Abstract
Recent molecular clock studies date the origin of Metazoa to 750–800 million years ago (Ma), roughly coinciding with evidence from geochemical proxies that oxygen levels rose from less than 0.1% present atmospheric level (PAL) to perhaps 1–3% PAL O2. A younger origin of Metazoa would require greatly increased substitution rates across many clades and many genes; while not impossible, this is less parsimonious. Yet the first fossil evidence for metazoans (the Doushantuo embryos) about 600 Ma is followed by the Ediacaran fossils after 580 Ma, the earliest undisputed bilaterians at 555 Ma, and an increase in the size and morphologic complexity of bilaterians around 542 Ma. This temporal framework suggests a missing 150–200 Myr of early metazoan history that encompasses many apparent novelties in the early evolution of the nervous system. This span includes two major glaciations, and complex marine geochemical changes including major changes in redox and other environmental changes. One possible resolution is that animals of these still unknown Cryogenian and early Ediacaran ecosystems were relatively simple, with highly conserved developmental genes involved in cell-type specification and simple patterning. In this model, complex nervous systems are a convergent phenomenon in bilaterian clades which occurred close to the time that larger metazoans appeared in the fossil record.
Keywords: metazoa, Cryogenian, Ediacaran, redox, molecular clock, innovation
1. Introduction
The origin and early divergence of animals occurred from the Cryogenian (ca 850–635 Ma) into the Ediacaran (635–542 Ma) and Cambrian periods, and the information from geological and fossil records provides useful constraints on hypotheses concerning the origin and early divergence of animals, and more specifically on early nervous system evolution. Molecular clock analyses provide estimates of divergence times between major clades, phylogenetic analyses establish the branching topology of these groups, and when synthesized with geological data the environmental context in which these events occurred can be inferred. In this contribution, I summarize recent work on metazoan phylogeny and molecular clock estimates. The Cryogenian and Ediacaran were intervals of pervasive environmental changes, many of them unlike anything that has occurred in the past 540 million years, but the global glaciations, changes in marine redox and possible extensive changes in oxygen levels in the oceans and atmosphere provide critical information on the environmental context of early metazoan life. Comparative studies of the development of living animals support conflicting interpretations of the probable morphology of animals during this time. The integration of geological and geochemical data with developmental information allows one to evaluate the plausibility of alternative models for the early history of animals.
2. Rate and topology of early metazoan divergences
The relationships between most animal groups have been satisfactorily established through molecular phylogenetics (figure 1). The bilaterians include the Lophotrochozoa plus Edcysozoa, together comprising the protostomes, and the Deuterostome clades. Most of the unresolved issues of metazoan phylogeny involve the basal metazoan groups of sponges, cnidarians, ctenophores and placozoans. Specifically, these include disagreements over whether sponges are a single monophyletic clade [2] or a suite of paraphyletic clades [3], controversial claims that ctenophores may lie at the base of Metazoa outside sponges [4,5], and uncertainty over the position of acoel flatworms and Xenoturbella [6]. These issues were analysed in a recent review [7]. None of these issues can be considered resolved, but their resolution is critical for understanding patterns of developmental evolution in the early history of metazoans, particularly whether neurons are shared character among ctenophores, cnidarians and bilaterians, or arose with ctenophores but were lost in sponges [4,5] or arose separately in ctenophores and the cnidarian plus bilaterian clade. These issues are also important for understanding the ecosystems of the late Neoproterozoic, and for interpreting the role of highly conserved genes.
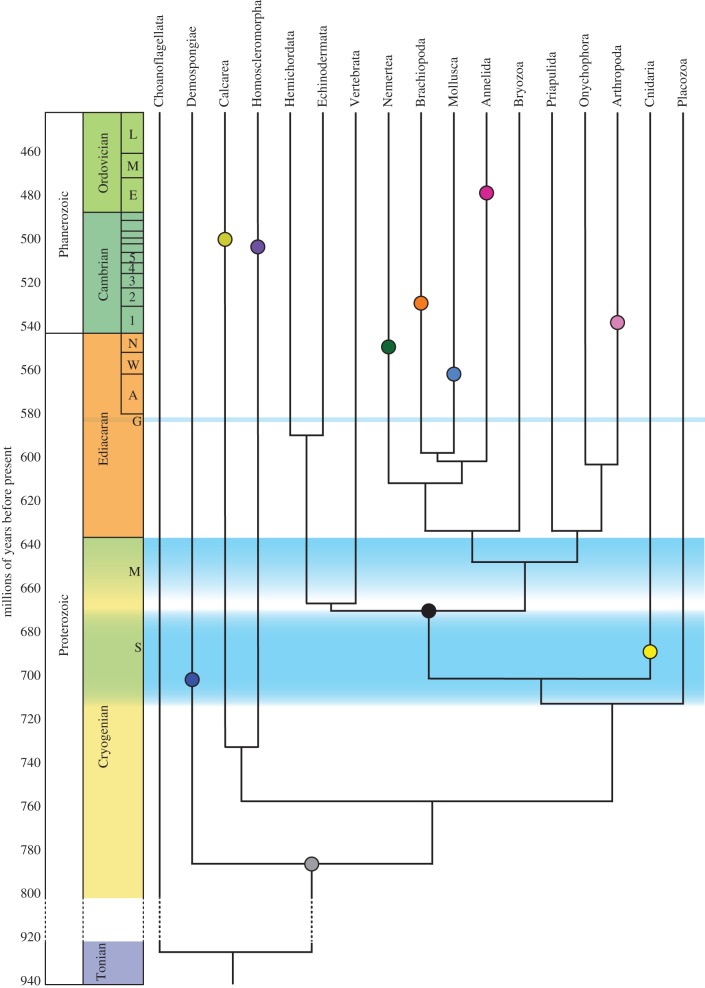
Figure 1.
A simplified topology of the metazoan radiation, based on the phylogeny and molecular clock results from reference [1]. The coloured circles are the estimated ages of the origin of the crown group of each clade, based on molecular clock estimates. The uncertainties on the divergence times are not shown on this diagram for clarity. These match closely with estimates from the fossil record, except for crown demosponges and crown cnidarians, which occur in the Cryogenian. The duration of the major glaciations are indicated: S, Sturtian; M, Marinoan; G, Gaskiers. (Online version in colour.)
Taking each of these three issues in turn, if sponges are paraphyletic this provides a much stronger inference that the last common ancestor (LCA) of metazoans must have been sponge-like. Alternatively, if sponges are a single clade, we cannot rely upon inferences about the morphology of these nodes to infer the morphological and developmental attributes of the most basal metazoans. Several recent studies suggest that sponges are likely to be monophyletic rather than paraphyletic [8,9]. Earlier studies indicated that resolving the phylogenetic position of ctenophores was complicated by considerable long-branch attraction [10], but more recent studies have attempt to resolve these difficulties and support a basal position for the clade [4,5,11], but have also been criticized [12,13]. In a recent study, long-branch attraction was found to have potentially been involved in the results of reference [4], and other potential problems affected reference [5]. Nonetheless, this new study evaluated these problems through increased taxon sampling and other improvements, and found support for ctenophores at the base of metazoans [14,15]. In contrast, a detailed study of opsins and phototransduction found good support for the classic phylogeny with uniting ctenophores, cnidarians and bilaterians [16]. Other analyses are in progress and I view the issue as currently unresolved. The phylogenetic position of ctenophores has important implications for the early history of metazoans in general, and for the nervous system in particular, and will be discussed in §4 and elsewhere in this issue [17]. Analyses placing acoel flatworms and Xenoturbella at the base of deuterostomes rather than below the last common protostome–deuterostome ancestor (LCPDA) were surprising, but even if this result is confirmed by further research, comparative analysis suggests that organisms morphologically similar to acoels likely occurred below the LCPDA [18].
Many molecular clock analyses have been conducted in the past several decades to estimate the divergence times of the metazoan clades. Figure 1 presents a simplified version of the analysis published in 2011 [1]. The study used concatenated sequences of several different housekeeping genes for 118 taxa in all major metazoan clades. Ctenophores were initially included but subsequently excluded because of analytical difficulties. The study included 24 different calibration points distributed across the tree, in contrast to the dominantly vertebrate calibration points used in previous studies, and employed a relaxed clock analysis. Further details can be found in reference [1]. Several aspects of the results are worth noting. First, the results suggest an origin of Metazoa during the Cryogenian, approximately 780–800 Ma, the LCA of cnidarians and bilaterians at approximately 700 Ma and of bilaterians approximately 688 Ma (all estimates have analytical uncertainties discussed in detail in reference [1]). Because so many taxa were included we were able to estimate the ages of the crown groups for the larger clades, and these largely fell during the late Ediacaran and Cambrian. The congruence between these estimates for the origin of the crown group and the actual appearances of the crown group in the fossil record provides additional confidence in these results. Second, if these results are reliable, they suggest crown-group cnidarians were present by approximately 700 Ma, raising the issue of why a nematocyst had evolved at such an early date. One reviewer of this manuscript argued that this was a non-issue, but nematocysts are clearly unique to the clade, despite recurrent attempts to link them to other groups [19]. They are involved in both predation and protection, often in specialized forms and are energetically costly elements to produce. Crown-group cnidarians with nematocysts at approximately 700 Ma indicates greater complexity to later Neoproterozoic ecosystems than has been previously appreciated. Third, these results imply that many metazoan lineages must have been present during the Ediacaran, not only sponges, cnidarians, but also stem groups of arthropods, priapulids, annelids, molluscs, brachiopods, vertebrates, echinoderms, hemichordates and others. As discussed further in §4, this does not require that such lineages had acquired the morphological characters of the crown groups (see also discussion in reference [11]). The only generally accepted fossil representative of any of these clades identified from the Ediacaran to date is Kimberella, from 555 Ma rocks in the White Sea of Russia, which could be a stem mollusc but is certainly a stem lophotrochozoan [20,21]. This molecular clock analysis should be re-done to assess the impact of a possible basal position for ctenophores, but such a topology would seem to require an earlier Cryogenian date for the origin of Metazoa. Moreover, the ctenophore-early hypothesis exacerbates a challenge posed by the inference of stem-group cnidarians at 700 Ma: both cnidarians and ctenophores are active and often voracious predators. In the absence of zooplankton, on what were they feeding?
One frequently proposed challenge to metazoan divergences that greatly predate evidence from the fossil record is the possibility of increased rate of molecular evolution across early metazoan lineages, bringing the molecular clock estimates into closer congruence with the fossil record. Several recent studies have examined this possibility, largely focusing on arthropods. One study rejected a rapid, late Ediacaran divergence of arthropod lineages, estimating the origin of the arthropod crown group at 706 Ma (with a very large uncertainty) [22], substantially older than our estimate of 560 Ma. However, this study used only eight calibration points. Another analysis used expressed sequence tags for 129 genes in 117 taxa, of which 101 were arthropods, to estimate divergences through the panarthropod clade. The estimated divergence of Panarthropoda was placed at approximately 600 Ma [23], which is not substantially different from our estimate of approximately 630 Ma. Another study inverted the question and concluded that rates of phenotypic evolution would have been four times and molecular rates five and half times greater to produce arthropod divergences largely consistent with the fossil record [24]. But even such elevated evolutionary rates would necessarily require much earlier divergences for bilaterian and basal metazoan clades. Some continue to reject the molecular clock evidence in favour of a fairly literal reading of the fossil record [25]. I long held this position [26], but I no longer view it as tenable. The analytical methods used in molecular clock studies, the calibration points and the quality of the data are such that even the studies strongly suggest divergences for the basal metazoan clades (sponges, cnidarians) in the Cryogenian. Although I will continue to use the results from reference [1] in the remainder of this contribution, I hardly view this study as the final word in molecular clock studies of early metazoan divergences. The acquisition of larger datasets and improved techniques should continue to refine our understanding of the tempo of early metazoan history. In particular, ctenophores were not included in these studies because of apparent long-branch attraction problems. New studies will be required estimate the pace of metazoan divergences with basal ctenophores.
Details of the Ediacaran–Cambrian explosion of animal life have been exhaustively reviewed elsewhere [1,18,27]. To summarize issues of direct relevance to the origin of nervous systems: likely early metazoan embryos have been described from the Doushantuo Formation in south China approximately 600 Ma [28], followed by the enigmatic macrofossils of the Ediacaran, ranging from 578 to 542 Ma. The fronds, discs and other forms of this assemblage probably represent a variety of clades but very few exhibit diagnostic metazoan characteristics or the presence of a gut, appendages, etc., with the exception of Kimberella mentioned previously. The Cambrian radiation begins about 542 Ma with a suite of small shelly fossils, most of which were probably of lophotrochozoan affinities. Most bilaterian groups appear somewhat later, although the pattern of first appearance of most clades appears to largely reflect preservational conditions. Several lines of evidence, including the absence of diagnostic bilaterian trace fossils through most of the Ediacaran, indicate that the Cambrian radiation accurately reflects the origin of these large, bilaterian bodyplans, if not the origin of the lineages themselves. I will return to this point in §4b.
In summary, integration of molecular clock and fossil data suggests that metazoans originated during the Cryogenian, perhaps 780 Ma, and diversified into many of the major clades through the Cryogenian and Ediacaran. The first widely accepted animals appeared in the fossil record by 555 Ma, although more controversial evidence extends back to about 600 Ma. Most clades of macroscopic bilaterians appear after 542 Ma. This implies a missing 150–200 Myr of early history of metazoans, a span encompassing most of the critical episodes in the evolution of nervous systems.
3. Environmental framework
Two extensive glacial episodes occurred during the Cryogenian period: the Sturtian glaciation (716–665 Ma) and the Marinoan glaciation (ca 650–635 Ma; figure 1) [29]. Geological evidence shows that these glaciations extended to sea level in near-equatorial latitudes. Overlying the glacial deposits are rocks described as ‘cap carbonates' with unusual sedimentary textures suggesting they were deposited during rapid deglaciation. Although the Sturtian and Marinoan glaciations are commonly described as though they involved persistent glaciation, we currently lack sufficient geologic and age control to exclude the possibility of shorter interglacial intervals within the Sturtian and Marinoan glacial episodes. One proposed explanation for these glacial episodes posits that essentially the entire Earth was enveloped in ice before build-up of carbon dioxide from volcanism triggered abrupt deglaciation [30,31]. Such a snowball Earth hypothesis has been very controversial, but it is important to emphasize that any explanation for these non-uniformitarian glacial deposits will involve circumstances that have not occurred in the past 550 Ma: widespread glaciations have not occurred at sea-level near the equator since the Marinoan. The deglaciation phase likely involved particularly extreme environmental conditions, including a rapid transition from glacial to greenhouse conditions, high alkalinity and high nutrient loading of the oceans as a consequence of the massive delivery to the oceans of the products of chemical weathering on the continents [32,33]. That multiple metazoan lineages persisted through these events is suggested by the molecular clock results reviewed in §2, but as yet we have only speculations as to how this was accomplished. The short-lived Gaskiers glaciation occurred about 580 Ma but may not have been as extensive as the Sturtian and Marinoan episodes. The Cryogenian and Ediacaran periods also included extensive geochemical changes, including some of the largest shifts in carbon isotopes of the past billion years, as well as changes in sulfur isotopes and trace element geochemistry [30,34].
Increased levels of oxygen have long been invoked as an explanation for the origin of animals (see review in [35]). Over the past several decades, geochemists have applied a variety of proxies to infer oxygen levels during the Cryogenian and Ediacaran, including carbon, sulfur and molybdenum isotopes as well as iron speciation. One challenge is that each technique preserves a record of the redox conditions in specific environments: for example, in deep-sea sediments, or the atmosphere. Consequently, some proxies may not be directly relevant for the primary environment of interest in the evolution of animals, which are oxygen levels in shallow seas. In the past few years, the introduction of new proxies such as nitrogen and chromium isotopes has provided important new data, although results from such studies are still being validated. As a consequence, results from older proxies such as carbon and sulfur isotopes are playing less of a role. Interpreting these studies also requires understanding the constraints oxygen levels may have played on early metazoan evolution. For decades, the consensus view was that oxygen levels of 1–10% of present atmospheric levels (PALs) were required for metazoan life and particularly for collagen biosynthesis. However, recent studies have radically revised this view, indicating that at least at small size, animals could have lived and reproduced at much lower oxygen levels [35–38]. Discussions about the drivers of this late Neoproterozoic oxygenation event have led many authors to conclude that the biological activity of organisms may have played an important role [18,36,37,39].
The results from geochemical proxies suggest an emerging model of redox condition through the late Neoproterozoic, with atmospheric oxygen levels less than 0.1% PAL O2 before approximately 800 Ma and partially oxygenated oceans from approximately 750 to 580 Ma with levels of approximately 1–3% PAL O2 [36,40,41]. During this time, the redox state of the oceans varied locally and regionally depending on the geological conditions, but overall the oceans were no more than 40% anoxic and 10% euxinic. Oxygen levels in shallow seas appear to have been highly variable after 600 Ma and probably did not stabilize until after approximately 560 Ma [32], and possibly not until the early Cambrian. The combination of glaciations and the low and fluctuating oxygen levels would have greatly influenced the course of early animal evolution. Recent studies suggest that low oxygen levels would not have precluded the origin of metazoans, as was once believed, but are quite likely to have limited their maximum body size [38,42,43]. The absence of evidence for macroscopic metazoans before 579 Ma, or bilaterians until after 555 Ma is consistent with a role for oxygen levels in controlling body size, rather the origin of clades.
Geologists have known for over a century of widespread exposure of the continents near the Ediacaran–Cambrian boundary. Known as the ‘great unconformity’ the prolonged erosion and denudation of the continents during this exposure may have generated a pulse of nutrients when the sea level rose during the early Cambrian, contributing to the pulse of biomineralization during the Cambrian explosion [44]. One of the striking aspects of the late Neoproterozoic was the extent of the environmental perturbations. Glaciations, shifts in ocean geochemistry and redox changes have occurred throughout the past several billion years of the Earth's history, but those of the late Neoproterozoic were unprecedented [45].
4. Implications for early evolution of nervous systems
Many models of the origin and early evolution of neurons and nervous systems have been proposed over the past several decades, often without reference to the temporal and environmental constraints imposed by integrating fossil, geological and molecular clock evidence. The temporal and environmental framework presented in §§2 & 3 places constraints on the likely morphologic and developmental attributes of animals during the Cryogenian and early Ediacaran, and thus on nervous systems. The absence of any fossil record of animals during the Cryogenian and early Ediacaran establishes that animals alive during this interval lacked a skeleton and were not easily fossilizable. The absence of trace fossils indicates that any mobile, benthic forms must have been less than 1 cm in maximum dimension (in some environments even smaller forms can be detected), and had a metabolic rate consistent with shallow marine oxygen levels of 1–3% PAL. In particular, three questions arise from the pattern of deep divergence of metazoan clades indicated by the molecular clock results: (i) What were the original roles of highly conserved genes in early metazoans? (ii) What changes in nervous system complexity were associated with the pronounced increase in body size of many bilaterian clades near the Cambrian boundary? (iii) Did nervous systems independently converge on more complex morphogenetic patterns? Here, I present a model for the early evolution of nervous systems that is consistent with this temporal and environmental framework.
(a). Phase 1
The origin of basal metazoan clades occurred during the Cryogenian and early Ediacaran, to about 600 Ma. There have been many reviews of the deep conservation of transcription factors and signalling pathways across metazoans, and we have done so recently [18,46,47]. A decade or more ago early comparative studies, often limited to bilaterians, inferred morphologically complex bilaterian LCAs with high conservation of developmental roles. On this basis, the bilaterian LCA was inferred to have eyes, segmentation, a gut, heart and considerable anterior/posterior and dorsal/ventral patterning. As comparative developmental studies were extended to cnidarians and sponges, the accompanying discovery that many critical components of developmental patterning were present early in metazoan history forced a revision to the early views of morphological conservation. Recent comparative studies suggest the following pattern of developmental and morphological novelties (based primarily on reference [46]), assuming that sponges are basal to metazoans: the sponge–cnidarian LCA (demosponge–cnidarian LCA if sponges are paraphyletic), the cnidarian–bilaterian LCA and the bilaterian LCA generally lacked the complicated morphogenetic developmental pathways observed in modern bilaterians. Rather, elements of these were co-opted from developmental regulatory circuits initially involved in less sophisticated patterning [1,18,47–50]. Transcription factors for cell-type specification were present, producing limited regional patterning of the body and a loosely controlled body plan. Feeding at this node was likely via proto-epithelial filter-feeding. Alternatively, for the ctenophore-early hypothesis a greater array of developmental tools could have been present at the base of Metazoa, with elements lost among sponges. In either case, body size of early metazoans must have been small with limited developmental patterning of the embryo and a lack of complex morphogenetic patterning. During this time, oxygen concentrations were low and unstable, probably placing primacy on opportunistic life strategies with easy dispersal. The basis of foodwebs for clades other than sponges is unclear. The cnidarian–bilaterian LCA (approx. 720 Ma) was able to form primary and orthogonal body axes and regional organization of structures in relation to body axes. This LCA possessed a mouth, gut and multifunctional cells. By approximately 650 Ma, the bilaterian LCA was more highly tuned for direct interaction with environment and exhibited both anterior/posterior and dorsal/ventral regional patterning and elaboration. Comparative studies strongly suggest that the bilaterian LCA possessed a suite of neurogenic capabilities [51], although the extent of a central nervous system remains a topic of discussion [52]. The capacity for feeding, movement and sensation were well developed.
(b). Phase 2
During the Ediacaran–Cambrian radiation (ECR) environmental conditions began to ameliorate, glacial activity ended, there was a decline in the magnitude of perturbations in carbon isotopes and an increase in oxygen levels in the deep and shallow oceans, especially after 555 Ma. Metazoan body sizes increased significantly, first with the appearance of the Ediacaran macrofauna after 579 Ma, and then with the appearance of larger-bodied bilaterians in the early Cambrian. Both these phases of the ECR were accompanied by increases in the complexity of developmental patterning. This increased morphogenetic patterning was often accomplished by co-option of developmental processes initially established for simpler processes. Ecological complexity increased rapidly, particularly near the Ediacaran–Cambrian boundary and positive feedback may have driven much of the ECR. Critically, the origin of many of the major metazoan clades is divorced from their acquisition of larger body size and of the distinctive bodyplans that characterize the modern clades. If this viewpoint is largely correct, then comparative studies of extant clades may provide only limited insights into the morphology and development of bilaterian clades prior to the late Ediacaran.
This scenario for the early history of animals is consistent with views in which multifunctional cells early in animal evolution gave rise to more specialized nerve cells, and the complexity of nervous systems increased slowly during the late Cryogenian and Ediacaran with the independent origins of nervous systems in cnidarians, ctenophores and bilaterians long after the origins of these clades [52–55]. Thus, sensory and nervous cells may have arisen early, but nervous systems were a later and independent novelty in different lineages, consistent with many earlier suggestions [13,54,56,57]. A reconstruction of the evolutionary history of ion channel genes established that ctenophores, cnidarians and bilaterians have experienced independent expansions of the major families of ion-channel proteins, as well as several rounds of gene loss, with no evidence for a rapid increase in complexity associated with the origin of nerves [53]. This study emphasized that the ion channel data are most consistent with the early origin of ‘very rudimentary’ nervous systems that persisted for a long time before the independent and convergent expansion of channel complexity. This long period of rudimentary nervous system development likely involved the deployment of genes now associated with the nervous system in proto-nervous system tissues, similar to Trichoplax today [13,53,58]. Such a model of early nervous system evolution is more consistent with the environmental data for the Cryogenian and early Ediacaran than is a model for an early, homologous nervous system shared across eumetazoans. Note that the model suggested here is independent of the controversy over the phylogenetic position of ctenophores relative to sponges and cnidarians.
Many evolutionary biologists instinctively link morphologic novelties to ecological opportunism and evolutionary success. This is hardly surprising as Simpson [59] and Mayr [60] explicitly made such claims. The discovery of deep conservation of developmentally significant gene and gene networks, as described in §2, led to similar expectations that novelties in developmental patterning would find quick phenotypic expression in similar ways, as these elements are employed in living bilaterians. As described in §2, the recognition that many of these signalling pathways and transcription factors were present in basal metazoans has led to a rejection of a straightforward interpretation of the ancestral role of these developmental components. Thus, we must distinguish between the origin of highly conserved genes, the formation of the developmental GRNs that control morphogenetic patterning, which has, in some cases, included co-option of genes previously employed in different contexts, and the origin of individuated phenotypic characters, or morphological novelties. Of particular relevance here is the need to distinguish between the origin of evolutionary novelty, defined as the formation of individuated, homologous characters [61], and innovation, the ecological or evolutionary success of such innovations. Morphological novelties do not necessarily lead to immediate evolutionary success if that is measured by an increase in taxic diversity. Macroevolutionary lags between the origin of the morphologic novelty and innovation have been well documented in the fossil record [18,62,63]. Similarly, inferring patterns of morphologic innovation from comparative developmental studies is hampered by the fact that the acquisition of a particular genetic novelty is not necessarily immediately expressed as a developmental innovation.
Acknowledgements
I appreciate useful discussions of these topics with E. Davidson, S. Tweedt, J. W. Valentine and members of the MIT NASA Astrobiology group, and reviews from an anonymous reviewer and Greg Edgecombe.
Competing interests
I delcare I have no competing interests.
Funding
This research was supported by a NASA Astrobiology Institute grant to D.H.E. through the MIT node.
References
- 1.Erwin DH, Laflamme M, Tweedt SM, Sperling EA, Pisani D, Peterson KJ. 2011. The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science 334, 1091–1097. ( 10.1126/science.1206375) [DOI] [PubMed] [Google Scholar]
- 2.Worheide G, Dohrmann M, Erpenbeck D, Larroux C, Maldonado M, Voigt O, Borchiellini C, Lavrov DV. 2012. Deep phylogeny and evolution of sponges (phylum Porifera). Adv. Mar. Biol. 61, 1–78. ( 10.1016/B978-0-12-387787-1.00007-6) [DOI] [PubMed] [Google Scholar]
- 3.Sperling EA, Peterson KJ, Pisani D. 2009. Phylogenetic-signal dissection of nuclear housekeeping genes supports the paraphyly of sponges and the monophyly of Eumetazoa. Mol. Biol. Evol. 26, 2261–2274. ( 10.1093/molbev/msp148) [DOI] [PubMed] [Google Scholar]
- 4.Ryan JF, et al. 2013. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342, 6164 ( 10.1126/science.1242592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moroz LL, et al. 2014. The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109–114. ( 10.1038/nature13400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Philippe H, Brinkmann H, Copley RR, Moroz LL, Nankano H, Poustka AJ, Wallberg A, Peterson KJ, Telford MJ. 2011. Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature 470, 255–258. ( 10.1038/nature09676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn CW, Giribet G, Edgecombe GD, Hejnol A. 2014. Animal phylogeny and its evolutionary implications. Annu. Rev. Ecol. Evol. Syst. 45, 371–395. ( 10.1146/annurev-ecolsys-120213-091627) [DOI] [Google Scholar]
- 8.Riesgo A, Farrar N, Windsor PJ, Giribet G, Leys SP. 2014. The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol. Biol. Evol. 31, 1102–1120. ( 10.1093/molbev/msu057) [DOI] [PubMed] [Google Scholar]
- 9.Nosenko T, et al. 2013. Deep metazoan phylogeny: when genes tell different stories. Mol. Phylogenet. Evol. 67, 223–233. ( 10.1016/j.ympev.2013.01.010) [DOI] [PubMed] [Google Scholar]
- 10.Philippe H, Brinkmann H, Lavrov DV, Littlewood DTJ, Manuel M, Worheide G, Baurain D. 2011. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 9, e1000602 ( 10.1371/journal.pbio.1000602) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borowiec ML, Lee EK, Chiu JC, Plachetzki DC. In press. Dissecting phylogenetic signal and accounting for bias in whole-genome data sets: a case study of the Metzoa. Mol. Biol. Evol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marlow H, Arendt D. 2014. Evolution: ctenophore genomes and the origin of neurons. Curr. Biol. 24, R757–R761. ( 10.1016/j.cub.2014.06.057) [DOI] [PubMed] [Google Scholar]
- 13.Jekely G, Paps J, Nielsen C. 2015. The phylogenetic position of ctenophores and the origins(s) of nervous systems. Evo. Devo 6, 1 ( 10.1186/2041-9139-6-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whelan NV, Kocot KM, Halanych KM. In press. Employing phylogenomics to resolve the relationships among cnidarians, ctenophores, sponges, placozoans, and bilaterians. Int. Comp. Biol. ( 10.1093/icb/icv037) [DOI] [PubMed] [Google Scholar]
- 15.Whelen NV, Kocot KM, Moroz LI, Halanych KM. 2015. Error, signal, and the placement of Ctenophora sister to all other animals. Proc. Natl Acad. Sci. USA 112, 5773–5778. ( 10.1073/pnas.1503453112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feuda R, Rota-Stabelli O, Oakley TH, Pisani D. 2014. The comb jelly opsins and the origins of animal phototransduction. Genome Biol. Evol. 6, 1964–1971. ( 10.1093/gbe/evu154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan JF, Chiodin M. 2015. Where is my mind? How sponges and placozoans may have lost neural cell types. Phil. Trans. R. Soc. B 370, 20150059 ( 10.1098/rstb.2015.0059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erwin DH, Valentine JW. 2013. The Cambrian explosion: the construction of animal biodiversity. Greenwood, CO: Roberts & Co. [Google Scholar]
- 19.Fautin DG. 2009. Structural diversity, systematics and evolution of Cnidae. Toxicon 54, 1054–1064. ( 10.1016/j.toxicon.2009.02.024) [DOI] [PubMed] [Google Scholar]
- 20.Fedonkin MA, Simonetta A, Ivantsov AY. 2007. New data on Kimberella, the Vendian mollusc-like organism (White Sea region, Russia): paleontological and evolutionary implications. In The rise and fall of the Ediacaran biota (eds Vickers-Rich P, Komarower P), pp. 157–179. London, UK: Geological Society. [Google Scholar]
- 21.Vinther J. 2015. The origins of molluscs. Palaeontology 58, 19–34. ( 10.1111/pala.12140) [DOI] [Google Scholar]
- 22.Wheat CW, Wahlberg N. 2013. Phylogenomic insights into the Cambrian explosion, the colonization of land and the evolution of flight in Arthropoda. Syst Biol. 62, 93–109. (doi:0.1093/sysbio/sys074) [DOI] [PubMed] [Google Scholar]
- 23.Rehm P, Borner J, Meusemann K, von Reumont BM, Simon S, Hadrys H, Misof B, Burmester T. 2011. Dating the arthropod tree based on large-scale transcriptome data. Mol. Phylogenet. Evol. 61, 880–887. ( 10.1016/j.ympev.2011.09.003) [DOI] [PubMed] [Google Scholar]
- 24.Lee MS, Soubrier J, Edgecombe GD. 2013. Rates of phenotypic and genomic evolution during the Cambrian explosion. Curr. Biol. 23, 1889–1895. ( 10.1016/j.cub.2013.07.055) [DOI] [PubMed] [Google Scholar]
- 25.Budd GE. 2015. Early animal evolution and the origins of nervous systems. Phil. Trans. R. Soc. B 370, 20150037 ( 10.1098/rstb.2015.0037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valentine JW, Jablonski D, Erwin DH. 1999. Fossils, molecules and embryos: new perspectives on the Cambrian explosion. Development 126, 851–859. [DOI] [PubMed] [Google Scholar]
- 27.Budd GE. 2008. The earliest fossil record of the animals and its significance. Phil. Trans. R. Soc. B 363, 1425–1434. ( 10.1098/rstb.2007.2232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao SH, Muscente AD, Chen L, Zhou CM, Schiffbauer JD, Wood AD, Polys NF, Yuan XL. 2014. The Weng'an biota and the Ediacaran radiation of multicellular eukaryotes. Nat. Sci. Rev. 1, 498–520. ( 10.1093/nsr/nwu061) [DOI] [Google Scholar]
- 29.Rooney AD, Macdonald FA, Strauss JV, Dudas FO, Hallmann C, Selby D. 2014. Re-Os geochronology and coupled Os-Sr isotope constraints on the Sturtian snowball Earth. Proc. Natl Acad. Sci. USA 111, 51–56. ( 10.1073/pnas.1317266110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halverson GP, Shields-Zhou G. 2011. Chemostratigraphy and the Neoproterozoic glaciations. In Memoir 36: The geological record of Neoproterozoic glaciations (eds Arnaud E, Halverson GP, Shields-Zhou G), pp. 51–66. London, UK: Geological Society. [Google Scholar]
- 31.Hoffman PF, Kaufman AJ, Halverson GP, Schrag DP. 1998. A Neoproterozoic snowball Earth. Science 281, 1342–1346. ( 10.1126/science.281.5381.1342) [DOI] [PubMed] [Google Scholar]
- 32.Higgins JA, Schrag DP. 2003. Aftermath of a snowball Earth. Geochem. Geophys. Geosyst. 4, 1028 ( 10.1029/2002GC000403) [DOI] [Google Scholar]
- 33.Planavsky NJ, Rouxel OJ, Bekker A, Lalonde SV, Konhauser KO, Reinhard CT, Lyons TW. 2010. The evolution of the marine phosphate reservoir. Nature 467, 1088–1090. ( 10.1038/nature09485) [DOI] [PubMed] [Google Scholar]
- 34.Johnston DT, Poulton SW, Dehler C, Porter S, Husson J, Canfield DE, Knoll AH. 2010. An emerging picture of Neoproterozoic ocean chemistry: insights from the Chuar Group, Grand Canyon, USA. Earth Planet. Sci. Lett. 290, 64–73. ( 10.1016/J.Epsl.2009.11.059) [DOI] [Google Scholar]
- 35.Mills DB, Ward LM, Jones C, Sweeten B, Forth M, Treusch AH, Canfield DE. 2014. Oxygen requirements of the earliest animals. Proc. Natl Acad. Sci. USA 111, 4168–4172. ( 10.1073/pnas.1400547111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lenton TM, Boyle RA, Poulton SW, Shields-Zhou G, Butterfield NJ. 2014. Co-evolution of eukaryotes and ocean oxygenation in the Neoproterozoic era. Nat. Geosci. 7, 257–265. ( 10.1038/ngeo2108) [DOI] [Google Scholar]
- 37.Butterfield NJ. 2009. Oxygen, animals and oceanic ventilation: an alternative view. Geobiology 7, 1–7. ( 10.1111/j.1472-4669.2009.00188.x) [DOI] [PubMed] [Google Scholar]
- 38.Sperling EA, Frieder CA, Raman AV, Girguis PR, Levin LA, Knoll AH. 2013. Oxygen, ecology and the Cambrian radiation of animals. Proc. Natl Acad. Sci. USA 110, 13 446–13 451. ( 10.1073/pnas.1312778110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mills DB, Canfield DE. 2014. Oxygen and animal evolution: did a rise of atmospheric oxygen ‘trigger’ the origin of animals? Bioessays 36, 1145–1155. ( 10.1002/bies.201400101) [DOI] [PubMed] [Google Scholar]
- 40.Ader M, Sansjofre P, Halverson GP, Busigny V, Trindade RIF, Kunzmann M, Nogueira AC. 2014. Ocean redox structure across the late Neoproterozoic oxygenation event: a nitrogen isotope perspective. Earth Planet. Sci. Lett. 396, 1–13. ( 10.1016/j.epsl.2014.03.042) [DOI] [Google Scholar]
- 41.Johnston DT, Poulton SW, Goldberg T, Sergeev VN, Podkovyrov V, Vorob'eva NG, Bekker A, Knoll AH. 2012. Late Ediacaran redox stability and metazoan evolution. Earth Planet. Sci. Lett. 335, 25–35. ( 10.1016/J.Epsl.2012.05.010) [DOI] [Google Scholar]
- 42.Knoll AH, Sperling EA. 2014. Oxygen and animals in Earth history. Proc. Natl Acad. Sci. USA 111, 3907–3908. ( 10.1073/pnas.1401745111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sperling EA, Halverson GP, Knoll AH, Macdonald FA, Johnston DT. 2013. A basin redox transect at the dawn of animal life. Earth Planet. Sci. Lett. 371, 143–155. ( 10.1016/J.Epsl.2013.04.003) [DOI] [Google Scholar]
- 44.Peters SE, Gaines RR. 2012. Formation of the 'Great Unconformity' as a trigger for the Cambrian explosion. Nature 484, 363–366. ( 10.1038/Nature10969) [DOI] [PubMed] [Google Scholar]
- 45.Erwin DH. 2015. Was the Ediacaran-Cambrian radiation a unique evolutionary event? Paleobiology 41, 1–15. ( 10.1017/pab.2014.2) [DOI] [Google Scholar]
- 46.Tweedt SM, Erwin DH. 2015. Origin of metazoan developmental toolkits and their expression in the fossil record. In Evolution of multicellularity (eds Ruiz-Trillo I, Nedelcu AM), pp. 47–78. Dordrecht, The Netherlands: Springer. [Google Scholar]
- 47.Davidson EH, Erwin DH. 2010. An integrated view of precambrian eumetazoan volution. Cold Spring Harb. Symp. Quant. Biol. 79, 65–80. ( 10.1101/sqb.2009.74.042) [DOI] [PubMed] [Google Scholar]
- 48.Davidson EH, Erwin DH. 2010. Evolutionary innovation and stability in animal gene networks. J. Exp. Zool. (Mol. Dev Evol.) 314B, 182–186. [DOI] [PubMed] [Google Scholar]
- 49.Erwin DH, Davidson EH. 2009. The evolution of hierarchical gene regulatory networks. Nat. Rev. Genet. 10, 141–148. ( 10.1038/nrg2499) [DOI] [PubMed] [Google Scholar]
- 50.Davidson EH, Erwin DH. 2006. Gene regulatory networks and the evolution of animal body plans. Science 311, 796–800. ( 10.1126/science.1113832) [DOI] [PubMed] [Google Scholar]
- 51.Hartenstein V, Stollewerk A. 2015. The evolution of early neurogenesis. Dev. Cell 32, 390–407. ( 10.1016/j.devcel.2015.02.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pani AM, Mullarkey EE, Aronowica J, Assimacopoulos S, Frove EA, Lowe CJ. 2012. Ancient deuterostome origins of vertebrate brain signaling centres. Nature 483, 289 ( 10.1038/Nature10838) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liebeskind BJ, Hillis DM, Zakon HH. 2015. Convergence of ion channel genome content in early animal evolution. Proc. Natl Acad. Sci. USA 112, E846–E851. ( 10.1073/pnas.1501195112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moroz LL. 2009. On the independent origins of complex brains and neurons. Brain Behav. Evol. 74, 177–190. ( 10.1159/000258665) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wenger Y, Galliot B. 2013. Punctuated emergences of genetic and phenotypic innovations in eumetazoan, bilaterian, euteleostome, and hominidae ancestors. Genome Biol. Evol. 5, 1949–1968. ( 10.1093/gbe/evt142) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keijzer F. 2015. Moving and sensing without input and output: early nervous systems and the origins of the animal sensorimotor organization. Biol. Philos. 30, 311–331. ( 10.1007/s10539-015-9483-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Northcutt RG. 2012. Evolution of centralized nervous systems: two schools of evolutionary thought. Proc. Natl Acad. Sci. USA 109(Suppl 1), 10 626–10 633. ( 10.1073/pnas.1201889109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith CL, Varoqueaux F, Kittelmann M, Azzam RN, Cooper B, Winters CA, Eitel M, Fasshauer D, Reese TS. 2014. Novel cell types, neurosecretory cells, and body plan of the early-diverging metazoan Trichoplax adhaerens. Curr. Biol. 24, 1565–1572. ( 10.1016/j.cub.2014.05.046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simpson GG. 1953. The major features of evolution. New York, NY: Columbia University Press. [Google Scholar]
- 60.Mayr E. 1960. The emergence of novelty. In The evolution of life (ed. Tax S.), pp. 349–380. Chicago, IL: University of Chicago Press. [Google Scholar]
- 61.Wagner GP. 2014. Homology, genes, and evolutionary innovation. Princeton, NJ: Princeton University Press. [Google Scholar]
- 62.Jablonski D, Bottjer DJ. 1990. The origin and diversification of major groups: environmental patterns and macroevolutionary lags. In Major evolutionary radiations (eds Taylor PD, Larwood GP), pp. 17–57. Oxford, UK: Clarendon Press. [Google Scholar]
- 63.Erwin DH. 2015. Novelty and innovation in the history of life. Curr. Biol. V. 25, R930–R940. [DOI] [PubMed] [Google Scholar]