for the development of bacterial biosensors for ecotoxicity testing. FEMS Microbiol Lett. 2001;197:159–165. doi: 10.1111/j.1574-6968.2001.tb10598.x. [DOI] [PubMed] [Google Scholar]
- Willardson BM, Wilkins JF, Rand TA, Schupp JM, Hill KK, Keim P, Jackson PJ. Development and testing of a bacterial biosensor for toluene-based environmental contaminants. Appl Environ Microbiol. 1998;64:1006–1012. doi: 10.1128/aem.64.3.1006-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson VS, Bobseine K, Gray LE. Development and characterization of a cell line that stably expresses an estrogen-responsive luciferase reporter for the detection of estrogen receptor agonist and antagonists. Toxicol Sci. 2004;81:69–77. doi: 10.1093/toxsci/kfh180. [DOI] [PubMed] [Google Scholar]
- Wilson VS, Bobseine K, Lambright CR, Gray LE. A novel cell line, MDA-kb2, that stably expresses an androgen- and glucocorticoid-responsive reporter for the detection of hormone receptor agonists and antagonists. Toxicol Sci. 2002;66:69–81. doi: 10.1093/toxsci/66.1.69. [DOI] [PubMed] [Google Scholar]
- Wolz J, Engwall M, Maletz S, Takner HO, van Bavel B, Kammann U, Klempt M, Weber R, Braunbeck T, Hollert H. Changes in toxicity and Ah receptor agonist activity of suspended particulate matter during flood events at the rivers Neckar and Rhine—a mass balance approach using in vitro methods and chemical analysis. Environ Sci Pollut Res. 2008;15:536–553. doi: 10.1007/s11356-008-0056-6. [DOI] [PubMed] [Google Scholar]
- Yagur-Kroll S, Belkin S. Upgrading bioluminescent bacterial bioreporter performance by splitting the lux operon. Anal Bioanal Chem. 2011;400:1071–1082. doi: 10.1007/s00216-010-4266-7. [DOI] [PubMed] [Google Scholar]