Abstract
Cnidarians, the sister group to bilaterians, have a simple diffuse nervous system. This morphological simplicity and their phylogenetic position make them a crucial group in the study of the evolution of the nervous system. The development of their nervous systems is of particular interest, as by uncovering the genetic programme that underlies it, and comparing it with the bilaterian developmental programme, it is possible to make assumptions about the genes and processes involved in the development of ancestral nervous systems. Recent advances in sequencing methods, genetic interference techniques and transgenic technology have enabled us to get a first glimpse into the molecular network underlying the development of a cnidarian nervous system—in particular the nervous system of the anthozoan Nematostella vectensis. It appears that much of the genetic network of the nervous system development is partly conserved between cnidarians and bilaterians, with Wnt and bone morphogenetic protein (BMP) signalling, and Sox genes playing a crucial part in the differentiation of neurons. However, cnidarians possess some specific characteristics, and further studies are necessary to elucidate the full regulatory network. The work on cnidarian neurogenesis further accentuates the need to study non-model organisms in order to gain insights into processes that shaped present-day lineages during the course of evolution.
Keywords: Cnidaria, nervous systems, neurogenesis, evolution, development
1. Why study cnidarian nervous systems?
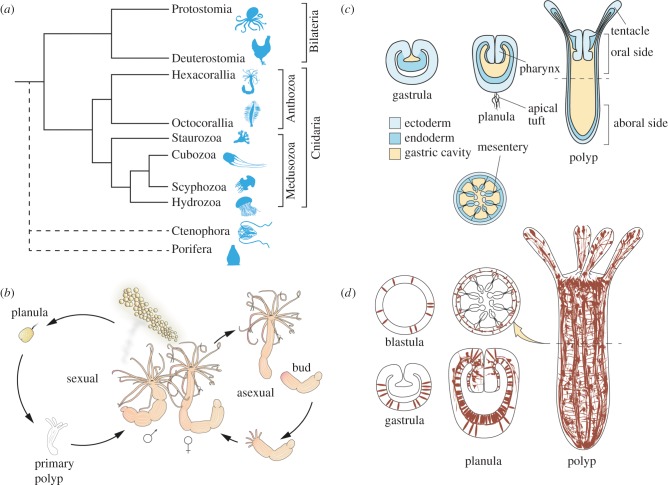
With the exception of Placozoa and Porifera, the nervous system is a defining characteristic of Metazoa, and its appearance was probably a crucial determinant in their diversification and their capability to conquer almost all ecological niches. Although the nervous system has been at the focus of attention for many years, and many aspects of its development and physiology are well understood, the knowledge about its evolutionary origins is still in its infancy. Most research on nervous systems has been carried out on standard model organisms, but their restricted phylogenetic representation makes it difficult to propose viable theories about the ancestral morphology and development of the nervous system. While the contentious phylogenetic positions of Porifera, Placozoa and Ctenophora are impacting on scenarios of the evolution of the nervous system (see also [1]), the Cnidaria have a robust position as a sister group to the Bilateria (figure 1a, [2,3]). Hence, the Cnidaria and the comparison with Bilateria are crucial for the reconstruction of a cnidarian–bilaterian ancestor and our understanding of the evolution of eumetazoan nervous systems. The cnidarians are divided into two major groups, the Anthozoa, consisting of Hexacorallia and Octocorallia, and the Medusozoa, which comprise Hydrozoa, Scyphozoa, Cubozoa and Staurozoa (figure 1a; [9]). The relatively simple morphology, underlined by an intricate gene repertoire, makes cnidarians an ideal system for studying the developmental and cellular processes that (i) led to the emergence of the nervous system and (ii) were involved in the adaptation of nervous systems to different environments and over long periods of time.
Figure 1.
(a) Phylogenetic relationships of cnidarians (after [4]). As the phylogenetic position of ctenophores and sponges is still not completely resolved, their lineages are marked with a dashed line [3,5–8]. The length of branches is for illustrative purposes only and does not represent time of divergence. (b) Life cycle of Nematostella vectensis, with both sexual and asexual reproduction. (c) Schematic of the body plan of N. vectensis throughout development. The lower panel represents a section through the polyp at the dashed line. (d) Schematic of the nervous system of N. vectensis throughout development. Neurons are depicted in brown. The schematic is based on several studies (see below), but it probably does not represent the whole neuronal population. All stages are showed as sections, except for the primary polyp. The section through the polyp (at the dashed line) shows the distribution of neurons in different layers.
Until recently, most of our knowledge on the development of cnidarian nervous systems came from Hydra. Hydra has been very helpful for getting insights into the mechanisms of neuronal differentiation during regeneration and homeostasis in the polyp (see below), however, it mainly propagates asexually by budding. The embryonic development occurs infrequently and is relatively derived, which makes it difficult to investigate and analyse the cellular and molecular differentiation processes during the initial formation of the nervous system.
Other species of the Medusozoa typically have a more complex life cycle, which involves a pelagic medusa stage and a sessile polyp stage. Medusae generally exhibit a more complex nervous system, with neural rings and eyes that are organized in rhopalia and statocysts. Processing and integration of information has been described in rhopalia [10,11], and the high concentration of neurites in the ecto- and endodermal nerve rings at the medusa bell appears to have a function in controlling swimming behaviour [12]. The more complex repertoire of sensory organs in the medusae allows for a more elaborate set of behaviours than found in the purely benthic polyps, and nerve rings and rhopalia may represent an independently evolved form of nervous system centralization [13].
In this review, we discuss cnidarian nervous systems with an emphasis on the recent findings in anthozoan starlet sea anemone Nematostella vectensis (figure 1b), because this system is amenable to functional studies investigating neurogenesis during embryogenesis. Nematostella became an important model system among cnidarians in the past decade [4,14–16]. This brackish water organism has been put forward among other anthozoans owing to its accessibility and amenability for experimental research. It is readily kept under laboratory conditions, spawning can be induced reproducibly, the genome has been sequenced, and gene knockdown methods and stable transgenics have been established, which were particularly insightful for our current understanding of neuronal development [17–19] (for review, see [15]). Nematostella has a surprisingly complex genome, including all major signalling pathways and most transcription factor families [18,20–24].
2. Structure of the Nematostella nervous system
The nervous system of Nematostella, as of other cnidarians, is comprised of two interconnected neuronal networks, one in the ectoderm and one in the endoderm. The principal cell types of cnidarian nervous systems are sensory cells, ganglion cells (the morphological equivalent of interneurons) and cnidocytes (stinging cells). Molecular analyses have revealed that the neuronal networks and the three main classes of neural cells comprise several subpopulations of neurons, which are marked, e.g. by the expression of different neuropeptides, and which can have different distributions along the body.
The search for a pan-neuronal marker for cnidarian neurons has been more difficult than expected. One of the candidate genes for a pan-neuronal marker is the homolog of Elav1, coding for an RNA-binding protein involved in neuronal differentiation [25]. In Nematostella, it indeed marks a large neuronal subpopulation [26,27], but is not a pan-neuronal marker (figure 2a,e), as it is part of a larger population marked by SoxB(2) [29] (see below).
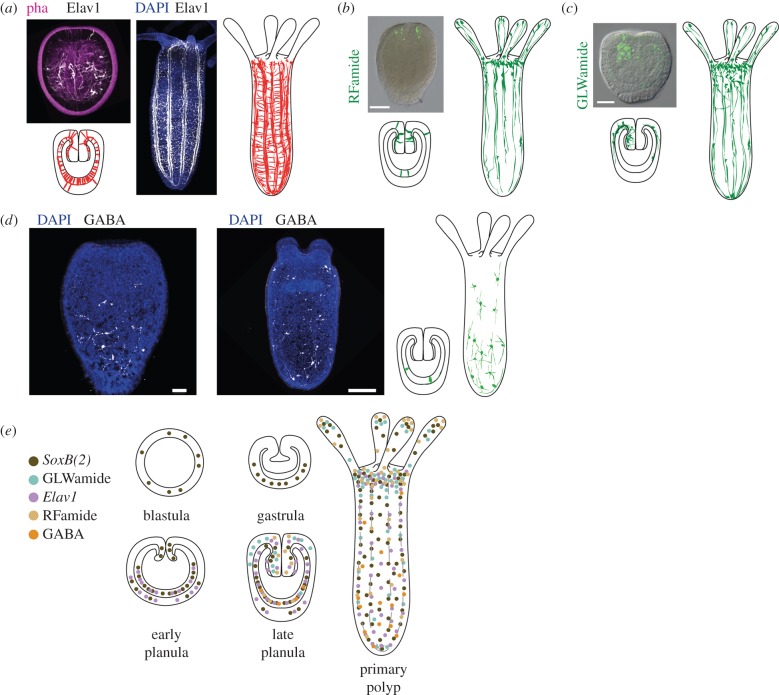
Figure 2.
(a) Distribution of Elav1-positive neurons in planula (left) and primary polyp (right). In the microscopic images (taken from reference [27]), Elav1-positive neurons are in white, phalloiding is in purple, and DAPI is in blue. In the schematic, Elav1-positive neurons are in red. (b) Distribution of RFamide-positive neurons in planula (left) and primary polyp (right). The inset is from reference [28]. RFamide-positive neurons are in green. (c) Distribution of GLWamide-positive neurons in planula (left) and primary polyp (right). The insert is from reference [28]. GLWamide-positive neurons are in green. (d) Distribution of GABA-positive neurons in planula larva and the primary polyp (GABA, white; DAPI, blue). The original data images are maximum projections of 20–30 single confocal images. (e) Distribution of different neuronal subpopulations during the development of N. vectensis. The schematic is based on the results of immunostaining with the antibody against the neuropeptide (GLWamide (turquoise) [28], RFamide (beige) [26,28], GABA (orange) (I.K. and U.T. 2015, unpublished data), or on the analysis of transgenic animals in which a fluorophore is under the control of the gene of interest promoter (SoxB(2) (brown) [29], Elav1 (purple) [27]). Scale bars, (b,c) 100 µm; (d) 50 µm (planula), 100 µm (primary polyp).
In the polyp, N. vectensis, the neuron density appears to be higher in the oral half of the animal, but previous suggestions of an oral nerve ring [26] could not be confirmed. Elav1-positive neurons form networks both in the ectoderm and in the endoderm (figure 2a,d). In the endoderm, however, many neurons follow the parietal muscles on either side of the eight mesenteries, forming prominent longitudinal tracts [27]. These tracts are connected via anastomoses of single neurons.
Two smaller neuronal subpopulations expressing specific neuropeptides, RFamide and GLWamide [30,31], are primarily found in the oral half of the young polyp. Both of them are found in all cnidarians and many bilaterians examined so far [26,28,32–34] (figure 2b,c,e). Individual RFamide-positive neurons appear in the tentacles (figure 2b,e) and may, as in Hydra and other hydrozoans, have a role in ectodermal sensory neurons.
The GLWamide-positive neuronal subpopulation has recently been shown to have a very interesting development. This population appears 2–3 days post fertilization (dpf), but, unlike the RFamide-positive neurons, the GLWamide-positive neurons initially appear on the one side of the developing pharynx in the planula [28]. Later, more GLWamide neurons are added in a radially symmetric pattern, similar to RFamide neurons, and the asymmetrical distribution of GLWamide-positive neurons becomes undetectable (figure 2c,e). However, we still have no insights into which special behaviour or physiological processes this (and maybe other) asymmetrical neuronal subpopulations might be involved in. It has been reported that at 4 dpf these subpopulations make up around 8% of the Elav1-positive neurons [28]. However, because Elav1 is not a pan-neuronal marker, the fraction of RFamide or GLWamide neurons of all neurons is unclear.
Both these neuropeptides mark neurons, which appear in both endoderm and ectoderm, but layer-specific neuronal populations have not yet been described. Recently, we discovered that the neurotransmitter gamma amino butyric acid (GABA) marks a population of neurons that is present only in the endoderm, with the tendency to be more concentrated in the aboral region (figure 2d,e; I.K. and U.T. 2015, unpublished data). Interestingly, the GABA-neurons do not appear to form connections to each other and seem embedded as individual neurons in the nervous system, raising questions about their role. The confined expression of individual neuronal markers reveals a hidden complexity of the Nematostella nervous system, with neuronal subpopulations that might be dedicated to different processes and/or behaviours.
When comparing Nematostella with polyps of other cnidarian species, we can see notable differences in the structure of neuronal subpopulations. For instance, in Hydra, RFamide marks mostly ectodermal sensory neurons of the hypostome and tentacles, but also ectodermal ganglion neurons of the peduncle, although they might also have at least a propriosensory function, because ultrastructural studies showed that they contain cilia [35]. In the planula larva of Clava multicornis, a hydrozoan, RFamide-positive neurons accumulate at the anterior end [36]. The difference between Nematostella and these species may reflect different constraints in their biology and shows that evolutionary interpretations of neuronal patterns have to be taken with caution.
Bioinformatic analysis of the Nematostella genome has shown that a large number of genes associated with chemical neurotransmission are present [37]. Based on these data, we can conclude that the number of neuronal subpopulations in Nematostella must be larger than the several described ones (see above), maybe numbering in dozens. However, in the absence of a pan-neuronal marker, limited number of antibodies and the lack of possibilities for double and triple stainings, it will be difficult to conclude which fraction of neurons express two or more neuropeptides. Also, one has to take into account the discrepancy of bioinformatic data with the empirical data (lack of serotonin orthologues in reference [37], with serotonin immunostaining in reference [26] in Nematostella). In order to tackle this problem, it is necessary to develop antibodies against cnidarian neurotransmitters and improve the immunostaining protocols.
3. Establishment of the nervous system in Nematostella vectensis
In most bilaterians, the neurogenic potential is unequally distributed in the ectodermal tissue. While sensory neurons can often be generated throughout most of the ectoderm, interneurons are typically generated only in the so-called neuroectoderm, the territory from which the central nervous system (CNS) develops. With some exceptions (e.g. hemichordates, acoel worms and flatworms), the specification of the neuroectoderm and CNS is a result of the formation of the dorsoventral (DV) axis by a gradient of BMP signalling [38]. While most cnidarians are considered radially symmetric, anthozoans form a second body axis, the directive axis, which depends on a gradient of BMP signalling [21,39,40]. However, this BMP signalling gradient has been detected only considerably later (at gastrula stage, [39,40]) than the occurrence of the first neural progenitor cells (NPCs; at mid-blastula stage [29]) and early neural differentiation is not biased along the directive axis [26,27,29,41]. Interestingly, the previously mentioned asymmetric distribution of early GLWamide-positive neurons (see above), together with the RFamide-positive population, depends on BMP signalling along the directive axis [28].
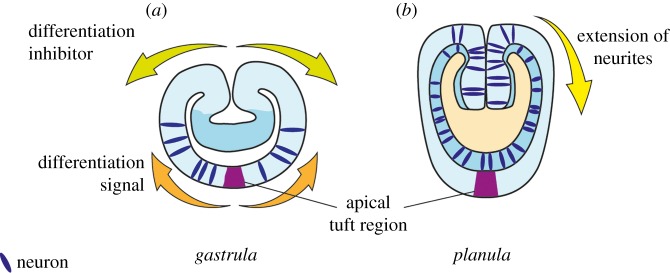
Recent observations suggest that in Nematostella neurogenesis commences at mid-to-late blastula stage in an aboral territory that spans approximately 75% of the body length (figure 3a). At early gastrulation, the oral cap is devoid of differentiating neurons, whereas after gastrulation, more neurons—including some specific subpopulations (RFamide, GLWamide neurons)—are born in the oral half (in and around the pharynx) and in the endoderm [26–28] (figure 3b). Notably, the most aboral region, the ‘apical organ’, often referred to as a sensory centre, remains free of Elav1 and SoxB(2) neural cell bodies [27,29]. It is unclear which signals prevent the early neurogenesis at the oral domain. Because the blastopore expresses various Wnt genes, and the oral–aboral axis of Nematostella is patterned by Wnt signalling [42,43], Wnt signalling might be involved in suppressing early neuronal differentiation at the oral pole. However, in contrast to this idea, recent work showed that manipulation of the Wnt pathway affects the development of oral RFamide and Elav1 neurons [28], suggesting a conserved role for Wnt signalling in promoting neurogenesis. This switch from aboral to more orally located neurogenesis during embryonic development might also indicate a shift from early on-site differentiation of neurons, to a somewhat more restricted neurogenic field. It would be interesting to investigate the potential migration patterns that the neural progenitors and/or neurons, born in this more restricted area, undergo, in order to establish the nervous system of an adult, both in the ecto- and endoderm.
Figure 3.
Early neurogenesis switch in N. vectensis. During very early development (blastula to ea rly gastrula; (a)), individual neurons start appearing in the aboral half, yet excluding future apical organ at the aboral pole. The location and morphology of the cells speak in favour of neuronal determination in situ, either guided by stochastic processes, or by the action of a yet-unknown gradient. Neurogenesis during later stages of embryonic development (b) is still poorly understood, but seems to have a more localized character. In the planula, many neurons are now born at the oral side and in the endoderm. It is still unclear whether and to what extent neuronal precursors migrate towards the aboral side of the embryo.
While in bilaterians neurons originate from the ectoderm, in Nematostella, both ectoderm and endoderm appear to be capable of producing neurons [27]. Endodermal neurons appear shortly after completion of gastrulation. By using transplantation experiments between transgenic Elav1::memOrange and wild-type embryos, Nakanishi et al. [27] have shown that the endoderm can produce neurons independently of the ectoderm. Whether the underlying genetic programme and developmental processes are the same in these two germ layers remains to be elucidated. Because nematocytes differentiate only in ectodermal tissue, one would expect distinct molecular mechanisms that ensure nematocyte and neuronal differentiation in the ectoderm, but neuronal differentiation only in the endoderm.
The differentiation of neurons as assessed by the formation of basal neurites begins at late gastrula stage and becomes more prominent at early planula stage. Analysis of the SoxB(2)::mOrange transgenic line, which broadly labels neural progenitors and their progeny, showed that neurites can extend in any direction from the onset of differentiation [29]. Interestingly, Elav1::mOrange-positive sensory cells, which constitute a subset of the SoxB(2)::mOrange cells, predominantly project in an aboral direction at early- and mid-planula stage. Later-born Elav1 neurons, however, preferentially project in transverse orientation. This is paralleled by the development of the mesenteries, and soon neuronal tracts run along the parietal muscle in the mesentery, with individual neurons situated in between and connecting them [27]. This change in the neurite projection pattern may indicate chemical cues that turn on and off during development in order to correctly orient the projections of neural subpopulations in the developing nervous system; however, they have not yet been identified. In fact, the expression patterns of candidate guidance molecules, such as Netrin or RGM, do not obviously relate to the observed changes in neurite projections of Elav1-positive neurons [24,39].
The development of the nervous system in Nematostella displays some striking differences to that in Hydra and other hydrozoans such as Clytia hemisphaerica and Hydractinia echinata. In these cnidarians, neurons, as well as nematocytes (cnidocytes), differentiate from multipotent interstitial stem cells (i-cells). i-cells predominantly reside between the ectodermal epithelial cells of the body column. Interestingly, the distribution of i-cells is virtually complementary to the density of neurons, which are highest at both extremities, i.e. in hypostome, tentacles and peduncle. i-cells become committed to become neurons either stochastically or by unknown signals. Neuronal progenitors then migrate orally or aborally to the site of differentiation, where they undergo a final mitosis and differentiate—probably by local cues—to a specific neuronal phenotype [44,45]. However, i-cells have only been found in hydrozoans and therefore are considered a specific feature of hydrozoans. In other cnidarians, e.g. the scyphozoan Aurelia aurita, neurons likely arise from epithelial cells or intermediate progenitors, more akin to the situation in Nematostella [46]. This variability further emphasizes the need to compare several species of one clade.
4. Developmental genetics of Nematostella neurons
The publication of several cnidarian genomes has shown that much of the molecular architecture underlying neurogenesis and neuron functioning is conserved between bilaterians and cnidarians [17,47–51]. In bilaterians, neurons are born from specialized cell populations, termed NPCs, which arise in an area of the ectoderm dedicated to developing the CNS. With cnidarians lacking a centralized system, and having both ectodermal and endodermal neurogenesis, it has been questioned whether this conserved molecular toolkit is employed in the same way.
Recent research on Wnt and BMP signalling during embryonic neurogenesis in Nematostella gives us some insight into the involvement of these conserved pathways in cnidarian neurogenesis [28]. The Wnt/β-catenin pathway is involved in neural patterning and neurogenesis in bilaterians [52,53]. Use of a β-catenin signalling inhibitor resulted in a severe reduction of RFamide, GLWamide and Elav1::mOrange neurons at planula stage, whereas ectopic activation of β-catenin increased the number of these neurons [28]. These observations suggest that β-catenin can positively regulate neural development in Nematostella. However, because the oral Wnt signalling centre of the blastopore is devoid of early neurogenesis, it is not clear whether Wnt/β-catenin signalling has a direct role in early neurogenesis or a general positive function in establishing neurogenic potential. Surprisingly, while in flies and vertebrates the gradient of BMP signalling along the DV axis has an anti-neuralizing effect and localizes the CNS [38], in Nematostella, BMP signalling appears to have no effect on neuronal differentiation at an early phase, but a proneural function in the later phase of embryonic neurogenesis [28]. Future research also on other bilaterian phyla will reveal whether Wnt signalling or BMP signalling (or both) has an ancestral role in neurogenesis.
Interesting insights into the conservation of regional patterning came from the analysis of the bilaterian head patterning genes six3/6, FoxQ2a and irx, which are early anterior brain markers. Strikingly, six3/6, FoxQ2a and irx are actually expressed at the aboral end of the Nematostella planula [54], suggesting a stunning conservation of regional patterning genes. Knockdown of Nematostella six3/6 reduced the number of DmrtB-expressing aboral neurons, but did not affect the expression of the broader neural marker RFamide, suggesting that the effect on the aboral neurons is rather a consequence of the mis-specification of the aboral domain. These observations are similar to loss-of-function studies in sea urchin and the beetle Tribolium castaneum [55,56], but different from the situation in vertebrates, where six3/6 is crucial for anterior brain development. Thus, general axial patterning genes might have been coopted for the induction of the anterior CNS in the vertebrate lineage.
The genetic network underlying the transition from the NPC to the post-mitotic neuron has been extensively studied in Drosophila [57] and mammals, especially mouse [58]. The determination of NPCs from the epithelial layer represents a textbook example of the lateral inhibition by the Notch/Delta system [59]. In a previously designated neuroepithelial field, NPCs, which are destined to become neurons, are singled out by the interaction of receptors, and by differential expression of the neurogenic programme in individual cells. The cells, which remain dividing progenitors, are inhibited from expressing this programme for the moment. It appears that, in Nematostella, the Notch/Delta system is also involved in determining the fate of neural lineage cells. By using pharmacological treatments with the γ-secretase inhibitor DAPT, it was shown that the Notch/Delta signalling influences the expression of neurogenic markers [60], e.g. the achaete-scute homologue (AshA) among others [61].
More surprising was the finding that the Notch/Delta function in neurogenesis is not conducted through the canonical pathway, i.e. involving suppressor of hairy (Su(H)), but a non-canonical, yet unidentified route [61]. This difference might indicate that the Notch pathway had a more general role in cell differentiation in the ancestor of cnidarians and bilaterians [62], and was coopted in slightly different ways in the neurogenic pathways of both lineages. Another explanation is that the non-canonical Notch signalling is the ancestral form of this signalling pathway. This is based on the fact that only bilaterians have the full complement of the Notch/Delta pathway [63]. However, because two key elements of canonical Notch signalling, Su(H) and mastermind, are present in Nematostella, this hypothesis still awaits confirmation through data from other, non-bilaterian phyla.
Downstream of Notch/Delta signalling, a specific set of proneural genes of the bHLH transcription factors become activated, in particular the achaete-scute (Ash) and atonal (ato) gene family, which regulate the transition of the progenitor cell into a neuron. At least one, AshA, is expressed in single cells of the aboral half of the early embryo and is directly involved in neurogenesis: knockdown leads to loss of specific neuronal markers, overexpression increases the number of RFamide+ and Elav1-precursor cells in the aboral half [41]. The data suggest that AshA does not have a pan-neuronal role, which would also fit the model in which Ash and ato promote neurogenesis of distinct neuronal populations [64], as in bilaterians. However, as the data on the members of the ato family are still scarce, owing to their unresolved phylogeny [65], this idea cannot be yet confirmed. The expression patterns of several ato genes coincide with the expression patterns of AshB and SoxB2, a gene also involved in neurogenesis in Bilateria [28]. Interestingly, one of the ato genes, Arp6, is expressed asymmetrically in the developing embryo and functional analysis suggests that it regulates the asymmetric distribution of GLWamide-positive neurons. Notably, in hydra, chemical inhibition of Notch signalling suggests a role in boundary formation during detachment of the bud and in tentacle formation during regeneration [66], thus, fundamentally different processes from neuronal differentiation. It is possible that Notch signalling has distinct roles in embryogenesis and adult polyps.
In mammals, neural progenitors still have the capability to divide and produce either other types of progenitors or different subpopulations of neurons. Pax6 and Sox2 are markers of these intermediate progenitors in the developing mammalian brain [58]. It is still not clear whether a neural progenitor population similar to this one exists in cnidarians. Until recently, it was also unclear whether cnidocytes and neurons, both members of the neuronal lineage, come from the same populations of progenitors. Here, Sox proteins might be key to this question. Sox proteins are indispensable in the determination and maintenance of embryonic stem cells in mammals, and later, during brain development, in the population of NPCs [67]. Members of SoxB1 and SoxB2 subgroups are especially important during neurogenesis [68,69]. Of the 14 Sox genes present in Nematostella and Hydra [17,24,47,70], one gene, SoxB(2) (termed SoxB2 in reference [24] and SoxBa in reference [71]), is expressed in single cells during gastrulation, consistent with a role in neuronal differentiation [24]. Using a SoxB(2) transgenic reporter line, Richards and Rentzsch showed that SoxB(2) marks a population of cells that gives rise to ganglion and sensory neurons and cnidocytes, thus representing a general neural progenitor population [29]. The knockdown of SoxB(2) strongly reduces the production of neurons and cnidocytes. Thus, this gene appears necessary for the differentiation of both cell types. The tracing of EdU-labelled dividing SoxB(2)-positive cells suggested that daughter cells of one neural progenitor in Nematostella can have different cell cycle characteristics [29], a feature that is reminiscent of asymmetric cell fate in Drosophila and mammals [72]. It also shows that this Sox gene has a conserved role in neurogenesis in cnidarians and bilaterians. In addition to SoxB(2), as mentioned above, another SoxB2 gene is involved in patterning the oral nervous system [28]. Taken together, these data strongly suggest that some key aspects of the neurogenic programme are conserved between cnidarians and bilaterians. Interestingly, Sox genes are also expressed in putative progenitors that give rise to neurosensory cells in the ctenophore Pleurobrachia pileus [73]. This further confirms the ancestral role that Sox genes have in the development and evolution of the nervous systems, but also brings into question the independent origin of nervous systems in ctenophores [5,74,75].
5. Evolutionary context
Taking into account more than 500 million years of independent evolution of the bilaterian and cnidarian lineage [76], surprisingly, many elements of the developmental neuronal network and genes governing neuronal structure and function are conserved. We conclude that the last common ancestor of Bilateria and Cnidaria was an animal with a well-established nervous system, in which neurons were born out of epithelial cells, which were singled out to become neurons by a cell-determining system (e.g. the Notch/Delta system) [61] and this mechanism was inherited from the common ancestor of sponges and eumetazoans [77], which may or may not have had a nervous system. These epithelial cells most probably underwent an asymmetric division, to produce a differentiated cell—a neuron, and presumably another epithelial cell or a neural progenitor. What also seems to be conserved is the early proneural gene network, which is involved in the production of neurons during the embryonic stages.
It has been shown that members of the Sox family of genes are involved in the patterning of the neural field and the production of the components of the neural lineage [28,29]. After the specification of the cell as a neural progenitor, other downstream proneural genes finish the differentiation process of the progenitor into a neuron [41]. More studies are needed in order to fully reconstruct the basic genetic network underlying cnidarian neurogenesis, but some obvious candidates exist. Members of the Pax family of transcription factors represent an interesting starting point, as they are crucial for mammalian neurogenesis (especially eye development), with Pax6 being one of the main markers of neural progenitors. The findings that jellyfish PaxB gene is involved in the eye development of the cubomedusa Tripedalia cystophora and that PaxB can rescue a Drosophila eye mutant [78] suggests a conserved role of PaxB in neuronal development. In line with this, Nematostella PaxA and PaxB are expressed in single cells, reminiscent of a pattern present in progenitors and/or neurons [24]. The finding of asymmetrical divisions in the SoxB(2)-positive progenitors also invites the investigation of cell polarity proteins (e.g. Par3, Par6), and their role in the determination of cell fate. With the development of advanced in vivo imaging techniques, this problem becomes more accessible.
All of the previous studies spanning more than 150 years of cnidarian research points at the common origin of the nervous systems in Bilateria and Cnidaria. However, there are also marked differences. The curious aspect of endodermal neurogenesis existing in Nematostella presents a puzzle. Is neurogenesis in both germ layers an ancestral trait, and bilaterians have lost it or was it independently gained in the cnidarian lineage? Further detailed analyses of the processes governing neurogenesis in both germ layers, and potential comparison with genomic elements expressed in the bilaterian endoderm, are necessary to resolve this question. The cnidocyte, the cnidarian-specific cell type, is also a part of the neuronal lineage, as it seems that it stems from the same progenitor pool. Interestingly, cnidocytes express one of the three subfamilies of the ether-à-go-go (EAG) family of voltage-gated K+ channels [79]. However, its morphology and function are markedly different from any neuronal type in the rest of the animal kingdom. While our knowledge of the molecular basis of neuronal physiology is still scarce, there has been some recent progress. Nematostella has a significantly expanded set of 20 voltage-gated K+ channels of the shaker family, yet their function in Nematostella is still unclear [80]. Further, the diversification of the EAG family of voltage-gated K+ channels into Eag, Erg and Elk subfamilies occurred in the cnidarian/bilaterian ancestor after divergence from ctenophores. All three subfamilies seem to have at least partially conserved molecular functions, when tested in vitro [79,81]. An interesting case is the voltage-gated Na+ (Nav) channels, which are responsible for the action potential of neurons in bilaterians. Both cnidarian and bilaterian Nav channels have evolved from an ancestral voltage-gated Ca2+ (Cav) channel. However, the selectivity filter differs significantly in cnidarians and bilaterians, suggesting that a key component of neuronal physiology has evolved independently in these two lineages [82,83].
6. Outlook
Evolutionary developmental biology (evo-devo) has experienced a renaissance in the past 10–15 years. This is mostly owing to the development of new functional techniques and the advancement of sequencing methods. With this, scientists could return to using non-model organisms to answer questions about the evolutionary origin of pathways, cells, organs and whole systems. Cnidarians have been particularly interesting in the studies of germ layers and the nervous system. However, there are still many unanswered questions. One of the main puzzle pieces still missing is the molecular signature of the cnidarian neural progenitors and neurons, and how it relates to the bilaterian ones. This could be addressed by using a combination of transgenic animals and transcriptome sequencing, in order to decipher the molecular fingerprint of different neuronal populations. The cellular processes of the establishment and maintenance of the nervous system are also still largely unknown. Furthermore, the molecular and cellular basis of the dynamics and the physiology of the diffuse nervous system under conditions of homeostasis and growth are still not well understood. Applying new in vivo imaging techniques will allow us to track transgenic progenitor cells in the developing embryo and polyp to provide further insights into the formation and function of the cnidarian nervous system.
Acknowledgements
We thank two anonymous reviewers for their comments and the members of the Technau laboratory for discussion.
Ethics
The use of animals to obtain data for this study (figure 2) was approved by the Austrian Ministry for Science, Research and Economy.
Authors' contributions
All authors wrote and approved the manuscript together. I.K. prepared the figures.
Competing interests
We declare we have no competing interests.
Funding
Research in the laboratory of U.T. has been supported by the Austrian Science Fund (FWF) (P24858). Research in the laboratory of F.R. has been supported by Sars Centre core budget. I.K. was the recipient of the FWF Lise Meitner Incoming Fellowship (M1680-B21). We also acknowledge the Core Imaging Facility of the Faculty of Life Sciences in Vienna for providing the confocal microscopes.
References
- 1.Moroz LL. 2015. Convergent evolution of neural systems in Ctenophores. J. Exp. Biol. 218, 598–611. ( 10.1242/jeb.110692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hejnol A, et al. 2009. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc. R. Soc. B 276, 4261–4270. ( 10.1098/rspb.2009.0896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pick KS, et al. 2010. Improved phylogenomic taxon sampling noticeably affects nonbilaterian relationships. Mol. Biol. Evol. 27, 1983–1987. ( 10.1093/molbev/msq089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Technau U, Steele RE. 2011. Evolutionary crossroads in developmental biology: Cnidaria. Development 138, 1447–1458. ( 10.1242/dev.048959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moroz LL, et al. 2014. The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109–114. ( 10.1038/nature13400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryan JF, et al. 2013. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342, 1242592 ( 10.1126/science.1242592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Philippe H, et al. 2009. Phylogenomics revives traditional views on deep animal relationships. Curr. Biol. 19, 706–712. ( 10.1016/j.cub.2009.02.052) [DOI] [PubMed] [Google Scholar]
- 8.Nosenko T, et al. 2013. Deep metazoan phylogeny: when different genes tell different stories. Mol. Phylogenet. Evol. 67, 223–233. ( 10.1016/j.ympev.2013.01.010) [DOI] [PubMed] [Google Scholar]
- 9.Collins AG, Schuchert P, Marques AC, Jankowski T, Medina M, Schierwater B. 2006. Medusozoan phylogeny and character evolution clarified by new large and small subunit rDNA data and an assessment of the utility of phylogenetic mixture models. Syst. Biol. 55, 97–115. ( 10.1080/10635150500433615) [DOI] [PubMed] [Google Scholar]
- 10.Garm A, Mori S. 2009. Multiple photoreceptor systems control the swim pacemaker activity in box jellyfish. J. Exp. Biol. 212, 3951–3960. ( 10.1242/jeb.031559) [DOI] [PubMed] [Google Scholar]
- 11.Skogh C, Garm A, Nilsson DE, Ekström P. 2006. Bilaterally symmetrical rhopalial nervous system of the box jellyfish Tripedalia cystophora. J. Morphol. 267, 1391–1405. ( 10.1002/jmor.10472) [DOI] [PubMed] [Google Scholar]
- 12.Mackie GO. 2004. Central neural circuitry in the jellyfish Aglantha: a model ‘simple nervous system’. Neurosignals 13, 5–19. ( 10.1159/000076155) [DOI] [PubMed] [Google Scholar]
- 13.Koizumi O, Hamada S, Minobe S, Hamaguchi-Hamada K, Kurumata-Shigeto M, Nakamura M, Namikawa H. 2015. The nerve ring in cnidarians: its presence and structure in hydrozoan medusae. Zoology (Jena) 118, 79–88. ( 10.1016/j.zool.2014.10.001) [DOI] [PubMed] [Google Scholar]
- 14.Darling JA, Reitzel AR, Burton PM, Mazza ME, Ryan JF, Sullivan JC, Finnerty JR. 2005. Rising starlet: the starlet sea anemone, Nematostella vectensis. Bioessays 27, 211–221. ( 10.1002/bies.20181) [DOI] [PubMed] [Google Scholar]
- 15.Genikhovich G, Technau U. 2009. The starlet sea anemone Nematostella vectensis: an anthozoan model organism for studies in comparative genomics and functional evolutionary developmental biology. Cold Spring Harb. Protoc 9, pdb.emo129. [DOI] [PubMed] [Google Scholar]
- 16.Technau U, Genikhovich G, Kraus JEM. 2015. Cnidaria. In Evolutionary developmental biology of invertebrates, vol. 1 (ed Wanninger A.), pp. 115–163. Berlin, Germany: Springer [Google Scholar]
- 17.Putnam NH, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86–94. ( 10.1126/science.1139158) [DOI] [PubMed] [Google Scholar]
- 18.Rentzsch F, Fritzenwanker JH, Scholz CB, Technau U. 2008. FGF signalling controls formation of the apical sensory organ in the cnidarian Nematostella vectensis. Development 135, 1761–1769. ( 10.1242/dev.020784) [DOI] [PubMed] [Google Scholar]
- 19.Renfer E, Amon-Hassenzahl A, Steinmetz PRH, Technau U. 2010. A muscle-specific transgenic reporter line of the sea anemone, Nematostella vectensis. Proc. Natl Acad. Sci. USA 107, 104–108. ( 10.1073/pnas.0909148107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Technau U, et al. 2005. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 21, 633–639. ( 10.1016/j.tig.2005.09.007) [DOI] [PubMed] [Google Scholar]
- 21.Saina M, Genikhovich G, Renfer E, Technau U. 2009. BMPs and chordin regulate patterning of the directive axis in a sea anemone. Proc. Natl Acad. Sci. USA 106, 18 592–18 597. ( 10.1073/pnas.0900151106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matus DQ, Magie CR, Pang K, Martindale MQ, Thomsen GH. 2008. The Hedgehog gene family of the cnidarian, Nematostella vectensis, and implications for understanding metazoan Hedgehog pathway evolution. Dev. Biol. 313, 501–518. ( 10.1016/j.ydbio.2007.09.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matus DQ, Pang K, Daly M, Martindale MQ. 2007. Expression of Pax gene family members in the anthozoan cnidarian, Nematostella vectensis. Evol. Dev. 9, 25–38. ( 10.1111/j.1525-142X.2006.00135.x) [DOI] [PubMed] [Google Scholar]
- 24.Magie CR, Pang K, Martindale MQ. 2005. Genomic inventory and expression of Sox and Fox genes in the cnidarian Nematostella vectensis. Dev. Genes Evol. 215, 618–630. ( 10.1007/s00427-005-0022-y) [DOI] [PubMed] [Google Scholar]
- 25.Akamatsu W, et al. 2005. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc. Natl Acad. Sci. USA 102, 4625–4630. ( 10.1073/pnas.0407523102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marlow HQ, Srivastava M, Matus DQ, Rokhsar D, Martindale MQ. 2009. Anatomy and development of the nervous system of Nematostella vectensis, an anthozoan cnidarian. Dev. Neurobiol. 69, 235–254. ( 10.1002/dneu.20698) [DOI] [PubMed] [Google Scholar]
- 27.Nakanishi N, Renfer E, Technau U, Rentzsch F. 2012. Nervous systems of the sea anemone Nematostella vectensis are generated by ectoderm and endoderm and shaped by distinct mechanisms. Development 139, 347–357. ( 10.1242/dev.071902) [DOI] [PubMed] [Google Scholar]
- 28.Watanabe H, Kuhn A, Fushiki M, Agata K, Özbek S, Fujisawa T, Holstein TW. 2014. Sequential actions of β-catenin and Bmp pattern the oral nerve net in Nematostella vectensis. Nat. Commun. 5, 5536 ( 10.1038/ncomms6536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards GS, Rentzsch F. 2014. Transgenic analysis of a SoxB gene reveals neural progenitor cells in the cnidarian Nematostella vectensis. Development 141, 4681–4689. ( 10.1242/dev.112029) [DOI] [PubMed] [Google Scholar]
- 30.Schmich J, Rudolf R, Trepel S, Leitz T. 1998. Immunohistochemical studies of GLWamides in Cnidaria. Cell Tissue Res. 294, 169–177. ( 10.1007/s004410051167) [DOI] [PubMed] [Google Scholar]
- 31.Takahashi T, Hatta M. 2011. The importance of GLWamide neuropeptides in cnidarian development and physiology. J. Amino Acids 2011, 424501 ( 10.4061/2011/424501) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimmelikhuijzen C, Leviev I, Carstensen K. 1996. Peptides in the nervous systems of cnidarians: structure, function, and biosynthesis. Int. Rev. Cytol. 167, 37–89. ( 10.1016/S0074-7696(08)61345-5) [DOI] [PubMed] [Google Scholar]
- 33.Grimmelikhuijzen C, Williamson M, Hansen G. 2002. Neuropeptides in cnidarians. Can. J. Zool. 80, 1690–1702. ( 10.1139/z02-137) [DOI] [Google Scholar]
- 34.Jékely G. 2013. Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc. Natl Acad. Sci. USA 110, 8702–8707. ( 10.1073/pnas.1221833110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westfall JA. 1973. Ultrastructural evidence for a granule-containing sensory-motor-interneuron in Hydra littoralis. J. Ultrastruct. Res. 42, 268–282. ( 10.1016/S0022-5320(73)90055-5) [DOI] [PubMed] [Google Scholar]
- 36.Piraino S, et al. 2011. Complex neural architecture in the diploblastic larva of Clava multicornis (Hydrozoa, Cnidaria). J. Comp. Neurol. 519, 1931–1951. ( 10.1002/cne.22614) [DOI] [PubMed] [Google Scholar]
- 37.Anctil M. 2009. Chemical transmission in the sea anemone Nematostella vectensis: a genomic perspective. Comp. Biochem. Physiol. D Genomics Proteomics 4, 268–289. ( 10.1016/j.cbd.2009.07.001) [DOI] [PubMed] [Google Scholar]
- 38.De Robertis EM. 2008. Evo-devo: variations on ancestral themes. Cell 132, 185–195. ( 10.1016/j.cell.2008.01.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leclère L, Rentzsch F. 2014. RGM regulates BMP-mediated secondary axis formation in the sea anemone Nematostella vectensis. Cell Rep. 9, 1921–1930. ( 10.1016/j.celrep.2014.11.009) [DOI] [PubMed] [Google Scholar]
- 40.Genikhovich G, et al. 2015. Axis patterning by BMPs: cnidarian network reveals evolutionary constraints. Cell Rep. 10, 1646–1654. ( 10.1016/j.celrep.2015.02.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Layden MJ, Boekhout M, Martindale MQ. 2012. Nematostella vectensis achaete-scute homolog NvashA regulates embryonic ectodermal neurogenesis and represents an ancient component of the metazoan neural specification pathway. Development 139, 1013–1022. ( 10.1242/dev.073221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kusserow A, et al. 2005. Unexpected complexity of the Wnt gene family in a sea anemone. Nature 433, 156–160. ( 10.1038/nature03158) [DOI] [PubMed] [Google Scholar]
- 43.Marlow H, Matus DQ, Martindale MQ. 2013. Ectopic activation of the canonical wnt signaling pathway affects ectodermal patterning along the primary axis during larval development in the anthozoan Nematostella vectensis. Dev. Biol. 380, 324–334. ( 10.1016/j.ydbio.2013.05.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Technau U, Holstein TW. 1996. Phenotypic maturation of neurons and continuous precursor migration in the formation of the peduncle nerve net in Hydra. Dev. Biol. 177, 599–615. ( 10.1006/dbio.1996.0189) [DOI] [PubMed] [Google Scholar]
- 45.Hager G, David CN. 1997. Pattern of differentiated nerve cells in hydra is determined by precursor migration. Development 124, 569–576. [DOI] [PubMed] [Google Scholar]
- 46.Nakanishi N, Yuan D, Jacobs DK, Hartenstein V. 2008. Early development, pattern, and reorganization of the planula nervous system in Aurelia (Cnidaria, Scyphozoa). Dev. Genes Evol. 218, 511–524. ( 10.1007/s00427-008-0239-7) [DOI] [PubMed] [Google Scholar]
- 47.Chapman JA, et al. 2010. The dynamic genome of Hydra. Nature 464, 592–596. ( 10.1038/nature08830) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shinzato C, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323. ( 10.1038/nature10249) [DOI] [PubMed] [Google Scholar]
- 49.Galliot B, Quiquand M, Ghila L, de Rosa R, Miljkovic-Licina M, Chera S. 2009. Origins of neurogenesis, a cnidarian view. Dev. Biol. 332, 2–24. ( 10.1016/j.ydbio.2009.05.563) [DOI] [PubMed] [Google Scholar]
- 50.Galliot B, Quiquand M. 2011. A two-step process in the emergence of neurogenesis. Eur. J. Neurosci. 34, 847–862. ( 10.1111/j.1460-9568.2011.07829.x) [DOI] [PubMed] [Google Scholar]
- 51.Watanabe H, Fujisawa T, Holstein TW. 2009. Cnidarians and the evolutionary origin of the nervous system. Dev. Growth Differ. 51, 167–183. ( 10.1111/j.1440-169X.2009.01103.x) [DOI] [PubMed] [Google Scholar]
- 52.Holland LZ, Carvalho JE, Escriva H, Laudet V, Schubert M, Shimeld SM, Yu J-K. 2013. Evolution of bilaterian central nervous systems: a single origin? EvoDevo. 4, 27 ( 10.1186/2041-9139-4-27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hartenstein V, Stollewerk A. 2015. The evolution of early neurogenesis. Dev. Cell. 32, 390–407. ( 10.1016/j.devcel.2015.02.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinigaglia C, Busengdal H, Leclère L, Technau U, Rentzsch F. 2013. The bilaterian head patterning gene six3/6 controls aboral domain development in a cnidarian. PLoS Biol. 11, e1001488 ( 10.1371/journal.pbio.1001488) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Posnien N, Koniszewski NDB, Hein HJ, Bucher G. 2011. Candidate gene screen in the red flour beetle Tribolium reveals six3 as ancient regulator of anterior median head and central complex development. PLoS Genet. 7, e1002416 ( 10.1371/journal.pgen.1002416) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei Z, Yaguchi J, Yaguchi S, Angerer RC, Angerer LM. 2009. The sea urchin animal pole domain is a Six3-dependent neurogenic patterning center. Development 136, 1179–1189. ( 10.1242/dev.032300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Homem CCF, Knoblich JA. 2012. Drosophila neuroblasts: a model for stem cell biology. Development 139, 4297–4310. ( 10.1242/dev.080515) [DOI] [PubMed] [Google Scholar]
- 58.Paridaen JTML, Huttner WB. 2014. Neurogenesis during development of the vertebrate central nervous system. EMBO Rep. 15, 351–364. ( 10.1002/embr.201438447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pierfelice T, Alberi L, Gaiano N. 2011. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron 69, 840–855. ( 10.1016/j.neuron.2011.02.031) [DOI] [PubMed] [Google Scholar]
- 60.Marlow H, Roettinger E, Boekhout M, Martindale MQ. 2012. Functional roles of Notch signaling in the cnidarian Nematostella vectensis. Dev. Biol. 362, 295–308. ( 10.1016/j.ydbio.2011.11.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Layden MJ, Martindale MQ. 2014. Non-canonical Notch signaling represents an ancestral mechanism to regulate neural differentiation. EvoDevo. 5, 30 ( 10.1186/2041-9139-5-30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richards GS, Degnan BM. 2012. The expression of Delta ligands in the sponge Amphimedon queenslandica suggests an ancient role for Notch signaling in metazoan development. EvoDevo. 3, 15 ( 10.1186/2041-9139-3-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gazave E, Lapébie P, Richards GS, Brunet F, Ereskovsky AV, Degnan BM, Borchiellini C, Vervoort M, Renard E. 2009. Origin and evolution of the Notch signalling pathway: an overview from eukaryotic genomes. BMC Evol. Biol. 9, 249 ( 10.1186/1471-2148-9-249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bertrand N, Castro DS, Guillemot F. 2002. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517–530. ( 10.1038/nrn874) [DOI] [PubMed] [Google Scholar]
- 65.Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan BM, Vervoort M. 2007. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol. Biol. 7, 33 ( 10.1186/1471-2148-7-33) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Münder S, et al. 2013. Notch-signalling is required for head regeneration and tentacle patterning in Hydra. Dev. Biol. 383, 146–157. ( 10.1016/j.ydbio.2013.08.022) [DOI] [PubMed] [Google Scholar]
- 67.Guth SIE, Wegner M. 2008. Having it both ways: Sox protein function between conservation and innovation. Cell Mol. Life Sci. 65, 3000–3018. ( 10.1007/s00018-008-8138-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bylund M, Andersson E, Novitch BG, Muhr J. 2003. Vertebrate neurogenesis is counteracted by Sox1–3 activity. Nat. Neurosci. 6, 1162–1168. ( 10.1038/nn1131) [DOI] [PubMed] [Google Scholar]
- 69.Sandberg M, Källström M, Muhr J. 2005. Sox21 promotes the progression of vertebrate neurogenesis. Nat. Neurosci. 8, 995–1001. ( 10.1038/nn1493) [DOI] [PubMed] [Google Scholar]
- 70.Jager M, Quéinnec E, Le Guyader H, Manuel M. 2011. Multiple Sox genes are expressed in stem cells or in differentiating neuro-sensory cells in the hydrozoan Clytia hemisphaerica. EvoDevo. 2, 12 ( 10.1186/2041-9139-2-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Royo JL, et al. 2011. Transphyletic conservation of developmental regulatory state in animal evolution. Proc. Natl Acad. Sci. USA 108, 14 186–14 191. ( 10.1073/pnas.1109037108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morin X, Bellaïche Y. 2011. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev. Cell. 21, 102–119. ( 10.1016/j.devcel.2011.06.012) [DOI] [PubMed] [Google Scholar]
- 73.Jager M, Queinnec E, Chiori R, Le Guyader H, Manuel M. 2008. Insights into the early evolution of SOX genes from expression analyses in a ctenophore. J. Exp. Zool. B, Mol. Dev. Evol. 310, 650–667. ( 10.1002/jez.b.21244) [DOI] [PubMed] [Google Scholar]
- 74.Marlow H, Arendt D. 2014. Evolution: ctenophore genomes and the origin of neurons. Curr. Biol. 24, R757–R761. ( 10.1016/j.cub.2014.06.057) [DOI] [PubMed] [Google Scholar]
- 75.Jékely G, Paps J, Nielsen C. 2015. The phylogenetic position of ctenophores and the origin(s) of nervous systems. EvoDevo. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Steele RE, David CN, Technau U. 2011. A genomic view of 500 million years of cnidarian evolution. Trends Genet. 27, 7–13. ( 10.1016/j.tig.2010.10.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Richards GS, Simionato E, Perron M, Adamska M, Vervoort M, Degnan BM. 2008. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Curr. Biol. 18, 1156–1161. ( 10.1016/j.cub.2008.06.074) [DOI] [PubMed] [Google Scholar]
- 78.Kozmik Z, Daube M, Frei E, Norman B, Kos L, Dishaw LJ, Noll M, Piatigorsky J. 2003. Role of Pax genes in eye evolution: a cnidarian PaxB gene uniting Pax2 and Pax6 functions. Dev. Cell. 5, 773–785. ( 10.1016/S1534-5807(03)00325-3) [DOI] [PubMed] [Google Scholar]
- 79.Li X, Martinson AS, Layden MJ, Diatta FH, Sberna AP, Simmons DK, Martindale MQ, Jegla TJ. 2015. Ether-à-go-go family voltage-gated K+ channels evolved in an ancestral metazoan and functionally diversified in a cnidarian–bilaterian ancestor. J. Exp. Biol. 218, 526–536. ( 10.1242/jeb.110080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jegla T, Marlow HQ, Chen B, Simmons DK, Jacobo SM, Martindale MQ. 2012. Expanded functional diversity of shaker K+ channels in cnidarians is driven by gene expansion. PLoS ONE 7, e51366 ( 10.1371/journal.pone.0051366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martinson AS, van Rossum DB, Diatta FH, Layden MJ, Rhodes SA, Martindale MQ, Jegla T. 2014. Functional evolution of Erg potassium channel gating reveals an ancient origin for IKr. Proc. Natl Acad. Sci. USA 111, 5712–5717. ( 10.1073/pnas.1321716111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moran Y, Barzilai MG, Liebeskind BJ, Zakon HH. 2015. Evolution of voltage-gated ion channels at the emergence of metazoa. J. Exp. Biol. 218, 515–525. ( 10.1242/jeb.110270) [DOI] [PubMed] [Google Scholar]
- 83.Gur Barzilai M, Reitzel AM, Kraus JEM, Gordon D, Technau U, Gurevitz M, Moran Y. 2012. Convergent evolution of sodium ion selectivity in metazoan neuronal signaling. Cell Rep. 2, 242–248. ( 10.1016/j.celrep.2012.06.016) [DOI] [PMC free article] [PubMed] [Google Scholar]