Summary
Networks are a powerful and flexible methodology for expressing biological knowledge for computation and communication. Network-encoded information can include systematic screens for molecular interactions, biological relationships curated from literature, and outputs from analysis of Big Data. NDEx, the Network Data Exchange (www.ndexbio.org), is an online commons where scientists can upload, share, and publicly distribute networks. Networks in NDEx receive globally unique accession IDs and can be stored for private use, shared in pre-publication collaboration, or released for public access. Standard and novel data formats are accommodated in a flexible storage model. Organizations can use NDEx as a distribution channel for networks they generate or curate. Developers of bioinformatic applications can store and query NDEx networks via a common programmatic interface. NDEx helps expand the role of networks in scientific discourse and facilitates the integration of networks as data in publications. It is a step towards an ecosystem in which networks bearing data, hypotheses, and findings flow easily between scientists.
Graphical abstract
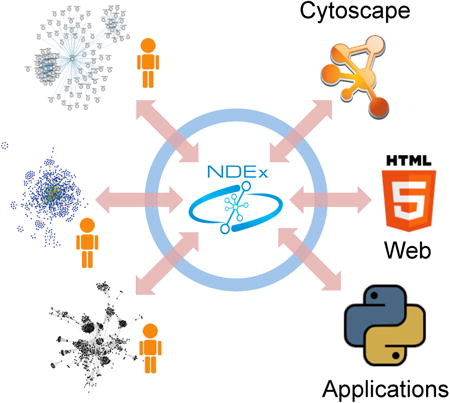
Networks are a precise and computable form in which biologists can express many kinds of information, including models of biological mechanisms, experimental facts, and relationships derived by systematic data analysis. When pathway diagrams evolved into repositories of small pathway networks (Croft et al., 2014; Kanehisa and Goto, 2000; Ogata et al., 1998), they became searchable resources and the basis for data interpretation and collaborative pathway editing applications (van Iersel et al., 2008). The emergence of repositories of large networks of molecular relationships (Franceschini et al., 2013; Orchard et al., 2014; Stark et al., 2011; Warde-Farley et al., 2010) in both simple and complex formats (Demir et al., 2010; Le Novere et al., 2006; Mi et al., 2010; OpenBEL, 2011) condensed collections of data into structured findings useful for hypothesis generation and computational prediction (Bandyopadhyay et al., 2006; Ingenuity; Vandin et al., 2012). In recent years, there has been rapid progress in the construction of networks inferred by the systematic processing of genome-scale information, providing an important avenue for interpretation and a counterpoint to literature curation (Califano et al., 2012; Chuang et al., 2007; Hofree et al., 2013). By providing a flexible computable medium for biological knowledge, networks are also becoming a critical element for new models of scientific publication, in which data and its derivatives are as important as text (CyNetShare, 2014).
NDEx, the Network Data Exchange, is an open-source (Data S1, Open Source) software framework that facilitates the sharing of networks of many types and formats, the publication of networks as data, and the use of networks in modular software. In comparison to repositories such as IntAct (Orchard et al., 2014) or KEGG (Kanehisa and Goto, 2000) where network content in specific formats is managed by the organization maintaining the resource, NDEx is a data commons where users manage the sharing and publication of their own networks (Data S1, Related Resources). It accommodates networks of any type, from pathway models and interaction maps to novel data-driven knowledge, handling diverse formats including SIF, XGMML, BioPAX3 and OpenBEL (Data S1, Network Formats). It promotes scientific publication and reproducibility by enabling the tracking of accession and provenance of networks. Finally, NDEx provides a flexible, programmatically accessible storage service that promotes modular software development and workflows in which networks output from one application can be input to another. NDEx does not perform biological analysis and visualization itself but instead enables interchange of networks between applications that do.
As a data commons, NDEx enables scientists and organizations that create accounts on the NDEx server to upload and save networks and to create communities of users, much like Google+ Circles or LinkedIn Groups. They can manage access to their networks, making them private, public, or shared with selected users and community groups, similar to shared document systems such as Google Docs or DropBox (Figure 1A) (Data S1, NDEx Basics). The shared networks preserve the distinct semantics of their original formats while standardizing the treatment of identifiers, citations, properties, and network topology (Figure 1B) (Data S1, Data Model). NDEx therefore differs in approach from WikiPathways (Pico et al., 2008), a pioneering collaborative platform for the curation of biological pathways, where all documents are publically edited and use a single format. Organizations that publish network content can use NDEx as a channel for distribution: networks from the NCI Pathway Interaction Database (Schaefer et al., 2009), Pathway Commons (Cerami et al., 2011), and the OpenBEL Consortium are among those available in NDEx (Data S1, Metrics).
Figure 1. Access Control, Network Data Structures, and Provenance History in NDEx.
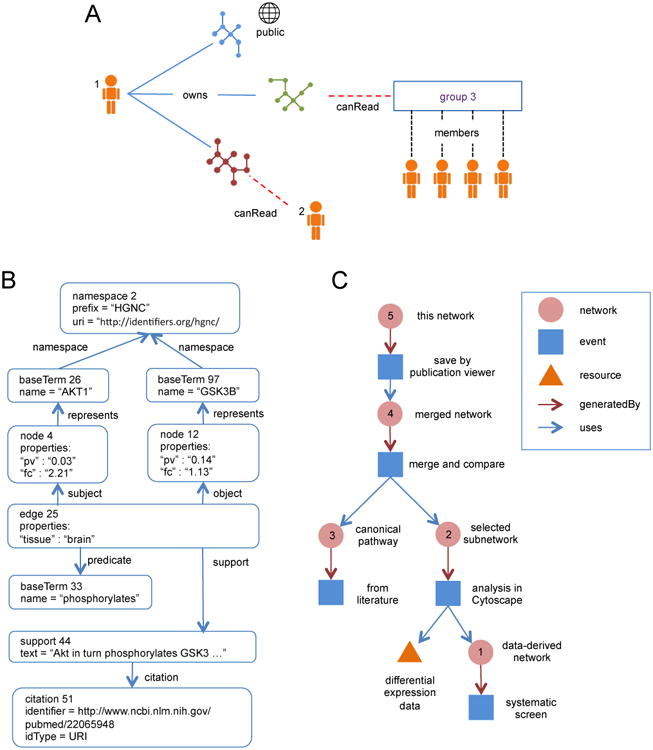
(A) Examples of access control relationships for networks in NDEx. User 1 owns the red, green, and blue networks. She shares the red network directly with user 2, the green network with the members of community group 3, and makes the blue network a public network available to any user or by anonymous query. (B) Example of one edge represented in the NDEx network data model (Supplemental Materials 10). Each box in the diagram is a network element, labeled with its type and id. Edge 25 connects nodes 4 and 12 by the subject and object relationships. The meaning of edge 25 is set by the predicate relationship to baseTerm 33, “phosphorylates”. BaseTerm objects define the vocabulary used by the network, and the primary meaning of node 4 is set by the represents relationship to baseTerm 26, “AKT1”. Node 12 represents baseTerm 97, “GSK3B”. Both baseTerm 97 and baseTerm 26 are associated with namespace 2, indicating that they are standard human gene symbols. Both nodes have user-defined properties “fc” and “pv” associated with them, used to record differential expression data that was mapped onto the network. Edge 25 has a user-defined property “tissue” = “brain” used by the authors to indicate the tissue context. The edge is also annotated with evidence text by support 44 associated with citation 51, the article from which the text was derived. (C) Abstract representation of the provenance history for network 5 in Figure 2. The provenance history records the workflow that led to the network as a tree structure of events, NDEx networks, and other resources.
To support publication of networks as data, it must be possible to unambiguously specify the identity of the network and trust that the content of a published network will remain constant. NDEx provides accession identifiers for every network, assigning a universal unique identifier (UUID) that distinguishes it from all other networks across all servers. The owner of an NDEx network can set its status to be read-only, preventing further edits. These features enable networks to be reliable, consistent references, suitable as inputs to further research.
When networks expressing data, hypotheses, and findings are both inputs and outputs of analysis and are referenced in publications, it becomes important to know how and when a network was created and which inputs and algorithms would be required to reproduce it. NDEx addresses these needs by including the “provenance history” (Figure 1C) (Data S1, Provenance) with each network. The provenance history captures the workflow leading to the current network by describing prior events, networks and other resources. The history grows as networks are created, modified, used, or copied. It incorporates concepts and vocabulary from ongoing work in provenance annotation (Ciccarese et al., 2013; DublinCore, 2012; PROV-O, 2013), but adopts a strategy where referenced resources are described rather than simply linked, preserving information in cases where the resources are later removed, altered, or become unavailable.
NDEx promotes the development of new network analysis algorithms and applications by expanding access to networks as inputs, by facilitating immediate sharing of network results and by providing a path to publication. An NDEx server stores and manages networks and all related information and can be accessed by applications via a web-based REST API (RElational State Transfer Application Programming Interface) (Fielding and Taylor, 2002). NDEx client libraries for this API have been created in the Java, Python, and R languages to facilitate easy use by scientists, though NDEx can be accessed via any language capable of Internet communications (Data S1, REST API). The NDEx website (www.ndexbio.org) is the most comprehensive example of an NDEx-enabled application that accesses the public NDEx server via the REST API. The website enables visitors to anonymously search, browse, and query networks and logged in users to upload content, manage groups and share networks. Simple analysis scripts (e.g., Python or R) can also query NDEx via the API to obtain input networks and then save analysis result networks directly to NDEx (Figure 2). This storage service model enables the researcher to focus on the core data analysis or algorithm rather than on the management, storage, and publication of networks.
Figure 2. NDEx Workflow.
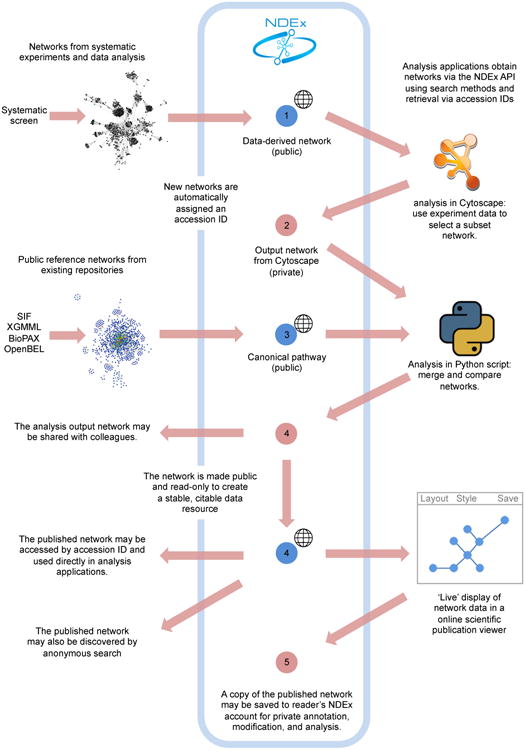
Example workflow in which network 1 is created by systematic analysis of genome scale data and stored in NDEx, network 2 is produced by a Cytoscape analysis that takes network 1 as an input, network 3 represents a canonical pathway uploaded from literature, and network 4 is the output of a bioinformatic script that operates on networks 2 and 3. Network 4 is made public and read-only and becomes part of a publication. Network 4 is viewed by readers of the publication using an NDEx-capable web application that enables them to directly act on the network data, such as saving a private copy to an NDEx account as network 5.
The rich Cytoscape biological analysis and visualization environment (Shannon et al., 2003) can also access NDEx via the REST API, enabling Cytoscape users to search, import and export networks. Under Cytoscape, a workflow might start by importing a transcriptional regulatory network from NDEx, after which the user could annotate the network with a differential mRNA expression dataset and process it to find subnetworks enriched for genes with significant changes in mRNA expression. The user could then export the subnetworks back to NDEx for review by collaborators or for use as inputs to further analyses. The CyNDEx App (Data S1, CyNDEx) implements access to NDEx, and upcoming releases of Cytoscape are expected to incorporate its functionality into the main application, making NDEx networks immediately available to users.
An important use of NDEx is to enable new models of scientific publication via network visualization applications in which live data structures replace static diagrams and supplemental files. Readers can immediately act on networks published via NDEx as data that can be dynamically visualized, inspected, and manipulated. For example, a biologist might save selected portions of a published network to their NDEx account to capture a mechanism of interest. Both the original and saved networks would be accessible to other NDEx-capable applications for analysis and visualization. This integration of viewing, annotation, sharing, and action can accelerate and enrich the process of scientific communication.
Finally, users can download and deploy the NDEx server software for private uses that would be impractical or unsupported on the shared public server. An NDEx server can be installed behind a firewall to handle cases where strong security is required, enabling storage of proprietary networks developed for the health sciences industry or those that incorporate patient information subject to privacy standards (e.g., HIPAA, the Health Insurance Portability and Accountability Act). A private NDEx can also be deployed on local servers or on a scientist's desktop for applications that store very large networks or perform frequent, large transactions. Applications can simultaneously access both public and private NDEx servers, or users can coordinate private NDEx instances with a public server using NDEx Sync (Data S1, NDEx Sync), a command line utility that can copy and update selected networks between servers.
In summary, NDEx provides distinctive capabilities as a data commons to further the use of biological networks in scientific discourse. It promotes the development of modular applications and the re-use of research products by creating a network exchange where the outputs from one project or application can readily become inputs to another. The NDEx platform is enabling new forms of publication and collaboration in which network information can be immediately analyzed, visualized, annotated, and shared.
Supplementary Material
NDEx, the Network Data Exchange is an online resource to enable new modes of collaboration and publication using biological networks. Scientists using NDEx can upload, share and distribute networks in diverse formats and sizes. Networks in NDEx can be private, shared for pre-publication collaboration, or publicly released. Bioinformatic applications can access NDEx via a common programmatic interface. These services can expand the role of networks in scientific discourse and enable new capabilities such as embedding ‘live’ networks directly in publications. NDEx is an important step towards an ecosystem in which networks bearing data, hypotheses, and findings flow easily between scientists.
Acknowledgments
NDEx is supported by F. Hoffmann-La Roche Ltd, Janssen Research and Development, LLC, Pfizer, Inc. and the National Cancer Institute under award U24 CA-184427.
Footnotes
Author Contributions: DP, JC, DW, RR, VR, and RP designed, implemented, tested, and documented the NDEx software. KO wrote the Network Publication Viewer. DP and TI wrote the paper. BD, CM, LH, SS, AS, JK, RD and MB provided valuable guidance, review, and technical support. TI, SS and DP conceived of the project, DP and TI directed its execution.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bandyopadhyay S, Kelley R, Ideker T. Discovering regulated networks during HIV-1 latency and reactivation. Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing. 2006:354–366. [PMC free article] [PubMed] [Google Scholar]
- Califano A, Butte AJ, Friend S, Ideker T, Schadt E. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nature genetics. 2012;44:841–847. doi: 10.1038/ng.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, Schultz N, Bader GD, Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic acids research. 2011;39:D685–690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarese P, Soiland-Reyes S, Belhajjame K, Gray AJG, Goble C, Clark T. PAV ontology: provenance, authoring and versioning. Journal of biomedical semantics. 2013;4:37. doi: 10.1186/2041-1480-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, et al. The Reactome pathway knowledgebase. Nucleic acids research. 2014;42:D472–477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CyNetShare (2014). http://cynetshare.ucsd.edu.
- Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D'Eustachio P, Schaefer C, Luciano J, et al. The BioPAX community standard for pathway data sharing. Nature biotechnology. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DublinCore. Dublin Core Metadata Element Set, Version 1.1. 2012 http://www.dublincore.org/documents/dces/
- Fielding RT, Taylor RN. Principled design of the modern Web architecture. ACM Transactions on Internet Technology (TOIT) 2002;2:115–150. [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofree M, Shen JP, Carter H, Gross A, Ideker T. Network-based stratification of tumor mutations. Nature methods (United States) 2013:1108–1115. doi: 10.1038/nmeth.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingenuity. Ingenuity Systems, Inc. http://www.ingenuity.com.
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novere N, Bornstein B, Broicher A, Courtot M, Donizelli M, Dharuri H, Li L, Sauro H, Schilstra M, Shapiro B, et al. BioModels Database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic acids research. 2006;34:D689–691. doi: 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic acids research. 2010;38:D204–210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic acids research. 1998;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OpenBEL. OpenBEL. 2011 http://www.openbel.org.
- Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, Campbell NH, Chavali G, Chen C, del-Toro N, et al. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic acids research. 2014;42:D358–363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS biology. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PROV-O. PROV-O: The PROV Ontology. 2013 http://www.w3.org/TR/prov-o/
- Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic acids research. 2009;37:D674–679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. The BioGRID Interaction Database: 2011 update. Nucleic acids research. 2011;39:D698–704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Iersel MP, Kelder T, Pico AR, Hanspers K, Coort S, Conklin BR, Evelo C. Presenting and exploring biological pathways with PathVisio. BMC Bioinformatics. 2008;9:399. doi: 10.1186/1471-2105-9-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandin F, Clay P, Upfal E, Raphael BJ. Discovery of mutated subnetworks associated with clinical data in cancer. Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing. 2012:55–66. [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic acids research. 2010;38:W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


