Abstract
Sex differences in behaviour extend to cognitive-like processes such as learning but the underlying dimorphisms in neural circuit development and organization that generate these behavioural differences are largely unknown.
Here we define at the single-cell level, from development, through neural circuit connectivity, to function, the neural basis of a sex-specific learning in the nematode C. elegans. We show that sexual conditioning, a form of associative learning, requires a pair of male-specific interneurons whose progenitors are fully differentiated glia. These neurons are born during sexual maturation and incorporated into pre-exisiting sex-shared circuits to couple chemotactic responses to reproductive priorities. Our findings reveal a general role for glia as neural progenitors across metazoan taxa and demonstrate that the addition of sex-specific neuron types to brain circuits during sexual maturation is an important mechanism for the generation of sexually dimorphic plasticity in learning.
During sexual maturation, the nervous system undergoes sexually dimorphic changes that couple behaviour to reproductive needs 1-5. Sex differences in behaviour extend beyond courtship and mating to cognitive-like processes such as learning that also enhance reproductive success 6,7. However, the precise dimorphisms in neural circuit development and organization that generate these differences in behavioural plasticity are largely unknown.
C. elegans is a sexually dimorphic species with hermaphrodites that can self-fertilise and males that can only cross-fertilise hermaphrodites. As a consequence, males display a repertoire of reproductive behaviours not performed by the hermaphrodite and these behaviours require sex differences in the nervous system that develop during sexual maturation 8,9. The best-described dimorphisms are the male-specific sensory and motor circuits required for mating. However, non-reproductive behaviours such as learning are also sexually dimorphic. Males, but not hermaphrodites, are capable of sexual conditioning, a process that involves associative learning and leads to a switch in chemotactic responses that facilitates effective mate finding 10. Sexual conditioning requires sex-shared and male-specific sensory neurons 10, but whether the circuit also contains sexually dimorphic interneurons for integration is not known.
The MCMs: a novel class of interneurons
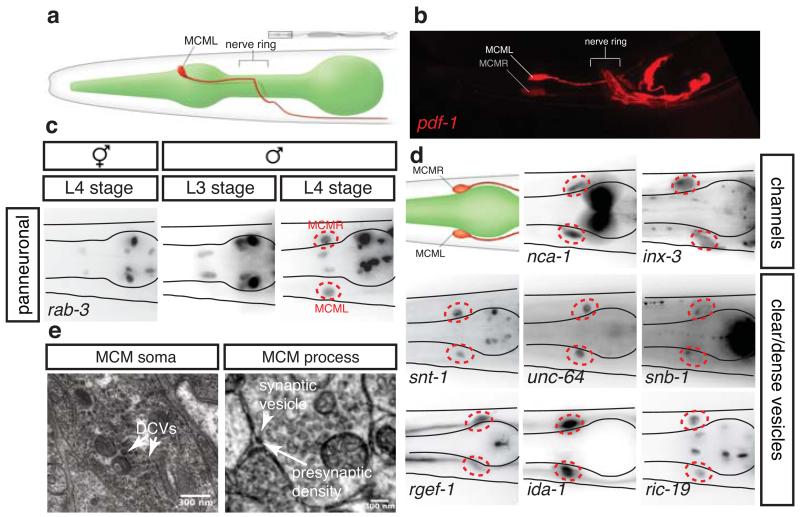
We identified a previously unnoticed bilateral pair of neurons in the head of males that are required for sexual conditioning. We have termed them the MCMs (for mystery cells of the male) (Fig. 1). The identification of the MCMs is surprising given that the C. elegans anatomy, neuronal connectivity and developmental lineage have been characterized extensively 8,9,12-15. We noted the presence of the MCMs when we were analyzing the expression pattern of a reporter transgene for the neuropeptide pdf-1. In males, but not in hermaphrodites, we observed pdf-1 expression in a bilateral pair of cells located dorsoanterior to the pharyngeal metacorpus, a region devoid of neuronal cell bodies in hermaphrodites (Fig. 1a-c). This region is comprised of glial and epithelial cells, some of which project anteriorly to the nose 14,16. Unlike any other cell in this region, we observed that the male’s pdf-1-positive cells send projections posteriorly into the nerve ring (Fig. 1b, and Extended Data Fig. 1a, b) The nerve ring is the main concentrated neuropil of the nematode brain, where most sensory neuron-interneuron synapses and neural processing occur 14. We found that the pdf-1-positive cells also express the panneuronal reporter rab-3, suggesting they may be neurons. rab-3 expression was first observed at the L4 stage, when sexual maturation begins (Fig. 1c). Through reporter gene analysis, and ultrastructural reconstruction, we found that the MCMs are indeed fully differentiated neurons. In addition to rab-3, these cells express neuronal gene batteries for both electrical and chemical communication (Fig. 1d, Extended Data Fig. 1c and Extended Data Table 1). These include components of the SNARE complex; Na+ and voltage-gated Ca2+ channel subunits; innexins; components of the machinery for neuropeptide secretion ric-19 and ida-1; and pdf-1. At the ultrastructural level, we observed dense-core vesicles in the MCM somata and projections, and clear-core vesicles at presynaptic densities (Fig. 1e). Together, these observations indicate that the MCMs are a novel class of male-specific interneurons.
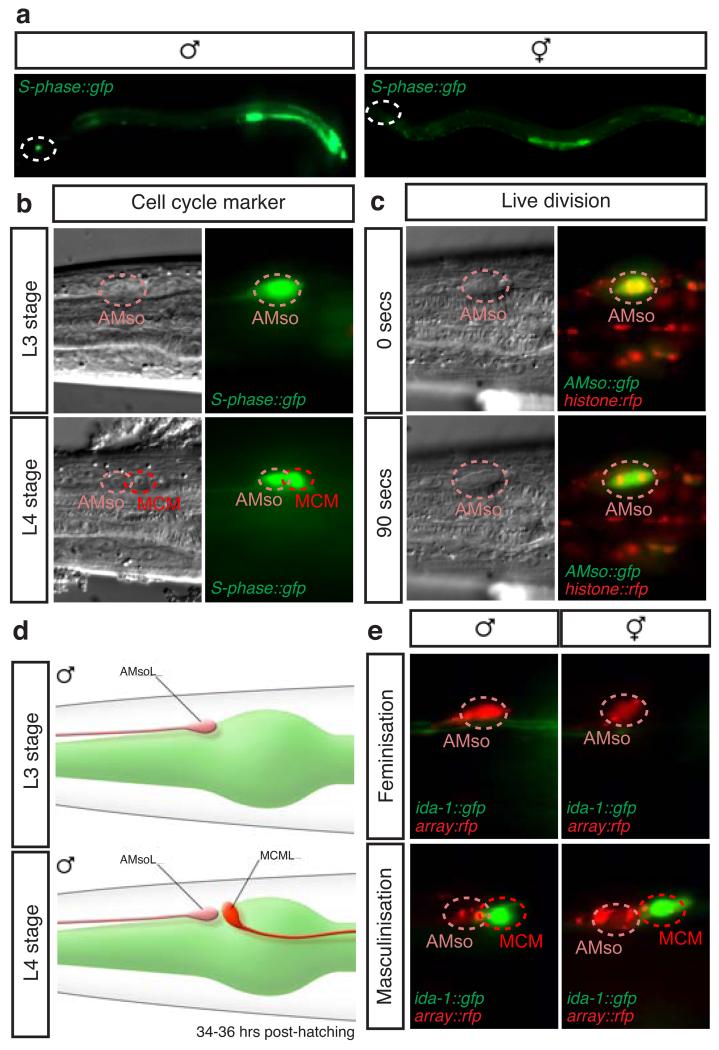
Figure 1. The MCMs are newly identified male-specific neurons.
Lateral views (a and b) and dorsal views (c and d) of animals oriented anterior to the left.
a, WormAtlas-style 11 diagram depicting the morphology and position, adjacent to the pharynx, of one of the bilateral pair of MCM neurons in the head of a male.
b, Confocal projection of pdf-1prom::rfp expression in the head of an adult male (same region as in a).
c, Expression of the pan-neuronal reporter transgene rab-3prom:rfp (Ras GTPase) in the head of an hermaphrodite and males at the third (L3) and fourth (L4) larval stages. The position of the MCMs is indicated with dashed red circles. R/L=Right/Left.
d, Expression in the MCMs in adult males of reporter transgenes for neuronal markers. snt-1 (synaptotagmin); unc-64 (syntaxin); snb-1 (synaptobrevin); rgef-1 (Ras exchange factor); nca-1 (NALCN Na+ channel subunit); inx-3 (gap junction innexin); ida-1 (tyrosine phosphatase-like receptor, phogrin); pdf-1 (neuropeptide pigment dispersing factor).
e, Electron micrographs of MCM showing dense-core vesicles (DCV) (left) and a synapse (right).
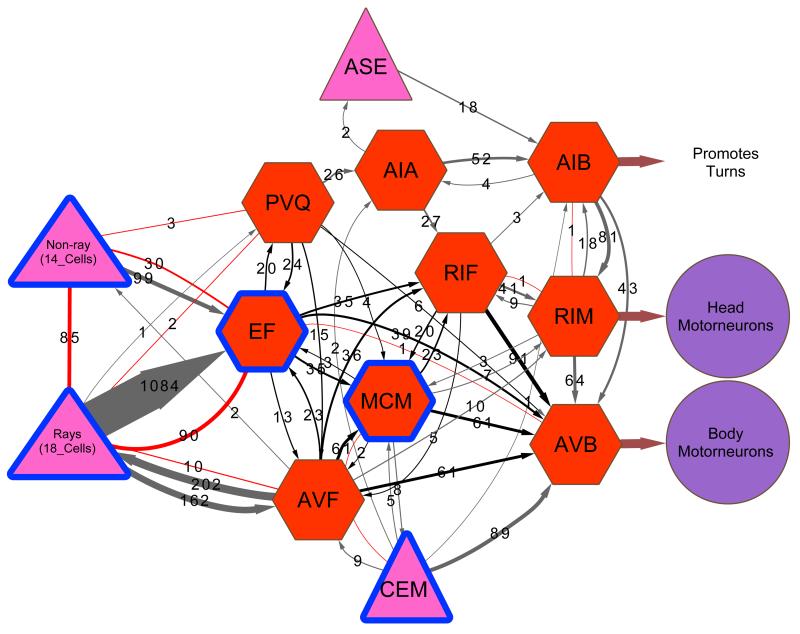
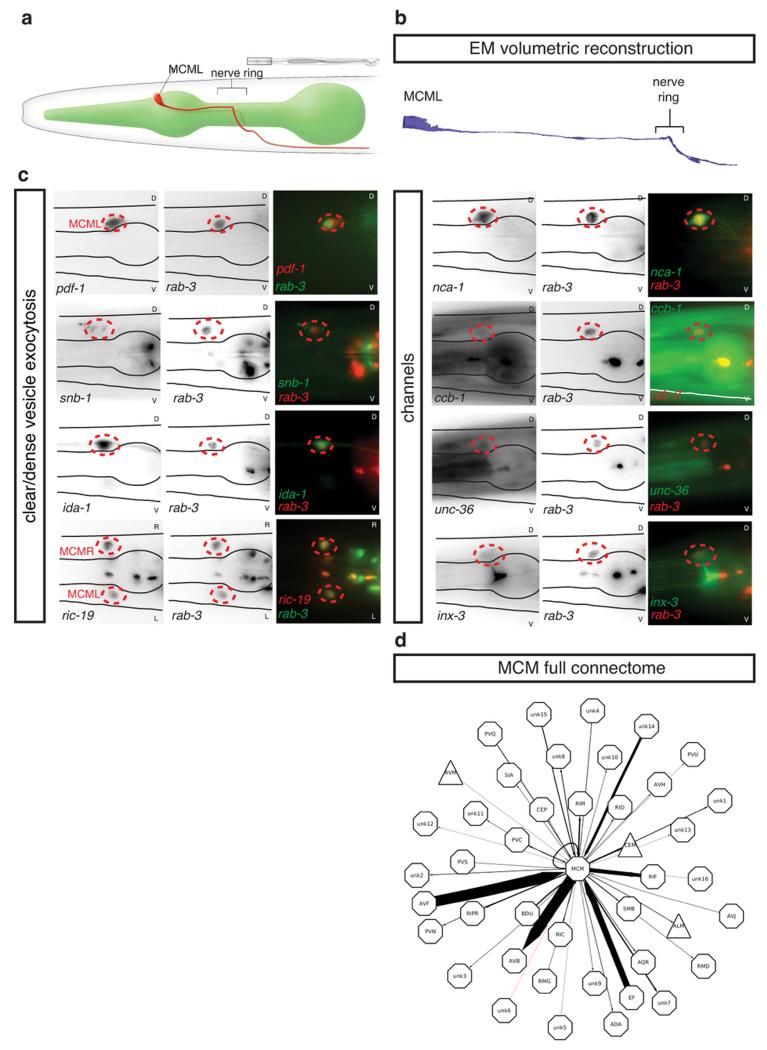
We identified the MCMs in electron micrographs of serial sections through the male head and established their entire neural connectivity (Fig. 2 and Extended Data Table 2). The cell bodies send processes posteriorly in the amphid bundles, from which they diverge to enter the nerve ring, and from which they exit and extend into the ventral nerve cord (Extended Data Fig. 1b). In the nerve ring and ventral nerve cord we identified synaptic interactions with 24 neuron classes (Extended Data Fig. 1e and Extended Data Table 2). The bulk of the synaptic input (65%) is from three interneuron classes, two of which are sex-shared (AVF and PVQ) and one of which is male-specific (EF). These interneurons receive extensive sensory inputs from the male copulatory circuits in the tail and extend processes through the ventral nerve cord and into the nerve ring where they connect to the MCMs both directly and through RIF (a sex-shared class of second-order interneurons that receive input from head chemosensory circuits) (Fig. 2). The largest synaptic output is onto the AVB pre-motor interneurons that drive forward locomotion. These outputs are both direct (41%) and indirect via RIF (14%). Thus, the presence of the MCMs creates a series of male-specific disynaptic feed-forward triplet motifs connecting male-specific EF and sex-shared AVF, PVQ and RIF to AVB (Fig. 2). Such triplet motifs recur frequently in the C. elegans nervous system 9,17. There is also a small amount of reciprocal interaction between the MCMs and the male-specific, pheromone-sensing CEM head sensory neurons. The major input from mate-sensing circuits and their output onto second- and third-order interneurons make the MCMs ideally placed for the integration of mate-experience into circuits regulating behavioural plasticity to sensory stimuli.
Figure 2. MCM connectivity.
a, Connectivity diagram of the MCMs showing the main inputs and outputs. The connections to the neurons known to regulate sexual conditioning (rays and CEMs), chemosensory plasticity (AIA, RIF), and salt sensation (ASE) are included. Grey and red connections indicate chemical and electrical synapses, respectively. The triplet motifs created by the MCMs are highlighted in black. The thickness of the lines is proportional to the anatomical strength of the connections, indicated in numbers (the number of EM serial sections of scored synaptic connectivity). Pink triangles, sensory neurons; red octagons, interneurons; purple circles, motor neurons; blue outline, male-specific.
The MCMs regulate sex-specific learning
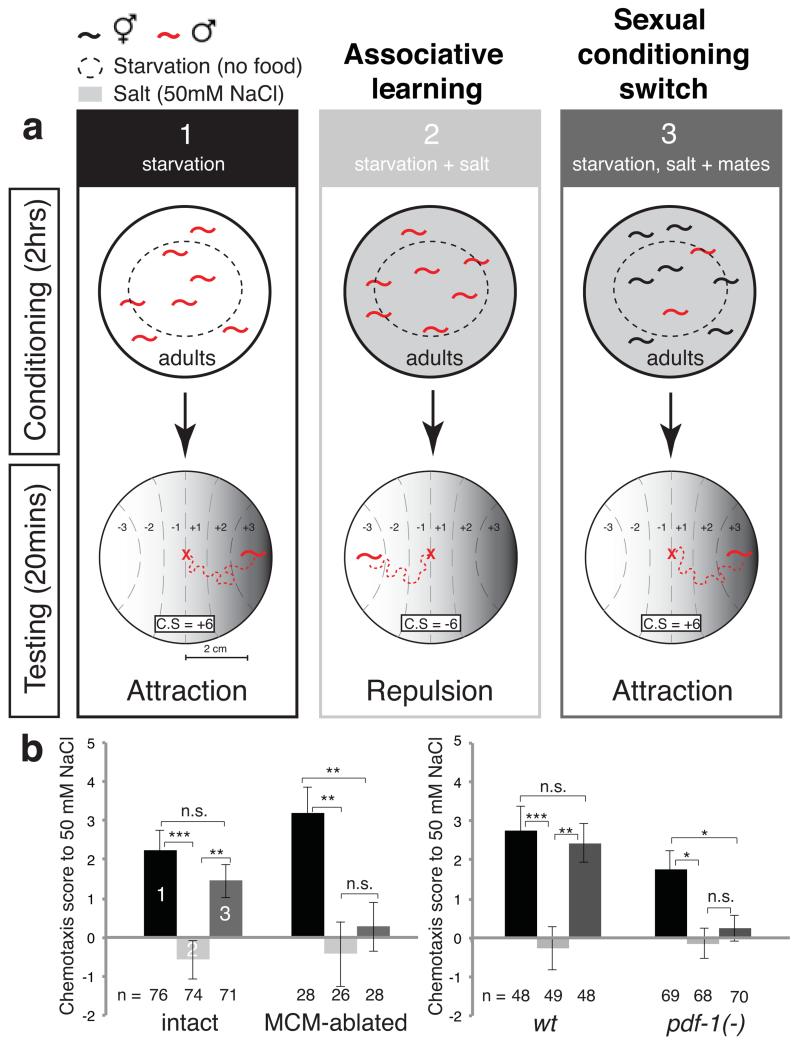
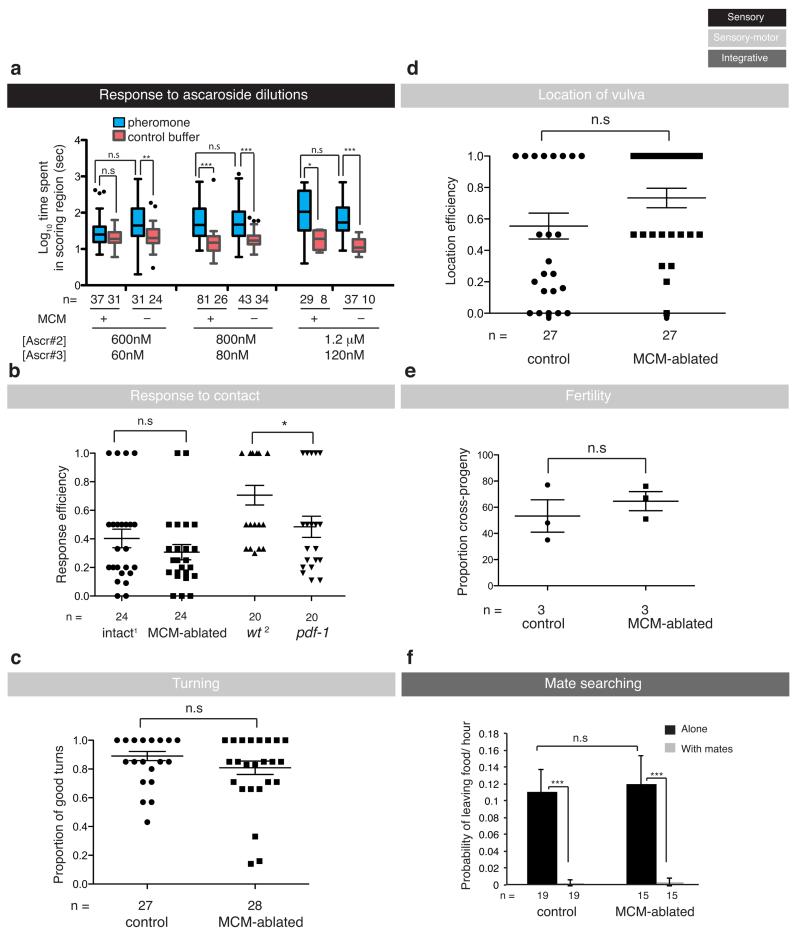
We found that the MCMs are required for a male-specific switch in chemosensory behaviour induced by sexual conditioning. Sexual conditioning is a mate-experience dependent process of behavioural modification that trumps the effects of starvation during chemosensory learning 10. This plasticity confers males with the ability to use recent environmental chemosensory cues as signals for effective mate finding and reflects the adult male prioritisation of sex over food 18,19. One chemosensory behaviour that is subject to sexual conditioning is salt-avoidance learning 10. C. elegans is normally attracted to salt but both males and hermaphrodites can learn to avoid salt when it is previously associated with an aversive stimulus such as starvation 20,21 (Fig. 3a). However, unlike hermaphrodites, males switch their behaviour and become attracted to salt if mates are present during previous conditioning 10 (Fig. 3a). Ablation of the MCMs at the late fourth larval (L4) stage with a laser microbeam resulted in males that failed to undergo a sexually conditioned switch, thus avoiding salt rather than being attracted to it, after conditioning with starvation, salt and mates (Fig. 3b). Importantly, MCM ablation did not disrupt salt attraction in non-conditioned males or salt avoidance learning after conditioning with salt and starvation in the absence of mates (Fig. 3b), indicating that the defects are specific to sexual conditioning and not due to inability to sense salt or starvation.
Figure 3. The MCMs are required for male-specific associative learning.
a, Diagram depicts the salt-avoidance learning and sexual conditioning assay. C.S, chemotaxis score.
b, Salt chemotaxis scores of previously conditioned intact and MCM-ablated males [inIs179(ida-1prom::gfp);him-8(e1489)], and wild-type [him-5(e1490)] and pdf-1(tm1996) mutant males. n= number of individual animals tested. Error bars indicate SEM. Mann-Whitney test was used for statistical analysis. ***P<0.001; **P<0.01; *P<0.05; n.s.= no statistically significant difference (p≥0.05);
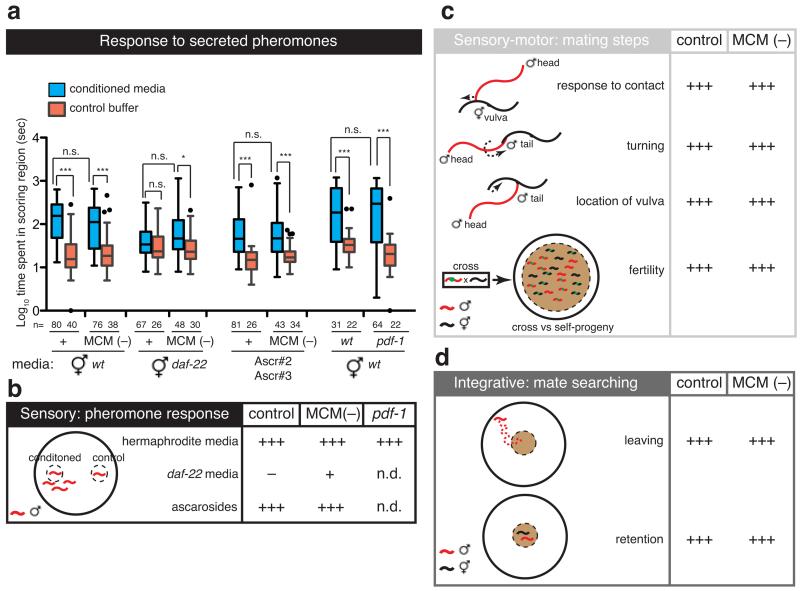
Sexual conditioning requires two types of mate inputs 10: a secreted pheromone synthesized by the enzyme DAF-22 and sensed through chemosensory amphid neurons and the male-specific CEM sensory neurons in the head 10,22, and a contact-dependent cue that is sensed through the male-specific ray neurons in the tail 10,23. MCM-ablated males responded efficiently to both daf-22-dependent secreted pheromones (ascarosides) (Fig. 4a, b and Extended Data Fig. 2a) and contact-dependent hermaphrodite cues that initiate the mating sequence and inhibit male food-leaving behaviour 23,24 (Fig. 4c, d and Extended Data Fig. 2b, f). These results indicate that the MCMs are not required for the sensory detection of mates, salt or starvation but that they specifically regulate the integration of these cues to produce behavioural plasticity in a context- and sex-specific manner.
Figure 4. MCM ablation does not affect other male-specific behaviours.
Efficiency of intact and MCM-ablated males [inIs179(ida-1prom::gfp);him-8(e1489) or otIs356(rab-3prom::rfp)him-5(e1490)], and wild-type [him-5(e1490)] and pdf-1(tm1996) mutant males in male-specific behaviours.
a, Response to wild-type or daf-22 (m130) hermaphrodite-conditioned media and purified ascaroside pheromones (80nM Ascr#3, 800nM Ascr#2). Graphs represent Tukey box plots of logarithmic transformations of the data; n= number of independent events (i.e. entry in scoring region); T-test with Bonferroni correction was used for statistical analysis. ***P<0.001; *P<0.05; n.s.= no statistically significant difference (p≥0.05).
b, Summary diagram of the data plotted in a. Statistical significance compared to control buffer is indicated by +++, P<0.001; +, P<0.05; − = no statistically significant difference. n.d., not determined.
c and d, Execution of mating sub-steps (c) and exploration in search of mates (d). +++ indicates performance level of intact, control males.
Having found a role for the MCMs in sexual conditioning, we next asked whether the MCMs are important regulators of other male-specific behaviours from the coordinated sensory-motor programmes of mating 25 to other integrative behaviours such as the decision to leave food in search of mates 23,26. MCM ablation did not cause defects in any other male-specific behaviour tested or overall fertility (Fig. 4c, d and Extended Data Fig. 2b-f), demonstrating a surprisingly specific role for the MCMs in sexual conditioning.
To establish whether sexual conditioning is regulated through neuromodulation, we tested pdf-1(tm996) mutants in the salt chemotaxis learning assay. The tm1996 deletion removes the minimal promoter and first exon of the pdf-1 gene and is likely to be a null 27,28. We found that pdf-1 mutant males, like MCM-ablated males, failed to undergo sexual conditioning and avoided salt after conditioning with starvation, salt and mates (Fig. 3b). Loss of pdf-1, like loss of MCMs, did not disrupt salt sensation in naïve animals or the ability to associate salt and starvation (Fig. 3b). pdf-1 mutants also responded to secreted and contact-dependent mate cues, although they were slightly, but significantly, less efficient than wild-type males at contact response (Fig. 4 a, b and Extended Data Fig. 2b) 23. Together, these results tentatively suggest that the MCMs may modulate the circuits that regulate chemosensory plasticity through neuropeptide secretion.
The MCMs arise from glia
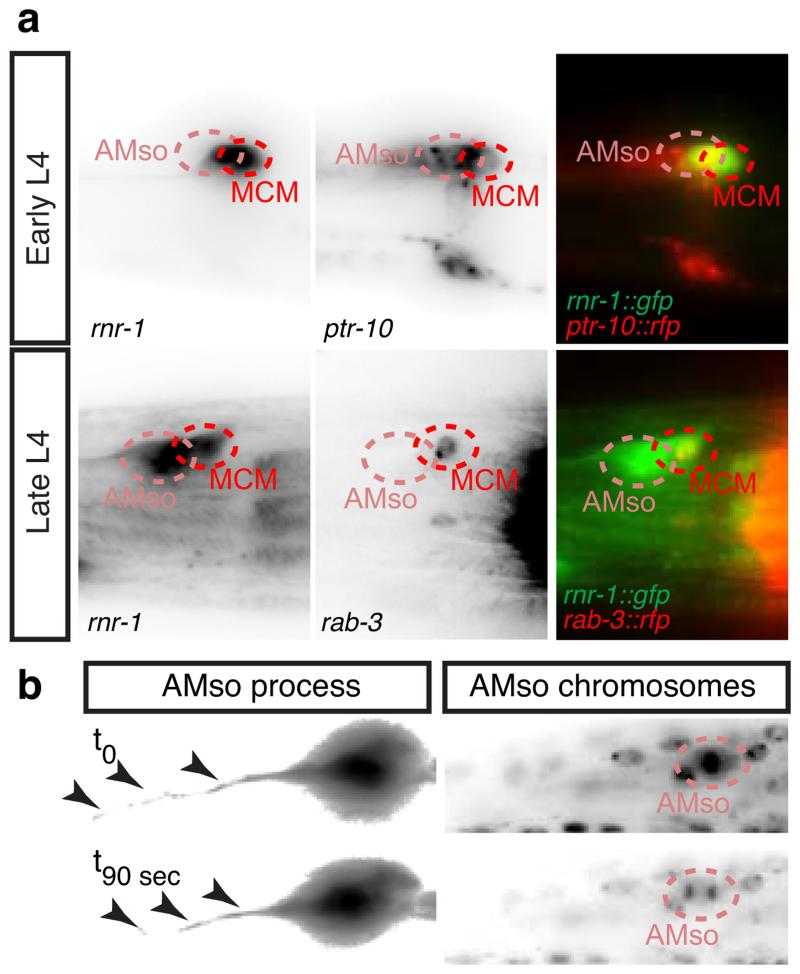
Next we sought to establish the developmental mechanisms by which these male-specific neurons arise. Unlike any other neurons in C. elegans and other invertebrates, which arise from epithelial or undifferentiated blast cells 8,12,13,29, we found that the MCMs arise from glial cells. Glia are specialized cells with projections that ensheath the cilia of the sensory neurons they are associated with, and provide structural and functional support to these neurons 30. Since neuronal gene expression in the MCMs begins during sexual maturation, we examined the expression of an rnr-1prom::gfp reporter, which labels cells in S-phase 10,31, to determine if the MCMs are born at this stage via an undescribed cell division in the male head. In the head region, we observed strong expression only in males (Fig. 5a). Expression was observed in a bilateral pair of cells at the late L3 stage, corresponding to the amphid socket (AMso) glial cells, and perduring in two bilateral pairs of cells at the early L4 stage, corresponding to the two AMso and the MCMs (Fig. 5b and Extended Data Fig.3a). The lack of expression in the hermaphrodite head is consistent with the lack of AMso glial cell division in this sex 13.
Figure 5. The MCMs originate from a male-specific cell division of the AMso glial cells.
Lateral views of animals oriented anterior to the left and dorsal to the top.
a and b, Expression of the S-phase reporter transgene rnr-1prom:gfp in whole animals at the L4 stage (a) and in the head of L3 and L4 males (b). The male-specific division in the head is indicated by dashed circles in a. The rest of expression corresponds to developing reproductive structures.
c, DIC and fluorescent images of the AMso cell body with RFP-labeled histones during cell division.
d, WormAtlas diagram depicting the morphology and position of the AMso and the MCM in the male head at the L3 and L4 stages.
e, Head of animals with sex-reversal genetic manipulations of AMso. Feminisation by expression of the grl-2prom:tra-2IC::SL2:mCherry transgene oleEx23. Masculinisation by expression of grl-2prom:fem-3::SL2:mCherry transgenes oleEx18 and oleEx24.
Ablation of one of the bilateral pair of AMso glial cells at the L3 stage in males resulted in a consistent loss of the MCM on the operated side (Extended Data Table 3). Importantly, the MCM was not lost when the AMso was ablated at the mid-L4 stage, after the MCM was born (Extended Data Table 3). Together this demonstrates that the AMso glial cells are the MCM progenitors.
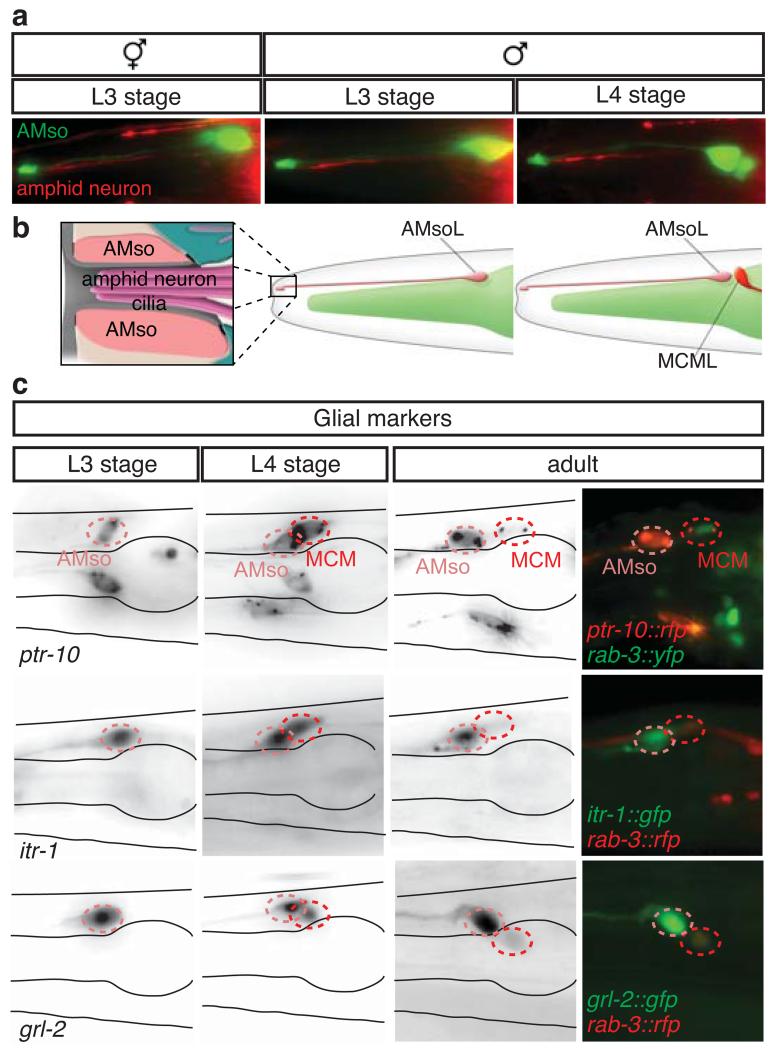
The AMso glial cells display a polarised morphology with a single projection running anteriorly along the A-P axis to the nose 16 (Fig. 5d and Fig. 6a, b). Live imaging of the AMso cell division in males revealed that the cleavage plane was perpendicular to the axis of AMso polarity and that the AMso projection was maintained throughout the division (Fig. 5c and Extended Data Fig. 3b). The projection was inherited by the self-renewing AMso glial daughter.
Figure 6. The male AMso cells are fully differentiated glia before and after the division that generates the MCM neuron.
Lateral views of animals oriented anterior to the left and dorsal to the top.
a, AMso glial cell (green, grl-2prom:gfp) and amphid sensory neuron dendrites (red, pdf-1prom:rfp) in an L3 hermaphrodite and L3 and L4 males.
b, Diagram of the AMso glial cell and the distal end of its projection to the nose, where it ensheaths the cilia of sensory dendrites.
c, Expression in AMso and perdurance in MCM of reporter transgenes for glial markers in L3, L4 and adult males. ptr-10 (Patch related receptor); itr-1 (IP3 receptor); grl-2 (hedgehog-like/Ground related).
AMso plasticity is intrinsically regulated
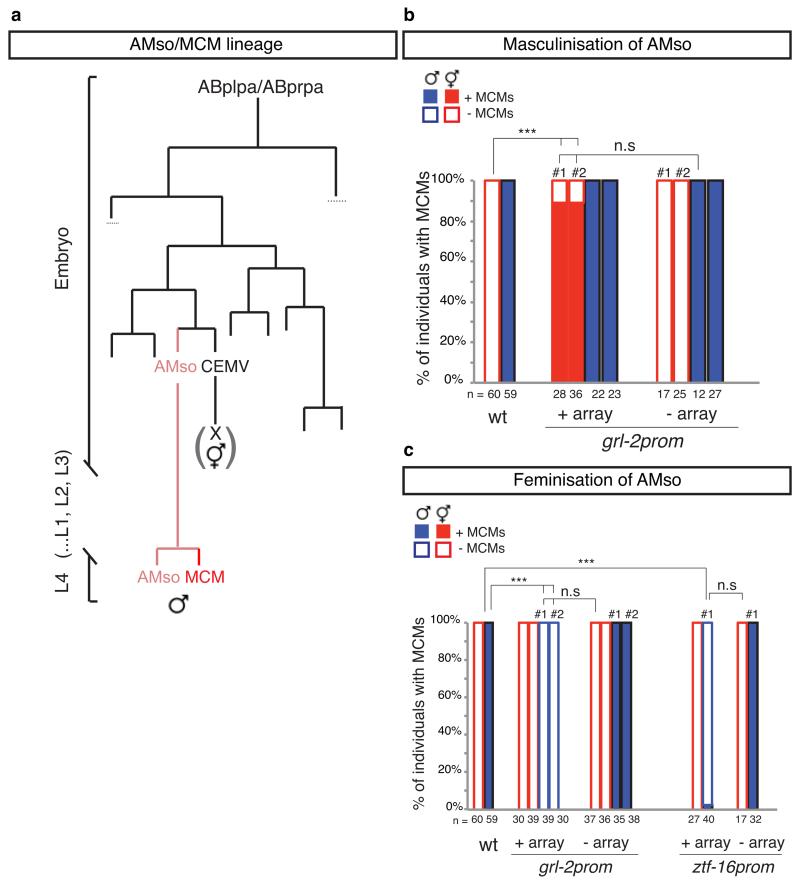
The AMso glial cells are born during embryogenesis from the same fully described lineages in both sexes (Extended Data Fig. 4a) 12, but retain their plasticity only in males, re-entering the cell cycle during sexual maturation. We found that the AMso developmental program occurred according to the genetic sex of the AMso cells and not according to that of the rest of their lineage or the animal. Sexual dimorphism in C. elegans is regulated by a genetic pathway that converges on the Ci/GLI-like zinc finger transcription factor tra-1, which inhibits male development and activates hermaphrodite development 32,33. Consistent with a role for the sex-determination pathway in MCM specification, 22/22 tra-1(e1488) sex-transformed XX pseudo-males produced MCMs (data not shown). The sister cells of the AMso are the male-specific CEM sensory neurons, which undergo cell death during embryogenesis in hermaphrodites 12 (Extended Data Fig. 4a). In order to establish whether sex-specific neurogenic competence of the AMso glial cells is specified extrinsically or intrinsically we expressed sex-reversing transgenes under cell-restricted promoters to uncouple the genetic sex of the AMso glial cells from that of the rest of the animal. We used the grl-2 and ztf-16 promoters, which drive expression in the AMso cells after they are born and one or two other non-overlapping cell types in the head in larval animals 34,35. Masculinisation of the AMso cells in hermaphrodites, by expression of a fem-3 transgene 36,37, resulted in AMso cell division and MCM neuronal differentiation (Fig. 5e, Extended Data Fig. 4b, c and Extended Data Table 4). Conversely, feminisation of AMso cells in males, by overexpression of a tra-2/C transgene 19,38, resulted in lack of AMso cell division and lack of MCMs (Fig. 5e, Extended Data Fig. 4b, c and Extended Data Table 4). Masculinising transgenes in males or feminising transgenes in hermaphodites had no effect on AMso development (Fig. 5e). These results indicate that the competence of the AMso glial cell to become a neural progenitor is specified intrinsically and extend the battery of sexual dimorphisms regulated by the sex-determination pathway to the intrinsic properties of glia.
The male AMso are fully differentiated glia
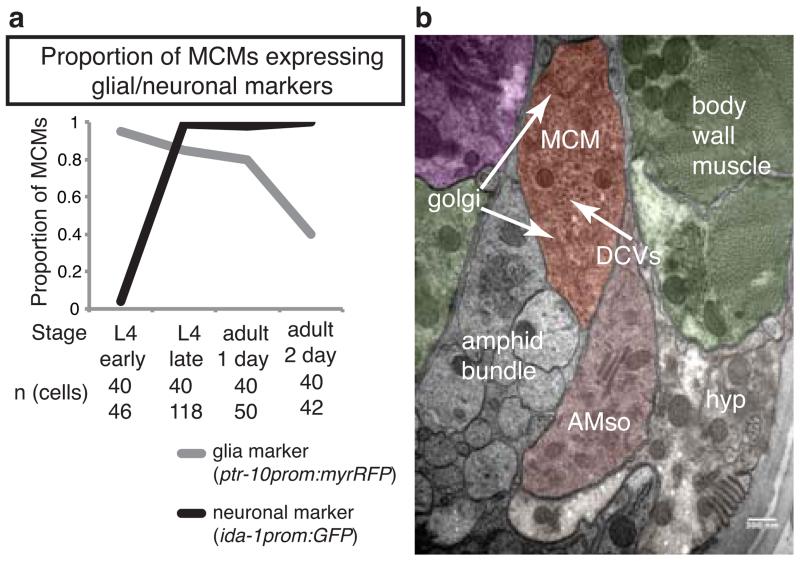
We found that the male AMso glial cells are fully differentiated specialized cells before they divide, indicating that the production of the MCMs from AMso involves a complete switch in terminal cell fate (Fig. 6). In hermaphrodites and adult males, the AMso cells are specialized glial cells that form a hole in the cuticle through which most amphid neurons contact the outside world 16, (Fig. 6a, b). Hermaphrodite AMso cells also express a battery of glial-cell markers 10,39. We observed that at the L3 stage, before their division, the male AMso cells display the same characteristics as the AMso of hermaphrodites and adult males: they project to the nose where they form a socket that ensheaths amphid neuron cilia (Fig. 6a, b), and they express AMso markers (Fig. 6c and Extended Data Table 5). Following AMso division, one daughter retains the AMso identity and the other becomes the MCM, which loses molecular and structural characteristics of glia (Extended Data Fig. 5) and, as described above, acquires a neuronal identity. Similar complete switches in terminal cell fates have been described as transdifferentiation events in C. elegans and other systems 12,40-42.
Discussion
The production of the MCMs from AMso glial cells through a division that results in self-renewal and neuronal differentiation is equivalent to a snapshot of the reiterative neuronal differentiation events arising from radial glia cell divisions during vertebrate neurogenesis 43. However, the extent to which vertebrate neural progenitors are fully differentiated glia remains unclear and in particular, it is not known whether the support and progenitor functions of glia can be integrated within one cell 44,45. Our results provide the first example of neurons arising from glia in a non-vertebrate organism and demonstrate that fully differentiated, functional glia can retain neural progenitor properties during normal development.
In C. elegans the best described transdifferentation event is the direct conversion of the rectal epithelial cell Y into a neuron 40,46. Other cell fate switches in C. elegans and other systems, which are not direct but require cell division, have also been suggested to be transdifferentation events 12,41,42. The complete cell fate swtich we observe suggests that the production of neurons from glia may be a process of natural transdifferentiation. It will be interesting to determine if direct and indirect transdifferentiaitons of this kind occur through similar molecular mechanisms.
Our findings provide a direct link between developmental and anatomical sexual dimorphism in higher-order processing areas of the brain and sexually dimorphic behavioural plasticity during learning. We have shown that sex differences in learning require the production, late in development, of a male-specific class of cephalic interneurons that were not known to exist. These interneurons are incorporated into pre-existing, sex-shared circuits allowing the male to change behavioural priorities according to its new reproductive needs. The MCMs function to confer salience to previous mate-experience during male navigation. Based on their connectivity, we suggest that these interneurons may function in other chemosensory-plasticity behaviours that may be subject to sexual conditioning. The production of more neurons in males is also a mechanism to generate cognitive sexual dimorphism in the song-learning system of songbirds, where neural progenitors have a glial identity 47. However, whether these neurons are a male-specific class and whether their progenitors are intrinsically sexually dimorphic is not known. Our findings indicate that the addition of sex-specific neurons is an effective way of remodeling brain circuits during sexual maturation and reveal a general, possibly ancient role for glia as neural progenitors to assemble circuits for higher order processing.
Methods
No statistical methods were used to predetermine sample size. No samples were excluded from analysis. For behavioural assays, investigators were blinded to the genotype or surgery of the animals during experiments and outcome assessment. Experiments testing response to secreted pheromones were randomized.
Strains
him-5(e1490) was used as the wild-type background strain.
BL5717, inIs179(ida-1prom::gfp);him-8(e1489) was used for MCM ablation and behavioural assays (data in Fig.3, Fig. 4 and Extended Data Fig. 2).
BAR4, otIs356(rab-3prom::rfp)him-5(e1490) was used for MCM ablation and behavioural assays (data in Fig. 4 and Extended Data Fig. 2)
PT2248, pdf-1(tm1996);him-5(e1490)
DR476, daf-22(m130)
CB369, unc-51(e369)
VT774, unc-36(e251);maIs103(rnr-1prom::gfp+unc-36 (+))
CB2823, tra-1(e1488)III;eDp6(III;f)
BAR63, stIs10116 [his-72prom::his-24::mCherry::let-858 3′UTR + unc-119(+)]; itIs37 [pie-1prom::mCherry::H2B::pie-1 3′UTR + unc-119(+)];him-omen5(e1490) Arrays for neuronal and amphid socket markers are indicated in Extended Data Table 1 and 5, respectively.
DNA constructs
Sex transformation constructs were made by PCR fusion of the regulatory sequences of grl-2 35 or ztf-16 34 directly upstream of the ATG of fem-3::SL2::mCherry or tra-2IC::SL2::mCherry 19,36-38. These contructs include the unc-54 3′ UTR. The grl-2 promoter included 862 bp upstream of the ATG start codon (this is a smaller region of the promoter from that used in 35). We checked transgenes driven by this promoter for embryonic expression and saw no expression before the birth of the AMso. The ztf-16 glia enhancer included the region −2536 to −4637 upstream of the ATG, as described in 34. The following arrays were generated:
| oleEx18[grl-2prom::fem-3::SL2mCherry (20ng/μl)+elt-2::GFP (40ng/μl)]; |
| oleEx24[grl-2prom::fem-3::SL2mCherry (8ng/μl)+elt-2::GFP (40ng/μl)]; |
| oleEx19[grl-2prom::tra-2IC::SL2mCherry (50ng/μl)+elt-2::GFP (30ng/μl)]; |
| oleEx23[grl-2prom::tra-2IC::SL2mCherry (10ng/μl)+elt-2::GFP (40ng/μl)]; |
| oleEx22[ztf-16enhancer:tra-2IC::SL2mCherry(5ng/μl)+elt-2:GFP(40ng/μl)]. |
Laser ablations
Standard protocol was used 48. L1, L3 or L4 males were mounted on a glass slide on a 5% agarose pad with 25mM NaN3 as anesthetic. For MCM candidate progenitor ablations the presence or absence of MCMs was identified with a rab-3 transgene (table S1). Epithelial cells were identified by morphology, position, and expression of the gaIs220 [col-93prom::HIS-24::mCherry + unc-119(+)] or syIs78[ajm-1prom::GFP+unc-119(+)] transgenes. Amphid socket cells were identified with glial markers (table S5).
For MCM ablation and behavioural assays the strains BL5717, inIs179(ida-1prom::GFP);him-8(e1489) and BAR4, otIs356(rab-3prom::rfp)him-5(e1490) were used. Animals were left to recover for 1 day and then assayed.
Behavioural assays
Sexual conditioning switch
Assays were performed as in 10 with some modifications. Assay plates and 50 mM NaCl conditioning plates were 5 cm in diameter and contained 4.5 ml of 2% agar, 5 mM potassium phosphate (pH 6.0), 1 mM CaCl2 and 1 mM MgSO4 (salt-conditioning plates also contained 50 mM NaCl). A radial gradient of salt was created in the assay plates by adding 10 μl of 50 mM NaCl 2 cm away from the center of the plate the night before the assay and then 5 μl three hours before the assay. Males and hermaphrodites were picked as L4s the night before the assay and transferred to single-sex plates with food. Animals were recovered from the food plates with wash buffer (1mM CaCl2, 1mM MgSO4, and 5mM pH 6.0 potassium phosphate) and centrifuged at 1,700 r.p.m. for 3 minutes. A total of three washes were performed to remove the food before placing the animals in the conditioning plates. Between 8 and 15 males were placed in each conditioning plate and 200 hermaphrodites were used for sexual conditioning. Animals were conditioned for 2 hrs. Ablated males were sexually conditioned in the same plate as control males, tested blind, and identified afterwards based on the absence of MCMs. Animals were tested individually on a radial gradient of salt by placing them in the center of the assay plate, 2 cm away from the source of NaCl. After 20 minutes, the tracks left by the animal were visualized and the chemotaxis score was calculated as in 10 by the sum of the scores of the regions by which the animal had travelled (Fig. 3a). At least five replicas were performed and on different days. Mann-Whitney test was used for statistical analysis.
Response to secreted mate pheromones
Assays were performed as in 23. Hermaphrodite-conditioned media was prepared by incubating wild-type or daf-22(m130) hermaphrodites in M9 buffer at a concentration of 1 hermaphrodite per 1 μl of M9 for 3 hrs. Animals were picked as L4s the night before. Assay plates were regular NGM plates seeded with 50 μl of OP50. A pheromone spot (1 μl of hermaphrodite-conditioned M9, or diutions of Ascr#2 and Ascr#3 in M9) and a control spot (1 μl of M9) were placed 3 mm a part. The two spots were interchanged every other trial to remove any bias. Upon drying, the spots left a visible rim on the food lawn. A population of 5 males (8 males for him-5 (e1490) and pdf-1(tm1996) trials) was placed 3 mm away from the spots and video-recorded for 20 minutes. Videos were scored blindly for time spent in each spot. Each entrance to a spot was considered an independent event if after leaving, the worm had moved at least 2 mm away from the spot. The average duration of events was calculated and compared between spots and populations. Mann-Whitney test with Bonferroni correction was used for statistical analysis. For clear visualization of the data in the plotted graphs, data was logarithmically transformed. This results in normalization of the distribution of the data. T-test with Bonferroni correction was used for statistical analysis of the transformed data. Comparison of arithmetic (raw data) or geometric (transformed data) means did not affect the interpretation of the data.
Seven independent population trials were performed for each condition except for daf-22(m130)-conditioned media (twelve), 600nM Ascr#2+60nM Ascr#3 (five) and 1200nM Ascr#2+120nM Ascr#3 (four). We selected three different ascaroside dilutions according to the response elicited in intact males: 600nM Ascr#2+60nM Ascr#3 (no robust and stochastic response), 800nM Ascr#2+80nM Ascr#3 (robust but low response), 1200nM Ascr#2+120nM Ascr#3 (robust and strong response) (Extended Data Figure 2).
Response efficiency to mate contact
Animals were isolated as L4s the night before. For the assay, single males were placed with 30 mates (hermaphrodites) in a 20 mm food lawn. Males were tested until they responded to a mate or for three minutes, whatever happened first. A male was scored as responding to mate contact if it placed its tail ventral down on the mate’s body and initiated the mating sequence by backing along the mate’s body to make a turn. The response efficiency was calculated by dividing a response by the total number of contacts made with the mate before responding. If a male did not respond within three minutes, it was scored as having 0 efficiency. Males were scored blindly. The Mann-Whitney test was used for statistical analysis.
Turning and location of vulva (lov) assays
Single males (isolated the night before as L4) were tested on a 10 μl-lawn of food (E. coli OP50) with 30 unc-51 hermaphrodites during 3 minutes. Upon response to contact, up to seven turns were scored as good or bad. Good turns: sharp ventral bend around the tip of the body of the mate, followed by uninterrupted scanning along the other side of the mate’s body. Bad turns: stutter (stops moving backwards before turning and moves forward before continuing with backward move); wide (tail positioned in a loose bend when turning around the tip of the mate); swim off (continues backing after reaching the end of the mate’s body and loses contact); or interrupted (after turning, the male does not continue backing along the mate’s body). The proportion of good turns out of total turns was scored.
For location of vulva efficiency, the number of encounters with the vulva until it was first located was scored. The assay finished when the male located the vulva or after three minutes, whatever happened first. Location efficiency was scored as 1/total number of encounters.
The Mann-Whitney test was used for statistical analysis.
Fertility assays
A single rab-3::rfp transgenic male was left to mate with two unc-51 hermaphrodites on a 15 μl-lawn of food (E. coli OP50) for 24 hours and then removed. The hermaphrodites were transferred to a fresh plate every 24 hours for a total of three days. The proportion of cross-progeny from total progeny was calculated for three ablated individuals and three controls. T-test was used for statistical analysis.
Food-leaving and retention assays
As described in 28.
Electron microscopy and serial reconstruction
Four samples were fixed by chemical fixation or high pressure freezing and freeze substitution as previously described 49. Ultrathin sections were cut using a RMC Powertome XL, collected onto grids, and imaged using either a Philips CM10 TEM or Zeiss Supra 40 FE-SEM. Sections were elastically aligned 50 and volumetrically reconstructed using TrakEM2 51 . The MCM cell bodies were identified in the EM sections based on position and morphology and in comparison to similar hermaphrodite sections. This identity was further confirmed by the identification of a posterior projection, as no other cell with its soma in the same region is known to project posteriorly. This was followed by serial tracing of the projections to establish their morphology and connectivity. Synaptic connectivity and skeleton diagrams were determined using Elegance 52 . Circuit diagrams of connectivity were generated using Cytoscape 53. We estimated the anatomical strength of synaptic connectivity between two neurons by summing the number of serial sections where we observed the ultrastructural morphology presynaptic components using the same criteria as 14
Extended Data
Extended Data Figure 1. The MCMs are newly identified male-specific neurons.
a, WormAtlas-style diagram depicting the morphology and position of one of the bilateral pair of MCM neurons in the head of a male worm and its projection within the nerve ring and along the ventral cord.
b, Volumetric reconstruction of the MCML cell body and projection based on tracing of serial EM sections.
c, Co-expression of transgenes for neuronal markers in the rab-3-positive cells identified as MCMs (indicated with dashed red circles). All photographs are lateral views of animals oriented anterior to the left and dorsal to the top except for ric-19, which are dorsal views. Transgenes are listed in table S1. pdf-1 (neuropeptide pigment dispersing factor); snb-1 (synaptobrevin); ida-1 (tyrosine phosphatse-like receptor, ortholog of mammalian phogrin); ric-19 (rab-2 effector); nca-1 (NALCN Na+ channel subunit); ccb-1 (voltage-gated Ca2+ channel subunit); unc-36 (voltage-gated Ca2+ channel subunit); inx-3 (gap junction innexin). R/L=Right/Left and D/V=Dorsal/Ventral.
d, Diagram of the neurons that directly connect to and from the MCMs. Triangles, sensory neurons; octagons, interneurons and unidentified neurons. The thickness of the arrows is proportional to the anatomical strength of the connections (Extended Data Table 2).
Extended Data Figure 2. The MCMs are not required for other male-specific behaviours.
a, Response of intact and MCM-ablated males (inIs179(ida-1prom::gfp);him-8(e1489) and otIs356(rab-3prom::rfp)him-5(e1490)) to dilutions of ascaroside pheromones (Ascr). Graphs represent Tukey box plots of logarithmic transformations of the data; n= number of independent events (i.e. entry in scoring region). T-test with Bonferroni correction was used for statistical analysis. ***P<0.001; **P<0.01; *P<0.05; n.s.= no statistically significant difference (p≥0.05).
b, Response efficiency to mate contact of intact, MCM-ablated and pdf-1(tm1996) mutant males measured as the proportion of responses out of total contacts with an hermaphrodite. 1 Intact animals were inIs179(ida-1prom::gfp);him-8(e1489). 2 Control animals were him-5(e1490). A response indicates that the male placed its tail ventral down on the mate’s body and backed along it to make a turn.
c and d, Proportion of good turns (c) and location of vulva efficiency (d) of intact and MCM-ablated males (inIs179(ida-1prom::gfp);him-8(e1489) and otIs356(rab-3prom::rfp)him-5(e1490)).
e, Fertility (measured as proportion of cross-progeny) of intact and MCM-ablated males (otIs356(rab-3prom::rfp)him-5(e1490)).
For b – e, n= number of individual animals tested. Error bars indicate SEM.
Mann-Whitney test was used for statistical analysis. *P<0.05; n.s.= no statistically significant difference (p≥0.05).
f, Mate-searching behavior, measured as PL values (probability of leaving food per hour) in the absence or presence of mates, of intact and MCM-ablated males (otIs356(rab-3prom::rfp)him-5(e1490)). n= number of individual animals tested. Two independent population assays were performed on different days. Maximum likelihood statistical analysis was used to compare PL values. Error bars indicate SEM. ***P<0.001; n.s.= no statistically significant difference (p≥0.05).
Extended Data Figure 3. The MCMs arise from a division of the AMso glial cell.
All photographs are lateral views of animals oriented anterior to the left and dorsal to the top.
a, Fluorescent photographs showing the two cells expressing rnr-1prom::gfp co-labeled with the glial marker ptr-10prom::rfp and the neuronal marker rab-3prom::rfp in the head of males at the early and late L4 stages. The AMso and MCM cell bodies are indicated with dashed lines.
b, Fluorescent images of the AMso cell body and its projection at two time points during cell division. Photos are overexposed for visualization of the projection, indicated by arrows. The chromosomes are labeled with a histone:rfp transgene, and the AMso cell body is indicated by dashed lines.
Extended Data Figure 4. AMso plasticity is regulated by AMso genetic sex.
a, Diagram of the left AMso and MCM lineage.
b and c, proportion of individuals with MCMs in control animals and animals expressing sex-reversing transgenes in AMso. b, AMso masculinization with grl-2prom::fem-3::SL2::mCherry transgenes (oleEx18 and oleEx24). c, AMso feminization with grl-2prom::tra-2IC::SL2::mCherry transgenes (oleEx19 and oleEx23) and ztf-16enhancer::tra-2IC::SL2::mCherry transgene oleEx22. MCM cell fate was identified with ida-1prom::gfp or rab-3prom::yfp reporter transgenes. In the head, the grl-2 promoter drives expression in AMso and the excretory duct and pore cells, and the ztf-16 glial enhancer drives expression in the AMso and amphid sheath glia. # indicates an independent transgenic array line for each manipulation. χ2 test was used for statistical analysis; ***P<0.001; n.s.= no statistical significant difference (p≥0.05); n= number of animals scored.
Extended Data Figure 5. The MCMs lose molecular and structural characteristics of glia after birth.
a, Proportion of MCMs with presence of the glial marker ptr-10prom::myrRfp or the neuronal marker ida-1prom::gfp at different stages after MCM birth.
b, Electron micrograph of a cross section of an adult male head showing the MCM and AMso cell body ultrastructure. Neighbouring tissues are color-coded according to WormAtlas (http://www.wormatlas.org/colorcode.htm). Purple (pharynx), muscle (green), hypodermis (light cream), AMso (amphid socket, pink). The dendrites of the amphid neurons (amphid bundle) are not colored.
Extended Data Table 1.
Reporter transgenes for neuronal markers tested for MCM expression
| protein/function | gene | array | MCM expression |
|---|---|---|---|
| Neuronal GTPase | rab-3 | otIs291 (rab-3prom:: NLS YFP+rol-6) and otIs356(rab-3prom::rfp) 1 | + 62/62 |
| Neuronal GEF | rgef-1 | evIs111 (rGEFprom:: GFP) 2 | + |
| Neuropeptide | pdf-1 | myEx696 (pdf-1prom::RFP+unc-122::GFP) 3 | + |
| Phogrin/Exocytosis | ida-1 | inIs179(ida-1prom::GFP) 4 | + 59/59 |
| Neuropeptide exocytosis | ric-19 | otEx6173 (ric-19prom6::NLS-TagRFP) (gift from Hobert lab) 5 | + |
| Syntaxin | unc-64 | otEx4553 (fosmid-based transcriptional reporter) (gift from Hobert lab) 5 | + |
| Synaptobrevin | snb-1 | otEx5422 (fosmid-based translational reporter) (gift from Hobert lab) | + |
| Synaptotagmin | snt-1 | otEx5924( fosmid-based transcriptional reporter) (gift from Hobert lab) 5 | + |
| Na2+ channel subunit | nca-1 | hpEx528 (nca-1prom::GFP) (gift from Zhen lab) | + |
| Ca2+ channel subunit | ccb-1 | sEX10836(T28F2.5prom::GFP+pCeh361) 6 | + |
| Ca2+ channel subunit | unc-36 | sEx12299(C50C3.9prom::GFP+pCeh361) 6 | + |
| Innexin | inx-3 | zwEx103 [inx-3prom::GFP +lin-15(+)] 7 | + |
| VGLUT | eat-4 | otIs388 (fosmid-based reporetr) 8 | − |
| Choline transporter | cho-1 | otIs323 (fosmid-based reporter) 9 | − |
| VGAT | unc-47 | otIs348[unc-47prom(300bp)::mCHOPTI::unc54-3’UTR), pha-1(+)] 9 | − |
| Intraflagellar transport (marker for ciliated neurons) | osm-6 | mnIs17(osm-6prom::osm-6:GFP) 10 | − |
| Fatty acid hydroxylase (marker for the XXX cell) | daf-9 | dhIs59(daf-9prom::GFP) 11 | − |
| Octopamine synthesis | tbh-1 | nIs107[tbh-1prom::GFP+lin-15(+)] 12 | − |
| Neuropeptide | flp-21 | nyIs80 (flp-21prom::GFP) 13 | − |
| Nematocin receptor | ntr-1 | lstEx33 [ntr-1prom::gfp] 14 | − |
| Ca2+ channel subunit | cca-1 | sEx14060(C54D2.5aprom::GFP+pCeh361) 6 | − |
| K+ channel subunit | unc-103 | rgEx[unc-103prom::YFP + pBx1] 15 | − |
| Innexin | inx-12 | zwEx112 [inx-12prom::GFP +lin-15(+)] 7 | − |
| Innexin | inx-13 | zwEx113 [inx-13prom::GFP +lin-15(+)] 7 | − |
| Innexin | inx-5 | zwEx105 [inx-5prom::GFP +lin-15(+)] 7 | − |
| Innexin | inx-12 | zwEx112 [inx-12prom::GFP +lin-15(+)] 7 | − |
+ indicates presence; − indicates absence. For integrated arrays, quantification is indicated. At least 15 animals were examined for each transgene.
Tursun, B. et al. Science (2011).
Altun, Z.F. et al. Development (2001).
Barrios, A. et al. Nature Neuroscience (2012).
Zahn, T. R. et al. (2000).
Stefanakis, N. et al. Neuron (2015).
McKay, S. J. et al. Cold Spring Harb. Symp. Quant. Biol. (2003).
Altun, Z.F. et al. Dev Dyn. 238, 1936 (2009).
Serrano-Saiz, E. et al. Cell (2013).
Kratsios, P. et al. Nature Neuroscience (2011).
Bell, L. R. et al. Genetics (2006).
Gerisch, B. et al. Dev. Cell (2001).
Alkema, M. J. et al. Neuron (2005).
Kim, K. & Li, C., J. Comp. Neurol. (2004).
Beets, I. et al. Science (2012).
Gruninger, T. R. et al. J. Neurosci. (2006).
Extended Data Table 2.
MCM connectivity
| type | presynaptic | postsynaptic | #synapses | #EM sections |
|---|---|---|---|---|
| chemical | AVF | MCM | 16 | 61 |
| chemical | AVJ | MCM | 1 | 2 |
| chemical | AVM | MCM | 1 | 1 |
| chemical | BDU | MCM | 3 | 7 |
| chemical | CEM | MCM | 2 | 5 |
| chemical | CEP | MCM | 1 | 1 |
| chemical | EF | MCM | 6 | 35 |
| chemical | MCM | ADA | 1 | 3 |
| chemical | MCM | ALM | 2 | 3 |
| chemical | MCM | AQR | 2 | 3 |
| chemical | MCM | AVB | 29 | 61 |
| chemical | MCM | AVF | 1 | 2 |
| chemical | MCM | AVH | 1 | 2 |
| chemical | MCM | BDU | 1 | 3 |
| electrical | MCM | CEM | 1 | 1 |
| chemical | MCM | CEM | 5 | 8 |
| chemical | MCM | CEP | 1 | 1 |
| chemical | MCM | EF | 2 | 3 |
| chemical | MCM | MCM | 2 | 6 |
| chemical | MCM | PVC | 1 | 2 |
| chemical | MCM | PVN | 3 | 3 |
| chemical | MCM | PVU | 1 | 1 |
| chemical | MCM | RIC | 2 | 3 |
| chemical | MCM | RID | 1 | 3 |
| chemical | MCM | RIF | 11 | 23 |
| chemical | MCM | RIM | 3 | 7 |
| chemical | MCM | RIPR | 3 | 6 |
| chemical | MCM | RMD | 1 | 2 |
| chemical | MCM | SIA | 1 | 2 |
| chemical | MCM | SMB | 2 | 3 |
| chemical | MCM | unk1* | 2 | 4 |
| chemical | MCM | unk10* | 1 | 2 |
| chemical | MCM | unk11* | 1 | 2 |
| chemical | MCM | unk12* | 1 | 1 |
| chemical | MCM | unk13* | 1 | 1 |
| chemical | MCM | unk2* | 1 | 3 |
| chemical | MCM | unk3* | 1 | 2 |
| electrical | MCM | unk6* | 1 | 1 |
| chemical | MCM | unk7* | 3 | 7 |
| chemical | MCM | unk8* | 4 | 4 |
| chemical | MCM | unk9* | 1 | 2 |
| chemical | PVC | MCM | 1 | 3 |
| chemical | PVQ | MCM | 1 | 4 |
| chemical | PVS | MCM | 1 | 2 |
| chemical | RIC | MCM | 1 | 1 |
| chemical | RID | MCM | 1 | 1 |
| chemical | RIF | MCM | 10 | 20 |
| chemical | RIM | MCM | 3 | 3 |
| chemical | RMG | MCM | 1 | 2 |
| chemical | SMB | MCM | 2 | 3 |
| chemical | unk14* | MCM | 3 | 15 |
| chemical | unk15* | MCM | 1 | 5 |
| chemical | unk16* | MCM | 1 | 1 |
| chemical | unk4* | MCM | 2 | 3 |
| chemical | unk5* | MCM | 1 | 1 |
unk refers to neurons whose identity has not been unambiguously confirmed in the EM serial sections
Extended Data Table 3.
Cell ablations of candidate MCM progenitors
| ablated cell | stage | animals with loss of MCM/total |
|---|---|---|
| AMso | L3 | 6/7 |
| AMso | L4 | 0/3 |
| H0 seam cell | L1 | 1/16 |
| H1 seam cell | L1 | 0/2 |
| H0+H1 | L1 | 0/1 |
| Hyp1 (lateral nucleus) | L1 | 0/4 |
| H0+Hyp1(lateral nucleus) | L1 | 0/1 |
Extended Data Table 4.
Mosaic analysis of sex-transformation arrays, scoring the presence of MCMs
| sex reversal | array | array in AMso and other cells* | array in other cells* only |
|---|---|---|---|
| Masculinised hermaphrodites | oleEx18 (grl-2prom::fem-3:SL2:mCherry) | 89% (n=28) | 0% (n=22) |
| Feminised males | oleEx19 (grl-2prom::tra-2IC:SL2:mCherry) | 0% (n=39) | 100% (n=7) |
| Feminised males | oleEx22 (ztf-16prom::tra-2IC:SL2:mCherry) | 2.5% (n=40) | 87.5%(n=16) |
Expression of oleEx18 (grl-2prom::fem-3:SL2:mCherry) in the head was observed in AMso, excretory duct and pore cells and sometimes in the pharynx and/or hypodermis. Expression of oleEx19 (grl-2prom::tra-2IC:SL2:mCherry) in the head was observed in AMso, excretory duct and pore cells and sometimes in the hypodermis. Expression of oleEx22 (ztf-16prom::tra-2IC:SL2:mCherry) in the head was observed in AMso and sometimes in the amphid sheath and/or a neuron in the nerve ring.
Extended Data Table 5.
Reporter transgenes for glial/AMso markers
| protein/function | gene | array | glial subtype expression | AMso expression |
|---|---|---|---|---|
| Patched (PTCHD3) related receptor | ptr-10 1 | nsIs108[ptr-10prom::myristyl-Rfp] | All glia | + |
| Hedgehog-like/Ground-related | grl-2 2 | sEx12852[T16G1.8prom::GFP+pCeh361] | AMso and PHso | + |
| Inositol trisphospate receptor |
itr-1 {Gower:2001ix, Heiman:2009bj} |
jwEx51(itr1promB::GFP+rol-6) and nsEX1153 [F16F9.3prom::mCherry+itr-1prom::CFP+rol-6(su1006)] | AMso | + |
| C2H2 zinc-finger transcription factor | ztf-16 3 | oleEx22[ztf-16enhancer::tra-2IC::SL2cherry+elt-2::GFP] | AMso, AMsh, PHso | + |
| Basic helix-loop-helix transcription factor | hlh-17 1 | leEx1713[hlh-17prom::GFP+unc-119(+)] | CEPsh | − |
| Caspr (Neurexin superfamily) | itx-1 4 | otEx[W03D8.6prom::GFP+rol-6] | OL and IL glia | − |
+ indicates presence; − indicates absence. At least 15 animals were examined for each transgene. AMso, amphid socket; PHso, phasmid socket; AMsh, amphid sheath; PHsh, phasmid sheath; CEPsh, cephalic sensilla sheath; OLL, outer labial; IL, inner labial.
Yoshimura, S. et al. Development (2008).
Hao, L. et al. BMC Genomics (2006).
Procko, C. et al. Genetics (2012).
Haklai-Topper, L. et al. Gene Expr. Patterns (2011).
Acknowledgements
We would like to acknowledge Maureen Barr, in whose laboratory A.B. discovered the MCMs; WormAtlas for illustrations (with permission); Travis Jarrell for contributions to the EM reconstruction; and Will Letton for the generation of strains and preliminary ablation studies. We thank Mike Boxem, Doug Portman, Howard Baylis, Laura Bianchi and Rene Garcia for strains and reagents; Mei Zhen, Oliver Hobert, Ines Carrera, Nikos Stefanakis and Shai Shaham, for unpublished reagents. Purified ascarosides were obtained from a gift of Frank Schroeder to the Barr lab. Additional strains were obtained from the CGC, which is funded by NIH grant P40 OD010440. We thank Luisa Cochella, Ines Carrera, Sophie Jarriault, and several of our close colleagues in CDB and NPP at University College London for discussions and comments on the manuscript; Chris Barnes for advice on statistical analysis.
This work was supported by a Master it! Scholarship Scheme (Malta and EU) to M.S., by NIH grant OD010943 to D.H.H., by Marie Curie CIG grant 618779 to R.J.P. and by a grant from The G. Harold and Leila Y. Mathers Charitable Foundation to S.W.E; S.J.C is supported by NIH grant 5T32GM007491; R.J.P. is a Wellcome Trust Research Career Development Fellow 095722/Z/11/Z; A.B. is supported by the Wellcome Trust Institutional Strategic Support Fund097815/Z/11/A.
Footnotes
The authors declare no competing financial interests.
References
- 1.Kimura K-I, Ote M, Tazawa T, Yamamoto D. Fruitless specifies sexually dimorphic neural circuitry in the Drosophila brain. Nature. 2005;438:229–233. doi: 10.1038/nature04229. [DOI] [PubMed] [Google Scholar]
- 2.Ruta V, et al. A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature. 2010;468:686–690. doi: 10.1038/nature09554. [DOI] [PubMed] [Google Scholar]
- 3.Yang CF, et al. Sexually dimorphic neurons in the ventromedial hypothalamus govern mating in both sexes and aggression in males. Cell. 2013;153:896–909. doi: 10.1016/j.cell.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nature Neuroscience. 2010;13:458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stowers L, Logan DW. Sexual dimorphism in olfactory signaling. Curr. Opin. Neurobiol. 2010;20:770–775. doi: 10.1016/j.conb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nottebohm F, Arnold AP. Sexual dimorphism in vocal control areas of the songbird brain. Science. 1976;194:211–213. doi: 10.1126/science.959852. [DOI] [PubMed] [Google Scholar]
- 7.Keleman K, et al. Dopamine neurons modulate pheromone responses in Drosophila courtship learning. Nature. 2012;489:145–149. doi: 10.1038/nature11345. [DOI] [PubMed] [Google Scholar]
- 8.Sulston JE, Albertson DG, Thomson JN. The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev. Biol. 1980;78:542–576. doi: 10.1016/0012-1606(80)90352-8. [DOI] [PubMed] [Google Scholar]
- 9.Jarrell TA, et al. The Connectome of a Decision-Making Neural Network. Science. 2012;337:437–444. doi: 10.1126/science.1221762. [DOI] [PubMed] [Google Scholar]
- 10.Sakai N, et al. A sexually conditioned switch of chemosensory behavior in C. elegans. PLoS ONE. 2013;8:e68676. doi: 10.1371/journal.pone.0068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WormAtlas (2002-2015) at < http://www.wormatlas.org>.
- 12.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 13.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 14.White JG, Southgate E, Thomson JN, Brenner S. The Structure of the Nervous System of the Nematode Caenorhabditis elegans. Philosophical Transactions of the Royal Society B: Biological Sciences. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 15.Hall DH, Russell RL. The posterior nervous system of the nematode Caenorhabditis elegans: serial reconstruction of identified neurons and complete pattern of synaptic interactions. J. Neurosci. 1991;11:1–22. doi: 10.1523/JNEUROSCI.11-01-00001.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward S, Thomson N, White JG, Brenner S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans.?2UU. J. Comp. Neurol. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- 17.Varshney LR, Chen BL, Paniagua E, Hall DH, Chklovskii DB. Structural properties of the Caenorhabditis elegans neuronal network. PLoS Comput. Biol. 2011;7:e1001066. doi: 10.1371/journal.pcbi.1001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrios A. Exploratory decisions of the Caenorhabditis elegans male: a conflict of two drives. Semin. Cell Dev. Biol. 2014;33:10–17. doi: 10.1016/j.semcdb.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Ryan DA, et al. Sex, Age, and Hunger Regulate Behavioral Prioritization through Dynamic Modulation of Chemoreceptor Expression. Curr. Biol. 2014;24:2509–2517. doi: 10.1016/j.cub.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saeki S, Yamamoto M, Iino Y. Plasticity of chemotaxis revealed by paired presentation of a chemoattractant and starvation in the nematode Caenorhabditis elegans. J. Exp. Biol. 2001;204:1757–1764. doi: 10.1242/jeb.204.10.1757. [DOI] [PubMed] [Google Scholar]
- 21.Vellai T, McCulloch D, Gems D, Kovács AL. Effects of sex and insulin/insulin-like growth factor-1 signaling on performance in an associative learning paradigm in Caenorhabditis elegans. Genetics. 2006;174:309–316. doi: 10.1534/genetics.106.061499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srinivasan J, et al. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature. 2008;454:1115–1118. doi: 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrios A, Nurrish S, Emmons SW. Sensory regulation of C. elegans male mate-searching behavior. Curr. Biol. 2008;18:1865–1871. doi: 10.1016/j.cub.2008.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barr MM, Sternberg PW. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401:386–389. doi: 10.1038/43913. [DOI] [PubMed] [Google Scholar]
- 25.Barr MM, García LR. Male mating behavior. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.78.1. doi:10.1895/wormbook.1.78.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipton J, Kleemann G, Ghosh R, Lints R, Emmons SW. Mate searching in Caenorhabditis elegans: a genetic model for sex drive in a simple invertebrate. J. Neurosci. 2004;24:7427–7434. doi: 10.1523/JNEUROSCI.1746-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janssen T, et al. Discovery and characterization of a conserved pigment dispersing factor-like neuropeptide pathway in Caenorhabditis elegans. Journal of Neurochemistry. 2009;111:228–241. doi: 10.1111/j.1471-4159.2009.06323.x. [DOI] [PubMed] [Google Scholar]
- 28.Barrios A, Ghosh R, Fang C, Emmons SW, Barr MM. PDF-1 neuropeptide signaling modulates a neural circuit for mate-searching behavior in C. elegans. Nature Neuroscience. 2012;15:1675–7682. doi: 10.1038/nn.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egger B, Chell JM, Brand AH. Insights into neural stem cell biology from flies. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2008;363:39–56. doi: 10.1098/rstb.2006.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oikonomou G, Shaham S. The Glia of Caenorhabditis elegans. Glia. 2011;59:1253–1263. doi: 10.1002/glia.21084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong Y, Roy R, Ambros V. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development. 1998;125:3585–3597. doi: 10.1242/dev.125.18.3585. [DOI] [PubMed] [Google Scholar]
- 32.Zarkower D. Somatic sex determination. WormBook. 2006:1–12. doi: 10.1895/wormbook.1.84.1. doi:10.1895/wormbook.1.84.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodgkin J. A genetic analysis of the sex-determining gene, tra-1, in the nematode Caenorhabditis elegans. Genes & Development. 1987;1:731–745. doi: 10.1101/gad.1.7.731. [DOI] [PubMed] [Google Scholar]
- 34.Procko C, Lu Y, Shaham S. Sensory Organ Remodeling in Caenorhabditis elegans Requires the Zinc-Finger Protein ZTF-16. Genetics. 2012;190:1405–1415. doi: 10.1534/genetics.111.137786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hao L, Johnsen R, Lauter G, Baillie D, Bürglin TR. Comprehensiveanalysis of gene expression patterns of hedgehog-related genes. BMC Genomics. 2006;7:280. doi: 10.1186/1471-2164-7-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White JQ, et al. The sensory circuitry for sexual attraction in C. elegans males. Curr. Biol. 2007;17:1847–1857. doi: 10.1016/j.cub.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 37.Lee K, Portman DS. Neural sex modifies the function of a C. elegans sensory circuit. Curr. Biol. 2007;17:1858–1863. doi: 10.1016/j.cub.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 38.Mowrey WR, Bennett JR, Portman DS. Distributed Effects of Biological Sex Define Sex-Typical Motor Behavior in Caenorhabditis elegans. J. Neurosci. 2014;34:1579–1591. doi: 10.1523/JNEUROSCI.4352-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.WormBase. at < http://www.wormbase.org/species/all/anatomy_term/WBbt:0003929#01--10 >.
- 40.Jarriault S, Schwab Y, Greenwald I. A Caenorhabditis elegans model for epithelial-neuronal transdifferentiation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3790–3795. doi: 10.1073/pnas.0712159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okada TS. Transdifferentiation. Flexibility in Cell Differentiation. Clarendon Press; 1991. [Google Scholar]
- 42.Eguchi G, Kodama R. Transdifferentiation. Current Opinion in Cell Biology. 1993;5:1023–1028. doi: 10.1016/0955-0674(93)90087-7. [DOI] [PubMed] [Google Scholar]
- 43.Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nature Neuroscience. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 44.Doetsch F. The glial identity of neural stem cells. Nature Neuroscience. 2003;6:1127–1134. doi: 10.1038/nn1144. [DOI] [PubMed] [Google Scholar]
- 45.Ninkovic J, Gotz M. Fate specification in the adult brain--lessons for eliciting neurogenesis from glial cells. Bioessays. 2013;35:242–252. doi: 10.1002/bies.201200108. [DOI] [PubMed] [Google Scholar]
- 46.Zuryn S, et al. Transdifferentiation. Sequential histone-modifying activities determine the robustness of transdifferentiation. Science. 2014;345:826–829. doi: 10.1126/science.1255885. [DOI] [PubMed] [Google Scholar]
- 47.Arnold AP. Developmental plasticity in neural circuits controlling birdsong: Sexual differentiation and the neural basis of learning. J. Neurobiol. 1992;23:1506–1528. doi: 10.1002/neu.480231010. [DOI] [PubMed] [Google Scholar]
- 48.Bargmann CI, Avery L. Laser killing of cells in Caenorhabditis elegans. Methods Cell Biol. 1995;48:225–250. doi: 10.1016/s0091-679x(08)61390-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hall DH, Hartwieg E, Nguyen KCQ. Modern electron microscopy methods for C. elegans. Methods Cell Biol. 2012;107:93–149. doi: 10.1016/B978-0-12-394620-1.00004-7. [DOI] [PubMed] [Google Scholar]
- 50.Saalfeld S, Fetter R, Cardona A, Tomancak P. Elastic volume reconstruction from series of ultra-thin microscopy sections. Nat Meth. 2012;9:717–720. doi: 10.1038/nmeth.2072. [DOI] [PubMed] [Google Scholar]
- 51.Cardona A, et al. TrakEM2 software for neural circuit reconstruction. PLoS ONE. 2012;7:e38011. doi: 10.1371/journal.pone.0038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu M, et al. Computer assisted assembly of connectomes from electron micrographs: application to Caenorhabditis elegans. PLoS ONE. 2013;8:e54050. doi: 10.1371/journal.pone.0054050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]