Abstract
The fitness landscape—the function that relates genotypes to fitness—and its role in directing evolution are a central object of evolutionary biology. However, its huge dimensionality precludes understanding of even the basic aspects of its shape. One way to approach it is to ask a simpler question: what are the properties of a function that assigns fitness to each possible variant at just one particular site—a single position fitness landscape—and how does it change in the course of evolution? Analyses of genomic data from multiple species and multiple individuals within a species have proved beyond reasonable doubt that fitness functions of positions throughout the genome do themselves change with time, thus shaping protein evolution. Here, I will briefly review the literature that addresses these dynamics, focusing on recent genome-scale analyses of fitness functions of amino acid sites, i.e. vectors of fitnesses of 20 individual amino acid variants at a given position of a protein. The set of amino acids that confer high fitness at a particular position changes with time, and the rate of this change is comparable with the rate at which a position evolves, implying that this process plays a major role in evolutionary dynamics. However, the causes of these changes remain largely unclear.
Keywords: epistasis, selection, proteins, macroevolution, microevolution
1. Introduction
Evolution is the change of genotype with time; adaptive evolution is associated with an increase of fitness. To understand evolution and adaptation, we need to know the fitness landscape—the function that relates genotype to fitness [1]. However, it is hard to study its shape: because the number of conceivable genotypes is huge, it is impossible to measure the fitness of each of them. A protein of length L has 20L possible variants, which is an immense number for realistic protein lengths (L > 100). Although probably the fitness conferred by the overwhelming majority of these variants is always zero, it is not clear how many viable variants exist, how they are distributed in the protein space, and how much they differ in fitness [2,3]. Complete fitness sub-landscapes—values of fitness associated with each possible combination of alleles at a subset of positions—have so far only been obtained for at most a handful of positions in experimental systems [4]. While such data are extremely helpful for informing our intuition, it is unlikely that they will ever be obtainable at genomic scale [5]. Experimental data show that real-life fitness landscapes are complex and that the large number of dimensions is biologically important [6].
Because fitness landscapes shape evolution and variation, information about their properties can be obtained from comparisons of genomes of different species, and of individuals within a species. Genomes of extant organisms, and reconstructed genomes of extinct ones, illuminate the subset of the landscape that is viable, although they cover an infinitesimal fraction of the genotype space and thus tell us little about the region of the landscape that is uninhabitable.
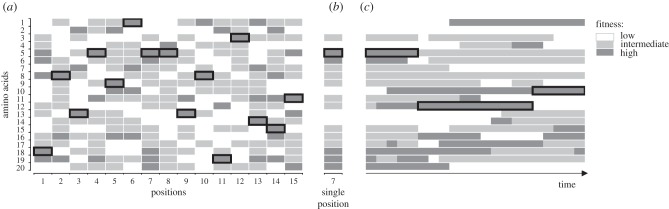
The most basic level of understanding of a fitness landscape of proteins pertains to individual amino acid positions (figure 1a). An amino acid position may be occupied by up to 20 different amino acids. The fitness values conferred by each of them comprise, for each genomic background (amino acids at other positions of the same protein, and the rest of the genome) and for each environment faced by the organism, a vector of length 20. This vector of amino acid propensities [8,9], or single position fitness landscape (SPFL), is a minimal meaningful cross-section of the complete fitness landscape (figure 1b). In the course of evolution, the SPFL may change (figure 1c) due to two reasons: changes elsewhere in the genome (i.e. when the complete fitness landscape remains invariant, but its different cross-section is considered), or environmental changes (i.e. when the complete fitness landscape changes).
Figure 1.
Single position fitness landscape (SPFL). Horizontal rows correspond to individual amino acids at a site. (a) At each moment of time, a protein can be described by the fitness values of all its one-step mutational neighbours (for simplicity, all amino acid variants are assumed to be accessible by mutation). The currently predominant amino acid at each site (surrounded by black rectangles) confers high fitness. (b) The SPFL of position 7. (c) The change of the SPFL with time; the fitness of individual amino acids at the position may increase, decrease or remain invariant owing to changes in the genomic background or of the environment. Fitness changes are modelled as a Poisson process, as in [7].
While this single-site approach obviously may provide only a very limited understanding of the properties of the complete fitness landscape, it is applicable at the genome scale, and much of the accumulated comparative genomics data is relevant to it. Here, I will briefly review some evidence that SPFLs of amino acid sites change with time, and some general characteristics of this change. I will mainly focus on the statistical evidence obtained from genome-level patterns of variation between and within species, thus omitting much other relevant lower-level data. In particular, experimental evidence for epistasis—changes in SPFL owing to changes in the genomic background—has been the subject of several recent reviews [10–13] and will mostly not be considered here. I will only address protein-coding sequences, thus omitting the extensive literature on (somewhat simpler and better understood) fitness landscapes of nucleic acids. Finally, I will consider the possible underlying causes of changes in SPFLs, asking whether we can distinguish between them.
2. Evidence for changes in position-specific landscapes
(a). Sustained positive selection and divergence
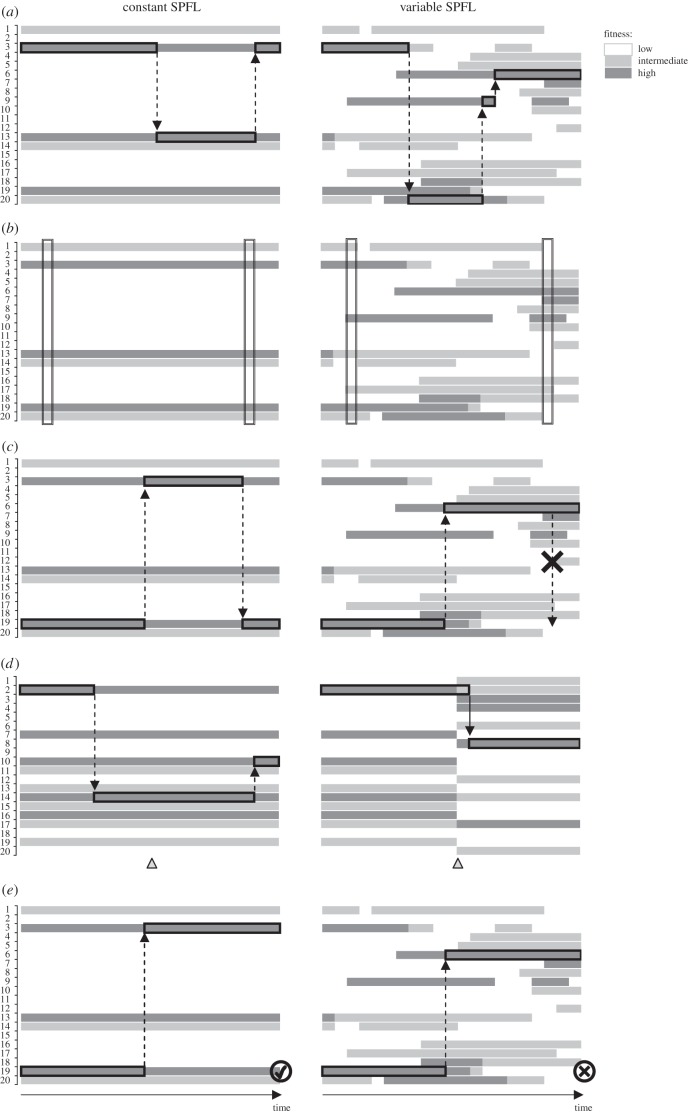
Statistical evidence for changes in amino acid propensities may be obtained from genomic patterns even without knowledge of the exact changes that are going on. A change of the SPFL may provoke evolution: after the previously optimal allele becomes suboptimal, a substitution to the new optimal variant is favoured. Therefore, evolution by itself is consistent with changes in the SPFL. However, it does not require them. Indeed, evolution at a site may proceed indefinitely if the fitness conferred by two or more most-fit variants is the same [14] (figure 2a), or substantially similar [14–16]. This evolution is not associated with a net long-term excess of beneficial over deleterious substitutions, and thus is not adaptive [16,17].
Figure 2.
Inferring changes in the SPFL. Left column, constant SPFL; right column, varying SPFL. (a) Abundance of positive selection and sustained sequence divergence. (b) Different sets of permitted variants at different time points (or in different species). (c) Reduction in the rate of reversals with time, owing to the ancestral variant being no longer fit. (d) Positive selection provoked by a change elsewhere in the genome (triangle). (e) Direct data on low (cross) or high (check mark) fitness of the ancestral variant. Broken lines, neutral substitutions; solid lines, positively selected substitutions. The currently predominant amino acid at each site is surrounded by a black rectangle.
Conversely, an influx of strongly beneficial substitutions at a site that is sustained for a long time implies that the SPFL changes. From comparisons of extant and reconstructed ancestral genomes, the corresponding positive fitness flux [16,17] may be revealed from strong positive selection favouring new variants (figure 2a). Numerous methods for detection of strong positive selection have been developed. Typically, these methods use patterns of within-species variation, perhaps together with between-species divergence, to infer the fraction of positively selected substitutions, and the strength of this selection [18–23]. Application of such methods has frequently revealed a substantial fraction of substitutions that have been fixed by rather strong positive selection [24,25]. For example, approximately 20–50% of the amino acid-changing substitutions were positively selected in the recent evolution of the Drosophila melanogaster lineage [18,21,22]. This value has remained nearly constant over the past approximately 60 million years since divergence from D. virilis [22], suggesting that positive selection was not a response to some transient event in the history of this lineage, but instead is a stationary process [16] that continues more or less uniformly throughout evolution. Most tests interpret SPFLs fluctuating at different timescales as positive selection [20,22,23,26,27].
Even in the absence of evidence for positive selection, SPFL changes may be inferred from sustained protein sequence divergence for evolutionarily long periods of time [28]. This divergence is inconsistent with constant SPFLs: under constant SPFLs, divergence is expected to reach its asymptotic level rather quickly—at the same timescale as neutrally diverging sequences [29]. By contrast, the amino acid sequence similarity between even the most anciently homologous proteins still continues to decrease 3.5 billion years after their origin from the last universal common ancestor [28].
Furthermore, under a constant SPFL, strong constraint observed at each time point (as inferred from the low ratio of non-synonymous to synonymous substitutions) implies that only a few amino acids are permitted at most amino acid positions. However, when multiple alignments of proteins from large numbers of species are considered, many different amino acids are observed at a typical site [30]. By itself, this does not necessarily mean that the SPFL has changed, as some of the amino acid variants may be permanently weakly deleterious, but still observable in some species [31]; however, other evidence supports SPFL dynamics as the main cause of the observed discrepancy [32].
(b). Differences in patterns of substitutions between clades
In theory, SPFLs can be inferred directly from rates of amino acid substitution, which in turn can be estimated from observed patterns of substitution in a maximum likelihood or Bayesian framework. If a position evolves under different SPFLs in two groups of species, the amino acid frequencies and the substitution matrices at this position will be different between these groups, and with sequence data from enough species from each group, these differences may be inferred with statistical tests. However, the inference of static, let alone variable, position-specific amino acid propensity vectors involves fitting a very large number of parameters [8,33,34] and may be statistically questionable [35]. Furthermore, there is a huge number of conceivable ways in which these vectors could change with time, making model specification insurmountable [36]. Still, this is an active direction of research.
Changes both in the rate of evolution and in the mode of selection at individual amino acid sites have been modelled and studied extensively. As early as 1970, Fitch & Markowitz [37,38] showed that different amino acid sites within a protein possess different levels of between-species variability and that variability increases with divergence of considered species (figure 2b); they interpreted this as evidence that the set of substitutions ‘acceptable’ at a site, and the set of variable sites, changes with time. This led to a productive ‘covarion’ model, in which sites are allowed to switch between variable and invariable [37,39,40]; and, more generally, to heterotachy (‘different speed’) models that allow arbitrary changes in the rate of evolution of a site with time [41–43]. Heterotachy implies variability of SPFLs: under a constant SPFL (and assuming that mutation rates do not change), all substitution rates would be constant with time, although perhaps different between different amino acid pairs. In the limit, a SPFL with a single peak gives rise to a completely invariable site, while a site with a ‘flat’ SPFL with equal fitness of all amino acid variants evolves neutrally. Models involving heterotachy generally receive high support from the data, implying that position-specific rates of amino acid evolution, and, by inference, relative fitness values associated with different amino acids, do change. Furthermore, not just heterotachy, but also heteropecilly (‘different variation’ [44])—variability of position-specific profiles of substitutions—can be inferred [44,45]. Heterotachy and heteropecilly models are often created in the context of phylogenetic inference and are not necessarily easily tractable in terms of SPFLs; in particular, they confound the mutation and fixation probabilities in a single substitution matrix, which complicates distinguishing changes in SPFLs from changes in position-specific mutation rates. Existing models also assume that the breakpoints—times at which the substitution matrix changes—are known a priori.
(c). Distribution of homoplasies
Changes in SPFLs may be inferred empirically from phylogenetic distributions of substitutions. Homoplasies—multiple substitutions that repeatedly give rise to the same derived amino acid variant at the considered site—are usually frequent [46–48] and particularly useful in this respect. If the fitness of a particular allele relative to other alleles differs between groups of species, the rate of substitutions giving rise to it is increased, and the rate of substitutions replacing it is reduced, in the clades where this allele is more favoured. Changing SPFLs have been inferred from the changes in the rate of homoplasies: in more closely related species, convergence is more likely, i.e. the same variant is more likely to arise twice independently, compared with more distant species [28,48,49]. Furthermore, the probability that a particular amino acid change A→B becomes reversed (B→A) during subsequent evolution of this lineage declines with the evolutionary time since the A→B change [50] (figure 2c). Patterns of polymorphism are also informative: at a site of past A→B replacement, the ancestral variant A is more frequently observed as polymorphism in an extant species if the A→B replacement happened recently, compared with the case when it happened a long time ago. Consideration of the substitutions and polymorphisms giving rise to a third variant (C) shows that this pattern arises from two codirectional forces: the fitness of the replaced variant A tends to decline in the course of subsequent evolution after the A→B replacement, and the fitness of the derived variant B tends to increase [50]. The latter trend was also observed in simulations of protein folding, where it was attributed to epistasis between amino acid sites [9]. All of these patterns mean that the difference in fitness between two amino acids differs between parts of the phylogenetic tree, and therefore, that the SPFL changes in the course of evolution.
(d). Correlated genetic changes
When sequence beyond a single position is considered, SPFL changes may be inferred when amino acid substitutions are known to have been facilitated by prior genetic changes (figure 2d). In this case, SPFL changes are owing to epistatic interactions between the considered site and its genomic background. When the SPFL at multiple sites is thus affected, this may result in a rapid burst of substitutions towards the newly advantageous variants at these sites: an ‘adaptive walk’ [36]. Such an increased rate of amino acid substitutions has been observed after prior amino acid changes elsewhere in the same [51–53] or a different [54] protein; after an insertion or deletion of several amino acids [55]; or after swapping a fraction of genes in the genome for their allelic variants [56]. For example, an insertion or deletion of a stretch of several amino acids leads to subsequent rapid accumulation of additional amino acid substitutions in the neighbouring segments of this protein, which is driven by positive selection [55]. As there are orders of magnitude more conceivable genetic interactions than have been considered by such analyses, the detected correlations probably reveal just the tip of the iceberg. Evolutionary models may provide a better fit to the rate of substitutions at a site when changes in protein structure introduced by substitutions at other sites are accounted for [57,58].
(e). Fitness effects of mutations
In the presence of large-scale data on fitness effects of mutations, SPFL changes may be revealed when an allele that is common in one species is deleterious in another. A negative effect of an amino acid variant on fitness can be revealed by reduced SNP or allele frequency, compared with the neutral expectation [22]; or from the known detrimental effect on the phenotype [59–62] (figure 2e). It has been estimated that in approximately 10% of all amino acid differences between humans and another species [59] or the ancestral variant [62], the non-human variant is pathogenic in humans. This fraction is similar in Drosophila [60]. High-throughput experimental data on functional effects of mutations may also be used to identify deleterious mutations, and, in combination with between-species comparisons, to infer SPFL changes, although this has only been done so far at the scale of one gene [63].
3. Rate of change
The above patterns can be used to estimate the rate at which the SPFL changes. It cannot be too slow, because few SPFL differences would otherwise be observed even between remote species. It is also not too fast, because SPFLs would otherwise be uncorrelated, even between closely related species. In Drosophila, the rate of change was found to be of the same order of magnitude as the neutral mutation rate [20]. The patterns observed in [28] and [30] suggest that, at an average site, approximately five amino acids out of 20 switch from ‘preferred’ to ‘unpreferred’ during the time it takes for one amino acid substitution to become fixed [64]. This estimate assumes binary fitness and a uniform rate of SPFL change both across sites [64] and in time. None of these assumptions holds. Still, the bulk of evidence shows unambiguously that the SPFL changes at timescales comparable with that of amino acid evolution (perhaps in addition to other, faster and/or slower, components).
4. Causes of change
What causes the changes in SPFL, and ultimately, adaptive evolution? Given infinite time and constant fitness landscape, the process of adaptation would ultimately come to a stop at a fitness peak (or, in the presence of deleterious substitutions, at a dynamic equilibrium in its vicinity). The fact that adaptation still proceeds, and apparently has not slowed down since the origin of life, implies that the complete fitness landscape is not constant, and/or that there has not been enough time for adaptation to converge on the fitness peak. At first sight, the second option seems implausible: surely billions of years of evolution would be enough for each amino acid site to become occupied by the amino acid conferring the highest fitness?
Indeed, this would be the case if the SPFL were independent of the rest of the genome [29]. However, this is not so. There is a progressive understanding of the importance of interactions within a genome, which are collectively referred to as epistasis [10–13]. Under epistasis, the SPFL is affected by genetic changes elsewhere in the genome. As evolution proceeds, such changes accumulate, and the SPFL deforms at a rate that depends on the number of interacting sites and on the rate of their evolution. This may result in ruggedness of the complete fitness landscape, restricting the number of accessible evolutionary paths towards the fitness peak and causing adaptation to take longer than on a non-epistatic landscape [28,29].
Thus, both environmental changes and epistasis may lead to changes in SPFL. Surprisingly, their relative importance is poorly understood. In the absence of solid evidence pointing one way or the other, evidence for a changing SPFL has been ascribed by different authors either to environmental changes (e.g. [23,26,36,65]) or to epistasis (e.g. [20,28,30,37]). Arguments have been put forward in support of both mechanisms. Environmental changes are usually invoked when the inferred fluctuations in selection are fast, and epistasis, when they are slow. However, to my knowledge, no systematic attempt has been made to distinguish between the two.
Relating genetic changes to underlying environmental changes is notoriously hard, with few unambiguous examples known [66,67]. On the other hand, most of the known examples of adaptation are in response to aspects of the environment that fluctuate [36]. The observed fluctuations of the environment that caused changes in allele distributions were generally rather rapid. However, the environment fluctuates on all timescales [66], and rare changes of major effect cannot, in general, be dismissed as a cause for the changes in SPFLs. In systems such as rapidly evolving pathogens, fluctuating selection is probably the predominant long-term mode of evolution.
In many cases, however, an environmental explanation for SPFL changes is implausible. It has been argued, for example, that the ancestral alleles found to be pathogenic in humans cannot be owing to adaptation of humans to a novel environment, as the phenotypes resulting from them are not reminiscent of ancestral ones [62]. Conversely, epistasis is expected to arise as an epiphenomenon of a wide range of biological processes [68], and its high prevalence is underscored by recently accumulating experimental data, including empirical data on interactions, limitations on the order in which substitutions may proceed, and repeatability of substitution paths in experimental evolution [11,13].
Although in rare cases it is possible to directly ascribe SPFL change to a change in the genomic background [59], many of the interactions are weak, non-specific and/or allosteric, complicating their inferences (reviewed in [12]). On the genome scale, experimental analysis of epistasis is still unfeasible. Still, some of the genomic evidence for SPFL changes—in particular, correlated changes at adjacent genomic sites—is only explainable by epistasis.
5. Conclusion
It is indisputable that the SPFLs of many amino acid sites change with time. The rate of this change appears to be high, so that it is comparable with the rate of protein evolution. Much of this change is owing to changes elsewhere in the genome; however, how much exactly is unclear. The available whole-genome analyses usually do not allow estimation of the relative contributions of epistasis and environmental fluctuations. Moreover, epistatic changes may ultimately be caused by changes in the outside environment: an environmental change may provoke a single change in the genome, which will in turn lead to a cascade of epistatic changes [20,36].
Estimating the fraction of substitutions in the genome that were caused by positive selection has been an important milestone in understanding the process of molecular evolution. While evidence for the existence of both weakly and strongly selected genomic changes was abundant, only the advent of whole-genome analyses has allowed estimation of the relative contribution of the two to evolution. The next natural milestone is understanding the mechanisms by which fitness changes and adaptive evolution proceeds. To what extent evolution involves traversing the complex fitness landscape, and to what extent it is a response to environmentally induced changes in the fitness landscape itself, remains a major open question.
Acknowledgements
I am grateful to Michael Lässig, Sergey Kryazhimskiy, Joshua Plotkin and two reviewers for comments that improved the manuscript. This work was performed in the Institute for Information Transmission Problems (Kharkevich Institute) of the Russian Academy of Sciences.
Competing interests
I declare I have no competing interests.
Funding
This work was supported by Russian Science Foundation grant no. 14-50-00150.
References
- 1.Wright S. 1932. The roles of mutation, inbreeding, crossbreeding and selection in evolution. In Proc. of the 6th Int. Congress of Genetics, Vol. 1, pp. 356–366. Brooklyn, NY: Brooklyn Botanic Garden. [Google Scholar]
- 2.Maynard Smith J. 1970. Natural selection and the concept of a protein space. Nature 225, 563–564. ( 10.1038/225563a0) [DOI] [PubMed] [Google Scholar]
- 3.Gavrilets S. 2004. Fitness landscapes and the origin of species. Princeton, NJ: Princeton University Press. [Google Scholar]
- 4.Podgornaia AI, Laub MT. 2015. Pervasive degeneracy and epistasis in a protein–protein interface. Science 347, 673–677. ( 10.1126/science.1257360) [DOI] [PubMed] [Google Scholar]
- 5.Weinreich DM. 2011. High-throughput identification of genetic interactions in HIV-1. Nat. Genet. 43, 398–400. ( 10.1038/ng.820) [DOI] [PubMed] [Google Scholar]
- 6.Weinreich DM, Lan Y, Wylie CS, Heckendorn RB. 2013. Should evolutionary geneticists worry about higher-order epistasis? Curr. Opin. Genet. Dev. 23, 700–707. ( 10.1016/j.gde.2013.10.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillespie JH. 1993. Substitution processes in molecular evolution. I. Uniform and clustered substitutions in a haploid model. Genetics 134, 971–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodrigue N, Philippe H, Lartillot N. 2010. Mutation-selection models of coding sequence evolution with site-heterogeneous amino acid fitness profiles. Proc. Natl Acad. Sci. USA 107, 4629–4634. ( 10.1073/pnas.0910915107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollock DD, Thiltgen G, Goldstein RA. 2012. Amino acid coevolution induces an evolutionary Stokes shift. Proc. Natl Acad. Sci. USA 109, E1352–E1359. ( 10.1073/pnas.1120084109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DePristo MA, Weinreich DM, Hartl DL. 2005. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat. Rev. Genet. 6, 678–687. ( 10.1038/nrg1672) [DOI] [PubMed] [Google Scholar]
- 11.De Visser JAGM, Krug J. 2014. Empirical fitness landscapes and the predictability of evolution. Nat. Rev. Genet. 15, 480–490. ( 10.1038/nrg3744) [DOI] [PubMed] [Google Scholar]
- 12.Ivankov DN, Finkelstein AV, Kondrashov FA. 2014. A structural perspective of compensatory evolution. Curr. Opin. Struct. Biol. 26, 104–112. ( 10.1016/j.sbi.2014.05.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondrashov DA, Kondrashov FA. 2015. Topological features of rugged fitness landscapes in sequence space. Trends Genet. 31, 24–33. ( 10.1016/j.tig.2014.09.009) [DOI] [PubMed] [Google Scholar]
- 14.Kimura M. 1983. The neutral theory of molecular evolution. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 15.Ohta T. 1992. The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst. 23, 263–286. ( 10.1146/annurev.es.23.110192.001403) [DOI] [Google Scholar]
- 16.Mustonen V, Lässig M. 2009. From fitness landscapes to seascapes: non-equilibrium dynamics of selection and adaptation. Trends Genet. 25, 111–119. ( 10.1016/j.tig.2009.01.002) [DOI] [PubMed] [Google Scholar]
- 17.Mustonen V, Lässig M. 2010. Fitness flux and ubiquity of adaptive evolution. Proc. Natl Acad. Sci. USA 107, 4248–4253. ( 10.1073/pnas.0907953107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith NGC, Eyre-Walker A. 2002. Adaptive protein evolution in Drosophila. Nature 415, 1022–1024. ( 10.1038/4151022a) [DOI] [PubMed] [Google Scholar]
- 19.Macpherson JM, Sella G, Davis JC, Petrov DA. 2007. Genomewide spatial correspondence between nonsynonymous divergence and neutral polymorphism reveals extensive adaptation in Drosophila. Genetics 177, 2083–2099. ( 10.1534/genetics.107.080226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mustonen V, Lässig M. 2007. Adaptations to fluctuating selection in Drosophila. Proc. Natl Acad. Sci. USA 104, 2277–2282. ( 10.1073/pnas.0607105104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eyre-Walker A, Keightley PD. 2009. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol. Biol. Evol. 26, 2097–2108. ( 10.1093/molbev/msp119) [DOI] [PubMed] [Google Scholar]
- 22.Bazykin GA, Kondrashov AS. 2011. Detecting past positive selection through ongoing negative selection. Genome Biol. Evol. 3, 1006–1013. ( 10.1093/gbe/evr086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benger E, Sella G. 2013. Modeling the effect of changing selective pressures on polymorphism and divergence. Theor. Popul. Biol. 85, 73–85. ( 10.1016/j.tpb.2012.10.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fay JC. 2011. Weighing the evidence for adaptation at the molecular level. Trends Genet. 27, 343–349. ( 10.1016/j.tig.2011.06.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellegren H. 2014. Genome sequencing and population genomics in non-model organisms. Trends Ecol. Evol. 29, 51–63. ( 10.1016/j.tree.2013.09.008) [DOI] [PubMed] [Google Scholar]
- 26.Huerta-Sanchez E, Durrett R, Bustamante CD. 2008. Population genetics of polymorphism and divergence under fluctuating selection. Genetics 178, 325–337. ( 10.1534/genetics.107.073361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gossmann TI, Waxman D, Eyre-Walker A. 2014. Fluctuating selection models and McDonald-Kreitman type analyses. PLoS ONE 9, e84540 ( 10.1371/journal.pone.0084540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Povolotskaya IS, Kondrashov FA. 2010. Sequence space and the ongoing expansion of the protein universe. Nature 465, 922–926. ( 10.1038/nature09105) [DOI] [PubMed] [Google Scholar]
- 29.Kondrashov AS, Povolotskaya IS, Ivankov DN, Kondrashov FA. 2010. Rate of sequence divergence under constant selection. Biol. Direct 5, 5 ( 10.1186/1745-6150-5-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breen MS, Kemena C, Vlasov PK, Notredame C, Kondrashov FA. 2012. Epistasis as the primary factor in molecular evolution. Nature 490, 535–538. ( 10.1038/nature11510) [DOI] [PubMed] [Google Scholar]
- 31.McCandlish DM, Rajon E, Shah P, Ding Y, Plotkin JB. 2013. The role of epistasis in protein evolution. Nature 497, E1–E2; discussion E2–E3 ( 10.1038/nature12219) [DOI] [PubMed] [Google Scholar]
- 32.Breen MS, Kemena C, Vlasov PK, Notredame C, Kondrashov FA. 2013. The role of epistasis in protein evolution (Reply). Nature 497, E2–E3. ( 10.1038/nature12220) [DOI] [Google Scholar]
- 33.Halpern AL, Bruno WJ. 1998. Evolutionary distances for protein-coding sequences: modeling site-specific residue frequencies. Mol. Biol. Evol. 15, 910–917. ( 10.1093/oxfordjournals.molbev.a025995) [DOI] [PubMed] [Google Scholar]
- 34.Lartillot N, Philippe H. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109. ( 10.1093/molbev/msh112) [DOI] [PubMed] [Google Scholar]
- 35.Rodrigue N. 2013. On the statistical interpretation of site-specific variables in phylogeny-based substitution models. Genetics 193, 557–564. ( 10.1534/genetics.112.145722) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillespie JH. 1991. The causes of molecular evolution. Oxford, UK: Oxford University Press. [Google Scholar]
- 37.Fitch WM, Markowitz E. 1970. An improved method for determining codon variability in a gene and its application to the rate of fixation of mutations in evolution. Biochem. Genet. 4, 579–593. ( 10.1007/BF00486096) [DOI] [PubMed] [Google Scholar]
- 38.Fitch WM. 1971. The nonidentity of invariable positions in the cytochromes c of different species. Biochem. Genet. 5, 231–241. ( 10.1007/BF00485794) [DOI] [PubMed] [Google Scholar]
- 39.Galtier N. 2001. Maximum-likelihood phylogenetic analysis under a covarion-like model. Mol. Biol. Evol. 18, 866–873. ( 10.1093/oxfordjournals.molbev.a003868) [DOI] [PubMed] [Google Scholar]
- 40.Wang H-C, Spencer M, Susko E, Roger AJ. 2007. Testing for covarion-like evolution in protein sequences. Mol. Biol. Evol. 24, 294–305. ( 10.1093/molbev/msl155) [DOI] [PubMed] [Google Scholar]
- 41.Lopez P, Casane D, Philippe H. 2002. Heterotachy, an important process of protein evolution. Mol. Biol. Evol. 19, 1–7. ( 10.1093/oxfordjournals.molbev.a003973) [DOI] [PubMed] [Google Scholar]
- 42.Yang Z, Nielsen R. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19, 908–917. ( 10.1093/oxfordjournals.molbev.a004148) [DOI] [PubMed] [Google Scholar]
- 43.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8, e1002764 ( 10.1371/journal.pgen.1002764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roure B, Philippe H. 2011. Site-specific time heterogeneity of the substitution process and its impact on phylogenetic inference. BMC Evol. Biol. 11, 17 ( 10.1186/1471-2148-11-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamuri AU, dos Reis M, Hay AJ, Goldstein RA. 2009. Identifying changes in selective constraints: host shifts in influenza. PLoS Comput. Biol. 5, e1000564 ( 10.1371/journal.pcbi.1000564) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bazykin GA, Kondrashov FA, Brudno M, Poliakov A, Dubchak I, Kondrashov AS. 2007. Extensive parallelism in protein evolution. Biol. Direct 2, 20 ( 10.1186/1745-6150-2-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kryazhimskiy S, Bazykin GA, Plotkin J, Dushoff J. 2008. Directionality in the evolution of influenza A haemagglutinin. Proc. R. Soc. B 275, 2455–2464. ( 10.1098/rspb.2008.0521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rogozin IB, Thomson K, Csürös M, Carmel L, Koonin EV. 2008. Homoplasy in genome-wide analysis of rare amino acid replacements: the molecular-evolutionary basis for Vavilov's law of homologous series. Biol. Direct 3, 7 ( 10.1186/1745-6150-3-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldstein RA, Pollard ST, Shah SD, Pollock DD. 2015. Nonadaptive amino acid convergence rates decrease over time. Mol. Biol. Evol. 32, 1373–1381. ( 10.1093/molbev/msv041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naumenko SA, Kondrashov AS, Bazykin GA. 2012. Fitness conferred by replaced amino acids declines with time. Biol. Lett. 8, 825–828. ( 10.1098/rsbl.2012.0356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bazykin GA, Kondrashov FA, Ogurtsov AY, Sunyaev S, Kondrashov AS. 2004. Positive selection at sites of multiple amino acid replacements since rat–mouse divergence. Nature 429, 558–562. ( 10.1038/nature02601) [DOI] [PubMed] [Google Scholar]
- 52.Callahan B, Neher RA, Bachtrog D, Andolfatto P, Shraiman BI. 2011. Correlated evolution of nearby residues in Drosophilid proteins. PLoS Genet. 7, e1001315 ( 10.1371/journal.pgen.1001315) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kryazhimskiy S, Dushoff J, Bazykin GA, Plotkin JB. 2011. Prevalence of epistasis in the evolution of influenza A surface proteins. PLoS Genet. 7, e1001301 ( 10.1371/journal.pgen.1001301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neverov AD, Kryazhimskiy S, Plotkin JB, Bazykin GA. 2014. Coordinated evolution of influenza A surface proteins. PLOS Genet. 11, e1005404 ( 10.1371/journal.pgen.1005404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leushkin EV, Bazykin GA, Kondrashov AS. 2012. Insertions and deletions trigger adaptive walks in Drosophila proteins. Proc. R. Soc. B 279, 3075–3082. ( 10.1098/rspb.2011.2571) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neverov AD, Lezhnina KV, Kondrashov AS, Bazykin GA. 2014. Intrasubtype reassortments cause adaptive amino acid replacements in H3N2 influenza genes. PLoS Genet. 10, e1004037 ( 10.1371/journal.pgen.1004037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson DM, Jones DT, Kishino H, Goldman N, Thorne JL. 2003. Protein evolution with dependence among codons due to tertiary structure. Mol. Biol. Evol. 20, 1692–1704. ( 10.1093/molbev/msg184) [DOI] [PubMed] [Google Scholar]
- 58.Rodrigue N, Philippe H. 2010. Mechanistic revisions of phenomenological modeling strategies in molecular evolution. Trends Genet. 26, 248–252. ( 10.1016/j.tig.2010.04.001) [DOI] [PubMed] [Google Scholar]
- 59.Kondrashov AS, Sunyaev S, Kondrashov FA. 2002. Dobzhansky–Muller incompatibilities in protein evolution. Proc. Natl Acad. Sci. USA 99, 14 878–14 883. ( 10.1073/pnas.232565499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kulathinal RJ, Bettencourt BR, Hartl DL. 2004. Compensated deleterious mutations in insect genomes. Science 306, 1553–1554. ( 10.1126/science.1100522) [DOI] [PubMed] [Google Scholar]
- 61.Azevedo L, Suriano G, van Asch B, Harding RM, Amorim A. 2006. Epistatic interactions: how strong in disease and evolution? Trends Genet. 22, 581–585. ( 10.1016/j.tig.2006.08.001) [DOI] [PubMed] [Google Scholar]
- 62.Soylemez O, Kondrashov FA. 2012. Estimating the rate of irreversibility in protein evolution. Genome Biol. Evol. 4, 1213–1222. ( 10.1093/gbe/evs096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Melamed D, Young DL, Miller CR, Fields S. 2015. Combining natural sequence variation with high throughput mutational data to reveal protein interaction sites. PLoS Genet. 11, e1004918 ( 10.1371/journal.pgen.1004918) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Usmanova DR, Ferretti L, Povolotskaya IS, Vlasov PK, Kondrashov FA. 2015. A model of substitution trajectories in sequence space and long-term protein evolution. Mol. Biol. Evol. 32, 542–554. ( 10.1093/molbev/msu318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takahata N, Ishii K, Matsuda H. 1975. Effect of temporal fluctuation of selection coefficient on gene frequency in a population. Proc. Natl Acad. Sci. USA 72, 4541–4545. ( 10.1073/pnas.72.11.4541) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bell G. 2010. Fluctuating selection: the perpetual renewal of adaptation in variable environments. Phil. Trans. R. Soc. B 365, 87–97. ( 10.1098/rstb.2009.0150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bergland AO, Behrman EL, O'Brien KR, Schmidt PS, Petrov DA. 2014. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 10, e1004775 ( 10.1371/journal.pgen.1004775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lehner B. 2011. Molecular mechanisms of epistasis within and between genes. Trends Genet. 27, 323–331. ( 10.1016/j.tig.2011.05.007) [DOI] [PubMed] [Google Scholar]