Abstract
Enhanced sensitivity to Wnts is an emerging hallmark of a subset of cancers, defined in part by mutations regulating the abundance of their receptors. Whether these mutations identify a clinical opportunity is an important question. Inhibition of Wnt secretion by blocking an essential post-translational modification, palmitoleation, provides a useful therapeutic intervention. We developed a novel potent, orally available PORCN inhibitor, ETC-1922159 (henceforth called ETC-159) that blocks the secretion and activity of all Wnts. ETC-159 is remarkably effective in treating RSPO-translocation bearing colorectal cancer (CRC) patient-derived xenografts. This is the first example of effective targeted therapy for this subset of CRC. Consistent with a central role of Wnt signaling in regulation of gene expression, inhibition of PORCN in RSPO3-translocated cancers causes a marked remodeling of the transcriptome, with loss of cell cycle, stem cell and proliferation genes, and an increase in differentiation markers. Inhibition of Wnt signaling by PORCN inhibition holds promise as differentiation therapy in genetically defined human cancers.
Introduction
Wnts are a family of 19 evolutionarily conserved cysteine rich morphogens that interact with at least 15 different receptors and co-receptors to regulate a multitude of developmental and homeostatic processes.1, 2 Wnts signal through both β-catenin dependent and β-catenin independent pathways. Dysregulation of Wnt signaling is thought to be causal in a subset of cancers due to mutations in either upstream or downstream components.3, 4 Mutations in downstream genes that result in stabilization of β-catenin protein have been well documented. More recently, cancer-associated mutations that alter the abundance of Wnt receptors Frizzled and lipoprotein-related receptor 5 or 6 (LRP5/6) have been reported, which adds to the complexity of Wnt signaling in cancer.5, 6, 7, 8, 9, 10, 11, 12 The E3 ubiquitin ligases ring-finger protein 43 (RNF43) and zinc and ring finger 3 (ZNRF3) negatively regulate Wnt signaling by ubiquitinating the Frizzled and LRP5/6 receptors, promoting their endocytosis and subsequent degradation.13, 14 The secreted Wnt agonists of the R-spondin family, RSPO1-4, in turn negatively regulate RNF43/ZNRF3. Gain of function gene fusions involving RSPO2 and RSPO3 lead to increased cell surface abundance of Frizzleds and LRP5/6 and consequently enhanced Wnt signaling.6, 9, 13 This is clinically relevant for a subset of patients with difficult to treat cancers. Chromosomal translocations fusing the regulatory sequences of EIF3a or PTPRK with RSPO2 and RPSO3 are found in 10% of APC wild-type colon cancers6, 7 and with varying frequencies (~1–11%) in ovarian, esophageal, lung and head and neck cancers.15 Thus, there appears to be a subset of cancers driven by enhanced cellular sensitivity to Wnts.
Wnt driven cancers can be targeted at several steps in the pathway.16, 17 One approach is to target the secretion of all Wnts by inhibiting the enzymatic activity of Porcupine (PORCN), an endoplasmic reticulum resident enzyme that post-translationally palmitoleates Wnts at a highly conserved serine residue.18, 19 This palmitoleation of Wnts is essential for their secretion and binding to the Frizzled receptors.20, 21, 22 Inhibition of PORCN enzymatic activity offers an approach to overcome the limitations of β-catenin inhibitors that can only block the canonical Wnt signaling pathway23, 24, 25, 26 or the anti-Frizzled antibodies that are limited in their ability to target all the Frizzled receptors.17
Here we describe the efficacy of a novel small molecule inhibitor of PORCN, ETC-159. ETC-159 has robust activity in multiple cancer models driven by high Wnt signaling. Most importantly, we have identified that ETC-159 is highly efficacious in molecularly defined colorectal cancers (CRCs) with R-spondin translocations. Consistent with a broad Wnt-dependent signaling network, CRCs with gain of function RSPO mutations respond to ETC-159 treatment with a rapid and marked shift in the transcriptome including a highly significant decrease in proliferation and stem cell markers, and an increase in differentiation genes. RSPO translocations are novel predictive biomarkers for identifying Wnt ligand-dependent cancers that are responsive to a new class of Wnt-pathway inhibitors.
Results
Identification of novel inhibitors of Wnt signaling
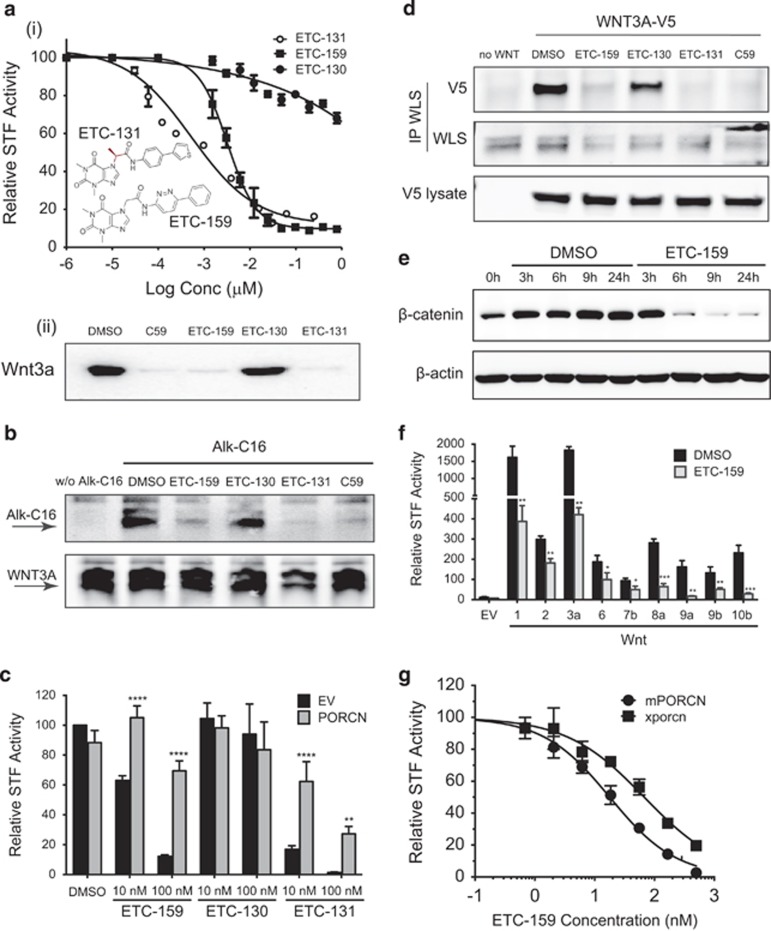
To identify potent inhibitors of Wnt secretion, we screened a library of ~225 755 small molecules using a multi-step cell-based screen.27 HEK293 cells with constitutive high Wnt/β-catenin signaling due to stable expression of WNT3A and harboring a luciferase-based Wnt/β-catenin reporter (Super 8xTOPFLASH) (STF3A cells) were incubated with small molecules for 24 h. Luciferase reporter activity was used as a measure of Wnt-pathway activity. To specifically identify inhibitors of Wnt secretion, potent compounds that were not cytotoxic were tested using a HEK293 cell line with an integrated STF reporter plasmid (STF cells) and exogenously supplied WNT3A conditioned medium. Compounds that selectively inhibited signaling in STF3A but not in STF cells with WNT3A conditioned medium and had an IC50 <1 μM were selected as potential PORCN inhibitors. This screen identified a novel pharmacophore that was further refined by reiterative structure/activity analysis into a compound named ETC-159 and its related derivatives (Figure 1a, inset) that are potent inhibitors of Wnt secretion. ETC-131 and ETC-159 inhibited β-catenin reporter activity in a dose-dependent manner with an IC50 of 0.5 nM and 2.9 nM, respectively (Figure 1a, i). ETC-131 showed stereo-specificity, as its enantiomer ETC-130 was four logs less potent, with an IC50 of 5.4 μM. ETC-159, ETC-131 and the chemically unrelated PORCN inhibitor Wnt-C59, but not ETC-130, effectively inhibited the secretion of WNT3A into culture media (Figure 1a, ii) but did not inhibit β-catenin signaling in STF cells supplemented with Wnt3A-conditioned medium (IC50 >10 μM) (Supplementary Figures 1A and B). These data confirm that ETC-159 and ETC-131 are potent and specific inhibitors of Wnt secretion.
Figure 1.
Development of novel PORCN inhibitors. (a) ETC-159 and ETC-131 inhibit Wnt/β-catenin reporter activity and secretion of Wnt3a (i) STF3A cells were treated with the indicated compounds and luciferase activity was measured after 24 h. Data represents mean±s.d. Inset: structure of ETC-159 and ETC-131. (ii) STF3A cells were treated with 100 nM of the indicated compounds and Wnt secretion was assessed by immunoblot of Wnt3A protein in culture supernatants. (b) ETC-159 and ETC-131 inhibit Wnt3A palmitoleation: HeLa cells transiently expressing Wnt3A-V5 were metabolically labeled for 16 h with alkyne-palmitate (Alk-C16) in the presence of 100 nM of the indicated compounds. Biotin-azide-clicked palmitate (upper panel), Wnt3a-V5 (lower panel). (c) PORCN overexpression rescues the inhibition of β-catenin reporter activity: HT1080 cells transfected with Wnt3a, PORCN expression plasmids, and Super 8xTOPFLASH reporter were treated as indicated for 16 h. The cells were harvested and luciferase activity was measured. Data represents mean±s.d. Data were analyzed using unpaired t-test and corrected for multiple comparisons. **P⩽0.01, ****P⩽0.0001 (d) ETC-159 and ETC-131 prevent interaction of Wnt with Wntless: HeLa cells transiently expressing Wnt3A-V5 were treated overnight with 100 nM of the indicated compounds before immunoprecipitation of endogenous Wntless. (e) ETC-159 promotes β-catenin degradation: mouse L cells stably expressing Wnt3a were trypsinizised and treated with DMSO or 100 nM ETC-159 before plating in cell culture dishes. The cells were harvested at indicated time points and total β-catenin levels were assessed by immunoblot. (f) ETC-159 inhibits Wnt/β-catenin reporter activity induced by diverse Wnts: HT1080 cells were transfected with the Super 8xTOPFLASH and indicated Wnt expression plasmid, and then treated with 100 nM ETC-159 for 24 h before luciferase assay. Bars represent the mean±s.d. Data were analyzed using unpaired t-test. *P⩽0.05, **P⩽0.01, ***P⩽0.001 (g) ETC-159 is a more potent inhibitor of mammalian PORCN: porcupine null HT1080 cells transfected with either a murine (1 ng) or Xenopus porcn (0.75 ng) expression plasmid and Super 8xTOPFLASH reporter were treated with indicated concentrations of ETC-159. The β-catenin reporter activity is shown as percentage of the DMSO treated control. Data represent the mean±s.d.
PORCN-mediated O-palmitoleation is indispensable for the secretion and function of all Wnts.21 To test if these novel compounds inhibit Wnt palmitoleation, Wnt3A-V5 was expressed in HeLa cells metabolically labeled with alkyne-palmitic acid.28,29 Alkyne-palmitoylated Wnt was detected by immunoprecipitation and click-chemistry coupling of azido-biotin to the alkyne moiety. Treatment of cells with either ETC-159, ETC-131 or Wnt-C59 prevented the incorporation of palmitate into Wnt3A (Figure 1b). Furthermore, overexpression of PORCN reversed the inhibitory effects of ETC-131 and ETC-159 (Figure 1c). These findings indicate that PORCN is the direct target of ETC-131 and ETC-159.
Wnt palmitoleation is required for its interaction with its carrier protein WLS.20 Treatment of cells with ETC-159, ETC-131 or Wnt-C59 but not an inactive enantiomer ETC-130 prevented this interaction (Figure 1d), explaining why PORCN inhibitors block Wnt secretion. Inhibition of PORCN also led to a reproducible decrease in WLS protein abundance, which may be a consequence of altered WLS expression or trafficking.30 ETC-159 treatment also caused decreased abundance of Wnt3a-stabilized β-catenin protein in both mouse L cells and HEK293 cells (Figure 1e and Supplementary Figure 1C). Confirming the central role of PORCN in the biogenesis of all Wnts, ETC-159 inhibited β-catenin signaling in response to multiple active Wnts (Figure 1f).31
Xenopus laevis Porcn is less sensitive than mammalian PORCN to inhibition by Wnt-C59.21 We therefore compared the activity of ETC-159 against Xenopus Porcn, which is 77% identical to mouse PORCN. HT1080 cells null for PORCN32 were reconstituted by transient expression of either mouse HA-Porcn-D or Xenopus HA-porcn expression plasmids at near-identical expression and activity levels as assessed by immunoblot and TOPFLASH assay (Supplementary Figures 1D and E). ETC-159 inhibited mouse PORCN with an IC50 of 18.1 nM, whereas the IC50 for Xenopus Porcn was approximately fourfold higher (70 nM) (Figure 1g). These differences in activity demonstrate its selectivity for mammalian PORCN and provide genetic evidence that PORCN is the molecular target of ETC-159.
ETC-159 is orally bioavailable and effectively inhibits the growth of mouse mammary tumor virus-Wnt1 tumors
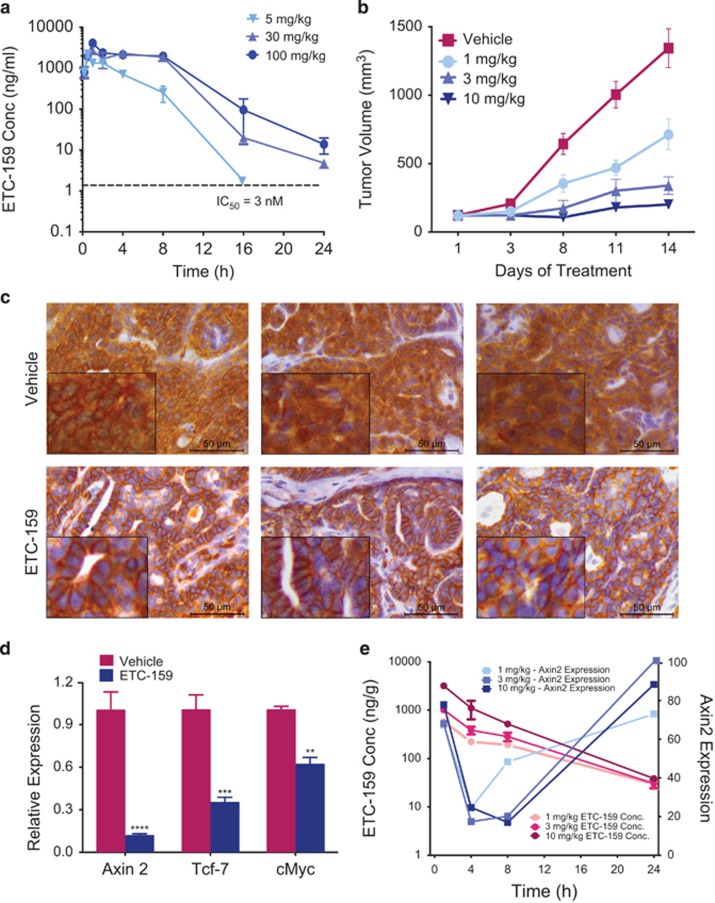
ETC-159 exhibits good oral pharmacokinetics in mice allowing preclinical evaluation via oral administration. After a single oral dose of 5 mg/kg, ETC-159 was rapidly absorbed into the blood with a Tmax of ~0.5 h and oral bioavailability of 100%. The plasma half-life was ~1.18 h and its concentration in the blood remained above the in vitro IC50 for at least 16 h (Figure 2a). Treatment of mice with increasing doses of ETC-159 led to a dose-related increase in exposure (Figure 2a). ETC-131 had poor oral bioavailability and was therefore used only for in vitro assays.
Figure 2.
ETC-159 is orally bioavailable and effectively inhibits the growth of mouse mammary tumor virus (MMTV)-Wnt1 tumors.(a) ETC-159 is orally bioavailable with a dose proportional increase in mouse plasma: Plasma levels of ETC-159 were measured following a single oral dose of 5, 30 or 100 mg/kg. The data is presented as mean±s.d. (b) Anti-tumor efficacy of ETC-159 in the MMTV-Wnt1 orthotopic-mouse model: BALB/c nude mice were implanted in the 4th fat-pad with a fragment of MMTV-Wnt1 tumor from a transgenic mouse. Following the development of palpable tumors, mice were randomized into four groups matched for tumor size and treated daily unblinded either with vehicle or with indicated doses of ETC-159. Data are presented as mean±s.e.m. n=8/group. (c) PORCN inhibition promotes nuclear and cytoplasmic exclusion of β-catenin: Tumors harvested at the end of the study were stained for β-catenin. Representative sections from three independent tumors of vehicle and ETC-159 treatment group are shown. Scale bar=50 μm. (d) Downregulation of Wnt target gene expression: Transcript abundance of select Wnt/β-catenin target genes was assessed in the vehicle and ETC-159 treated tumors harvested 6 h after the last dose. n=8/group. Data were analyzed using unpaired t-test. **P⩽0.01, ***P⩽0.001, ****P⩽0.0001 (e) Robust but transient pharmacodynamic response of Axin2 to ETC-159: Axin2 mRNA and ETC-159 drug concentration were measured in MMTV-Wnt1 tumors at the indicated times following 1, 3 or 10 mg/kg dose. Data are presented as mean±s.d. n= 3/group.
To identify the doses of ETC-159 that are well tolerated, non-tumor bearing BALB/c nude mice were treated daily with 10, 50 and 100 mg/kg ETC-159 for 7 days and were then observed for an additional 7 days off treatment. There was no significant weight loss or visible signs of toxicity at these doses (Supplementary Figure 2A). Histological analysis of tissues collected from various intestinal compartments revealed normal architecture in treated mice from all groups (Supplementary Figure 2B).
The anti-tumor efficacy of ETC-159 was first tested using a well-established Wnt dependent, murine mammary cancer model, mouse mammary tumor virus-Wnt1. Mice carrying an mouse mammary tumor virus LTR-Wnt1 transgene have marked overexpression of Wnt1 in the mammary gland, driving hyperplasia and an eventual development of adenocarcinomas.33 Tumor fragments from mouse mammary tumor virus-Wnt1 cancers were orthotopically transplanted into the fourth mammary fat-pad of BALB/c nude mice. ETC-159 inhibited tumor growth by 52% and 78% at 1 and 3 mg/kg by once daily gavage, respectively. Tumor growth inhibition of 94% was obtained at 10 mg/kg/day (Figure 2b). Importantly, at these doses of ETC-159 there were no signs of toxicity and little effect on body weight (Supplementary Figure 2C).
To confirm that ETC-159 was inhibiting Wnt/β-catenin signaling in vivo, we examined the tumors from control and treated groups. Control tumors had abundant nuclear and cytoplasmic β-catenin staining, whereas in treated tumors β-catenin re-localized to the membrane (Figure 2c and Supplementary Figure 2D). The decrease in cytoplasmic and nuclear abundance of β-catenin was accompanied by a treatment-induced decrease in expression of the β-catenin target genes Axin2, Tcf7 and c-Myc (Figure 2d).
To assess drug penetration into tumor tissue, tumors were harvested at the indicated times following a single oral dose of 1, 3 or 10 mg/kg ETC-159. Drug concentrations in the tumors were initially high and fell as predicted by the plasma concentration. There was a maximal 80% inhibition of AXIN2 expression in the tumors from mice treated with 3 or 10 mg/kg between 4 and 8 h after treatment (Figure 2e). Notably, AXIN2 expression returned to normal before the next dose. As these doses were highly effective in blocking tumor growth, it suggests that intermittent Wnt-pathway inhibition is sufficient to obtain good anti-tumor efficacy.
ETC-159 effectively inhibits Wnt autocrine signaling and growth of teratocarcinomas
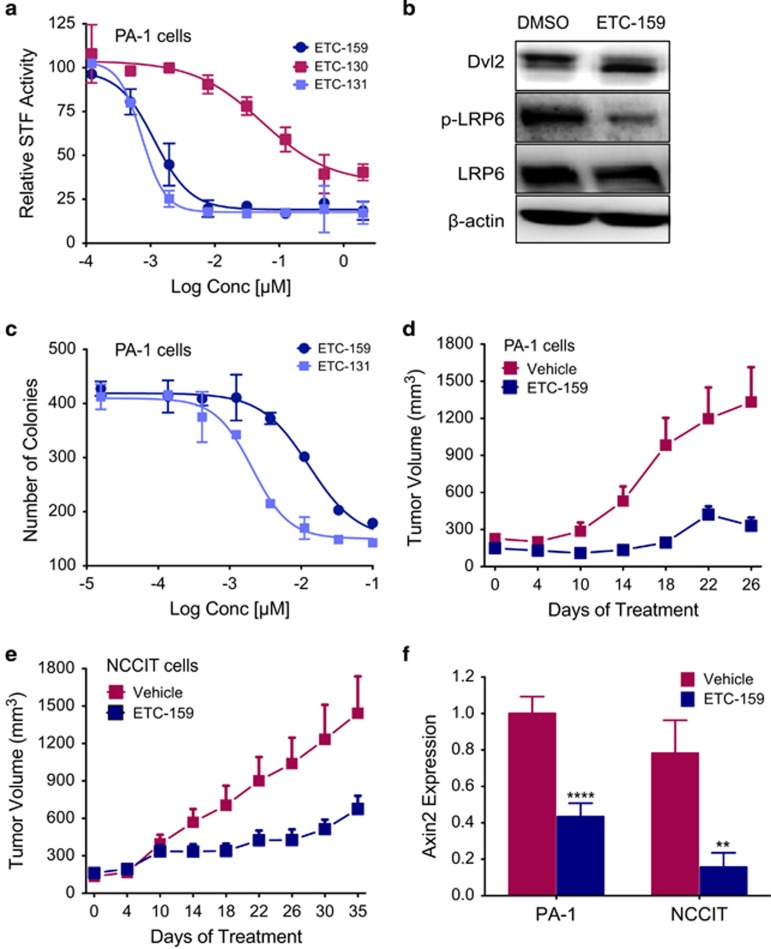
We next tested human cancer cell lines for their sensitivity to PORCN inhibitors. The human teratocarcinoma cell lines PA-1 and NCCIT have high autocrine Wnt signaling and express multiple Wnts, and the Wnt target gene AXIN2.34 Consistent with this, PA-1 cells transfected with the STF β-catenin reporter plasmid showed robust reporter activity, and both ETC-159 and ETC-131 potently inhibited the endogenous Wnt/β-catenin signaling in a dose-dependent manner (Figure 3a).
Figure 3.
ETC-159 inhibits Wnt autocrine signaling and growth of teratocarcinomas. (a) Specific inhibition of autocrine Wnt signaling in human teratocarcinomas cells: PA-1 cells transfected with Super 8xTOPFLASH reporter were treated with indicated concentrations of the compounds for 24 h before luciferase assay. Data represents mean±s.d. relative to the DMSO control. (b) ETC-159 decreases downstream activation of LRP6 and Dvl2: western blot analysis of PA-1 teratocarcinoma cells treated with 100 nM ETC-159 for 24 h. (c) Inhibition of anchorage-independent growth with ETC-159 and ETC-131: PA-1 cells were plated in soft agar and treated with indicated concentrations of ETC-159 and ETC-131. Total number of colonies was scored after 2–3 weeks. Each data point represents an average count of two wells. (d) ETC-159 prevents the growth of PA-1 teratocarcinomas: 3 weeks after inoculation with PA-1 cells, mice were divided into two treatment groups matched for tumor volume (average tumor volume ~150 mm3/group) and treated daily unblinded with vehicle or 30 mg/kg ETC-159. Data represents group mean±s.e.m. n=10 tumors/group. (e) ETC-159 inhibits the growth of NCCIT teratocarcinomas: As in d. n=9 tumors/group. (f) ETC-159 decreases Axin2 mRNA in PA-1 and NCCIT tumors: Tumors were harvested 4 h after the last dose and Axin2 expression was measured using qRT-PCR. Data represents mean±s.d. n=8 tumors/group. Data were analyzed using unpaired t-test, **P⩽0.01, ****P⩽0.0001.
Binding of Wnts to their receptors Frizzled and LRP5/6 stimulates phosphorylation of both LRP6 and Disheveled (Dvl). In agreement with the high autocrine Wnt signaling, PA-1 cells had readily detectable phosphorylated LRP6 and showed an electrophoretic mobility shift of Dvl2, indicative of phosphorylation. ETC-159 significantly reduced the phosphorylation of both Dvl2 and LRP6 (Figure 3b). ETC-159 and ETC-131 also potently inhibited PA-1 colony formation in soft agar with an IC50 of 35 nM and 7.7 nM, respectively (Figure 3c). ETC-159 treatment of athymic nude mice bearing PA-1 or NCCIT xenografts reduced tumor growth significantly, confirming the requirement for autocrine Wnt signaling in vivo (Figures 3d and e). Consistent with inhibition of Wnt signaling, ETC-159 treatment also significantly reduced AXIN2 expression in the tumors (Figure 3f). These findings confirm the efficacy of ETC-159 against human cancers.
ETC-159 effectively inhibits the growth and induces differentiation of colon cancers with RSPO translocations
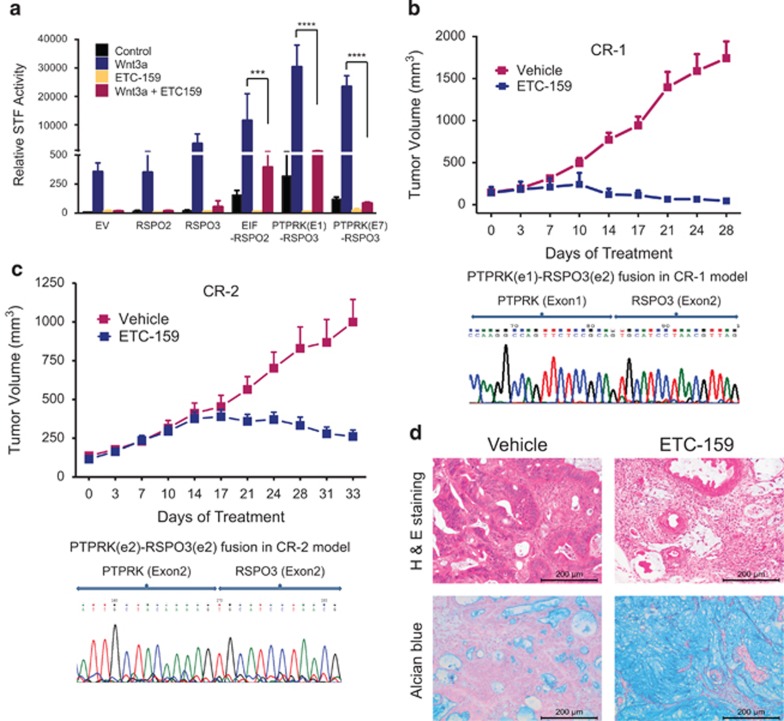
We sought genetically defined human cancers that might benefit from treatment with PORCN inhibitors. R-spondins are secreted Wnt-pathway agonists, and cancer-associated chromosome translocations that drive RSPO2 or RSPO3 expression have recently been described.6 We tested if high R-spondin expression drives sensitivity to PORCN inhibitors. Both RSPO2 and RSPO3 markedly potentiated Wnt/β-catenin reporter activity, with RSPO3 ~1000-fold more potent than RSPO2. Remarkably all the three RSPO fusion constructs; EIF-RSPO2, PTPRK(E1)-RSPO3 and PTPRK(E7)-RSPO3 further enhanced Wnt/β-catenin signaling by 20–60 fold (Figure 4a). Even in the absence of exogenous Wnt3a expression, the fusion constructs upregulated Wnt/β-catenin signaling >10 fold, most likely due to the presence of endogenous Wnt activity. Importantly, ETC-159 potently inhibited all RSPO2 and RSPO3-induced signaling, confirming they required the presence of active Wnts to drive signaling.
Figure 4.
ETC-159 prevents growth of colorectal tumors with RSPO fusions: (a) ETC-159 inhibits Wnt signaling induced by the RSPO fusion proteins: HEK293 cells transfected with the indicated RSPO expression plasmids and STF reporter were treated with DMSO or 100 nM ETC-159 for 16 h before analysis. The graph shows mean±s.d. and is representative of three independent experiments. Data were analyzed using unpaired t-test and corrected for multiple comparisons. ***P⩽0.001, ****P⩽0.0001. (b and c) Anti-tumor efficacy of ETC-159 in CRC patient-derived xenografts: female BALB/c nude mice with established subcutaneous tumors from two independent patient-derived xenografts CR-1 (b) and CR-2 (c) were treated daily with vehicle or 75 mg/kg ETC-159. The graph shows group means±s.e.m., n=12 tumors/group. Groups were matched for tumor size before unblinded treatment. The sequence chromatograms of the PTPRK-RSPO fusions in the tumors are shown below the respective graphs (d) ETC-159 promotes differentiation of colorectal tumors: tumors harvested at the end of the study were stained with hematoxylin and eosin (upper panel) or Alcian blue (lower panel). Representative sections from tumors of vehicle and ETC-159-treated group are shown. Scale bar=200 μm.
We therefore tested the efficacy of ETC-159 in two patient-derived colon cancer xenografts with confirmed R-spondin fusion genes; CR-1 with the (PTPRK(e1)-RSPO3(e2) and CR-2 with PTPRK(e2)-RSPO3(e2) fusions (Figures 4b and c). Tumor fragments were implanted into the flanks of BALB/c nude mice. Following development of palpable tumors, the mice were administered ETC-159 or vehicle by gavage once daily. Both patient-derived colon cancer xenografts models showed treatment-related growth inhibition following a variable period of slow response (Figures 4b and c and Supplementary Figures 3A and B). Most importantly, PORCN inhibitor therapy in the CRCs with RSPO translocations led to differentiation. Histologic analysis of tumors treated for 28–30 days showed a near complete loss of adenocarcinoma and markedly increased acidic polysaccharides including mucopolysaccharides as detected with Alcian blue staining consistent with mucinous differentiation (Figure 4d).
ETC-159 induces global remodeling of gene expression in colon cancers with RSPO translocations
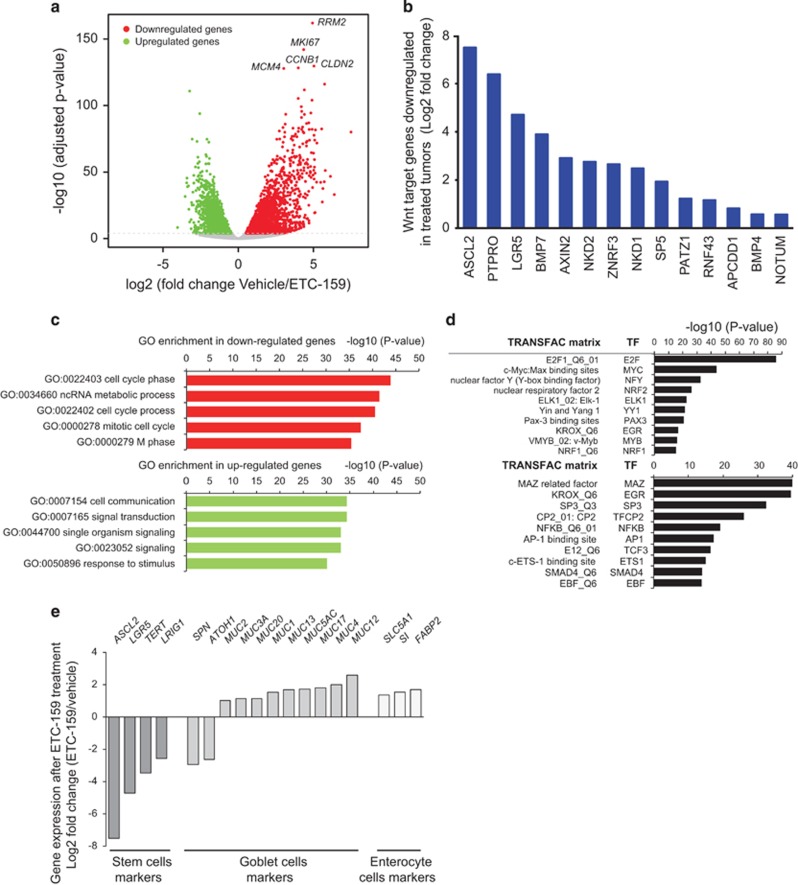
To identify the consequences of Wnt-pathway inhibition, we performed RNAseq on CR-1 patient-derived colon cancer xenograft tumors following 3 days of treatment with vehicle or ETC-159. ETC-159 treatment caused a highly significant remodeling of gene expression, with downregulation of 2744 (2420 protein-coding) genes and upregulation of 2518 (2252 protein-coding) genes (Benjamini–Hochberg adjusted P-value <0.0001, Figure 5a and Supplementary Table 1). Confirming the effect on Wnt signaling, the expression of multiple well-established β-catenin target genes was significantly reduced (Figure 5b). Importantly, other downregulated genes were significantly enriched for genes involved in cell cycle, mitosis and DNA replication, consistent with a substantial block in proliferation (Figure 5c and Supplementary Table 2). Indeed, the five most significantly downregulated genes, ribonucleotide reductase (RRM2), Ki-67 (MKI67), MCM4, cyclin B1 (CCNB1), and claudin2 (CLDN2) are known to be overexpressed in CRC and are clinical markers for cancer progression (Supplementary Table 1).35 We then investigated whether differentially expressed genes (upregulated and downregulated) were enriched for common genomic signatures using GATHER,36 which interrogates the TRANSFAC (TRANScription FACtor) database37 for over-representation of common transcription factor-binding sites. This analysis identified transcription factors (Figure 5d), including variants of the binding site of E2F transcription factor family (important regulators of cell cycle, DNA synthesis and mitosis), as well as other transcription factors that co-regulate E2F (for example, MYC38 and NYF/NRF139) or act as downstream effectors of E2F (for example, MYB40). In contrast, ETC-159 treatment for 3 days led to an upregulation of genes involved in inter/intra signal transduction, cell differentiation, cell communication and response to stimulus (Figures 5a, c and e and Supplementary Table 2). Consistent with the histologically observed differentiation, markers of differentiated intestinal cells were significantly enriched (P=0.0005, hypergeometric test) for genes showing increased expression following treatment (Supplementary Table 3). Importantly, intestinal stem cell markers including ASCL2, LGR5 and TERT were significantly reduced upon treatment with ETC-159 (P=0.0005, hypergeometric test) (Figure 5e and Supplementary Figure 3C). In summary, we found widespread transcriptional changes in genes related to cell cycle, cell proliferation, cell differentiation, and intestinal stem cell markers after treatment of RSPO3-driven CRC with ETC-159. In a subset of mice, short-term treatment (as brief as 6 days) lead to durable suppression of tumor growth (data not shown). These results suggest that suppression of Wnt/β-catenin signaling with ETC-159 induces irreversible cellular differentiation thus preventing regrowth of these tumors. These data establish that RSPO translocations are bona fide predictive biomarkers for PORCN inhibitors, and PORCN inhibition induces differentiation of RSPO-driven tumors.
Figure 5.
Global remodeling of gene expression in ETC-159 treated colon cancers with RSPO translocations. (a) Differential expression of genes in CRCs: The volcano plot shows the fold change in gene expression (x axis) vs the Benjamini–Hochberg adjusted P-value between vehicle and ETC-159 treated CR-1 tumors. n=4/group. Dotted line, Benjamini–Hochberg adjusted P-value=0.0001. (b) Downregulation of Wnt/β-catenin target genes in ETC-159 treated CRCs: the bar graph shows the fold changes in expression of Wnt/β-catenin target genes in ETC-159 treated CR-1 tumors. (c) Functional enrichment analysis of the genes downregulated and upregulated after ETC-159 treatment. False Discovery Rate <5%. (d) Analysis of over-representation of transcription factor-binding sites in the genes downregulated after ETC-159 treatment (top panel) and upregulated after ETC-159 treatment (bottom panel). False Discovery Rate <5%. (e) Functional enrichment analysis of the genes upregulated after ETC-159 treatment. False Discovery Rate <5%.
Treatment with ETC-159 prevents regrowth of tumors
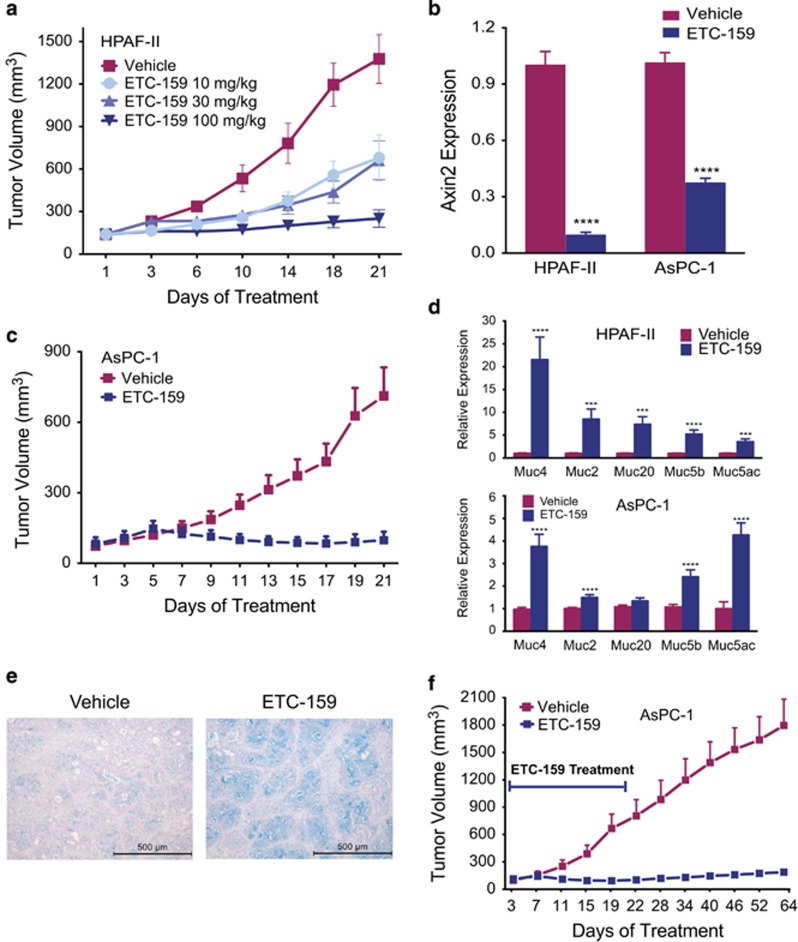
We wished to extend the results with ETC-159 to other cancers with Wnt-sensitizing mutations. We identified an RNF43 p.S720X mutation in the AsPC-1 pancreatic cell line (Supplementary Figure 4A), and c.826_827delCT in MCAS, an ovarian cell line and confirmed the presence of a p.E174X mutation in the HPAF-II pancreatic cancer cell line. These RNF43 mutations also predict sensitivity of autocrine signaling to ETC-159 as assessed by effects on AXIN2 (Supplementary Figure 4B), colony formation in soft agar (Supplementary Figures 4C–E) and low-density plating (Supplementary Figure 4F). ETC-159 decreased HPAF-II tumor growth in a dose-dependent manner, with tumor growth inhibition of 91% at 100 mg/kg and no loss of body weight (Figure 6a, Supplementary Figure 4G). At 30 mg/kg we found significant downregulation of AXIN2 expression (Figure 6b). AsPC-1 tumor xenografts were also highly sensitive in vivo to ETC-159 (Figure 6c). Consistent with what was observed in RSPO3-translocated CRCs, the ETC-159 treated RNF43 mutant pancreatic tumors showed signs of differentiation, with increased expression of mucin genes (Figure 6d) and markedly increased Alcian blue staining (Figure 6e). Mice with bilateral AsPC-1 flank xenografts were treated for 21 days and then observed for an additional 6 weeks (Figure 6f, 10 mice, 20 tumors per arm). Notably, not a single tumor re-grew during the observation period. Similar results were obtained with pancreatic patient-derived xenografts with RNF43 mutations (data not shown). These results suggest that suppression of Wnt/β-catenin signaling with ETC-159 in genetically defined tumors induces irreversible cellular differentiation thus preventing regrowth of these tumors.
Figure 6.
Treatment with ETC-159 prevents growth of RNF43 mutant pancreatic tumors and induces differentiation. (a) Three weeks after inoculation with HPAF-II cells, the mice were divided into four treatment groups matched for tumor volume (average tumor volume of ~150 mm3) and treated daily unblinded with vehicle or indicated doses of ETC-159. The graph shows group mean±s.e.m., n=9/group. (b) AXIN2 levels are decreased in ETC-159 treated HPAF-II and AsPC-1 tumors: HPAF-II and AsPC-1 tumors were harvested 6 h after the last dose and AXIN2 expression was measured by qRT-PCR. ****P⩽0.0001, n=6/group (HPAF-II) and n =16/group (AsPC-1). (c) ETC-159 treatment prevents the growth of AsPC-1 xenografts: 3 weeks after inoculation of AsPC-1 cells, mice were randomized into two matched groups and treated twice daily unblinded with 15 mg/kg/dose ETC-159 or vehicle. Data is plotted as group mean±s.d., n=16/group. (d) Increased expression of mucins with ETC-159 treatment: Mucin gene expression was assessed using qRT-PCR. Data were analyzed using unpaired t-test, ***P⩽0.001, ****P⩽0.0001, n=6/group (HPAF-II) and n=16/group (AsPC-1). (e) Representative images of Alcian blue stained tumor sections from vehicle and ETC-159 treated HPAF-II tumors. Scale bar=500 μm. (f) ETC-159 treatment prevents the growth of AsPC-1 xenografts after stopping treatment: following establishment of matched size AsPC-1 tumors, the mice were treated twice daily unblinded with 15 mg/kg/dose ETC-159 or vehicle for 21 days before the treatment was stopped. Tumor volumes were measured for up to 6 weeks after stopping the treatment. The graph shows group means±s.d., n=20/group.
Discussion
Precision therapy in cancer requires both knowledge of individual driver lesions, and having an appropriate targeted intervention for that specific lesion. Our study shows that genetically defined human CRCs with RSPO2/3 translocations are highly sensitive to the novel PORCN inhibitor ETC-159. This establishes that the over-expressed RSPO works wholly or at least significantly through the Wnt pathway, and that inhibiting production of Wnts can be an effective therapy for these cancers. The development of potent upstream Wnt inhibitors, coupled with these readily detectable predictive biomarkers, is an important step forward in the treatment of selected patients. Notably, the response of these Wnt-dependent cancers to PORCN inhibition appears to be terminal differentiation, consistent with a role for Wnts in maintaining these cancers in an undifferentiated state.
A number of agents are in development to block Wnt signaling upstream, by inhibiting Wnt biogenesis, or by blocking the interaction of Wnts with their receptors.17, 41, 42, 43 Antibody and recombinant protein-based therapeutics have advantages of high affinity and specificity but have long half-lives and low off-rates, characteristics that may contribute to toxicity. Small molecule PORCN inhibitors are highly effective in preventing Wnt secretion and overcome the limitations of antibody and protein-based therapeutics. At therapeutic doses, PORCN inhibitors do not produce overt intestinal or skin toxicity in mouse models, perhaps owing to differences in tissue penetration or dosing schedules.21, 42 However, given the well-established role of Wnts in bone metabolism it will be important to assess the effect of PORCN inhibitors on bone metabolism in clinical studies.
The efficacy of PORCN inhibitors in CRCs has not been demonstrated previously. CRCs often have downstream activation of Wnt/β-catenin signaling due to APC or β-catenin mutations, which should render them insensitive to PORCN inhibition. Molecules that prevent the interaction of β-catenin with co-activators such as CREB binding protein (CREBBP/CBP) may prove efficacious in the CRCs with APC or β-catenin mutations. One such molecule PRI-724 is in phase I clinical trials, however, its efficacy in CRCs needs to be established.44 The identification of a subset of CRC with RSPO overexpression confirms the importance of Wnt signaling in CRC.6 RSPO overexpression is postulated to drive cancers via sensitization to locally expressed Wnts (recently reviewed by Madan et al.43). Our data confirm that transcriptional activation after RSPO overexpression requires active Wnts. The highly significant therapeutic effect of ETC-159 on the patient-derived CRC xenografts suggests that the major function of over-expressed RSPOs in cancer is to make cells highly responsive to Wnts. The most striking consequence of ETC-159 treatment of both the pancreatic and the colorectal models was the cellular differentiation of the tumors, which was accompanied by increased expression of various mucins. Upstream inhibition of Wnt signaling may be a form of tumor differentiation therapy that prevents tumor regrowth rather than causing tumor death.9, 17 Consistent with this hypothesis, there was massive remodeling of the transcriptome in RSPO-overexpressing CRC xenografts within 3 days of treatment. The marked decrease in expression of proliferation, cell cycle and intestinal stem cell maintenance genes, and an increase in differentiation markers suggests that a high Wnt signal holds these cells in a proliferative, but undifferentiated state. Loss of Wnts appears to both turn-off proliferation, and allows differentiation. Notably, our transcriptional analysis did not identify a stress response nor a DNA damage response. Whether the transcriptome alterations are simply a consequence of interrupted β-catenin signaling, or are due to a combination of inhibition of multiple canonical (β-catenin) and non-β-catenin pathways will require further study.
In summary, we demonstrate that ETC-159 has remarkable efficacy in preclinical models of genetically defined cancers. In US alone there are ~130 000 new cases of CRC every year. An estimated 9% patients have RSPO translocations,6 suggesting ~12 000 of these patients might benefit from a PORCN inhibitor. The efficacy of ETC-159 in preventing growth of CRCs with RSPO fusions strongly suggests that additional RSPO fusion-bearing cancers will also be highly responsive to treatment with PORCN inhibitors. Furthermore, ~4–18% of patients with ovarian, endometrial and gastric cancer patients have RSPO translocations and could potentially benefit from the use of ETC-159. The safety and efficacy of ETC-159 warrants its evaluation in patients with Wnt driven cancers.
Material and methods
Reagents
The following plasmids and antibodies were gifts from various research groups, Super 8x TOPFLASH reporter (STF) from Randy Moon, pGK-WNT3A from Karl Willert, pMKIT-3xHA-mPORCN-D from Tatsuhiko Kadowaki, human PORCN (isoform B) from Charles Murtaugh, RSPO3 from Wanjin Hong and R-spondin fusion constructs from Genentech, WNT3A antibody from Shinji Takada.
Wnt secretion and TOPFLASH assays
HEK293 cells stably transfected with STF reporter and pPGK-WNT3A plasmid (STF3A cells) were treated with varying concentrations of compounds. HT1080 cells were transfected in 24-well plates with 50 ng Wnt, 100 ng mCherry and 550 ng STF plasmids. For PORCN rescue experiments, 100 ng 3xHA-mPORCN-D was added. For testing the sensitivity of mouse and Xenopus porcupine, PORCN null HT1080 cells were transfected with 1 ng of PORCN plasmids. After 24 h of transfection, the cells were lysed in 0.6% NP40 in PBS containing protease inhibitors. STF reporter activity was measured using firefly luciferase substrate (Promega, Madison, WI, USA) and was normalized to the cell viability, determined using LDH assay.27 For Wnt secretion, STF3A cells were treated with ETC-159 diluted in 1% fetal bovine serum-containing media. Wnt3A-conditioned medium was obtained from L cells stably expressing Wnt3A (American Type Cell Culture: CRL-2647).
Metabolic labeling with Alk-C16 and click chemistry and Wnt-WLS interaction
HeLa cells transfected with V5-tagged WNT3A were cultured in DMEM containing fatty acid-free BSA and ω-alkynyl palmitic acid (Alk-C16). Following overnight incubation with the compounds, cells were lysed, WNT3A was immunoprecipitated and subjected to click labeling and detection as described.21 HeLa cells transfected with WNT3A-V5 were treated with 0.1% DMSO or indicated compounds for 16 h. Following immunoprecipitation of WLS, the proteins were analyzed as previously described.21
Soft-agar colony assay
For colony formation assays cells were plated in 24-well culture plates. 1500 cells/well mixed with 0.35% agar and complete media were layered on top of 0.5% agar supplemented with complete growth media. An additional 500 μl of medium with or without ETC-159 was added to each well. After 2–3 weeks, colonies were stained with 5 mg/ml MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) and counted with a Gelcount instrument (Oxford Optronix, Abingdon, UK).
Immunoblot
Cells were lysed using 50 mM Tris-HCl, 150 mM NaCl, 1% sodium deoxycholate, 0.25 mM EDTA (pH 8.0), 1% Triton X-100, 0.2% sodium fluoride and protease inhibitor cocktail (Sigma, St Louis, MO, USA). Dvl2 (cat# 3224 S), pLRP6 (cat# 2568 S), LRP6 (cat# 3395 S), anti-rabbit IgG-HRP (cat# P0448), anti-mouse IgG-HRP (cat# P0447) antibodies were obtained from Cell Signaling Technology (Danvers, MA, USA). Immunoblots on PVDF were developed using SuperSignal West Dura substrate (Thermo Scientific, Rockford, IL, USA). The images were captured using the LAS-3000 Life Science Imager (Fujifilm; Tokyo, Japan).
Animal care
BALB/c nude, NCr nude or NOD-scid-gamma mice were purchased from the InVivos, Singapore or Jackson Laboratories (Bar Harbor, ME, USA). The Duke-NUS Institutional Animal Care and Use Committee or BRC Institutional Animal Care and Use Committee approved all animal studies. Animals were housed in standard cages and were allowed access ad libitum to food and water.
Tumor implantation and treatment of mice
PA-1, NCCIT, AsPC-1 and HPAF-II cells were obtained from American Type Cell Culture. All cell lines were mycoplasma-free. In all, 5–10 × 106 cells resuspended in 50% matrigel were injected subcutaneously into flanks of BALB/c nude, NCr nude or NOD-scid-gamma mice. For human xenograft models, patient-derived solid tissue fragments were subcutaneously implanted in BALB/c nude mice. All groups were matched for tumor size with equal variance before treatment. ETC-159 formulated in 50% PEG400 (vol/vol) in water was administered by oral gavage at a dosing volume of 10 μl/g body weight. Tumors were measured as described.21
Immunohistochemistry
Tumors were processed for β-catenin immunohistochemistry as described.21 For staining mucins, sections were incubated with 3% acetic acid for 3 min, stained with Alcian blue (pH 2.5) for 30 min and counterstained with neutral fast red.
RNA isolation and qRT-PCR
Total RNA isolated from the cell lines or tumors using RNAeasy kit (Qiagen, Hilden, Germany) was reverse transcribed with iScript reverse transcriptase (BioRAD, Hercules, CA, USA). Real time quantitative PCR (qPCR) was performed with SsoFast EvaGreen assay from BioRad. HPRT and ACTB were used as housekeeping genes (Supplementary Table 4).
Sample preparation for pharmacokinetic and Liquid chromatography–mass spectrometry/mass spectrometry analysis
For pharmacokinetic analysis, plasma mixed with 50 ng/ml carbamazepine and extraction solvent (70% acetronitrile and 0.1% formic acid) was incubated for 10 min at –20 °C. After vigorous shaking for 30 min and centrifugation, samples were resolved on the Kinetex C18 column (Phenominex, Torrance, CA, USA). Mass spectrometry parameters for ETC-159: multiple reaction monitoring m/z 392.0→m/z 212; collision energy, 29 V; declustering potential, 50 V and collision cell exit, 12 V. Mass spectrometry parameters for carbamazepine: multiple reaction monitoring m/z 237.1→m/z 194.1; collision energy, 27 V; DP, 100 V and CXP; 26 V. Quantitation was carried out using the multiple reaction monitoring of the transitions. The lower limit of quantification of ETC-159 was 1 ng/ml. Pharmacokinetic parameters were calculated by the non-compartmental method45 using Phoenix WinNonlin 6.3 software (Pharsight, Princeton, NJ, USA).
RNA-seq analysis
RNA-seq libraries were prepared using the Illumina TruSeq stranded Total RNA protocol with subsequent PolyA enrichment. Paired-end reads (100 bp) from the RNA-seq libraries were aligned against the human genome (hg19/GRCh37) and Ensembl annotated transcripts (build 75) using Tophat version 2.0.946 allowing two mismatches per mate. Read counts were summarized at the level of individual genes using HTseq-count47 considering only reads that mapped unambiguously to the transcriptome. Differential expression analysis was carried out using DESeq.48 Only genes showing expression lower than the 60th quantile were filtered out. Genes showing changes in their expression at the significance level of Benjamini–Hochberg adjusted P-value <0.0001 were defined as being differentially expressed after correction for multiple testing. Enrichment for manually curated markers of differentiation cell types and stem cell genes were performed using a series of hypergeometric tests. In this, only those genes expressed in the set of samples were used as background. The results of these enrichments remain the same, regardless of the filtering criteria used before the differential expression analysis or whether the set of reads used was pre-filtered using Xenome.49
Data analysis
Data was analyzed using Prism v5.0 (GraphPad, La Jolla, CA, USA) and R. Significance for all tests was set at P⩽0.05 unless otherwise stated.
Acknowledgments
We acknowledge the assistance of members of the Virshup lab including Jamal Aliyev, Naushad Moti and Edison and members of Experimental Therapeutics Centre including Shermaine Q Y Lim, Sifang Wang, Yu Wang, Vivien Wei Wen Cheong, and Grace Ruiting Lin. We acknowledge Ivana Mihalek for help with the alignment of PORCN sequences and Ralph Bunte, DVM, for his expert advice with histological analysis. We also acknowledge the assistance of the vivarium staff including Hock Lee. This research is supported in part by the National Research Foundation Singapore and administered by the Singapore Ministry of Health's National Medical Research Council under the STAR Award Program to DMV. The research in Experimental Therapeutics Centre is supported by the Agency for Science, Technology and Research (A*STAR), Singapore. EP and NH acknowledge the support of the MRC Clinical Sciences Centre, Imperial College, London.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Many of the authors are named in the patent regarding ETC-159.
Supplementary Material
References
- Chien AJ, Conrad WH, Moon RT. A Wnt survival guide: from flies to human disease. J Investig Dermatol 2009; 129: 1614–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R, Varmus H. Three decades of Wnts: a personal perspective on how a scientific field developed. EMBO J 2012; 31: 2670–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer 2013; 13: 11–26. [DOI] [PubMed] [Google Scholar]
- Yu J, Virshup DM. Updating the Wnt pathways. Biosci Rep 2014; 34. doi:10.1042/BSR20140119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol 2012; 13: 767–779. [DOI] [PubMed] [Google Scholar]
- Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB et al. Recurrent R-spondin fusions in colon cancer. Nature 2012; 488: 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinmura K, Kahyo T, Kato H, Igarashi H, Matsuura S, Nakamura S et al. RSPO fusion transcripts in colorectal cancer in Japanese population. Mol Biol Rep 2014; 41: 5375–5384. [DOI] [PubMed] [Google Scholar]
- Giannakis M, Hodis E, Jasmine Mu X, Yamauchi M, Rosenbluh J, Cibulskis K et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet 2014; 46: 1264–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Hao H-X, Growney JD, Woolfenden S, Bottiglio C, Ng N et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci USA 2013; 110: 12649–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryland GL, Hunter SM, Doyle MA, Rowley SM, Christie M, Allan PE et al. RNF43 is a tumour suppressor gene mutated in mucinous tumours of the ovary. J Pathol 2013; 229: 469–476. [DOI] [PubMed] [Google Scholar]
- Ong CK, Subimerb C, Pairojkul C, Wongkham S, Cutcutache I, Yu W et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet 2012; 44: 690–693. [DOI] [PubMed] [Google Scholar]
- Wu J, Jiao Y, Dal Molin M, Maitra A, de Wilde RF, Wood LD et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc Natl Acad Sci USA 2011; 108: 21188–21193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H-X, Xie Y, Zhang Y, Charlat O, Oster E, Avello M et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 2012; 485: 195–200. [DOI] [PubMed] [Google Scholar]
- Koo B-K, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 2012; 488: 665–669. [DOI] [PubMed] [Google Scholar]
- Cardona GM, Bell K, Portale J, Gaffney D, Moy C, Platero S et alIdentification of R-Spondin Fusions in Various Types of Human Cancer. Proceedings of the 105th Annual Meeting of the American Association for Cancer Research; 5–9 April 2014; San Diego, CA, USA. Abstract no. 2408..
- Kahn M. Can we safely target the WNT pathway? Nat Rev Drug Discov 2014; 13: 513–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney A, Axelrod F, Bond CJ, Cain J, Chartier C, Donigan L et al. Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc Natl Acad Sci USA 2012; 109: 11717–11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L, Clevers H. The Unusual Case of Porcupine. Science 2012; 337: 922–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios-Esteves J, Resh MD. Stearoyl CoA Desaturase Is Required to Produce Active, Lipid-Modified Wnt Proteins. Cell Rep 2013; 4: 1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs GS, Yu J, Canning CA, Veltri CA, Covey TM, Cheong JK et al. WLS-dependent secretion of WNT3A requires Ser209 acylation and vacuolar acidification. J Cell Sci 2010; 123: 3357–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proffitt KD, Madan B, Ke Z, Pendharkar V, Ding L, Lee MA et al. Pharmacological inhibition of the Wnt acyltransferase PORCN prevents growth of WNT-driven mammary cancer. Cancer Res 2013; 73: 502–507. [DOI] [PubMed] [Google Scholar]
- Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural Basis of Wnt Recognition by Frizzled. Science 2012; 337: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi-Yanaga F, Kahn M. Targeting Wnt signaling: can we safely eradicate cancer stem cells? Clin Cancer Res 2010; 16: 3153–3162. [DOI] [PubMed] [Google Scholar]
- Lee SB, Gong Y-D, Park YI, Dong M-S. 2,3,6-Trisubstituted quinoxaline derivative, a small molecule inhibitor of the Wnt/beta-catenin signaling pathway, suppresses cell proliferation and enhances radiosensitivity in A549/Wnt2 cells. Biochem Biophys Res Commun 2013; 431: 746–752. [DOI] [PubMed] [Google Scholar]
- Huang S-MA, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 2009; 461: 614–620. [DOI] [PubMed] [Google Scholar]
- Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M et al. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription. Proc Natl Acad Sci USA 2004; 101: 12682–12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs GS, Schmitt AA, Canning CA, Alok A, ICC Low, Banerjee N et al. Modulation of Wnt/β-catenin signaling and proliferation by a ferrous iron chelator with therapeutic efficacy in genetically engineered mouse models of cancer. Oncogene 2012; 31: 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannoush RN, Arenas-Ramirez N. Imaging the lipidome: omega-alkynyl fatty acids for detection and cellular visualization of lipid-modified proteins. ACS Chem Biol 2009; 4: 581–587. [DOI] [PubMed] [Google Scholar]
- Gao X, Arenas-Ramirez N, Scales SJ, Hannoush RN. Membrane targeting of palmitoylated Wnt and Hedgehog revealed by chemical probes. FEBS Lett 2011; 585: 2501–2506. [DOI] [PubMed] [Google Scholar]
- Fu J, Jiang M, Mirando AJ, Yu H-MI, Hsu W. Reciprocal regulation of Wnt and Gpr177/mouse Wntless is required for embryonic axis formation. Proc Natl Acad Sci USA 2009; 106: 18598–18603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najdi R, Proffitt K, Sprowl S, Kaur S, Yu J, Covey TM et al. A uniform human Wnt expression library reveals a shared secretory pathway and unique signaling activities. Differentiation 2012; 84: 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proffitt KD, Virshup DM. Precise regulation of porcupine activity is required for physiological Wnt signaling. J Biol Chem 2012; 287: 34167–34178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T, Varmus HE. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell 1988; 55: 619–625. [DOI] [PubMed] [Google Scholar]
- DeAlmeida VI, Miao L, Ernst JA, Koeppen H, Polakis P, Rubinfeld B. The soluble wnt receptor Frizzled8CRD-hFc inhibits the growth of teratocarcinomas in vivo. Cancer Res 2007; 67: 5371–5379. [DOI] [PubMed] [Google Scholar]
- Dhawan P, Ahmad R, Chaturvedi R, Smith JJ, Midha R, Mittal MK et al. Claudin-2 expression increases tumorigenicity of colon cancer cells: role of epidermal growth factor receptor activation. Oncogene 2011; 30: 3234–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics 2006; 22: 2926–2933. [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gößling E, Haubrock M, Hehl R et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 2003; 31: 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005; 435: 839–843. [DOI] [PubMed] [Google Scholar]
- Gurtner A, Fuschi P, Martelli F, Manni I, Artuso S, Simonte G et al. Transcription factor NF-Y induces apoptosis in cells expressing wild-type p53 through E2F1 upregulation and p53 activation. Cancer Res 2010; 70: 9711–9720. [DOI] [PubMed] [Google Scholar]
- Lam EW, Watson RJ. An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J 1993; 12: 2705–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Moon J, Dodge ME, Pan X, Zhang L, Hanson JM et al. The development of highly potent inhibitors for porcupine. J Med Chem 2013; 56: 2700–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Pan S, Hsieh MH, Ng N, Sun F, Wang T et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc Natl Acad Sci USA 2013; 110: 20224–20229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan B, Virshup DM. Targeting Wnts at the Source-New Mechanisms, New Biomarkers, New Drugs. Mol Cancer Ther 2015; 14: 1087–1094. [DOI] [PubMed] [Google Scholar]
- Lenz H-J, Kahn M. Safely targeting cancer stem cells via selective catenin coactivator antagonism. Cancer Sci 2014; 105: 1087–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibaldi M, Perrier D. Noncompartmental analysis based on statistical moment theory. Pharmacokinetics 1982; 2: 409–417. [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010; 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T, Wazny J, Bromage A, Tymms M, Sooraj D, Williams ED et al. Xenome—a tool for classifying reads from xenograft samples. Bioinformatics 2012; 28: i172–i178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.