Abstract
Background
Corynebacterium glutamicum is generally regarded as a safe microorganism and is used to produce many biochemicals, including l-glutamate. 5-Aminolevulinic acid (ALA) is an l-glutamate derived non-protein amino acid, and is widely applied in fields such as medicine and agriculture.
Results
The products of the gltX, hemA, and hemL genes participate in the synthesis of ALA from l-glutamate. Their annotated C. glutamicum homologs were shown to be functional using heterologous complementation and overexpression techniques. Coexpression of hemA and hemL in native host led to the accumulation of ALA, suggesting the potential of C. glutamicum to produce ALA for research and commercial purposes. To improve ALA production, we constructed recombinant C. glutamicum strains expressing hemA and hemL derived from different organisms. Transcriptome analysis indicated that the dissolved oxygen level and Fe2+ concentration had major effects on ALA synthesis. The downstream pathway of heme biosynthesis was inhibited using small molecules or introducing genetic modifications. Small-scale flask cultures of engineered C. glutamicum produced 1.79 g/L of ALA.
Conclusion
Functional characterization of the key enzymes indicated complex regulation of the heme biosynthetic pathway in C. glutamicum. Systematic analysis and molecular genetic engineering of C. glutamicum may facilitate its development as a system for large-scale synthesis of ALA.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-015-0364-8) contains supplementary material, which is available to authorized users.
Keywords: Corynebacterium glutamicum, 5-Aminolevulinic acid, Heme biosynthesis, Metabolic engineering
Background
Corynebacterium glutamicum, which occurs naturally in soil, is a Gram-positive, nonpathogenic, biotin auxotroph bacterium that is used for large-scale industrial production of the flavor enhancer l-glutamate and several other amino acids [1]. Recent studies demonstrate the potential of C. glutamicum to produce a variety of other commercially interesting compounds such as organic acids, diamines, and biofuels [2]. Because of its importance to industrial biotechnology, C. glutamicum serves as a prominent model organism for studying prokaryotic metabolism and its regulation as well as providing a subject for applying the tools and concepts of synthetic biology [3].
The iron-containing tetrapyrrole heme is a cofactor of the protein components of the electron transport chain that drives aerobic and anaerobic respiration [4]. Certain central metabolic pathways and enzymes require heme for activity, although excess heme is toxic because of its reactive nature [5]. The mechanism of the biosynthesis of heme from its first precursor, 5-aminolevulinic acid (ALA), is highly conserved among organisms. However, the biosynthesis of ALA is regulated at different levels depending on species and may be subject to feedback inhibition by heme [6]. The key genes gltX, hemA, and hemL, which participate in the synthesis of ALA from glutamate, were identified in the genome of C. glutamicum using in silico techniques, including sequence alignments and the identification of domains shared with those of their functionally verified counterparts (Fig. 1). However, the sequences of these C. glutamicum genes are only 31.53, 25.32, and 47.05 % identical to those of GluRS, HemA, and HemL, respectively, of Escherichia coli, and the functions of the former are unknown.
Fig. 1.
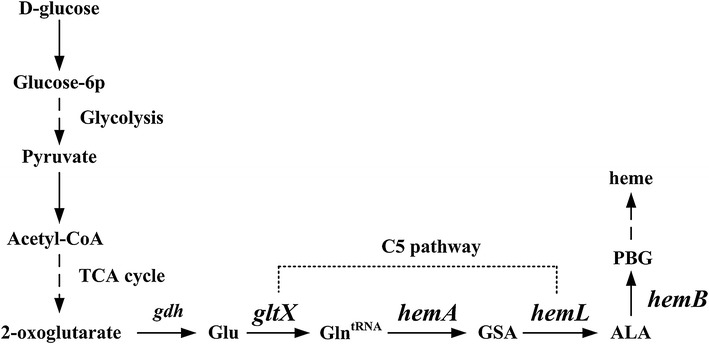
The ALA biosynthesis pathway in C. glutamicum. The enzymes encoded by the corresponding genes are: gdh glutamate dehydrogenase; gltX glutamyl-tRNA synthetase; hemA glutamyl-tRNA reductase; hemL glutamate-1-semialdehyde aminotransferase; hemB 5-aminolevulinic acid dehydratase. Glu glutamate; Gln tRNA Glutamyl-tRNA; GSA glutamate 1-semialdehyde aminotransferase; ALA 5-aminolevulinic acid; PBG porphobilinogen
5-Aminolevulinic acid is used for photodynamic therapy (PDT) in gastroenterology, urology, and dermatology and is used as well as a photosensitizer in photodynamic diagnosis [7]. Moreover, the application of low concentrations of ALA for agricultural purposes increases the tolerance of plants to low temperatures and high salt concentrations and is a biodegradable herbicide and insecticide [8]. Because the chemical synthesis of ALA is complicated and generates relatively low yields, the focus of researchers is on using microbial cell factories to synthesize ALA, because such systems are environmentally safe, economical, and sustainable [9]. For example, microbes such as Rhodobacter sphaeroides and E. coli were engineered to produce ALA [10]. However, most of these studies focused on the condensation reaction of succinyl-CoA and glycine that is catalyzed by ALA synthase (C4 pathway) [11]. The C5 biosynthetic pathway was recently engineered in E. coli to achieve yields of 4.13 g/L using batch fermentation [12]. The synthesis of ALA from glucose by the C5 pathway is an important advantage over the C4 pathway. The heme pathway was further optimized, and its regulatory factors such as small RNA ryhB were studied as well [13, 14].
Because l-glutamate is a precursor of ALA, and glutamate-producing C. glutamicum is generally regarded as safe, we reasoned that it might serve as an ideal host for the production of ALA. For this purpose, we conducted heterologous complementation and overexpression experiments, and demonstrate here, for the first time to our knowledge, the function of the key C. glutamicum enzymes involved in heme biosynthesis. Moreover, an engineered C. glutamicum strain produced ALA at yields of 1.79 g/L.
Results
Functional identification of the key genes involved in ALA biosynthesis
5-Aminolevulinic acid is a direct derivative of the tricarboxylic acid (TCA) cycle in the C5 pathway of C. glutamicum, which was predicted by analysis of the KEGG database or in previous studies [15]. However, there is no biochemical evidence, to our knowledge, that demonstrates the role of any component of the pathway. Three C. glutamicum genes, gltX (NP_600515.1), hemA (NP_599664.2), and hemL (NP_599684.1) were predicted to encode the enzymes involved in ALA biosynthesis. However, the predicted amino acid sequences of these genes are only 31.53, 25.32, and 47.05 % identical to their respective cognate counterparts in E. coil, indicating they need functional verification. C. glutamicum gltX gene was predicted to encode a glutamyl-tRNA synthetase (GluRS). Conserved Domain Search showed that the conserved HIGH and KMSKS motifs also existed in the catalytic domain of C. glutamicum GluRS (Additional file 1: Figure S1). GluRS from C. glutamicum probably belonged to the Class I synthetase that aminoacylates the 3′ hydroxyl group of the cognate tRNA [16]. Glutamyl-tRNA reductase (HemA) was predicted to be encoded by hemA. The reactive sulfhydryl group (Cys-50) of E. coli HemA requires Mg2+ to attack the α-carbonyl group of glutamyl-tRNA [17], and Cys-51 of C. glutamicum HemA is presumably similar to Cys-50 of the active site of E. coli HemA. Gly-197, Arg-222, Asp-285, and Pro-289 are conserved within the NADPH binding domain and may bind NADPH. The predicated HemL sequence contains a pyridoxal 5′-phosphate binding domain, which may participate in amino acid transfer [18]. These features suggest a functional relationship to their E. coli counterparts.
To verify these assumptions, the three genes from C. glutamicum were subcloned into pUC19 (designated pGX, pHA, and pHL, respectively) and tested for complementation of the respective E. coli mutants. The E. coli mutants JP1449, SASX41B, and GE1377 harboring a defective gltX, hemA, or hemL mutant, respectively, were unable to grow in minimal medium containing glucose but grew on solid medium when transformed with the plasmids encoding the cognate C. glutamicum genes. Further, the recombinant strains cultivated in LB medium (Fig. 2a–c) grew to higher optical densities, measured at 600 nm (OD600), than the controls, verifying the anaplerotic functions of these three genes. Moreover, adding ALA to cultures of the control strains increased cell growth to the levels of the recombinants, suggesting that ALA is a key intermediate that supports the growth of the mutants. To further characterize these genes, pGX, pHA, and pHL were used to transform wild-type E. coli DH5α to generate the E. coli strains PDGX, PDHA, and PDHL, which were cultivated in modified minimal medium containing 20 g/L glucose, and analyzed for ALA production. Although OD600 values of PDGX, PDHA, and PDHL were lower than that of wild-type E. coli DH5α, ALA production by each strain was higher than that of each control (Fig. 2d). These results suggest that these putative gltX, hemA, and hemL genes encode glutamyl-tRNA synthetase, glutamyl-tRNA reductase, and glutamate-1-semialdehyde aminotransferase, respectively.
Fig. 2.
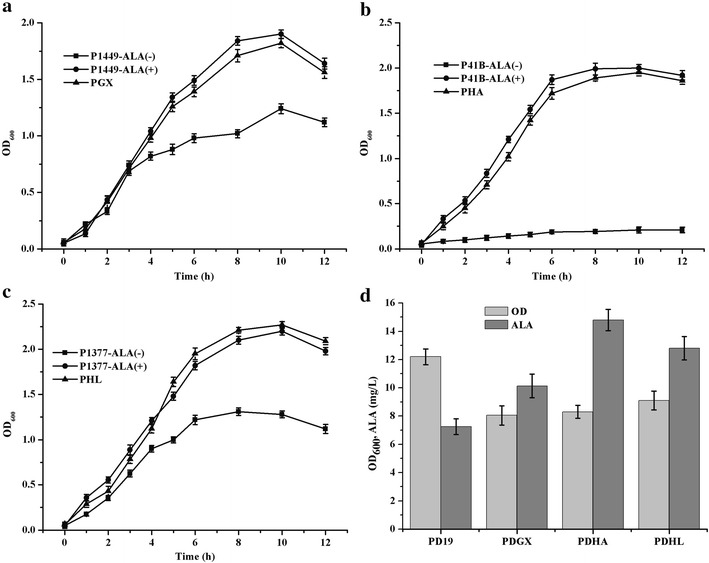
The complementation and ALA accumulation experiments in recombinant E. coli. a The ALA auxotrophic E. coli JP1449 containing pGX. b The ALA auxotrophic E. coli SASX41B containing pHA. c The ALA auxotrophic E. coli GE1377containing pHL. d The ALA accumulation in recombinant E. coli DH5α expressing gltX, hemA and hemL from C. glutamicum. Complementation experiments using the ALA auxotrophic strains were grown in LB medium supplemented with 50 μg/mL ALA (ALA+) or without ALA (ALA−). The wild type E. coli DH5α was respectively transformed with pUC19, pGX, pHA and pHL, generating the strain PD19, PDGX, PDHA and PDHL. Samples were taken and measured at 24 h with an interval of 4 h. The results were the average of three individual experiments
The shuttle vector pECXK99E was used to overexpress C. glutamicumgltX, hemA, and hemL (PEGX, PEHA, and PEHL, respectively) in their native host. Only PEHA produced increased levels of ALA (Table 1). These data indicate that the conversion of glutamyl-tRNA to glutamate-1-semialdehyde catalyzed by HemA is likely a rate-limiting step. Coexpression of hemL and hemA significantly improved ALA production, which was not increased by further overexpression of gltX, suggesting that a complex mechanism regulates the activity of the heme biosynthetic pathway in C. glutamicum (Table 1).
Table 1.
ALA accumulation in recombinant C. glutamicum expressing endogenous gltX, hemA and hemL
| C. glutamicum strains | Expressed genes | Cell biomass (OD600) | Glutamate (g/L) | ALA accumulation (mg/L) | PBG accumulation (mg/L) | Heme accumulation (mg/L) |
|---|---|---|---|---|---|---|
| PECX | – | 18.84 ± 0.76 | 1.9 ± 0.14 | 25.44 ± 1.56 | 1.26 ± 0.26 | 0.12 ± 0.05 |
| PEGX | gltX | 18.62 ± 0.98 | 1.85 ± 0.17 | 20.88 ± 1.34 | 1.27 ± 0.25 | 0.14 ± 0.04 |
| PEHA | hemA | 19.11 ± 1.56 | 1.95 ± 0.15 | 56.89 ± 9.21 | 3.16 ± 0.23 | 0.56 ± 0.12 |
| PEHL | hemL | 18.24 ± 0.87 | 2.05 ± 0.24 | 24.93 ± 2.53 | 1.21 ± 0.41 | 0.13 ± 0.08 |
| CGAL | hemA, hemL | 17.63 ± 0.46 | 3.55 ± 0.49 | 79.84 ± 6.56 | 9.46 ± 0.54 | 0.82 ± 0.28 |
| PALX | hemA, hemL, gltX | 17.86 ± 0.64 | 2.75 ± 0.31 | 64.55 ± 3.61 | 11.12 ± 0.32 | 1.05 ± 0.23 |
Samples were taken and measured until 65 h and cultivation was performed at 180 rpm
40 g/L glucose was added initially
Results are the average of three independent experiments
Engineering C. glutamicum to enhance ALA biosynthesis
Overexpression of endogenous hemA and hemL in C. glutamicum improved ALA production as well as that of its precursor, glutamate (Table 1), suggesting the potential use of C. glutamicum to produce ALA. To improve ALA production further, we employed genes required for its synthesis from other organisms (Table 2). We generated recombinant C. glutamicum strains designated CEAL, SCAL, SEAL, PSEC, and PSEE that coexpressed hemA with hemL and gltX from different organisms (Table 2). All five strains produced ALA, and the yield of SEAL was the highest (425 mg/L). Further, the acetate and lactate concentrations were low and will benefit ALA production by C. glutamicum (Table 2). The cells and media turned red after approximately 8 h, indicating the formation of specific intermediates (data not shown). Analysis of downstream metabolites revealed the accumulation of porphobilinogen (PBG) and heme. The relative increases in the concentrations of ALA corresponded to those of PBG and heme, except that strains PSEC and PSEE produced PBG and heme but not ALA (Table 2). The expression of gltX in E. coli increases the expression of hemB (encoding 5-aminolevulinic acid dehydratase, ALAD), which may contribute to the synthesis of more porphyrin derivatives or heme [12]. However, the mechanism in C. glutamicum is not clear. Taken together, the data reveal that the activity of the Salmonella arizona mutant hemAM was active in C. glutamicum and that its coexpression with hemL greatly increased ALA accumulation. Since ALA, as the precursor of heme biosynthetic pathway, accumulated in the engineered strain, the accumulation of downstream metabolites may also increase. As aforementioned, the heme biosynthetic pathway was complicatedly regulated and was hard to be regulated. In order to improve ALA production, some useful strategies should be explored and applied.
Table 2.
Engineering the ALA production in C. glutamicum using the gltX, hemA and hemL from different sources
| C. glutamicum strains | Expressed genes | Cell biomass (OD600) | ALA (mg/L) | Lactate (g/L) | Acetate (g/L) | PBG (mg/L) | Heme (mg/L) |
|---|---|---|---|---|---|---|---|
| PECX | – | 18.84 ± 0.76 | 25.44 ± 1.56 | 7.45 ± 0.46 | 32.41 ± 1.27 | 1.26 ± 0.26 | 0.12 ± 0.05 |
| CEAL | hemA (C. glutamicum) and hemL (E. coli) | 18.07 ± 0.84 | 128.13 ± 8.94 | 4.25 ± 0.19 | 26.24 ± 2.44 | 21.34 ± 2.47 | 1.54 ± 0.28 |
| SCAL | hemA M (S. arizona) and hemL (C. glutamicum) | 17.75 ± 0.72 | 83.47 ± 7.33 | 0.19 ± 0.07 | 4.23 ± 0.59 | 11.26 ± 1.24 | 0.98 ± 0.18 |
| SEAL | hemA M (S. arizona) and hemL (E. coli) | 18.33 ± 0.88 | 425.11 ± 15.69 | 0.04 ± 0.008 | 0.28 ± 0.03 | 97.87 ± 4.02 | 3.18 ± 0.54 |
| PSEC | hemA M (S. arizona), hemL (E. coli) and gltX (C. glutamicum) | 16.81 ± 0.74 | 251.36 ± 10.11 | 0.17 ± 0.03 | 1.45 ± 0.12 | 117.92 ± 2.49 | 4.15 ± 0.75 |
| PSEE | hemA M (S. arizona), hemL (E. coli) and gltX (E. coli) | 16.45 ± 0.78 | 276.21 ± 9.26 | 0.14 ± 0.05 | 1.12 ± 0.34 | 121.38 ± 2.53 | 4.22 ± 0.62 |
Fermentations were performed at 180 rpm for 144 h, and the initial glucose concentration was 40 g/L
Results are the means ± standard deviations in three individual experiments
Transcriptional analysis to identify key factors that affect ALA synthesis
SEAL cells and the culture medium were red, indicating the presence of porphyrins and heme, which may affect metabolism. It is hypothesized therefore that specific sets of genes were differentially regulated in the SEAL strain to restore metabolic homeostasis. To optimize culture conditions to increase ALA synthesis, we compared the gene expression levels of the SEAL cells with those of the control strain PECX. Total RNA was isolated when the cultivation medium turned red (after approximately 8 h). Among the nearly 3000 genes analyzed, most were down-regulated and were identified as those involved in glucose transport, glycolysis, respiratory chain, or ATP synthesis (Additional file 1: Table S2). Specifically, the levels of ldh and ackA mRNAs required for the synthesis of lactate and acetate, respectively, were down-regulated. Consistent with these data, the SEAL strain exhibited reduced glycolytic activity indicated by slower acidification of the medium, which correlated with delayed consumption of glucose and decreased secretion of lactate and acetate (Table 2). Moreover, the down-regulation of the transcription of genes hemH for heme synthesis, sufC and sufD for Fe-S cluster synthesis which are also involved in Fe2+ transporters, combined with the up-regulation of the transcription of genes involved in oxidative stress and protein repair may represent strategies employed by cells to avoid toxicity caused by excess concentrations of heme and Fe2+. Down-regulated electron flow through the electron transport chain, which is possibly associated with the reduced synthesis of heme and the Fe-S cluster, suggests that a high respiratory rate may adversely affect cell metabolism.
The transcriptional data suggest the likelihood that the potentially toxic mechanism of iron is connected with the high level of dissolved oxygen (DO). To address this question, we determined Fe2+ concentrations and DO levels and found that ALA production increased in proportion to the decrease in the concentration of Fe2+, and decreased DO levels were associated with increased ALA production (Table 3). When we reduced the rotation of culture flasks to 120 rpm and reduced the concentration of FeSO4·7H2O to 0.01 mg/L, ALA production increased to 830 mg/L (Table 3), indicating that low DO and Fe2+ levels increased ALA production. However, when the culture flasks were rotated at 80 rpm, ALA production decreased, suggesting that the level of DO that enhanced ALA production must be sufficiently high to support cell growth. Therefore, in the following experiments, we used 0.01 mg/L FeSO4·7H2O and rotated the culture flasks at 120 rpm, which were optimal for producing ALA.
Table 3.
ALA production in C. glutamicum SEAL containing various concentrations of Fe2+ at 180, 120 and 80 rpm
| The concentration of FeSO4·7H2O (mg/L) | ALA accumulation (mg/L) | ||
|---|---|---|---|
| 180 rpm | 120 rpm | 80 rpm | |
| 10 | 528.2 ± 17.88 | 648.2 ± 16.25 | 364.4 ± 10.32 |
| 0.2 | 573.1 ± 16.34 | 652.1 ± 17.27 | 382.3 ± 13.45 |
| 0.1 | 612.4 ± 18.32 | 702.4 ± 18.49 | 402.5 ± 14.28 |
| 0.05 | 664.5 ± 19.48 | 738.3 ± 19.24 | 446.2 ± 13.89 |
| 0.01 | 678.3 ± 15.27 | 830.2 ± 21.24 | 482.6 ± 14.12 |
| 0 | 548.7 ± 14.58 | 692.7 ± 18.44 | 418.6 ± 16.02 |
Inhibition of ALA downstream pathway using different inhibitors
Levulinic acid (LA) is a competitive inhibitor of the downstream metabolic pathway of ALA and is used to enhance ALA production [19]. For the first time to our knowledge, we used the other inhibitors such as maleic acid (MA), phthalic acid (PA), and 2-nitrobenzoic acid (NA) for this purpose. These inhibitors did not influence growth but did affect the accumulation of ALA and PBG (Fig. 3). Addition of LA, MA and PA improved ALA production with the decrease of PBG accumulation. However, NA did not significantly increase the production of ALA (data not shown). ALA production gradually increased when cultures were treated with concentrations of LA from 2.5 to 15 mM. ALA production was significantly decreased in the presence of 30 mM LA. Further, PBG production decreased in the presence of 2.5–30 mM LA. In contrast, low concentrations of MA and PA increased ALA production, which reached 1289 and 1507 mg/L in the presence of 0.3 mM MA and 0.1 mM of PA, respectively. Compared with the control, the PBG accumulation was also decreased, and exhibited different behaviors in compare with the presence of LA. These results suggest that the inhibition of downstream metabolic pathway of ALA affected cell metabolism and that this strategy increases ALA production.
Fig. 3.
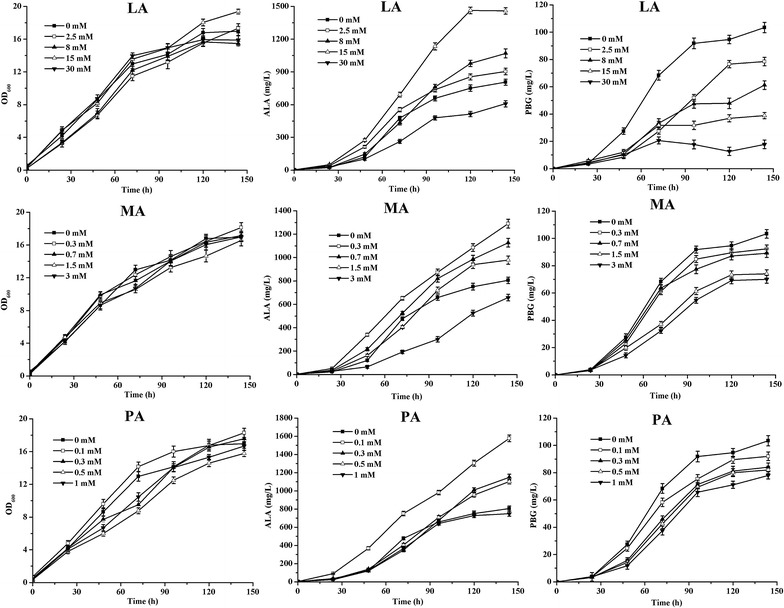
Effects on the growth, ALA and PBG accumulation in C. glutamicum SEAL with addition of LA, MA and PA. 40 g/L glucose was added initially as the sole carbon source. The results are the average of three individual experiments
Molecular genetic modifications that down-regulate ALAD activity
Because inhibition of the downstream heme biosynthesis increased the production of ALA, and hemB (encoding ALAD) is required for viability, we added a degradation tag into the C-terminus of ALAD to increase its rate of degradation. The predicted amino acid sequence of C. glutamicumhemB (NP_599678.1) showed 46.9 % identity to its E. coli counterpart; however, the function of the protein encoded by C. glutamicum hemB is unknown. Corynebacterial SsrA tags vary in their three C-terminal amino acid residues and are designated ASV (AAEKSQRDYAASV) and AAV (AAEKSQRDYAAAV) [20]. The ASV tag was successfully integrated into the C-terminus of ALAD, but the AAV tag was failed probably due to its strong degradation. The plasmids pSEAL that expressed hemAM (S. arizona) and hemL (E. coli) were used to transform C. glutamicum that expressed ALAD with the C-terminal ASV tag (SEAL1). Cells that expressed SEAL1 produced increased concentrations of ALA (1.79 g/L), and the concentrations of PBG in the medium were reduced (Fig. 4), suggesting that the predicated hemB gene encodes ALAD and that the degradation of ALAD increased ALA production.
Fig. 4.
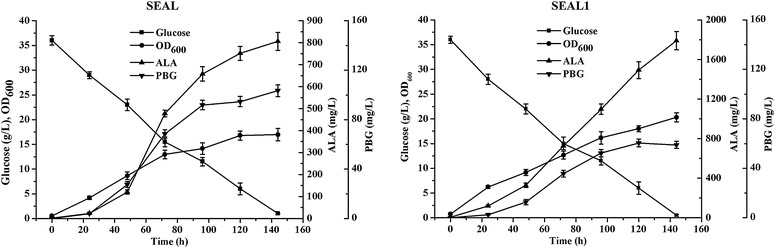
The growth, ALA and PBG accumulation in C. glutamicum SEAL and SEAL1. SEAL contains the plasmid pSEAL, and SEAL1 contains the plasmid pSEAL and the ASV tag in the C-terminus of ALAD. 40 g/L glucose was added initially as the sole carbon source. The results are the average of three independent experiments
Discussion
In the present study, we determined the functions of key C. glutamicum genes predicted to encode enzymes that mediate heme biosynthesis. Previous studies determined that HemA is a major control point for ALA biosynthesis [12]. However, our results indicate that the relative activity of HemA produced by C. glutamicum is not high. The HemA activity of S. typhimurium is high; however, the enzyme is not stable in the presence of heme [21, 22]. Therefore, the stability of HemA was increased by inserting two Lys residues (AAGAAG) between Thr-2 and Leu-3 at its N-terminus [21]. We show here that expression of HemAM from S. arizona with E. coli HemL by the recombinant strain designated SEAL produced the highest level of ALA, which indicates that these enzymes synergize, likely because of the formation of a tight complex. However, the glutamate was also detected in the culture medium.
Several lines of evidence indicate that heme plays a major role in the regulation of tetrapyrrole biosynthesis. For example, in the presence of an excess of intracellular heme, the activity of GluRS and the level of HemA are down-regulated and decrease the production of ALA [5]. Moreover, excess heme is toxic and perturbs the central metabolism of S. aureus [5]. However, we conclude that the low rates of growth, glucose consumption, and ALA synthesis may be explained by the intracellular accumulation of inhibitory concentrations of porphyrins and heme.
Downstream products such as protoporphyrin IX are potentially toxic in the presence of oxygen, leading to the generation of reactive oxygen species [23]. Further, the addition of iron drives ALA for porphyrins and heme production and causes the cultures to become red [24]. Higher iron concentrations influence cell growth and regulate heme synthesis [12]. Iron is an important cofactor involved in the formation of iron-sulphur (Fe-S) clusters and the activity of heme, which are components of oxygen-binding and regulatory proteins as well as those of the electron transport chain [25]. When the iron concentration is in excess, it may trigger the Fenton reaction and cause oxidative stress [26]. However, the suf operon is specifically adapted to synthesize Fe-S clusters under iron-limiting conditions [27], and, according to the transcriptome analysis of SEAL performed here, the synthesis of the Fe-S cluster will likely decrease when iron is in excess. Moreover, when the iron concentration decreased, heme biosynthesis was likely reduced, which may relieve feedback inhibition of the expression of GluRS and HemA to the benefit of ALA synthesis. It has been demonstrated in vertebrate that the homeostatic mechanisms that regulate iron and oxygen interact, and the balance between the contents of protoheme and iron to form active heme is critical [28]; however, their homeostatic concentrations in bacteria are unknown. It is interesting to note that low concentrations of Fe2+ and DO reduced the inhibitory effects of porphyrins and heme on ALA synthesis by C. glutamicum. Toxicity caused by nonspecific redox cycling occurs between heme and oxygen when excess heme accumulates within the cell membrane [29], and the reduced rates of respiration and heme biosynthesis may mitigate this toxicity. We hypothesize therefore that low levels of DO and Fe2+ may compensate for the excess iron available to form Fe-S clusters and to synthesize heme. Such a mechanism may adjust to down-regulate heme synthesis and electron transport under these conditions. The data presented are consistent with this hypothesis, because they indicate that reducing the levels of stressors and heme represents an efficient strategy to increase ALA production.
A hemB mutant of E. coli does not produce higher levels of ALA compared with wild-type [30], while d-glucose and d-xylose, which were used to inhibit the expression of ALAD, are usually metabolized [11]. Therefore, only LA is typically used to increase ALA production by R. sphaeroides [19, 31, 32], Chlorella vulgaris [33], and E. coli [34]. For example, 15 mmol/L LA inhibits ALAD activity by 60 % at pH 6.5 [19]. Studies of compounds with different inhibitory activities reveal that MA and PA increased ALA production and decreased the production of PBG [35]. These inhibitors interact at distinct steps of the reaction mechanism [36], and different classes of inhibitors exhibit significant differences in influencing the kinetics of the enzyme reactions that produce ALA [35]. PA may serve as a more desirable ALAD inhibitor, because PA is relatively inexpensive and is a highly efficient inhibitor.
In E. coli, protein degradation occurs, in part, through the tmRNA system. The C-terminal fusion of SsrA guides proteins to endogenous ClpXP, ClpAP, or FtsH proteases for rapid degradation. Variants of the E. coli SsrA tag are used to modify the degradation of proteins [37]. Moreover, ssrA is highly conserved, and a homologue is annotated in the C. glutamicum genome [38]. The SsrA tag is used to monitor dynamic gene expression patterns in C. glutamicum [20]. Here, the addition of the SsrA tag significantly decreased the production of PBG and increased ALA production in the absence of other inhibitors, indicating that ALAD is a key regulatory point in the heme biosynthetic pathway and that genetic modification of ALAD will be useful for improving ALA production.
Conclusion
In summary, we identified several key genes involved in heme biosynthesis and demonstrated their function in complementation and overexpression experiments. Recombinant C. glutamicum strains that produce ALA were constructed by expressing hemA and hemL from different organisms. Using a strategy that included transcriptome analysis and genomic modifications, we engineered C. glutamicum (strain SEAL1) to produce 1.79 g/L of ALA in a small-scale flask culture. ALA synthesis will likely be improved by further optimizing culture conditions using a rigorously controlled fermenter.
Methods
Strains, primers and plasmids
All strains, plasmids, and oligonucleotides used in this study are summarized in the additional files (see Additional file 1: Tables S3 and Table S4). The gltX, hemA, and hemL genes were amplified from the wild-type C. glutamicum ATCC 13032 genome using primers gltx-F, gltx-R and hema-F, hema-R, and heml-F, and heml-R, respectively. The three amplicons were ligated to pUC19, which was digested with SalI (Fermentas, China) according to the method of Gibson assembly that included T5 exonuclease (Epicentre, USA), Phusion DNA polymerase (New England Biolabs, USA), and Taq DNA ligase (New England Biolabs, USA) [39]. Moreover, gltX, hemA, and hemL were cloned from the genome of C. glutamicum ATCC 13032 using the primers cggltx-F, cggltx-R and cghema-F, cgdhema-R, and cgdheml-F, cgheml-R. To enhance the stability of HemA, hemA was cloned from the S. arizona genome, which encodes the same predicted HemA amino acid sequence of S. typhimurium. For this purpose, the primers with mutations (sthemAM-F and sthemAM-R) were used to insert two codons (AAGAAG) encoding Lys between Thr-2 and Leu-3 at the N-terminus [22]. One hemL gene was amplified using the primers echeml-F and echeml-R designed according to the sequence of the wild-type E. coli MG1655 genome, and hemL was cloned using primers cgheml-F and cgheml-R designed according to the sequence of the C. glutamicum ATCC 13032 genome. These amplicons contained 30-bp overlaps and were ligated individually or together into pECXK99E that was cleaved with KpnI (Fermentas, China) using the Gibson assembly method. The gltX genes were cloned from the genomes of E. coli MG1655 and C. glutamicum ATCC 13032 using the primers mgltx-F, mgltx-R and cgltx-F, cgltx-R and then ligated into pSEAL digested with PstI using Gibson assembly. E. coli DH5α served as the host for molecular cloning and construction of plasmids. The inducible suicide vector pKJL, which was constructed from pK18mobsacB, was used to add ASV and AAV tags to the C-terminus of ALAD encoded by the C. glutamicum ATCC 13032 genome [40]. The sequences of the plasmids were verified by the BioSune Company (Shanghai, China). Transformation of ALA auxotrophs and E. coli DH5α was achieved using the CaCl2 procedure, whereas C. glutamicum was electrophoretically transformed [41]. C. glutamicum strain ATCC 13032 was used as parental strain for ALA production.
Media and culture conditions
LB medium (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl, pH 7.2) and BHIS medium (2.5 g/L beef extract, 5 g/L tryptone, 5 g/L NaCl, 18.5 g/L brain heart infusion, and 91 g/L sorbitol) were for molecular genetic procedures. Kanamycin (50 μg/mL) and ampicillin (100 μg/mL) were added as selective agents as required. E. coli was cultured using modified minimal medium [12] (16 g/L (NH4)2SO4, 3 g/L KH2PO4, 16 g/L Na2HPO4·12H2O, 1 g/L MgSO4·7H2O, 0.01 g/L MnSO4·7H2O, 2 g/L yeast extract, and 20 g/L glucose. C. glutamicum was cultured using a modified minimal medium (CGXII) containing glucose as the sole carbon source. CGXII contains (per liter) 20 g of (NH4)2SO4, 5 g of urea, 1 g of KH2PO4, 1 g of K2HPO4, 0.25 g of MgSO4·7H2O, 42 g of MOPS (3-morpholinopropanesulfonic acid), 10 mg of CaCl2, 10 mg of FeSO4·7H2O, 10 mg of MnSO4·H2O, 1 mg of ZnSO4·7H2O, 0.2 mg CuSO4, 0.02 mg NiCl2·6H2O, 0.02 g citrate sodium, and pH 7.0. To induce the expression of plasmid genes, isopropyl-β-d-thiogalactopyranoside was added to the cultures at a final concentration of 0.25 mM.
ALA was produced at 30 °C in 300 mL using ordinary flasks containing 50 mL modified CGXII. Starter cultures contained 5 ml LB with glucose were routinely inoculated with a single colony from a freshly streaked agar plate, the culture flasks were rotated at 180 rpm for 14 h at 30 °C, and the cells were harvested, and washed with sterile 0.9 % NaCl and CGXII containing 5.0 μg/L biotin. The resuspended cells were used to inoculate a second culture containing 5.0 μg/L biotin, and the initial OD600 was adjusted to approximately 0.5. After approximately 12 h incubation at 30 °C at 170 rpm, the cells in the exponential growth phase were harvested and washed sequentially with sterile 0.9 % NaCl solution and CGXII containing 1.0 μg/L biotin and inoculated into the main culture that was adjusted to approximately 0.8 OD600. The initial pH was adjusted to 7.0 (optimum), then maintained at approximately pH 6.5 using 4 M NaOH. The inhibitors LA, MA, PA, and NA were added at approximately 8 h.
Substrate, product, and RNA-Seq analysis
For quantification of extracellular glucose, glutamate, and ALA, aliquots of the culture were withdrawn and the cells were removed by centrifugation (12,000×g, 10 min). The OD600 was determined using a spectrophotometer (Shimadzu, Japan), and the supernatant was analyzed for glucose and glutamate using a SBA-40C biosensor (developed by Biology Institute of Shandong Academy of Sciences) equipped with glucose and glutamate oxidase immobilized on membranes. To determine the concentrations of organic acids, high-performance liquid chromatography (HPLC) was used (Shimadzu). The supernatant was filtered through a 0.22-mm syringe filter. The HPLC system was equipped with a HPX-87H column (300 mm × 7.8 mm, Bio-Rad, USA) and a differential refractive index (RI) detector (Shimadzu RID-10A). The mobile phase (0.5 mM H2SO4) was delivered at 0.6 mL/min at 65 °C. To measure ALA and PBG concentrations, we used modified Ehrlich’s reagent and measured absorbance at 554 nm [42]. For heme measurements, a fluorescence assay was used [43]. RNA-Seq was performed according to the Illumina mRNA Sequencing Sample Preparation Guide (Illumina), and sequenced using an Illumina HiSeq sequencer (Illumina) at BGI Tech Company (Shenzhen, China). The raw sequencing data were analyzed using the software included with the system (Illumina).
Authors’ contributions
XLY planned and conducted the experiments, analyzed and interpreted data, and wrote the manuscript; HYJ and WJL participated in the research; QSQ and QW supervised the research and finalized the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was funded by grants from the National Natural Science Foundation of China (31370085), the National High-Tech Research and Development Plan of China (2012AA022104) and the Independent Innovation and Achievements Transformation of Shandong Province (201422CX02602).
Competing interests
The authors declare that they have no competing interests.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional file
10.1186/s12934-015-0364-8 Figure S1. Multiple-sequence alignment of GluRS (A), HemA (B), and HemL (C) from E. coli and C. glutamicum. Table S1. Strains and plasmids used in this study. Table S2. Primers used in this study. Table S3. Down-regulated genes (-) and up-regulated genes (+) were identified using comparative transcriptome analysis of the C. glutamicum strains SEAL and PECX.
Contributor Information
Xiaoli Yu, Email: yuxl0814@163.com.
Haiying Jin, Email: 1534785275@qq.com.
Wenjing Liu, Email: 15138753767@126.com.
Qian Wang, Phone: +86-531-88362897, Email: qiqi20011983@gmail.com.
Qingsheng Qi, Phone: +86-531-88365628, Email: qiqingsheng@sdu.edu.cn.
References
- 1.Kinoshita S, Udaka S, Shimono M. Studies on the amino acid fermentation. Part 1. Production of l-glutamic acid by various microorganisms. J Gen Appl Microbiol. 2004;50:331–343. [PubMed] [Google Scholar]
- 2.Becker J, Wittmann C. Bio-based production of chemicals, materials and fuels-Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol. 2012;23:631–640. doi: 10.1016/j.copbio.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Woo HM, Park JB. Recent progress in development of synthetic biology platforms and metabolic engineering of Corynebacterium glutamicum. J Biotechnol. 2014;180:43–51. doi: 10.1016/j.jbiotec.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Nobles CL, Maresso AW. The theft of host heme by Gram-positive pathogenic bacteria. Metallomics. 2011;3:788–796. doi: 10.1039/c1mt00047k. [DOI] [PubMed] [Google Scholar]
- 5.Mike LA, Dutter BF, Stauff DL, Moore JL, Vitkoe NP, Aranmolate O, Kehl-Fie TE, Sullivan S, Reid PR, DuBois JL, Richardson AR, Caprioli RM, Sulikowski GA, Skaar EP. Activation of heme biosynthesis by a small molecule that is toxic to fermenting Staphylococcus aureus. Proc Natl Acad Sci. 2013;110:8206–8211. doi: 10.1073/pnas.1303674110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levicán G, Katz A, Armas M, Núñez H, Orellana O. Regulation of a glutamyl-tRNA synthetase by the heme status. Proc Natl Acad Sci. 2007;104:3135–3140. doi: 10.1073/pnas.0611611104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondo Y, Murayama Y, Konishi H, Morimura R, Komatsu S, Shiozaki A, Kuriu Y, Ikoma H, Kubota T, Nakanishi M, Ichikawa D, Fujiwara H, Okamota K, Sakakura C, Takahashi K, Inoue K, Nakajima M, Otsuji E. Fluorescent detection of peritoneal metastasis in human colorectal cancer using 5-aminolevulinic acid. Int J Oncol. 2014;45:41–46. doi: 10.3892/ijo.2014.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z, Li H, Zhou W, Takeuchi Y, Yoneyama K. Effect of 5-aminolevulinic acid on development and salt tolerance of potato (Solanum tuberosum L.) microtubers in vitro. Plant Growth Regul. 2006;49:27–34. [Google Scholar]
- 9.Liu SL, Zhang GM, Li XK, Zhang J. Microbial production and applications of 5-aminolevulinic acid. Appl Microbiol Biotechnol. 2014;98:7349–7357. doi: 10.1007/s00253-014-5925-y. [DOI] [PubMed] [Google Scholar]
- 10.Neidle EL, Kaplan S. Expression of the Rhodobacter sphaeroideshemA and hemT genes, encoding two 5-aminolevulinic acid synthase isozymes. J Bacteriol. 1993;175:2292–2303. doi: 10.1128/jb.175.8.2292-2303.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin J, Fu W, Cen P. Characterization of 5-aminolevulinate synthase from Agrobacterium radiobacter, screening new inhibitors for 5-aminolevulinate dehydratase from Escherichia coli and their potential use for high 5-aminolevulinate production. Bioresour Technol. 2009;100:2293–2297. doi: 10.1016/j.biortech.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Kang Z, Wang Y, Gu PF, Wang Q, Qi QS. Engineering Escherichia coli for efficient production of 5-aminolevulinic acid from glucose. Metab Eng. 2011;13:492–498. doi: 10.1016/j.ymben.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Li FF, Wang Y, Gong K, Wang Q, Liang QF, Qi QS. Constitutive expression of RyhB regulates the heme biosynthesis pathway and increases the 5-aminolevulinic acid accumulation in Escherichia coli. FEMS Microbiol Lett. 2014;350:209–215. doi: 10.1111/1574-6968.12322. [DOI] [PubMed] [Google Scholar]
- 14.Zhang JL, Kang Z, Chen J, Du GC. Optimization of the heme biosynthesis pathway for the production of 5-aminolevulinic acid in Escherichia coli. Sci Rep. 2015;5:8584. doi: 10.1038/srep08584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bott M, Niebisch A. The respiratory chain of Corynebacterium glutamicum. J Bacteriol. 2003;104:129–153. doi: 10.1016/s0168-1656(03)00144-5. [DOI] [PubMed] [Google Scholar]
- 16.Franklund CV, Goldberg JB. Cloning of the glutamyl-tRNA synthetase (gltX) gene from Pseudomonas aeruginosa. J Bacteriol. 1999;181:3582–3586. doi: 10.1128/jb.181.11.3582-3586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schauer S, Chaturvedi S, Randau L, Moser J, Kitabatake M, Lorenz S, Verkamp E, Schubert WD, Nakayashik T, Murai M, Wall K, Thomann HU, Heinz DW, Inokuchi H, Söll D, Jahn D. Escherichia coli glutamyl-tRNA reductase. J Biol Chem. 2002;277:48657–48663. doi: 10.1074/jbc.M206924200. [DOI] [PubMed] [Google Scholar]
- 18.Grimm B. Primary structure of a key enzyme in plant tetrapyrrole synthesis: glutamate semialdehyde aminotransferase. Proc Natl Acad Sci. 1990;87:4169–4173. doi: 10.1073/pnas.87.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasaki K, Watanabe M, Nishio N. Inhibition of 5-aminolevulinic acid (ALA) dehydratase by undissociated levulinic acid during ALA extracellular formation by Rhodobacter sphaeroides. Biotechnol Lett. 1997;19:421–424. doi: 10.1023/A:1018331824331. [DOI] [Google Scholar]
- 20.Hentschel E, Will C, Mustafi N, Burkovski A, Rehm N, Frunzke J. Destabilized eYFP variants for dynamic gene expression studies in Corynebacterium glutamicum. Microbial Biotechnol. 2012;6:196–201. doi: 10.1111/j.1751-7915.2012.00360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Wilson S, Elliott T. A mutant HemA protein with positive charge close to the N terminus is stabilized again stheme-regulated proteolysis in Salmonella typhimurium. J Bacteriol. 1999;181:6033–6041. doi: 10.1128/jb.181.19.6033-6041.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Elliott M, Elliott T. Conditional stability of the HemA protein (glutamyl-tRNA reductase) regulates heme biosynthesis in Salmonella typhimurium. J Bacteriol. 1999;181:1211–1219. doi: 10.1128/jb.181.4.1211-1219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kochevar IE. Mechanisms of drug photosensitization. Photochem Photobiol. 1987;45:891–895. doi: 10.1111/j.1751-1097.1987.tb07899.x. [DOI] [PubMed] [Google Scholar]
- 24.Kang Z, Zhang J, Zhou J, Qi Q, Du G, Chen J. Recent advances in microbial production of δ-aminolevulinic acid and vitamin B12. Biotechnol Adv. 2012;30:1533–1542. doi: 10.1016/j.biotechadv.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Schultz IJ, Chen C, Paw BH, Hamza I. Iron and porphyrin trafficking in heme biogenesis. J Biol Chem. 2010;285:26753–26759. doi: 10.1074/jbc.R110.119503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fränzel B, Frese C, Penkova M, Metzler-Nolte N, Bandow JE, Wolters DA. Corynebacterium glutamicum exhibits a membrane-related response to a small ferrocene-conjugated antimicrobial peptide. J Biol Inorg Chem. 2010;15:1293–1303. doi: 10.1007/s00775-010-0689-z. [DOI] [PubMed] [Google Scholar]
- 27.Outten FW, Djaman O, Storz G. A suf operon requirement for Fe–S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol. 2004;52:861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 28.Graça-Souza AV, Maya-Monteiro C, Paiva-Silva GO, Braz GR, Paes MC, Sorgine MH, Oliveira MF, Oliveira PL. Adaptations against heme toxicity in blood-feeding arthropods. Insect Biochem Mol Biol. 2006;36:322–335. doi: 10.1016/j.ibmb.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 29.Wakeman CA, Stauff DL, Zhang Y, Skaar EP. Differential activation of Staphylococcus aureus heme detoxification machinery by heme analogues. J Bacteriol. 2014;196:1335–1342. doi: 10.1128/JB.01067-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie L, Eiteman MA, Altman E. Production of 5-aminolevulinic acid by an Escherichia coli aminolevulinate dehydratase mutant that overproduces Rhodobacter sphaeroides aminolevulinate synthase. Biotechnol Lett. 2003;25:1751–1755. doi: 10.1023/A:1026035912038. [DOI] [PubMed] [Google Scholar]
- 31.Ishii K, Hiraishi A, Arai T, Kitamura H. Light-dependent porphyrin production by suspended and immobilized cells of Rhodobacter sphaeroides. J Ferment Bioeng. 1990;69:26–32. doi: 10.1016/0922-338X(90)90159-T. [DOI] [Google Scholar]
- 32.Nishikawa S, Watanabe K, Tanaka T, Miyachi N, Hotta Y, Murooka Y. Rhodobacter sphaeroides mutant which accumulate 5-aminolevulinic acid under aerobic and dark conditions. J Biosci Bioeng. 1999;87:798–804. doi: 10.1016/S1389-1723(99)80156-X. [DOI] [PubMed] [Google Scholar]
- 33.Ano A, Funahashi H, Nakao K, Nishizawa Y. Effects of levulinic acid on 5-aminolevulinic acid production in heterotrophic cultures of Chlorella regularis YA-603. J Biosci Bioeng. 2000;89:176–180. doi: 10.1016/S1389-1723(00)88733-2. [DOI] [PubMed] [Google Scholar]
- 34.Choi C, Hong BS, Sung HC, Lee HS, Kim JH. Optimization of extracellular 5-aminolevulinic acid production from Escherichia coli transformed with ALA synthase gene of Bradyrhizobium japonicum. Biotechnol Lett. 1999;21:551–554. doi: 10.1023/A:1005520007230. [DOI] [Google Scholar]
- 35.Jarret C, Stauffer F, Henz ME, Marty M, Lüönd RM, Bobálová J, Schürmann P, Neier R. Inhibition of Escherichia coli porphobilinogen synthase using analogs of postulated intermediates. Chem Biol. 2000;7:185–196. doi: 10.1016/S1074-5521(00)00089-2. [DOI] [PubMed] [Google Scholar]
- 36.Tschudy DP, Hess RA, Frykholm BC. Inhibition of delta-aminolevulinic acid dehydrase by 4,6-dioxoheptanoic acid. J Biol Chem. 1981;256:9915–9923. [PubMed] [Google Scholar]
- 37.Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Krämer R, Linke B, McHardy AC, Meyer F, Möckel B, Pfefferle W, Pühler A, Rey DA, Rückert C, Rupp O, Sahm H, Wendisch VF, Wiegräbe I, Tauch A. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J Biotechnol. 2003;104:5–25. doi: 10.1016/S0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- 39.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 40.Jiang LY, Chen SG, Zhang YY, Liu JZ. Metabolic evolution of Corynebacterium glutamicum for increased production of l-ornithine. BMC Biotechnol. 2013;13:47. doi: 10.1186/1472-6750-13-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tauch A, Kirchner O, Löffler B, Götker S, Pöhler A, Kalinowski J. Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr Microbiol. 2002;45:362–367. doi: 10.1007/s00284-002-3728-3. [DOI] [PubMed] [Google Scholar]
- 42.Burnham BF. δ-Aminolevulinicacid synthase (Rhodopseudomonas sphaeroides) Methods Enzymol. 1970;17:195–204. doi: 10.1016/0076-6879(71)17179-0. [DOI] [Google Scholar]
- 43.Sinclair PR, Gorman N, Jacobs JM. Measurement of heme concentration. Curr Protoc Toxicol. 2001;8(8):3. doi: 10.1002/0471140856.tx0803s00. [DOI] [PubMed] [Google Scholar]
- 44.Verkamp E, Chelm BK. Isolation, nucleotide sequence, and preliminary characterization of the Escherichia coli K-12 hemA Gene. J Bacteriol. 1989;171:4728–4735. doi: 10.1128/jb.171.9.4728-4735.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ilag LL, Jahn D, Eggertsson G, Söll D. The Escherichia colihemL gene encodes glutamate 1-semialdehyde aminotransferase. J Bacteriol. 1991;173:3408. doi: 10.1128/jb.173.11.3408-3413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]


