Abstract
Understanding the mechanisms of evolution of brain pathways for complex behaviours is still in its infancy. Making further advances requires a deeper understanding of brain homologies, novelties and analogies. It also requires an understanding of how adaptive genetic modifications lead to restructuring of the brain. Recent advances in genomic and molecular biology techniques applied to brain research have provided exciting insights into how complex behaviours are shaped by selection of novel brain pathways and functions of the nervous system. Here, we review and further develop some insights to a new hypothesis on one mechanism that may contribute to nervous system evolution, in particular by brain pathway duplication. Like gene duplication, we propose that whole brain pathways can duplicate and the duplicated pathway diverge to take on new functions. We suggest that one mechanism of brain pathway duplication could be through gene duplication, although other mechanisms are possible. We focus on brain pathways for vocal learning and spoken language in song-learning birds and humans as example systems. This view presents a new framework for future research in our understanding of brain evolution and novel behavioural traits.
Keywords: brain pathway, duplication, parrots, song systems, brain evolution, speech
1. Introduction
The evolution of brain pathways for generation of complex behavioural traits remains an enigmatic and fundamental question in biology. Therefore, examining the proximate and ultimate mechanisms driving changes in brain structure and function provides an exciting opportunity to understand the evolution of complex behavioural traits. In this regard, various hypotheses have been proposed to explain evolution of complex behavioural traits, including increases in brain or brain region size relative to body size, increases in total number of neurons or neuron density, and presence versus absence of particular neural networks that control a specific type of behaviour [1–5]. Some such changes may have occurred with the emergence of the telencephalon during the invertebrate to vertebrate transition, indicating that the central nervous system has been an important target of selection [4,6–8]. However, current empirical evidence for such models and theories are few and wanting.
Another fundamental problem in explaining the evolution of complex behaviours and brain pathways is understanding the contributing cellular and molecular mechanisms. One overall hypothesis is that significant changes in the brain can be generated by novel gene functions owing to gene duplications or expansion of gene regulatory networks [7,9–12]. One of the duplicated genes may then acquire a mutation in coding or regulatory sequences, which enables it to acquire a new function that then undergoes selection, a process known as neofunctionalization [12–15]. Other hypotheses posit that existing genes are modified, including changes in coding sequence, cis-regulatory motifs [16,17] or new alternative mRNA splice variants [18–21], a process known as subfunctionalization [12–14]. However, the origin and evolution of such molecular changes in the evolution of the nervous system and behavioural complexity are not well understood.
Here, we review and expand upon an underappreciated theory of evolution of brain complexity, namely by brain region or whole brain pathway duplication from pre-existing brain circuits. We propose hypotheses on cellular and molecular mechanisms for brain region and pathway duplication, including by gene duplication. We believe that such mechanisms may form a cornerstone of evolution of brain and behaviour complexity, which enable adaptations to new environments and social situations.
2. Theories on brain region and brain pathway evolution for brain complexity
Comparative neurobiology studies indicate that many primitive features of brain organization have been preserved to varying degrees in extant species [22]. The brain also has evolved in a mosaic pattern, with some regions changing dramatically, while others have remained little changed through the course of evolution [23]. While it is still unclear how brains evolve, past theories posit that brain evolution could be understood by examining how brains develop embryonically and how such development can be modified [22]. It is thought that the early embryonic state of the brain across species represents a more similar and thus ancestral state, and that during development, brain cells, regions and pathways diverge towards lineage- or species-specific states. This is one way in which homologous brain regions can become diverse in adults across species. Based on this view, the vertebrate brain is proposed to consist of three basic divisions, with the spinal cord and brainstem (hindbrain, midbrain and thalamus) having more conserved organization, and the telencephalon more divergent organization [24]. In turn, the telencephalon consists of three major subdivisions, with the pallidum and striatum having more conserved organization and the pallium or cortex a more divergent organization. The pallium is largely layered in mammals, and mostly nuclear in birds, reptiles and other vertebrates, but with divergences among them [24,25].
With these fundamental principles, one can argue that divergences may occur in many forms leading to more behavioural complexity, including: (i) larger brain-to-body size ratios endowing those animals with more advanced abilities [3]; (ii) novel connectivity within a pre-existing brain circuit that enhances that particular circuit's function for complex behaviours [26,27]; and (iii) the de novo presence of a specific brain region or circuit that controls a newly evolved behaviour, as has been proposed for the evolution of brain pathways for vocal learning and spoken language [2,28,29]. It is this latter theory that requires greater explanation.
A long proposed explanation for generating increased cortical complexity is that a single region gradually differentiates into two or more areas [30–35]. This could occur by expansion of an existing region and then selectively partitioning part of the older region to the new function, while the other part maintains the old function [36]. Allman and Kaas also proposed that development of the brain could be altered owing to a gene mutation so that a given area is duplicated [33,37]. The duplication event would modify the function of either one or both of the ancestral and duplicated areas to take on a new function. Duplication itself may modify the selection pressure on both structures, thereby allowing the individual structures to use the once single functional space in a mechanism reminiscent of adaptive radiation [38]. More recently, Feenders et al. [39] suggested that whole brain pathways could duplicate, followed by divergence of one of the duplicated copies. This idea was proposed as a mechanism to explain what they called the Motor Theory of Vocal Learning Origin, which we review next.
(a). The motor theory of vocal learning origin and brain pathway duplication
Vocal learning, a critical component of spoken language acquisition, is the ability to modify acoustic and/or syntactic features of sounds produced, including vocal imitation and improvization. Vocal learning is a rare trait, so far discovered in five distantly related groups of mammals (humans, bats, elephants, cetaceans (dolphins and whales) and pinnipeds (seals and sea lions)) and three distantly related groups of birds (parrots, songbirds and hummingbirds) [1,40–42]. In the past few decades, significant advances have been made in guiding our understanding of the evolution and mechanisms of brain pathways for vocal learning in birds and humans [2,40,42–49] (figure 1).
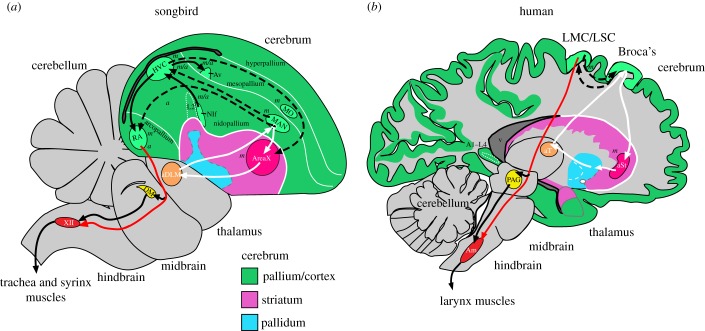
Figure 1.
Brain pathways controlling song in songbirds and spoken language in humans. (a) Vocal learning song pathway of songbirds. (b) Spoken language pathway of humans. Black arrows, posterior vocal motor pathway; white arrows, anterior vocal learning pathway; dashed arrows, connections between the two pathways; red arrows, specialized direct projection from forebrain to brainstem vocal MN in vocal learners. Italicized letters indicate that these regions mainly show motor (m), auditory (a), equally both motor and auditory (m/a) neural activity or activity-dependent gene expression in awake animals. Adapted from [2,50]. Not all connections are shown, for simplicity. Some connections in the human brain are proposed based on known connectivity of adjacent brain regions in non-human primates. A1–L4, primary auditory cortex—layer 4; Am, nucleus ambiguous; aSt, anterior striatum; Av, avalanche; aDLM, anterior dorsolateral nucleus of the thalamus; DM, dorsal medial nucleus of the midbrain; HVC, a vocal nucleus (no abbreviation); L2, field L2; LMC, laryngeal motor cortex; LSC, laryngeal somatosensory cortex; NIf, interfacial nucleus of the nidopallium; MAN, magnocellular nucleus of the anterior nidopallium; MN, motor neurons; MO, oval nucleus of the anterior mesopallium; PAG, peri-aqueductal gray; RA, robust nucleus of the arcopallium; v, ventricle space.
The independently evolved lineages of vocal learning birds and humans share distinct forebrain pathways that control the acquisition and production of learned vocalizations [2]. Within these pathways, all three avian lineages contain seven cerebral (telencephalic) vocal nuclei and several thalamic nuclei. These nuclei, best characterized in songbirds and parrots, are distributed between two subpathways (figure 1a): (i) the vocal production, or posterior, pathway that influences the production of learned song—which includes an arcopallium nucleus (songbird RA (robust nucleus of the arcopallium), parrot AAC (central nucleus of the anterior arcopallium), hummingbird VA (vocal nucleus of the arcopallium)), analogous to the laryngeal motor cortex (LMC) in humans (figure 1b) that makes a specialized direct projection to brainstem vocal motor neurons (MN), which in turn controls the vocal organs, the syrinx (birds) and larynx (humans); and (ii) the vocal learning, or anterior, pathway that is primarily responsible for vocal imitation and plasticity, which forms a pallial–basal ganglia–thalamic loop, analogous to such loops in the mammalian brain that presumably include Broca's speech area in humans. The song and speech regions in both these pathways are embedded in or adjacent to non-vocal motor brain areas [39,50]. These non-vocal motor regions are present in other vertebrate species examined thus far, and are thought to be involved in the production and learning of non-vocal motor behaviours. Based on these findings, Feenders et al. [39] proposed a motor theory of vocal learning origin, which stated that ‘cerebral brain pathways for vocal learning in distantly related animals evolved independently as specializations of a pre-existing motor system inherited from their common ancestor’ ([39], p.1). This was a more general theory of the motor theory of language origin [51], but with specific brain regions identified and a proposed mechanism. The motor theory of vocal learning origin suggested that the last common ancestor of birds and mammals had a motor forebrain pathway, including a motor cortex or pallium region. This is because although the motor pallial domain in mammals consists of six layers of cells (layered) and nuclear subdivisions in birds and reptiles (clustered), they function similarly and developed from the same embryonic primordium. This is supported by results obtained from activity-dependent gene expression and differential gene expression experiments, which show that the avian pallium has a functional columnar organization similar to the mammalian pallial domain [39,52–54]. Further, the mammalian non-vocal motor descending pathway and the pre-motor pathway share similar connectivity patterns in avian posterior and anterior motor pathways, respectively, suggesting the presence of a pre-existing motor system shared by both groups and their most recent ancestor [1,39,55,56].
The proposed mechanism of evolution of vocal learning pathways was by brain pathway duplication [39]. In this regard, it was hypothesized that parallel forebrain motor learning pathways with auditory, somatosensory or other sensory input, normally replicate multiple times during embryonic development and connect to different brainstem and spinal cord neurons to control different muscle groups. In vocal learners, this forebrain pathway is hypothesized to replicate one more time and then connect to the brainstem circuits that control vocalizations and respiration. Then the new vocal learning pathway would diverge to form novel connections and functions relative to the adjacent non-vocal motor pathways. Under this duplication hypothesis, the vocal learning pathways share a deep homology with the surrounding motor pathways, but convergence in the independent lineages of vocal learners.
Several alternative hypotheses have been proposed for evolution of vocal learning pathways, including that the pathways in humans and song-learning birds originated out of either a pre-existing auditory pathway [57,58], a non-motor cognitive region [59,60], a combined auditory–motor pathway [61], or completely de novo [62]. In support of an auditory origin hypothesis, the songbird posterior vocal motor pathway is also partly adjacent to the auditory pathway and has some parallel connections with the descending auditory system [58]. However, such an anatomical position is not present in hummingbirds, parrots, or humans [1,2]. With the exception of the completely de novo hypothesis, even if the vocal learning pathway arose from a non-motor pathway, the hypothesis of pathway duplication could still apply.
If the duplication hypothesis were true, then one would expect to find most genes expressed in vocal learning pathways to be similar to the pathway from which they were duplicated. Further, one would expect to find divergent molecular changes in neural connectivity genes associated with the unique connections found in vocal learning pathways. These ideas were recently tested in a high-throughput gene expression study using a novel computational approach that determines homologous and convergent specialized anatomical gene expression profiles from thousands of samples and genes from multiple species [50]. Using comparative microarray gene expression profiling of approximately 3000–7000 genes in vocal learning and vocal non-learning avian and primate species, Pfenning et al. [50] found that the song and speech brain pathway regions of vocal learning birds and humans have gene expression profiles that more closely match motor and premotor cortex and striatal pathway regions adjacent to them than they do to auditory, somatosensory or other brain regions (figure 1). These results corroborated some earlier single gene expression, developmental, functional, and connectivity studies [24,52,63–67]. Combined, the findings support the idea that the similarities are owing to homology and not convergence. Further, Pfenning et al. [50] found divergent changes in expression of genes that control neural connectivity in the avian song and the human speech regions from the surrounding motor areas, but that were convergent among the vocal learning birds and humans. There were also convergent changes in some genes involved in neural development, neuroprotection and synaptic communication functions. The brain expression characterization of these genes [50,68] led to the discovery of a further apparent duplication in the parrot brain [56], as described next.
(b). Parrots contain a song system within a song system
Parrots surpass other vocal learning avian species in their ability to imitate human speech and also rival non-human primates in their display of advanced cognitive skills and ability for tool use [2,69–74]. From 1981 until recently in 2015 (approx. 34 years), the neural pathways for vocal learning had been studied in only one parrot species, the budgerigar (Melopsittacus undulatus) [55,75–79]. From these studies, it was apparent that the budgerigar song system shows some differences from the songbird and hummingbird song systems [55,75,76,79,80]. The posterior song system of songbirds and parrots (and presumably hummingbirds) receives auditory input from the posterior auditory pathway (e.g. auditory Field L), but in parrots it receives additional auditory input from a small part of the nucleus basorostralis (B), the remainder of which is somatosensory [81]. Neural tracing and singing-regulated immediate early gene studies revealed some differences in connectivity and position or shape of song nuclei in parrots, but no clear differences were noted in the presence or absence of song system structures [55,75,76,78–80].
Recently, partly based on the high-throughput gene expression [50,68] and other findings, we led a study [56] characterizing gene expression profiles that are specialized in avian and/or human song/speech vocal learning circuits (e.g. PVALB, SLIT1, FOXP1, NR2A, GLUR1, NADPH-d, AR, mENK, TH, CGRP-LI) to understand the organization of the song system in diverse groups of parrots representing all the three superfamilies, Strigopoidea, Cacatuoidea and Psittacoidea [82]. We found that the parrot pallial (cortical) song nuclei had core regions that differed in gene expression from surrounding shell regions, and both in turn differed from the surrounding cortical motor areas (figure 2) [56]. Surprisingly, a subset of the genes (including PVALB) had moderate specialized expression in the immediate surrounding non-vocal motor areas. Both the core and shell song regions were functionally active in the production of learned vocalizations, as revealed by vocalizing-driven immediate early gene expression (EGR-1, C-FOS and DUSP-1), whereas the surrounding brain regions were active in production of non-singing motor behaviour, as revealed by rhythmic controlled hopping-driven gene expression [39,56,79,85,86].
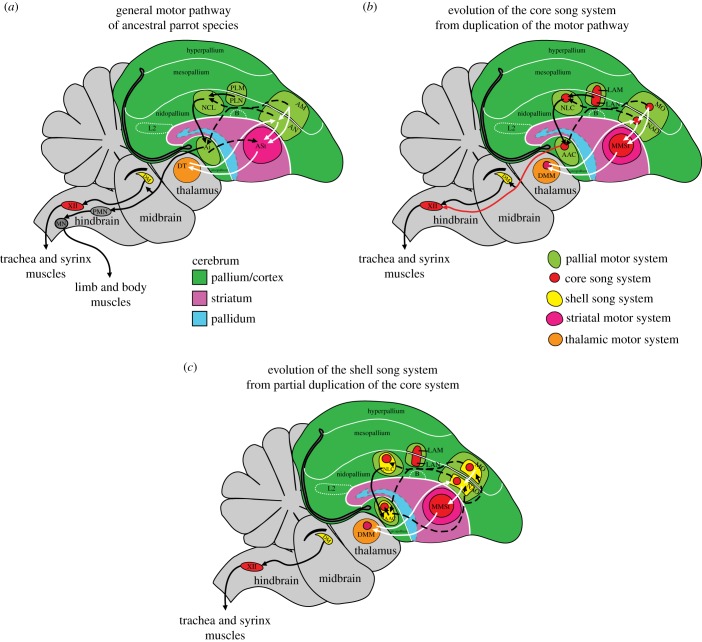
Figure 2.
Hypothesis of evolution of song system in parrots owing to sequential pathway duplications. (a) The parrot ancestral motor pathway (light green) with the posterior motor connections (in black arrows) and the anterior motor connections (in white arrows). (b) The parrot core song system (red), proposed to have evolved out of the pre-existing motor pathway through duplication. (c) The parrot shell song system (yellow), proposed to have evolved out of a partial duplication of the core song system. Black arrows, posterior vocal motor pathway; white arrows, anterior vocal motor pathway; dashed arrows, connections between the two pathways; red arrow, specialized direct projection from forebrain to brainstem vocal MN in vocal learners. Connectivity based on summaries in [39,55,56,76,83,84]. See fig. 20 from Chakraborty et al. [56] for additional connections of the core and shell song pathways. Not all connections are shown for simplicity, including reciprocal connections and additional thalamic projections. AAC, central nucleus of the anterior arcopallium; Ai, intermediate arcopallium; AM, anterior mesopallium; AN, anterior nidopallium; ASt, anterior striatum; B, basorostralis; DM, dorsal medial nucleus of the midbrain; DMM, magnocellular nucleus of the dorsomedial thalamus; DT, dorsal thalamus; L2, L4, auditory areas; NAO, oval nucleus of the anterior nidopallium; NCL, nidopallium caudal lateral; NLC, central nucleus of the lateral nidopallium; PMN, premotor neurons; LAN, lateral nucleus of the anterior nidopallium; LAM, lateral nucleus of the anterior mesopallium; MMSt, magnocellular nucleus of the medial striatum; MO, oval nucleus of the anterior mesopallium; PLM, posterior lateral mesopallium; PLN, posterior lateral nidopallium; XII, 12th motor nucleus.
The connectivity of the core and shell systems were similar to each other, but with some significant differences. One fundamental difference was that the core system made the rare direct projection to brainstem vocal MN (via the AAC core nucleus), whereas the parallel shell song system made mostly intra-cortical projections (via AAC shell; figure 2b,c). The direct projection to the brainstem vocal MN is considered critical for the evolution of vocal learning and spoken language, as it is either absent or very sparse in vocal non-learners [2,42,87–92]. There were sparse connections between the parrot cores and shells within and among each song nucleus.
The presence of these song nuclei in the kea, the most distantly related to the other extant parrot species [82,93], indicates that parrots evolved the core and shell song systems over 29 Ma before the kea split from the other parrot lineages. The kea shell system, however, was less well differentiated in terms of its gene expression specializations. There were also large species differences in relative sizes of the core and shell regions, where the shell had a significant log-linear relationship with their brain section size, but the core did not. This meant that shell regions were relatively larger in species with bigger brains such as the gold and blue macaw, and the African Grey and Amazon parrots that are considered to have more advanced communication and cognitive capacities.
The fact that the shell system AAC nucleus does not have the direct projection to the vocal MN (which is restricted to the core region of AAC), but may be correlated with more complex vocal learning behaviour, indicates that such direct projections may not be the only means to increase learned motor behavioural complexity over innate motor behaviours. We speculate that it is possible that the direct projection may not be required for the ability to imitate complex vocalizations, but strictly for the production of those learned vocalizations. Clearly, further studies will be required to explicitly test this hypothesis in parrots.
The gene expression specializations and neural connectivity of the parrot core song nuclei were most similar to the song nuclei of songbirds and hummingbirds, whereas the shells were unique to parrots. The shell specializations appear to be restricted to the cortical parrot song nuclei, as no shells have yet been found for the striatal or thalamic song nuclei. The songbird and hummingbird species examined to date do not have parallel vocal motor shell regions for any of their nuclei.
These findings indicate that the core and shell are two parallel, partially independent systems, performing similar and some unique functions for vocalizations. They support a partial brain pathway duplication hypothesis of brain evolution. In particular, we suggest that the core song system evolved convergently in parrots, songbirds and hummingbirds as a duplication event in each lineage from the surrounding motor areas (figure 2a,b). Thereafter, the parrot core cortical song nuclei underwent a further partial duplication event to evolve the shell song system (figure 2c). The shell song system went on to evolve specializations that allow more complex vocal communication abilities in parrots compared to other avian vocal learners. This dual system evolved early in the parrot lineage, and has lasted and expanded for millions of years in different species. In addition, changes in the regulation of some genes that may allow greater vocal–motor–auditory integration in vocal learning systems could have influenced changes in the surrounding motor areas to allow greater auditory–motor entrainment and synchronizing of body movements to the rhythm of music for dance in parrots [94–96].
It would be exciting to determine if similar duplications of brain pathways have occurred in humans, not only in the speech pathways but also for other advanced motor movements such as dancing [97–99]. In the human brain, areas 44 and 45 constitute Broca's area, the ventrolatreal frontal region critical for spoken language acquisition and production [100]. Petrides et al. [101,102] showed that there is a comparable area 44-like region involved in orofacial musculature functions in macaque monkeys. What is still unknown is whether area 44 in the human and the macaque brain share common ancestry since there is lack of an outgroup comparison so far. It is tempting to speculate that Broca's area could constitute one or more duplications of a more ancestral area 44, with divergent specializations for learned vocalizations and thus spoken language.
3. Some alternative hypotheses to the motor theory and brain pathway duplications
Expanding upon the alternative hypotheses, it is possible that the core and shell systems in parrots arose simultaneously, in which case the shell would not be a duplication of the core. However, for this alternative, it would be difficult to explain why the parrot core pathway is more similar to the songbird and hummingbird song systems, than an apparently evolutionary older state (i.e. appearing first). The shell song system of parrots does not appear to be a functionally differentiated region of the core system, since the core system still exists in them and other vocal learners.
A second alternative is that the shell is an independent duplication of the adjacent motor pathway, as the shell has a more similar gene expression profile to the adjacent motor pathway than the core. However, if this were the case, it would be hard to explain how evolution of the shell is more independent of the core, considering that the shell song nuclei surround the cores and are not located elsewhere in the motor pathway and that both core and shell are interconnected without notable major connections to the surrounding motor areas.
A third alternative is that the parrot shell and core song nuclei, as well as the songbird, hummingbird and human song/speech region analogues, are all specialized transformations of an existing motor pathway, i.e. subfunctionalizations, rather than duplications from it, i.e. neofunctionalizations. This would mean that the vocal learning species lost non-vocal motor learning pathway neurons to gain vocal motor pathway neurons. However, there is as yet no evidence for loss of non-vocal motor or other functions in vocal learning species, but rather gain of functions even beyond vocal learning (such as learning how to dance) [94,96,97,99] and increases in relative brain-to-body sizes [103].
A fourth alternative is that vocal learning pathway neurons migrate into their adult locations from developing non-adjacent and even non-motor brain areas, and then by adjacent association they adopt some of the motor learning pathway phenotypes. Although this alternative is theoretically possible, one would expect that the vocal learning pathway cells would have some vestigial properties of their non-motor origin. Thus far, the evidence has not revealed such an alternative origin, although only in songbirds does the HVC (a letter-based name) motor pathway nucleus share some secondary profiles in gene expression with the human secondary auditory cortex [50].
A bigger challenge to the motor theory and duplication hypotheses might at first glance be derived from a recently proposed ‘continuum hypothesis' of vocal learning. Based on findings that mice have a rudimentary forebrain circuit involved in modulating vocalizations but with a very sparse direct projection to brainstem vocal MN, buried within a motor region that also controls other behaviours, and that mice and non-human primates have at least a very limited ability to modify their vocalizations based on auditory experience, Arriaga & Jarvis [104] and Petkov & Jarvis [2] proposed a continuum hypothesis of vocal learning. In this hypothesis, vocal learning is considered to range from complex (e.g. humans and parrots), moderate (some songbirds), to limited or none (mice and non-human primates) [105–108]. In this model, there would be a rudimentary pre-existing forebrain vocal pathway within the vertebrate motor learning pathway, but in the more complex vocal learners these vocal pathway neurons independently expanded and segregated out of the motor learning pathway. However, it is also possible that initially the motor learning pathway duplicates within the non-vocal motor learning circuits to form a limited vocal learning circuit, which subsequently evolves enhanced functions and moves outside of to become adjacent to the motor learning circuit.
Resolving these hypotheses will require more comparative research. The nuances for limited vocal plasticity and an associated neural circuit in mice and non-human primates are also still an ongoing debate that requires further investigation [104–111]. Thus far, of all the hypotheses proposed, we believe the existing data most support the motor origin and duplication hypothesis for vocal learning pathways.
4. Other examples of duplicated morphological structures and structural subdivisions in the evolution of functional complexity
If brain pathway evolution by duplication were possible for vocal learning circuits, then it could be one broad mechanism of brain evolution. The presence of the well-known parallel cortical–basal ganglia–thalamic–cortical loops through the anterior forebrain of mammals and birds is consistent with such an idea. These parallel loops could be replicates of a basic motor learning pathway design. Since all of the cortex is connected with all of the basal ganglia and thalamus [112], when a cortex region is duplicated, one would expect to see a concomitant duplication in the connected basal ganglia and thalamic regions [22,33]. However, our finding of only cortical shell song nuclei duplications thus far in parrots indicates that it could be possible that the duplicated cortical regions co-innervate the non-duplicated striatal and thalamic regions (figure 2c). Such flexibility would allow for greater diversity in neural circuit evolution balanced with the constraint of limited cranial space to accommodate increases in brain size owing to duplications.
Studies of non-human primate motor cortex are consistent with the idea of duplicated brain components. For example, studies using retrograde transneuronal transport of the rabies virus from single muscles of rhesus macaque monkeys to identify layer 5 cortico-motoneuronal (CM) cells in the primary motor cortex (M1) have shown that the M1 region has two subdivisions [113]. The rostral subdivision of macaque M1 has been proposed to be an ‘older’ region as it contains fewer CM cells that make indirect projections to spinal cord MN and is present in most mammals requiring the indirect use of the spinal cord to influence motor output. The caudal subdivision of macaque M1 is proposed to be ‘newer’ as it contains the more rarely found CM cells that make direct projections to MN in the spinal cord and brainstem, including controlling shoulder, elbow and finger muscles to produce highly skilled motor actions. Based on these and other findings, it is generally proposed that the direct CM system of M1 is a recently evolved brain structure that conferred evolutionarily novel functions in the motor system in primates, including independent voluntary control of finger movements, which are more advanced in primates compared with non-primates [114–120]. Assuming that the evidence continues to support differences between species, one hypothesis, like the one we propose for vocal learning pathways, is that the newer caudal M1 region is a duplication of the older rostral M1 region, but with a divergent connection of the CM cells from cortical layer 5.
Examples of morphological duplications or subdivisions to enhance complexity also exist outside of the nervous system. Many animals, such as annelids, have repeated parallel body segments or specialized limb types among species, where the replicated parts are thought to be owing to a repeating developmental genomic programme [121,122]. A striking example of independently evolved morphological duplications is the diversification of the adductor mandibular muscles in teleost fish jaws, which have independently subdivided several times during tetraodontiform evolution [123–125]. Most of these divisions have been incomplete, which suggests that some parts were subfunctionalized instead of duplicated. The duplicated adductor mandibulae muscles continue to maintain similar morphological characteristics, but with increased morphological complexity associated with their functional complexity resulting in finer motor control for feeding [125]. Overall, structural duplications have been proposed to be one mechanism that allows for morphological decoupling [126]. Making structures, such as brain circuits, functionally independent of one another may provide increased complexity and opportunity for modification and diversification [126–128]. In this regard, brain evolution by brain region duplication, brain pathway duplication or structure subdivision may follow a general mechanism of morphological evolution to enhance functional complexity. Testing these hypotheses will be best informed by deciphering the cellular and molecular mechanisms for the development of additional, parallel circuits in the brain.
5. Proposed cellular and molecular mechanisms for evolution of brain pathway duplications
During development, neural stem cell/progenitor cells that give rise to forebrain circuits derive mainly from stem cells in the ventricular zone [129–131]. The daughter cells travel to their positions either by radial migration perpendicularly away from the ventricle (such as excitatory neurons within layers of the mammalian cortex), and/or by tangential migration parallel to the ventricle (such as inhibitory neurons that migrate from the basal ganglia into the cortex) [132–134]. Their local brain region identity is thought to be controlled by patterning transcription factors, such as the homeobox (Hox) genes [135,136]. Once daughter cells reach their target location in the brain, they find their connecting partners in a process that requires cell adhesion and axon guidance genes [137,138]. Given these principles, we propose that one possible mechanism for the evolution and development of duplicated segmented brain circuits is that there is a set of transcription factors that not only control the position of such circuits, but also the number of replicates of that circuit. A genetic change in such transcription factors could result in a new parallel circuit, such as that for vocal learning. Thereafter, changes in axon guidance genes in the new circuit would control divergence in connectivity relative to the older circuit. This begs the question of what kind of genetic change would this be?
We propose that gene duplication could be one such mechanism. Gene duplications have been found to influence the development and function of many organs and tissues, including brains, eyes and wings [11,139–147]. As proposed originally by Ohno [13,14], many consider gene duplication to be one of the most important factors in evolution, including neofunctionalization, subfunctionalization and evolutionary innovations. Gene duplication allows the old gene copy to maintain its function and the new copy to evolve new functions. Even theories on gene evolution through gene duplication have influenced the theories on brain evolution by morphological duplication [139,141]. The concept of neofunctionalization of genes [14] and subfunctionalization of genes [148] match those proposed for structural duplications [128,149,150].
One of the most well-studied and significant examples of duplicated genes controlling duplicated, repeated or segmented morphological structures are the Hox genes. These are transcription factors that control the anterior–posterior body plan axis and are situated in the genome in the same order as the body plan they control [136,151,152]. They are duplicated to different degrees in different animal lineages, with greater complexity and more anatomical segments correlated with more duplications [153]. Many invertebrates and Amphioxus possess one Hox gene cluster, whereas the remaining vertebrates have four Hox gene clusters, in part owing to two whole-genome duplication events that occurred early in vertebrate evolution [151,154–158]. Within the brain, the Hox genes and the greater Hox gene transcription factor superfamily (including OTX, EMX, DMBX, GBX and EN) are involved in brain division and subdivision segmentation [135]. They do so by controlling regional neuronal identity, stem cell progenitors, cell migration and cell death [159,160].
We propose that one possible mechanism for brain pathway duplication could be a local duplication of Hox superfamily genes in the genome segments that control forebrain development. One prediction of this hypothesis is that one should find such genes uniquely duplicated in vocal learning species that control brain development. Recently, based on comparative genomic analyses across the bird family tree, unique gene duplications were found in the songbird lineage and some of these genes had enriched or nearly exclusive expression in the song learning nuclei [161]. It remains to be determined, however, if any of these transcription factors belong to the Hox gene family.
We caution that we are not suggesting a one-to-one relationship of gene duplication with morphological duplication. There are many examples of gene duplications resulting in modifications of existing structures and functions. An example relevant to the topic of vocal learning and cognition is the Slit-Robo GTPase 2 gene (SRGAP2), which has undergone two partial duplications (SRGAP2B and SRGAP2C) uniquely in humans relative to other mammals [26,27,162]. The duplicated copies act as competitive inhibitors to slow cortical dendritic development of already existing brain pathways, which in turn allow greater neural plasticity into adulthood. SRGAP2 modulates activity of the ROBO axon guidance receptors, which are in turn activated by the SLIT family of protein ligands to modulate axonal/dendritic migration and branching in various brain regions [163–167]. Intriguingly, the SLIT1 ligand is uniquely downregulated in the song production nucleus RA analogue of vocal learning birds (songbird RA, parrot AAC and hummingbird VA) [56,68] and the analogous human LMC [50], which would mean that there could be a synergistic effect of the duplicated SRGAP2 GTPase and lower SLIT1 levels in the duplicated vocal motor pathways in humans. Another recent example of partial duplication includes another GTPase, the ARHGAP11B gene, which arose from ARHGAP11A in humans after separation from the chimpanzee lineage [168]. The duplicated copy of the ARHGAP11A gene causes cortical area expansion, and this expansion causes folding, which we surmise could be owing to the duplication.
Advances in genetic technologies have also allowed scientists to test some hypotheses on duplicating or eliminating neural structures genetically. For example, ectopic visual responsive eyes were induced in Drosophila with the addition of an extra copy of one transcription factor, Pax6, expressed during development in another part of the body [169]. Another study showed that electroporating an extra copy of the fibroblast growth factor 8 (FGF8) gene locally in the posterior cortical primordium of mouse embryos causes a partial duplication of the primary somatosensory cortex, with concomitant input from the thalamus to its layer 4 cortical cells, as shown by the presence of ectopic somatosensory barrel fields [170]. In vertebrates, the expression of the Hox1a gene marks the earliest stages of regionalization of the developing hindbrain. Mice mutant for the Hoxa1 gene lack the developing rhombomere 2 (r2) brain region, but the r2 neurons escape apoptosis and develop within r3 and r4, to still incorporate into appropriate circuits to drive the rhythm of breathing [171]. This suggests to us that Hox1a is needed for development of a separate, repeated rhombomere region, but that other factors are sufficient to develop the associated circuit within another circuit. An example of loss of gene function and functional redundancy leading to duplication of structure is the Mauthner (M) cells, a pair of reticulospinal neurons that control escape behaviour in zebra fishes [139,172–174]. During the escape response, if the threatening stimulus arrives from the left side, the left M cell fires, and its action potential travels to the right side so that the fish swerves to the right side owing to the contraction of the muscles on the right side to avoid the threat. Mutation of the notch1a/deadly seven (des) in zebra fish results in the development of extra M cells in r4 [175]. All extra copies of the M cells are responsive to the escape stimuli, suggesting that when duplication of the cells takes place, they receive the appropriate sensory information and respond in a normal way indicating adaptive plasticity of the escape-response circuit.
Other plausible hypotheses of molecular mechanisms that could lead to brain pathway duplication include: (i) changes in splice variants of a gene [176], which we propose could switch on and off at different developmental times to control the generation of parallel circuits; (ii) changes in the cis-regulatory elements of genes, which we propose again would change the reiterative use of a gene network in parallel developing circuits; and (iii) loss of function in a gene that may normally inhibit development of some circuits.
Overall, the various hypotheses may be tested with recent advances in genomics, transcriptomics and gene manipulations, using complete genome sequences from multiple species with and without the brain pathways of interest [93]. Until then, the existing evidence supports the possibility that brain pathway evolution through brain pathway duplication could be one mechanism to generate higher-order complexity in highly evolved animals.
6. Conclusion
In this review, we discussed new evidence from studies in birds, primates and other species that suggests that brain pathways for a novel convergent trait, vocal learning, possibly evolved by duplication from adjacent motor learning pathways. The continuum hypothesis of a pre-existing vocal learning pathway that was independently enhanced in vocal learners is an alternative, but could be compatible with the duplication hypothesis if the duplication occurs within an existing pathway, as seen with Hox1a r2 manipulations. Whether by duplication or enhancement, the pathways have diverged from their adjacent brain regions by specializations of genes involved in neural connectivity. These divergences may have been heavily selected upon for immediate and substantial phenotypic benefits. Despite these divergences, the vocal learning circuits share many properties with the adjacent motor pathways. The findings of the parrot core and shell song system lead us to wonder if humans evolved consecutive or simultaneous multiple duplications of a vocal learning pathway leading to more advanced spoken language abilities. Moreover, findings from studies outside of the vocal learning systems indicate that brain region or pathway duplication could be a general mechanism of brain evolution.
Answers to these questions can now be determined through comparative neurobiology and comparative genomics research. With the recent availability of genomes across the avian [93,177,178] and eventually primate [179,180] family trees, it becomes possible to discover candidate genes. They can then be studied with advanced technologies, such as transcriptomics and genome editing tools, including CRISPR-Cas9, RNAi, TALENs, Cre-Lox systems and more. The theoretical framework presented here will help guide use of these technologies.
Acknowledgements
The authors thank members of the Jarvis Lab (Matt Biegler, Greg Gedman, Ha Na Choe, Lindsey Catlin, Joshua Robinson and Jonathan Chabout) for critical discussions on the paper.
Authors' contributions
E.D.J. and M.C. conceived and wrote the paper.
Competing interests
We have no competing interests.
Funding
The authors were supported by funds from the Howard Hughes Medical Institute.
References
- 1.Jarvis ED. 2004. Learned birdsong and the neurobiology of human language. Ann. NY Acad. Sci. 1016, 749–777. ( 10.1196/annals.1298.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petkov CI, Jarvis ED. 2012. Birds, primates, and spoken language origins: behavioral phenotypes and neurobiological substrates. Front. Evol. Neurosci. 4, 12 ( 10.3389/fnevo.2012.00012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merker B. 2012. The vocal learning constellation: imitation, ritual culture, encephalization. In Music, language, and human evolution (ed. Bannan N.), pp. 215–259 Oxford, UK: Oxford University Press. [Google Scholar]
- 4.Williams RW, Herrup K. 1988. The control of neuron number. Annu. Rev. Neurosci. 11, 423–453. ( 10.1146/annurev.ne.11.030188.002231) [DOI] [PubMed] [Google Scholar]
- 5.Joseph-Harrigan W, Commons LM. 2014. The stage of development of a species predicts the number of neurons. Behav. Dev. Bull. 19, 12–19. ( 10.1037/h0101077) [DOI] [Google Scholar]
- 6.Holland LZ, Holland ND. 1999. Chordate origins of the vertebrate central nervous system. Curr. Opin. Neurobiol. 9, 596–602. ( 10.1016/S0959-4388(99)00003-3) [DOI] [PubMed] [Google Scholar]
- 7.Holland LZ, Short S. 2008. Gene duplication, co-option and recruitment during the origin of the vertebrate brain from the invertebrate chordate brain. Brain Behav. Evol. 72, 91–105. ( 10.1159/000151470) [DOI] [PubMed] [Google Scholar]
- 8.Chen JY. 2008. Early crest animals and the insight they provide into the evolutionary origin of craniates. Genesis 46, 623–639. ( 10.1002/dvg.20445) [DOI] [PubMed] [Google Scholar]
- 9.Emes RD, Grant SG. 2012. Evolution of synapse complexity and diversity. Annu. Rev. Neurosci. 35, 111–131. ( 10.1146/annurev-neuro-062111-150433) [DOI] [PubMed] [Google Scholar]
- 10.Davidson EH, Erwin DH. 2006. Gene regulatory networks and the evolution of animal body plans. Science 311, 796–800. ( 10.1126/science.1113832) [DOI] [PubMed] [Google Scholar]
- 11.Taylor JS, Raes J. 2004. Duplication and divergence: the evolution of new genes and old ideas. Annu. Rev. Genet. 38, 615–643. ( 10.1146/annurev.genet.38.072902.092831) [DOI] [PubMed] [Google Scholar]
- 12.Prince VE, Pickett FB. 2002. Splitting pairs: the diverging fates of duplicated genes. Nat. Rev. Genet. 3, 827–837. ( 10.1038/nrg928) [DOI] [PubMed] [Google Scholar]
- 13.Ohno S. 1967. Sex chromosomes and sex-linked genes. Berlin, Germany: Springer. [Google Scholar]
- 14.Ohno S. 1970. Evolution by gene duplication. New York, NY: Springer. [Google Scholar]
- 15.Zhang J. 2003. Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298. ( 10.1016/S0169-5347(03)00033-8) [DOI] [Google Scholar]
- 16.Johnson R, Samuel J, Ng CK, Jauch R, Stanton LW, Wood IC. 2009. Evolution of the vertebrate gene regulatory network controlled by the transcriptional repressor REST. Mol. Biol. Evol. 26, 1491–1507. ( 10.1093/molbev/msp058) [DOI] [PubMed] [Google Scholar]
- 17.Ono H, Kozmik Z, Yu JK, Wada H. 2014. A novel N-terminal motif is responsible for the evolution of neural crest-specific gene-regulatory activity in vertebrate FoxD3. Dev. Biol. 385, 396–404. ( 10.1016/j.ydbio.2013.11.010) [DOI] [PubMed] [Google Scholar]
- 18.Goymer P. 2007. Alternative splicing switches on the brain. Nat. Rev. Neurosci. 8, 576 ( 10.1038/nrn2200) [DOI] [Google Scholar]
- 19.Chen M, Manley JL. 2009. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 10, 741–754. ( 10.1038/nrm2777) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irimia M, et al. 2011. Stepwise assembly of the Nova-regulated alternative splicing network in the vertebrate brain. Proc. Natl Acad. Sci. USA 108, 5319–5324. ( 10.1073/pnas.1012333108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gueroussov S, Gonatopoulos-Pournatzis T, Irimia M, Raj B, Lin ZY, Gingras AC, Blencowe BJ. 2015. RNA splicing. An alternative splicing event amplifies evolutionary differences between vertebrates. Science 349, 868–873. ( 10.1126/science.aaa8381) [DOI] [PubMed] [Google Scholar]
- 22.Kaas JH. 1989. The evolution of complex sensory systems in mammals. J. Exp. Biol. 146, 165–176. [DOI] [PubMed] [Google Scholar]
- 23.Striedter GF. 2005. Principles of brain evolution. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 24.Jarvis ED, et al. 2005. Avian brains and a new understanding of vertebrate brain evolution. Nat. Rev. Neurosci. 6, 151–159. ( 10.1038/nrn1606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Northcutt RG. 1984. The evolution of the vertebrate central nervous system: patterns and processes. Am. Zool. 24, 701–716. ( 10.1093/icb/24.3.701) [DOI] [Google Scholar]
- 26.Charrier C, et al. 2012. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell 149, 923–935. ( 10.1016/j.cell.2012.03.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dennis MY, et al. 2012. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell 149, 912–922. ( 10.1016/j.cell.2012.03.033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fodor JA. 1983. The modularity of mind. Cambridge, MA: MIT Press. [Google Scholar]
- 29.Pinker S. 1994. The language instinct: how the mind creates language. New York, NY: W. Morrow. [Google Scholar]
- 30.Brodmann K. 1909. Vergleichende Lokalisationslehre der Grosshirnrinde. Leipzig, Germany: Barth. [Google Scholar]
- 31.Diamond IT, Hall WC. 1969. Evolution of neocortex. Science 164, 251–262. ( 10.1126/science.164.3877.251) [DOI] [PubMed] [Google Scholar]
- 32.von Economo C. 1929. The cytoarchitectonics of the human cortex. Oxford, UK: Oxford University Press. [Google Scholar]
- 33.Kaas JH. 1982. The segregation of function in the nervous system: why do sensory systems have so many subdivisions? In Contributions to Sensory Physiology, vol. 7 (ed. Neff WP.), pp. 201–240 New York, NY: Academic Press. [Google Scholar]
- 34.Ebbesson SOE. 1984. Evolution and ontogeny of neural circuits. Behav. Brain Sci. 7, 321–331. ( 10.1017/S0140525X00018379) [DOI] [Google Scholar]
- 35.Lende RA. 1969. A comparative approach to neocortex: localization in Monotremes, marsupials and insectivores. Ann. NY Acad. Sci. 167, 262–276. ( 10.1111/J.1749-6632.1969.Tb20449.X) [DOI] [Google Scholar]
- 36.Finlay BL, Cheung D, Darlington RB. 2005. Developmental constraints on or developmental structure in brain evolution? In Attention and performance XXI ‘process of change in brain and cognitive development’ (eds Munakata Y, Johnson M), pp. 131–162. Oxford, UK: Oxford University Press. [Google Scholar]
- 37.Allman JK, Kaas JH. 1971. A representation of the visual field in the caudal third of the middle temporal gyrus of the owl monkey (Aotus trivirgatus). Brain Res. 31, 85–105. ( 10.1016/0006-8993(71)90635-4) [DOI] [PubMed] [Google Scholar]
- 38.Hughes AL. 1994. The evolution of functionally novel proteins after gene duplication. Proc. R. Soc. Lond. B 256, 119–124. ( 10.1098/rspb.1994.0058) [DOI] [PubMed] [Google Scholar]
- 39.Feenders G, Liedvogel M, Rivas M, Zapka M, Horita H, Hara E, Wada K, Mouritsen H, Jarvis ED. 2008. Molecular mapping of movement-associated areas in the avian brain: a motor theory for vocal learning origin. PLoS ONE 3, e1768 ( 10.1371/journal.pone.0001768) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nottebohm F. 1972. The origins of vocal learning. Am. Nat. 106, 116–140. ( 10.1086/282756) [DOI] [Google Scholar]
- 41.Janik VM, Slater PJB. 1997. Vocal learning in mammals. Adv. Study Behav. 26, 59–99. ( 10.1016/S0065-3454(08)60377-0) [DOI] [Google Scholar]
- 42.Fitch WT, Jarvis ED. 2013. Birdsong and other animals models for human speech, song, and vocal learning. In Language, music, and the brain (ed. Arbib MA.), pp. 499–539. Cambridge, MA: MIT Press. [Google Scholar]
- 43.Brainard MS, Doupe AJ. 2013. Translating birdsong: songbirds as a model for basic and applied medical research. Annu. Rev. Neurosci. 36, 489–517. ( 10.1146/annurev-neuro-060909-152826) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Margoliash D. 1997. Functional organization of forebrain pathways for song production and perception. J. Neurobiol. 33, 671–693. () [DOI] [PubMed] [Google Scholar]
- 45.Fee MS, Goldberg JH. 2011. A hypothesis for basal ganglia-dependent reinforcement learning in the songbird. Neuroscience 198, 152–170. ( 10.1016/j.neuroscience.2011.09.069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mooney R. 1999. Sensitive periods and circuits for learned birdsong. Curr. Opin. Neurobiol. 9, 121–127. ( 10.1016/S0959-4388(99)80015-4) [DOI] [PubMed] [Google Scholar]
- 47.Mooney R. 2009. Neural mechanisms for learned birdsong. Learn. Mem. 16, 655–669. ( 10.1101/lm.1065209) [DOI] [PubMed] [Google Scholar]
- 48.Simonyan K, Horwitz B. 2011. Laryngeal motor cortex and control of speech in humans. Neuroscientist 17, 197–208. ( 10.1177/1073858410386727) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simonyan K, Horwitz B, Jarvis ED. 2012. Dopamine regulation of human speech and bird song: a critical review. Brain Lang. 122, 142–150. ( 10.1016/j.bandl.2011.12.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfenning AR, et al. 2014. Convergent transcriptional specializations in the brains of humans and song-learning birds. Science 346, 1256846 ( 10.1126/science.1256846) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Allott R. 1992. The motor theory of language: origin and function. In Language origin: a multidisciplinary approach (eds Wind J, Bichakjian BH, Nocentini A, Chiarelli B), pp. 105–119. Dordrecht, The Netherlands: Kluwer. [Google Scholar]
- 52.Jarvis ED, et al. 2013. Global view of the functional molecular organization of the avian cerebrum: mirror images and functional columns. J. Comp. Neurol. 521, 3614–3665. ( 10.1002/cne.23404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Brzozowska-Prechtl A, Karten HJ. 2010. Laminar and columnar auditory cortex in avian brain. Proc. Natl Acad. Sci. USA 107, 12 676–12 681. ( 10.1073/pnas.1006645107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shimizu T, Patton TB, Husband SA. 2010. Avian visual behavior and the organization of the telencephalon. Brain Behav. Evol. 75, 204–217. ( 10.1159/000314283) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Durand SE, Heaton JT, Amateau SK, Brauth SE. 1997. Vocal control pathways through the anterior forebrain of a parrot (Melopsittacus undulatus). J. Comp. Neurol. 377, 179–206. () [DOI] [PubMed] [Google Scholar]
- 56.Chakraborty M, et al. 2015. Core and shell song systems unique to the parrot brain. PLoS ONE 10, e0118496 ( 10.1371/journal.pone.0118496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Margoliash D, Fortune ES, Sutter ML, Yu AC, Wren-Hardin BD, Dave A. 1994. Distributed representation in the song system of oscines: evolutionary implications and functional consequences. Brain Behav. Evol. 44, 247–264. ( 10.1159/000113580) [DOI] [PubMed] [Google Scholar]
- 58.Mello CV, Vates GE, Okuhata S, Nottebohm F. 1998. Descending auditory pathways in the adult male zebra finch (Taeniopygia guttata). J. Comp. Neurol. 395, 137–160. () [DOI] [PubMed] [Google Scholar]
- 59.Edelman DB, Baars BJ, Seth AK. 2005. Identifying hallmarks of consciousness in non-mammalian species. Conscious. Cogn. 14, 169–187. ( 10.1016/j.concog.2004.09.001) [DOI] [PubMed] [Google Scholar]
- 60.Rendell L, Whitehead H. 2001. Culture in whales and dolphins. Behav. Brain Sci. 24, 309–324; discussion 324–382 ( 10.1017/S0140525X0100396X) [DOI] [PubMed] [Google Scholar]
- 61.Okanoya K. 2007. Language evolution and an emergent property. Curr. Opin. Neurobiol. 17, 271–276. ( 10.1016/j.conb.2007.03.011) [DOI] [PubMed] [Google Scholar]
- 62.Fisher SE, Marcus GF. 2006. The eloquent ape: genes, brains and the evolution of language. Nat. Rev. Genet. 7, 9–20. ( 10.1038/nrg1747) [DOI] [PubMed] [Google Scholar]
- 63.Dugas-Ford J, Rowell JJ, Ragsdale CW. 2012. Cell-type homologies and the origins of the neocortex. Proc. Natl Acad. Sci. USA 109, 16 974–16 979. ( 10.1073/pnas.1204773109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang C, et al. 2010. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc. Natl Acad. Sci. USA 107, 1518–1523. ( 10.1073/pnas.0913939107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reiner A, et al. 2004. Revised nomenclature for avian telencephalon and some related brainstem nuclei. J. Comp. Neurol. 473, 377–414. ( 10.1002/cne.20118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reiner A, Perkel DJ, Mello CV, Jarvis ED. 2004. Songbirds and the revised avian brain nomenclature. Ann. NY Acad. Sci. 1016, 77–108. ( 10.1196/annals.1298.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen CC, Winkler CM, Pfenning AR, Jarvis ED. 2013. Molecular profiling of the developing avian telencephalon: regional timing and brain subdivision continuities. J. Comp. Neurol. 521, 3666–3701. ( 10.1002/cne.23406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang R, Chen CC, Hara E, Rivas MV, Roulhac PL, Howard JT, Chakraborty M, Audet J, Jarvis ED. 2014. Convergent differential regulation of SLIT-ROBO axon guidance genes in the brains of vocal learners. J. Comp. Neurol. 523, 892–906. ( 10.1002/cne.23719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pepperberg I. 1999. The Alex studies: cognitive and communicative abilities of grey parrots. Cambridge, MA: Harvard University Press. [Google Scholar]
- 70.Pepperberg IM. 2010. Vocal learning in grey parrots: a brief review of perception, production, and cross-species comparisons. Brain Lang. 115, 81–91. ( 10.1016/j.bandl.2009.11.002) [DOI] [PubMed] [Google Scholar]
- 71.Auersperg AM, Oswald N, Domanegg M, Gajdon GK, Bugnyar T. 2014. Unrewarded object combinations in captive parrots. Anim. Behav. Cogn. 1, 470–488. ( 10.12966/abc.11.05.2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Striedter GF. 2013. Bird brains and tool use: beyond instrumental conditioning. Brain Behav. Evol. 82, 55–67. ( 10.1159/000352003) [DOI] [PubMed] [Google Scholar]
- 73.Emery NJ. 2006. Cognitive ornithology: the evolution of avian intelligence. Phil. Trans. R. Soc. B 361, 23–43. ( 10.1098/rstb.2005.1736) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Auersperg AM, von Bayern AM, Gajdon GK, Huber L, Kacelnik A. 2011. Flexibility in problem solving and tool use of kea and New Caledonian crows in a multi access box paradigm. PLoS ONE 6, e20231 ( 10.1371/journal.pone.0020231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paton JA, Manogue KR, Nottebohm F. 1981. Bilateral organization of the vocal control pathway in the budgerigar, Melopsittacus undulatus. J. Neurosci. 1, 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Striedter GF. 1994. The vocal control pathways in budgerigars differ from those in songbirds. J. Comp. Neurol. 343, 35–56. ( 10.1002/cne.903430104) [DOI] [PubMed] [Google Scholar]
- 77.Striedter GF, Lei K. 2006. Vocal performance and plasticity functions are segregated into dorsal and ventral subdivisions of a single nucleus in budgerigars (Melopsittacus undulatus). Poster number 818.10/X22. Society for Neuroscience, Atlanta, GA, USA.
- 78.Brauth SE, Liang W, Roberts TF. 2001. Projections of the oval nucleus of the hyperstriatum ventrale in the budgerigar: relationships with the auditory system. J. Comp. Neurol. 432, 481–511. ( 10.1002/cne.1115) [DOI] [PubMed] [Google Scholar]
- 79.Jarvis ED, Mello CV. 2000. Molecular mapping of brain areas involved in parrot vocal communication. J. Comp. Neurol. 419, 1–31. () [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jarvis ED, Ribeiro S, da Silva ML, Ventura D, Vielliard J, Mello CV. 2000. Behaviourally driven gene expression reveals song nuclei in hummingbird brain. Nature 406, 628–632. ( 10.1038/35020570) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Farabaugh SM, Wild JM. 1997. Reciprocal connections between primary and secondary auditory pathways in the telencephalon of the budgerigar (Melopsittacus undulatus). Brain Res. 747, 18–25. ( 10.1016/S0006-8993(96)01143-2) [DOI] [PubMed] [Google Scholar]
- 82.Joseph L, Toon A, Schirtzinger EE, Wright TF, Schodde R. 2012. A revised nomenclature and classification for family-group taxa of parrots (Psittaciformes). Zootaxa 3205, 26–40. [Google Scholar]
- 83.Veenman CL, Wild JM, Reiner A. 1995. Organization of the avian ‘corticostriatal’ projection system: a retrograde and anterograde pathway tracing study in pigeons. J. Comp. Neurol. 354, 87–126. ( 10.1002/cne.903540108) [DOI] [PubMed] [Google Scholar]
- 84.Atoji Y, Wild JM. 2012. Afferent and efferent projections of the mesopallium in the pigeon (Columba livia). J. Comp. Neurol. 520, 717–741. ( 10.1002/cne.22763) [DOI] [PubMed] [Google Scholar]
- 85.Horita H, Kobayashi M, Liu WC, Oka K, Jarvis ED, Wada K. 2012. Specialized motor-driven dusp1 expression in the song systems of multiple lineages of vocal learning birds. PLoS ONE 7, e42173 ( 10.1371/journal.pone.0042173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jarvis ED. 2007. Neural systems for vocal learning in birds and humans: a synopsis. J. Ornithol. 143, S35–S44. ( 10.1007/s10336-007-0243-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kuypers HGJM. 1958. Some projections from the peri-central cortex to the pons and lower brain stem in monkey and chimpanzee. J. Comp. Neurol. 100, 221–255. ( 10.1002/cne.901100205) [DOI] [PubMed] [Google Scholar]
- 88.Jurgens U. 2002. Neural pathways underlying vocal control. Neurosci. Biobehav. Rev. 26, 235–258. ( 10.1016/S0149-7634(01)00068-9) [DOI] [PubMed] [Google Scholar]
- 89.Simonyan K, Jurgens U. 2003. Efferent subcortical projections of the laryngeal motorcortex in the rhesus monkey. Brain Res. 974, 43–59. ( 10.1016/S0006-8993(03)02548-4) [DOI] [PubMed] [Google Scholar]
- 90.Fitch WT, Huber L, Bugnyar T. 2010. Social cognition and the evolution of language: constructing cognitive phylogenies. Neuron 65, 795–814. ( 10.1016/j.neuron.2010.03.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fitch WT. 2000. The evolution of speech: a comparative review. Trends Cogn. Sci. 4, 258–267. ( 10.1016/S1364-6613(00)01494-7) [DOI] [PubMed] [Google Scholar]
- 92.Arriaga G, Zhou EP, Jarvis ED. 2012. Of mice, birds, and men: the mouse ultrasonic song system has some features similar to humans and song learning birds. PLoS ONE 7, e46610 ( 10.1371/journal.pone.0046610) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jarvis ED, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346, 1320–1331. ( 10.1126/science.1253451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schachner A, Brady TF, Pepperberg IM, Hauser MD. 2009. Spontaneous motor entrainment to music in multiple vocal mimicking species. Curr. Biol. 19, 831–836. ( 10.1016/j.cub.2009.03.061) [DOI] [PubMed] [Google Scholar]
- 95.Schachner A. 2010. Auditory-motor entrainment in vocal mimicking species: additional ontogenetic and phylogenetic factors. Commun. Integr. Biol. 3, 290–293. ( 10.4161/cib.3.3.11708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Patel AD, Iversen JR, Bregman MR, Schulz I. 2009. Experimental evidence for synchronization to a musical beat in a nonhuman animal. Curr. Biol. 19, 827–830. ( 10.1016/j.cub.2009.03.038) [DOI] [PubMed] [Google Scholar]
- 97.Nettl B. 2000. An ethnomusicologist contemplates universals in musical sound and musical culture. In The origins of music (eds Wallin NL, Merker B, Brown S), pp. 463–472. Cambridge, MA: MIT Press. [Google Scholar]
- 98.Fitch WT. 2015. Four principles of bio-musicology. Phil. Trans. R. Soc. B 370, 20140091 ( 10.1098/rstb.2014.0091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Karpati FJ, Giacosa C, Foster NE, Penhune VB, Hyde KL. 2015. Dance and the brain: a review. Ann. NY Acad. Sci. 1337, 140–146. ( 10.1111/nyas.12632) [DOI] [PubMed] [Google Scholar]
- 100.Amunts K, Schleicher A, Burgel U, Mohlberg H, Uylings HB, Zilles K. 1999. Broca's region revisited: cytoarchitecture and intersubject variability. J. Comp. Neurol. 412, 319–341. () [DOI] [PubMed] [Google Scholar]
- 101.Petrides M, Cadoret G, Mackey S. 2005. Orofacial somatomotor responses in the macaque monkey homologue of Broca's area. Nature 435, 1235–1238. ( 10.1038/nature03628) [DOI] [PubMed] [Google Scholar]
- 102.Petrides M, Pandya DN. 2009. Distinct parietal and temporal pathways to the homologues of Broca's area in the monkey. PLoS Biol. 7, e1000170 ( 10.1371/journal.pbio.1000170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Iwaniuk AN, Dean KM, Nelson JE. 2005. Interspecific allometry of the brain and brain regions in parrots (psittaciformes): comparisons with other birds and primates. Brain Behav. Evol. 65, 40–59. ( 10.1159/000081110) [DOI] [PubMed] [Google Scholar]
- 104.Arriaga G, Jarvis ED. 2013. Mouse vocal communication system: are ultrasounds learned or innate? Brain Lang. 124, 96–116. ( 10.1016/j.bandl.2012.10.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hammerschmidt K, Reisinger E, Westekemper K, Ehrenreich L, Strenzke N, Fischer J. 2012. Mice do not require auditory input for the normal development of their ultrasonic vocalizations. BMC Neurosci. 13, 1471–2202. ( 10.1186/1471-2202-13-40) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hammerschmidt K, Whelan G, Eichele G, Fischer J. 2015. Mice lacking the cerebral cortex develop normal song: insights into the foundations of vocal learning. Sci. Rep. 5, 8808 ( 10.1038/srep08808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Portfors CV, Perkel DJ. 2014. The role of ultrasonic vocalizations in mouse communication. Curr. Opin. Neurobiol. 28, 115–120. ( 10.1016/j.conb.2014.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kikusui T, Nakanishi K, Nakagawa R, Nagasawa M, Mogi K, Okanoya K. 2011. Cross fostering experiments suggest that mice songs are innate. PLoS ONE 6, e17721 ( 10.1371/journal.pone.0017721) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hauser MD, Chomsky N, Fitch WT. 2002. The faculty of language: what is it, who has it, and how did it evolve? Science 298, 1569–1579. ( 10.1126/science.298.5598.1569) [DOI] [PubMed] [Google Scholar]
- 110.Hauser MD, Evans CS, Marler P. 1993. The role of articulation in the production of rhesus monkey, Macaca mulatta, vocalizations. Anim. Behav. 45, 423–433. ( 10.1006/anbe.1993.1054) [DOI] [Google Scholar]
- 111.Takahashi DY, Fenley AR, Teramoto Y, Narayanan DZ, Borjon JI, Holmes P, Ghazanfar AA. 2015. Language development. The developmental dynamics of marmoset monkey vocal production. Science 349, 734–738. ( 10.1126/science.aab1058) [DOI] [PubMed] [Google Scholar]
- 112.Swanson L. 2000. What is the brain? Trends Neurosci. 23, 519–527. ( 10.1016/S0166-2236(00)01639-8) [DOI] [PubMed] [Google Scholar]
- 113.Rathelot JA, Strick PL. 2009. Subdivisions of primary motor cortex based on cortico-motoneuronal cells. Proc. Natl Acad. Sci. USA 106, 918–923. ( 10.1073/pnas.0808362106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Maier MA, Armand J, Kirkwood PA, Yang HW, Davis JN, Lemon RN. 2002. Differences in the corticospinal projection from primary motor cortex and supplementary motor area to macaque upper limb motoneurons: an anatomical and electrophysiological study. Cereb. Cortex 12, 281–296. ( 10.1093/cercor/12.3.281) [DOI] [PubMed] [Google Scholar]
- 115.Schmidlin E, Brochier T, Maier MA, Kirkwood PA, Lemon RN. 2008. Pronounced reduction of digit motor responses evoked from macaque ventral premotor cortex after reversible inactivation of the primary motor cortex hand area. J. Neurosci. 28, 5772–5783. ( 10.1523/JNEUROSCI.0944-08.2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dum RP, Strick PL. 2005. Frontal lobe inputs to the digit representations of the motor areas on the lateral surface of the hemisphere. J. Neurosci. 25, 1375–1386. ( 10.1523/JNEUROSCI.3902-04.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lemon RN, Kirkwood PA, Maier MA, Nakajima K, Nathan P. 2004. Direct and indirect pathways for corticospinal control of upper limb motoneurons in the primate. Prog. Brain Res. 143, 263–279. ( 10.1016/S0079-6123(03)43026-4) [DOI] [PubMed] [Google Scholar]
- 118.Nakajima K, Maier MA, Kirkwood PA, Lemon RN. 2000. Striking differences in transmission of corticospinal excitation to upper limb motoneurons in two primate species. J. Neurophysiol. 84, 698–709. [DOI] [PubMed] [Google Scholar]
- 119.Bortoff GA, Strick PL. 1993. Corticospinal terminations in two new-world primates: further evidence that corticomotoneuronal connections provide part of the neural substrate for manual dexterity. J. Neurosci. 13, 5105–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lemon RN. 2008. Descending pathways in motor control. Annu. Rev. Neurosci. 31, 195–218. ( 10.1146/annurev.neuro.31.060407.125547) [DOI] [PubMed] [Google Scholar]
- 121.Carroll SB, Grenier K, Weatherbee SD. 2005. From DNA to diversity: molecular genetics and the evolution of animal design. Oxford, UK: Blackwell. [Google Scholar]
- 122.Tusscher Ten KHWJ. 2013. Mechanisms and constraints shaping the evolution of body plan segmentation. Eur. Phys. J. E. Soft Matter 36, 54 ( 10.1140/epje/i2013-13054-7) [DOI] [PubMed] [Google Scholar]
- 123.Friel JP, Wainwright PC. 1997. A model system of structural duplication: homologies of adductor mandibulae muscles in tetraodontiform fishes. Syst. Biol. 46, 441–463. ( 10.1093/sysbio/46.3.441) [DOI] [Google Scholar]
- 124.Friel JP, Wainwright PC. 1998. Evolution of motor patterns in tetraodontiform fishes: does muscle duplication lead to functional diversification? Brain Behav. Evol. 52, 159–170. ( 10.1159/000006560) [DOI] [PubMed] [Google Scholar]
- 125.Friel JP, Wainwright PC. 1999. Evolution of complexity in motor patterns and jaw musculature of tetraodontiform fishes. J. Exp. Biol. 202, 867–880. [DOI] [PubMed] [Google Scholar]
- 126.Schaefer SA, Lauder GV. 1996. Testing historical hypotheses of morphological change: biomechanical decoupling in loricarioid catfishes. Evolution 50, 1661–1675. ( 10.2307/2410902) [DOI] [PubMed] [Google Scholar]
- 127.Lauder GV. 1993. Design of the aquatic vertebrate skull: major patterns and their evolutionary interpretations. In The skull (eds Hanken J, Hall BK), pp. 113–149. Chicago, IL: University of Chicago Press. [Google Scholar]
- 128.Lauder GV. 1990. Functional morphology and systematics: studying functional patterns in an historial context. Ann. Rev. Ecol. Syst. 21, 317–340. ( 10.1146/annurev.es.21.110190.001533) [DOI] [Google Scholar]
- 129.Gage FH. 2002. Neurogenesis in the adult brain. J. Neurosci. 22, 612–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Alvarez-Buylla A, Lois C. 1995. Neuronal stem cells in the brain of adult vertebrates. Stem Cells 13, 263–272. ( 10.1002/stem.5530130307) [DOI] [PubMed] [Google Scholar]
- 131.Hatten ME. 1999. Central nervous system neuronal migration. Annu. Rev. Neurosci. 22, 511–539. ( 10.1146/annurev.neuro.22.1.511) [DOI] [PubMed] [Google Scholar]
- 132.Nomura T, Hattori M, Osumi N. 2009. Reelin, radial fibers and cortical evolution: insights from comparative analysis of the mammalian and avian telencephalon. Dev. Growth Differ. 51, 287–297. ( 10.1111/j.1440-169X.2008.01073.x) [DOI] [PubMed] [Google Scholar]
- 133.Tanaka DH, Oiwa R, Sasaki E, Nakajima K. 2011. Changes in cortical interneuron migration contribute to the evolution of the neocortex. Proc. Natl Acad. Sci. USA 108, 8015–8020. ( 10.1073/pnas.1102153108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Suzuki IK, Kawasaki T, Gojobori T, Hirata T. 2012. The temporal sequence of the mammalian neocortical neurogenetic program drives mediolateral pattern in the chick pallium. Dev. Cell 22, 863–870. ( 10.1016/j.devcel.2012.01.004) [DOI] [PubMed] [Google Scholar]
- 135.Holland PW, Takahashi T. 2005. The evolution of homeobox genes: implications for the study of brain development. Brain Res. Bull. 66, 484–490. ( 10.1016/j.brainresbull.2005.06.003) [DOI] [PubMed] [Google Scholar]
- 136.Mallo M, Alonso CR. 2013. The regulation of Hox gene expression during animal development. Development 140, 3951–3963. ( 10.1242/dev.068346) [DOI] [PubMed] [Google Scholar]
- 137.Brose K, Tessier-Lavigne M. 2000. Slit proteins: key regulators of axon guidance, axonal branching, and cell migration. Curr. Opin. Neurobiol. 10, 95–102. ( 10.1016/S0959-4388(99)00066-5) [DOI] [PubMed] [Google Scholar]
- 138.Marín O, Valiente M, Ge X, Tsai L. 2010. Guiding neuronal cell migrations. Cold Spring Harb. Perspect. Biol. 2, a001834 ( 10.1101/cshperspect.a001834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hurley I, Hale ME, Prince VE. 2005. Duplication events and the evolution of segmental identity. Evol. Dev. 7, 556–567. ( 10.1111/j.1525-142X.2005.05059.x) [DOI] [PubMed] [Google Scholar]
- 140.Innan H, Kondrashov F. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11, 97–108. ( 10.1038/nrg2689) [DOI] [PubMed] [Google Scholar]
- 141.Oakley TH, Rivera AS. 2008. Genomics and the evolutionary origins of nervous system complexity. Curr. Opin. Genet. Dev. 18, 479–492. ( 10.1016/j.gde.2008.12.002) [DOI] [PubMed] [Google Scholar]
- 142.Teichmann S, Patel NH. 2008. Genomes and evolution: multidimensional approaches to understanding diversity. Curr. Opin. Genet. Dev. 18, 469–471. ( 10.1016/j.gde.2008.11.003) [DOI] [PubMed] [Google Scholar]
- 143.Rivera AS, Pankey MS, Plachetzki DC, Villacorta C, Syme AE, Serb JM, Omilian AR, Oakley TH. 2010. Gene duplication and the origins of morphological complexity in pancrustacean eyes, a genomic approach. BMC Evol. Biol. 10, 123 ( 10.1186/1471-2148-10-123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Baker CR, Hanson-Smith V, Johnson AD. 2013. Following gene duplication, paralog interference constrains transcriptional circuit evolution. Science 342, 104–108. ( 10.1126/science.1240810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Magadum S, Banerjee U, Murugan P, Gangapur D, Ravikesavan R. 2013. Gene duplication as a major force in evolution. J. Genet. 92, 155–161. ( 10.1007/s12041-013-0212-8) [DOI] [PubMed] [Google Scholar]
- 146.Shukla V, Habib F, Kulkarni A, Ratnaparkhi GS. 2014. Gene duplication, lineage-specific expansion, and subfunctionalization in the MADF-BESS family patterns the Drosophila wing hinge. Genetics 196, 481–496. ( 10.1534/genetics.113.160531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Larhammar D, Nordström K, Larsson TA. 2009. Evolution of vertebrate rod and cone phototransduction genes. Phil. Trans. R. Soc. B 364, 2867–2880. ( 10.1098/rstb.2009.0077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liem KF. 1973. Evolutionary strategies and morphological innovations: cichlid pharyngeal jaws. Syst. Zool. 22, 425–441. ( 10.2307/2412950) [DOI] [Google Scholar]
- 149.Lauder GV. 1981. Intraspecific functional repertoires in the feeding mechanism of the characoid fishes Lebiasina, Hoplias and Chalceus. Copeia, 1981, 154–168. ( 10.2307/1444050) [DOI] [Google Scholar]
- 150.Lauder GV. 1982. Historical biology and the problem of design. J. Theor. Biol. 97, 57–67. ( 10.1016/0022-5193(82)90276-4) [DOI] [PubMed] [Google Scholar]
- 151.Ruddle FH, Bartels JL, Bentley KL, Kappen C, Murtha MT, Pendleton JW. 1994. Evolution of Hox genes. Annu. Rev. Genet. 28, 423–442. ( 10.1146/annurev.ge.28.120194.002231) [DOI] [PubMed] [Google Scholar]
- 152.Pearson JC, Lemons D, McGinnis W. 2005. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 6, 893–904. ( 10.1038/nrg1726) [DOI] [PubMed] [Google Scholar]
- 153.Wagner GP, Amemiya C, Ruddle F. 2003. Hox cluster duplications and the opportunity for evolutionary novelties. Proc. Natl Acad. Sci. USA 100, 14 603–14 606. ( 10.1073/pnas.2536656100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Garcia-Fernandez J, Holland PW. 1994. Archetypical organization of the amphioxus Hox gene cluster. Nature 370, 563–566. ( 10.1038/370563a0) [DOI] [PubMed] [Google Scholar]
- 155.Powers TP, Amemiya CT. 2004. Evidence for a Hox14 paralog group in vertebrates. Curr. Biol. 14, R183–R184. ( 10.1016/j.cub.2004.02.015) [DOI] [PubMed] [Google Scholar]
- 156.Holland PW. 1997. Vertebrate evolution: something fishy about Hox genes. Curr. Biol. 7, R570–R572. ( 10.1016/S0960-9822(06)00284-3) [DOI] [PubMed] [Google Scholar]
- 157.Holland PW, Garcia-Fernandez J. 1996. Hox genes and chordate evolution. Dev. Biol. 173, 382–395. ( 10.1006/dbio.1996.0034) [DOI] [PubMed] [Google Scholar]
- 158.Schughart K, Kappen C, Ruddle FH. 1989. Duplication of large genomic regions during the evolution of vertebrate homeobox genes. Proc. Natl Acad. Sci. USA 86, 7067–7071. ( 10.1073/pnas.86.18.7067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Reichert H, Bello B. 2010. Hox genes and brain development in Drosophila. Adv. Exp. Med. Biol. 689, 145–153. ( 10.1007/978-1-4419-6673-5_11) [DOI] [PubMed] [Google Scholar]
- 160.Tumpel S, Wiedemann LM, Krumlauf R. 2009. Hox genes and segmentation of the vertebrate hindbrain. Curr. Top. Dev. Biol. 88, 103–137. ( 10.1016/S0070-2153(09)88004-6) [DOI] [PubMed] [Google Scholar]
- 161.Wirthlin M, Lovell PV, Jarvis ED, Mello CV. 2014. Comparative genomics reveals molecular features unique to the songbird lineage. BMC Genomics 15, 1082 ( 10.1186/1471-2164-15-1082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tyler-Smith C, Xue YL. 2012. Sibling rivalry among paralogs promotes evolution of the human brain. Cell 149, 737–739. ( 10.1016/J.Cell.2012.04.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bagri A, Marin O, Plump AS, Mak J, Pleasure SJ, Rubenstein JL, Tessier-Lavigne M. 2002. Slit proteins prevent midline crossing and determine the dorsoventral position of major axonal pathways in the mammalian forebrain. Neuron 33, 233–248. ( 10.1016/S0896-6273(02)00561-5) [DOI] [PubMed] [Google Scholar]
- 164.Brose K, Bland KS, Wang KH, Arnott D, Henzel W, Goodman CS, Tessier-Lavigne M, Kidd T. 1999. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell 96, 795–806. ( 10.1016/S0092-8674(00)80590-5) [DOI] [PubMed] [Google Scholar]
- 165.Ma L, Tessier-Lavigne M. 2007. Dual branch-promoting and branch-repelling actions of Slit/Robo signaling on peripheral and central branches of developing sensory axons. J. Neurosci. 27, 6843–6851. ( 10.1523/JNEUROSCI.1479-07.2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ypsilanti AR, Zagar Y, Chedotal A. 2010. Moving away from the midline: new developments for Slit and Robo. Development 137, 1939–1952. ( 10.1242/dev.044511) [DOI] [PubMed] [Google Scholar]
- 167.Long H, et al. 2004. Conserved roles for Slit and Robo proteins in midline commissural axon guidance. Neuron 42, 213–223. ( 10.1016/S0896-6273(04)00179-5) [DOI] [PubMed] [Google Scholar]
- 168.Florio M, et al. 2015. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347, 1465–1470. ( 10.1126/science.aaa1975) [DOI] [PubMed] [Google Scholar]
- 169.Gehring WJ. 2001. The genetic control of eye development and its implications for the evolution of the various eye-types. Zoology 104, 171–183. ( 10.1078/0944-2006-00022) [DOI] [PubMed] [Google Scholar]
- 170.Fukuchi-Shimogori T, Grove EA. 2001. Neocortex patterning by the secreted signaling molecule FGF8. Science 294, 1071–1074. ( 10.1126/science.1064252) [DOI] [PubMed] [Google Scholar]
- 171.del Toro ED, Borday V, Davenne M, Neun R, Rijli FM, Champagnat J. 2001. Generation of a novel functional neuronal circuit in Hoxa1 mutant mice. J. Neurosci. 21, 5637–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Liu KS, Fetcho JR. 1999. Laser ablations reveal functional relationships of segmental hindbrain neurons in zebrafish. Neuron 23, 325–335. ( 10.1016/S0896-6273(00)80783-7) [DOI] [PubMed] [Google Scholar]
- 173.Zottoli SJ, Faber DS. 2000. The Mauthner cell: what has it taught us? Neuroscientist 6, 26–38. ( 10.1177/107385840000600111) [DOI] [Google Scholar]
- 174.Eaton RC, Lee RK, Foreman MB. 2001. The Mauthner cell and other identified neurons of the brainstem escape network of fish. Prog. Neurobiol. 63, 467–485. ( 10.1016/S0301-0082(00)00047-2) [DOI] [PubMed] [Google Scholar]
- 175.Liu KS, Gray M, Otto SJ, Fetcho JR, Beattie CE. 2003. Mutations in deadly seven/notch1a reveal developmental plasticity in the escape response circuit. J. Neurosci. 23, 8159–8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Johnson MB, et al. 2009. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron 62, 494–509. ( 10.1016/j.neuron.2009.03.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang G. 2015. Genomics: bird sequencing project takes off. Nature 522, 34 ( 10.1038/522034d) [DOI] [PubMed] [Google Scholar]
- 178.Koepfli KP, Paten B, Genome KCOS, O'Brien SJ. 2015. The genome 10K project: a way forward. Annu. Rev. Anim. Biosci. 3, 57–111. ( 10.1146/annurev-animal-090414-014900) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Scally A, et al. 2012. Insights into hominid evolution from the gorilla genome sequence. Nature 483, 169–175. ( 10.1038/nature10842) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xue Y, et al. 2015. Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science 348, 242–245. ( 10.1126/science.aaa3952) [DOI] [PMC free article] [PubMed] [Google Scholar]