Abstract
Accurate and defendable taxonomic identification of microalgae strains is vital for culture collections, industry and academia; particularly when addressing issues of intellectual property. We demonstrate the remarkable effectiveness of Matrix Assisted Laser Desorption Ionisation Time of Flight Mass Spectrometry (MALDI-TOF-MS) biotyping to deliver rapid and accurate strain separation, even in situations where standard molecular tools prove ineffective. Highly distinctive MALDI spectra were obtained for thirty two biotechnologically interesting Dunaliella strains plus strains of Arthrospira, Chlorella, Isochrysis, Tetraselmis and a range of culturable co-occurring bacteria. Spectra were directly compared with genomic DNA sequences (internal transcribed spacer, ITS). Within individual Dunaliella isolates MALDI discriminated between strains with identical ITS sequences, thereby emphasising and enhancing knowledge of the diversity within microalgae culture collections. Further, MALDI spectra did not vary with culture age or growth stage during the course of the experiment; therefore MALDI presents stable and accurate strain-specific signature spectra. Bacterial contamination did not affect MALDI’s discriminating power. Biotyping by MALDI-TOF-MS will prove effective in situations wherein precise strain identification is vital, for example in cases involving intellectual property disputes and in monitoring and safeguarding biosecurity. MALDI should be accepted as a biotyping tool to complement and enhance standard molecular taxonomy for microalgae.
The search for, and exploitation of marine and aquatic organisms for biotechnology applications - so called blue biotechnology1 - has risen to the forefront of the global research agenda over the past decade. Algae and cyanobacteria are intrinsic to the success of this sector2,3 and phycological research has benefitted greatly from the buoyant funding landscape this has created. Algae and cyanobacteria are some of the main biological protagonists that drive many global biogeochemical cycles, being directly responsible for over half of current planetary productivity and geologically for many fossil-fuel resources4. Their high conversion of solar energy into biomass, capacity to grow in saline or hypersaline environments, ability to metabolise industrial and domestic waste streams (including CO2 flue gases and wastewater), combined with the diverse and arguably untapped range of biochemicals they can synthesise5,6 makes algae and cyanobacteria attractive bioprospecting targets7,8. In this regard, culture collections that curate microalgae and cyanobacteria, whether public or private, have proven to be fruitful and convenient biological reservoirs for bioprospecting8,9.
Industry interests, often focused on intellectual property rights, are core to the boom in blue biotechnology10,11 with investment strategies often hinging on protection of novel or high performing strains, despite the fact that legal issues remain regarding the patenting of microbes in their wild-type versus genetically modified or even synthetic forms12,13. Added to this are concerns relating to biosecurity and traceability of industrial or genetically transformed strains, particularly if grown in open outdoor culture systems or in the event of an unwanted environmental discharge14. With this in mind, the need for rapid, accurate and defendable taxonomic identification of microalgae and cyanobacteria strains is paramount for culture collections, industry and academia; particularly when addressing issues of intellectual property and biosecurity.
A variety of approaches have been applied, successfully and otherwise, to characterise microalgae and cyanobacteria; including morphology and ultrastructure studies, genetic studies such as examining the secondary structures of the small subunit ribosomal RNA, the internal transcribed spacer (ITS) region of ribosomal DNA, Amplified Fragment Length Polymorphisms and metagenomics15,16,17,18,19. It has been estimated that there are well in excess of one thousand species of green algae alone20. Dunaliella species produce biochemicals with applications ranging from pharmaceuticals, high value pigments, and biofuels and platform chemicals21,22,23,24,25. Although the genus Dunaliella was introduced in 190526, due to the ecological, morphological and biochemical plasticity of this alga (particularly in relation to cell size, pigmentation and other nominally defining characters) the taxonomy of the genus has not been clearly resolved17. However, genetic barcoding approaches have identified twenty eight species separated into two main ecological groups, freshwater and marine species, with both groups having their own subdivisions27,28,29,30,31. In general, ITS rDNA sequencing has been accepted as a powerful tool for distinguishing microalgae species32 although the tufA gene has recently been advanced as a more appropriate DNA biomarker for chlorophytes18. ITS biotyping has been used for phylogeny of Dunaliella strains by several investigators15,33,34, with the most recent and comprehensive work undertaken by Assunçäo and co-investigators27,35 using ITS2. Analysis of ITS1, ITS2 and ITS2 secondary structure has revealed high intraspecific variation within Dunaliella15,33,34,36,37. ITS2 is thought to be an excellent marker for molecular phylogenetic studies, especially at lower taxonomic levels38.
An alternative approach to DNA-based strain separation that has very rarely been applied to microalgae and cyanobacteria is proteomic-based chemotaxonomic biotyping using MALDI-TOF mass spectrometry. Using mass spectrometers for characterising microorganisms began in 197539. Thereafter, the methods were rapidly optimised and expanded to include clinical and environmental species. MALDI-TOF-MS biotyping has successfully been used to identify bacteria, yeast, fungi, and even higher eukaryotes including insects40, molluscs41 and fish42. It has also been used in clinical and environmental contexts to identify and differentiate between strains of the pathogenic, microalga Prototheca43,44 and the bloom forming cyanobacterium Oscillatoria45. However, the potential for the approach to be applied to blue biotechnology has not hitherto been explored. As the molecular basis for the differences in MALDI-TOF-MS spectra are generally unknown the differences cannot be assumed to reflect phylogeny. The power of the method is in the potential capacity to discriminate between closely related organisms. In this context, we have undertaken a rigorous evaluation of the capacity of biotyping by MALDI-TOF-MS to function as both a complementary and standalone tool to differentiate between closely related strains of the commercially important chlorophyte microalgal genus Dunaliella.
Results
ITS sequence analysis
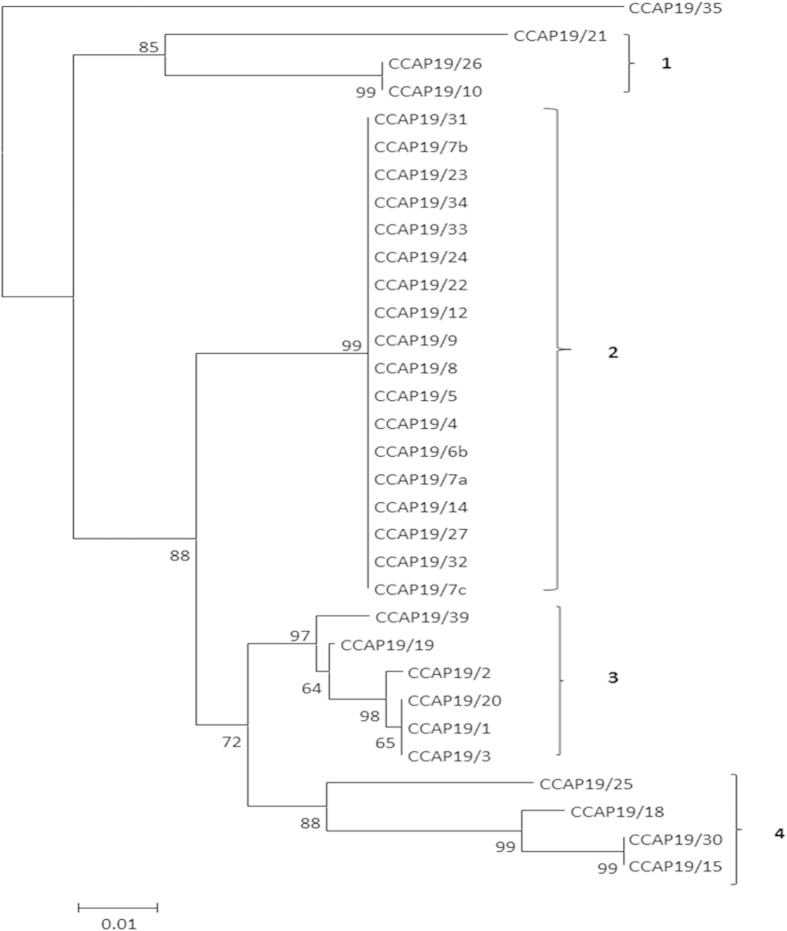
The thirty two Dunaliella isolates investigated here were previously classified and deposited with the Culture Collection of Algae and Protozoa (CCAP) on the basis of their gross phenotype (cell shape and size) and, in some cases using limited biochemical properties such as carotenoid production. In this study by using ITS4 and ITS5 primers, 729 bp of rDNA including ITS1, ITS2, and the 5.8S rRNA gene were amplified and sequenced. Three isolates, CCAP 19/10, 19/21, and 19/26, contained introns in their sequences. The introns were excluded during the construction of the phylogram presented in Fig. 1. Four major clades were observed encompassing thirty one of the thirty two available strains. The one exception was D. acidophila (CCAP 19/35), which stands as a distinctive strain. Clade 1 consisted of two strains of D. parva (CCAP 19/10 and 19/26) and one strain (CCAP 19/21) not previously designated to species level. Whilst not included in the phylogram construction, CCAP 19/10 and 19/26 have identical introns that imply that they are isolates of the same species. Clade 2 comprised eighteen isolates of which six have not previously been designated to species level. The remaining twelve isolates have been classified as eight distinct species (Table 1). In contrast, clade 2 is the only clade in which all isolates have identical ITS sequences (Fig. 1). Clade 3 consisted of three D. salina, one D. parva, one D. peircei isolates and a strain not previously designated to species level. Clade 4 contained three strains deposited as D. salina and one strain not previously designated to species level. Further analysis of the ITS2 sequence and structure supports the finding of the sequence only tree (Supplementary Figure 1).
Figure 1.
ITS phylogram of all thirty two Dunaliella isolates identified by strain number (see Table 1 for assigned species names) available from the Culture Collection of Algae and Protozoa (CCAP). Four distinctive phylogenetic clades, labelled 1-4, were identified containing between three and eighteen isolates. The exception was D. acidophila that had no significant nucleotide similarity with any other CCAP isolate.
Table 1. The thirty two Culture Collection of Algae and Protozoa Dunaliella isolates used in this study including their respective ITS barcode accession numbers.
| Name assigned by depositor | CCAP strain no. | Origin | ITS rDNA GenBank accession |
|---|---|---|---|
| D. peircei | CCAP 19/2 | USA | KJ094608 |
| D. parva | CCAP 19/9 | England | KJ094607 |
| CCAP 19/10 | Israel | KJ094617 | |
| CCAP 19/1 | Romania | KJ094618 | |
| CCAP 19/26 | Israel | KJ094630 | |
| D. tertiolecta | CCAP 19/6B | Norway | KJ094612 |
| CCAP 19/24 | Unknown | KJ094628 | |
| CCAP 19/27 | Unknown | KJ094631 | |
| D. quartolecta | CCAP 19/8 | England | KJ094616 |
| D. minuta | CCAP 19/5 | France | KJ094611 |
| D. bioculata | CCAP 19/4 | Soviet Union | KJ094610 |
| D. primolecta | CCAP 19/34 | England | KJ094636 |
| D. salina | CCAP 19/3 | Soviet Union | KJ094609 |
| CCAP 19/18 | Australia | KJ094622 | |
| CCAP 19/20 | Unknown | KJ094624 | |
| CCAP 19/25 | Mexico | KJ094629 | |
| CCAP 19/30 | Israel | KJ094632 | |
| CCAP 19/31 | Unknown | KJ094633 | |
| CCAP 19/39 | Spain | KJ094637 | |
| D. acidophila | CCAP 19/35 | Italy | KP005451 |
| D. polymorpha | CCAP 19/7A | England | KJ094613 |
| CCAP 19/7B | England | KJ094614 | |
| CCAP 19/7C | England | KJ094615 | |
| Dunaliella sp. | CCAP 19/22 | Unknown | KJ094619 |
| CCAP 19/33 | England | KJ094620 | |
| CCAP 19/15 | Israel | KJ094621 | |
| CCAP 19/21 | Egypt | KJ094623 | |
| CCAP 19/23 | Unknown | KJ094625 | |
| CCAP 19/14 | Israel | KJ094626 | |
| CCAP 19/12 | Israel | KJ094627 | |
| CCAP 19/19 | Australia | KJ094634 | |
| CCAP 19/32 | USA | KJ094635 |
MALDI-TOF analysis of microalgae and cyanobacteria isolates
A twenty day time point experiment using CCAP 19/7C and 19/27 demonstrated no significant difference between the general spectral pattern of the two isolates with time despite the cultures having been sampled during phases of logarithmic and linear growth, early and late stationary growth phases and early culture senescence (see Supplementary Figure 2). The only differences observed were in the intensity of the major peaks rather than their mass to charge ratio by MALDI-TOF. The MALDI-TOF-MS biotyping approach was therefore applicable throughout the full culture cycle.
Good quality spectra were obtained for numerous bacterial species that were co-occurring with two D. polymorpha isolates, two of which are presented in supplementary Fig. 4. There was no major overlap of peaks when overlaid with the spectra for D. polymorpha. Similarly, treatment of the cultures with antibiotics did not significantly change the D. polymorpha spectra.
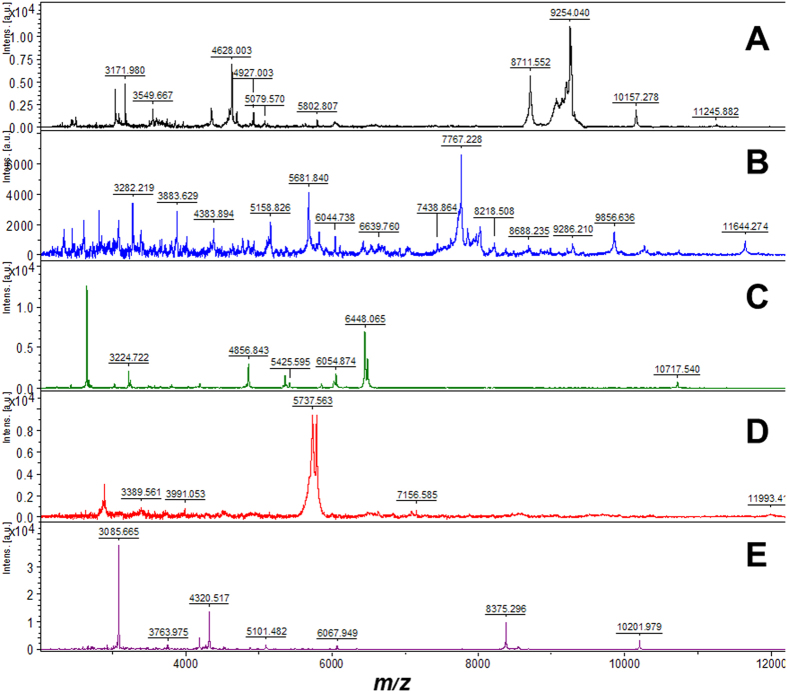
An initial experiment was conducted to validate the efficacy of biotyping by MALDI-TOF-MS to discriminate between distantly related microalgae and cyanobacteria genera using four eukaryote species; the chlorophytes Dunaliella polymorpha, Chlorella vulgaris and Tetraselmis suecica, the haptophyte Isochrysis galbana; and one prokaryote species; the cyanophyte Arthrospira platensis (Fig. 2). Each tested species generated highly distinct mass spectra, therefore demonstrating little chance of spectral overlap between separate genera.
Figure 2.
Comparison of the MALDI-TOF mass spectra of: A) Dunaliella polymorpha (CCAP 19/7B); B) Arthrospira platensis (CCMP 1295); C) Chlorella vulgaris (CCAP 211/63); D) Isochrysis galbana (CCAP 927/1); and E) Tetraselmis suecica (Seasalter Shellfish Ltd). Each mass spectrum was markedly different demonstrating that biotyping by MALDI-TOF-MS can easily distinguish between microalgae and cyanobacteria genera.
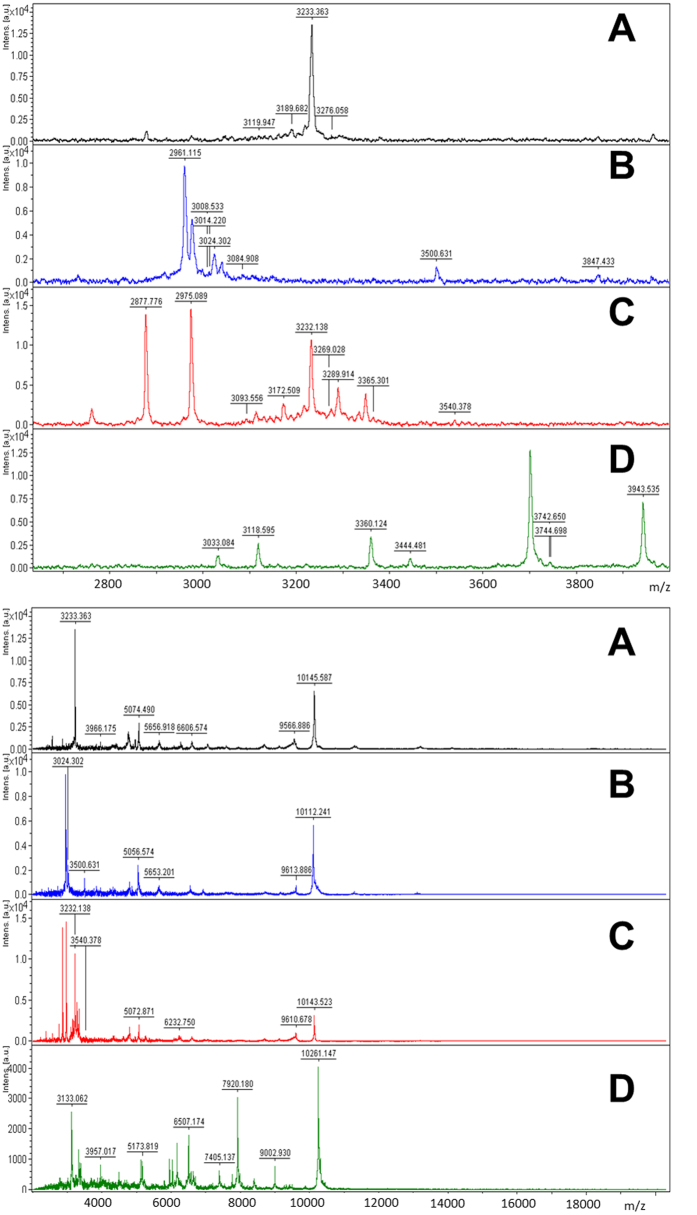
To determine whether biotyping by MALDI-TOF-MS may provide enhanced between-strain discrimination compared with ITS sequencing alone, the MALDI spectra of each ITS generated clade were analysed. MALDI-TOF-MS biotyping confirmed the output of ITS sequencing highlighting that D. acidophila (CCAP 19/35) is the most distantly related of the CCAP Dunaliella isolates. The spectrum is presented in Fig. 3 alongside clade 1 (CCAP 19/10, 19/21 and 19/26) for comparison. CCAP 19/21 has not previously been identified to species level, whereas both CCAP 19/10 and 19/26 were deposited as D. parva and their ITS rDNA sequences are identical. Peaks at m/z 2878, 3233, 5072, 6606, 10143 and 13211 are common between the D. parva isolates which suggests that they are indeed the same species. Their mass spectra also show strong similarities to that of CCAP 19/21. CCAP 19/21 and 19/26 have a peak at m/z 9611 ± 2, whereas CCAP 19/10 and 19/21 have a peak at m/z 5654 ± 2.
Figure 3.
MALDI-TOF mass spectra of the three Dunaliella isolates assigned to clade 1 in the ITS rDNA dendrogram (Fig. 1): A) CCAP 19/10; B) CCAP 19/21; and C) CCAP 19/26. Additionally, the spectrum for D. acidophila (CCAP 19/35) which had no significant ITS nucleotide sequence similarity with any other CCAP isolate is presented in panel D. Peaks in the m/z range of 2620-3500 for CCAP 19/10, 19/21 and 19/26 and m/z 2600-4000 for CCAP 19/35 are presented in the upper panels and peaks in the m/z range of 2000-15000 are presented in the lower panels. CCAP 19/10 and 19/26 isolates were deposited as D. parva and have identical ITS rDNA sequences yet the peaks at m/z 2975 and 3289 in CCAP 19/26 are absent in CCAP 19/10 and 21. However the peak at m/z 2961 is only present in the CCAP 19/21 spectrum. CCAP 19/10 and 19/26 also have common peaks at m/z 2877 and 3232.
Of particular interest were the mass spectra (summarised in Table 2) corresponding to the eighteen isolates comprising clade 2 that yielded identical ITS rDNA sequences. Nine major peaks were identified within clade 2 occurring at m/z 3045, 4627, 5803, 7588, 8711, 9254, 10157, 13019 and 18101 respectively. The mass spectra of eight of the isolates (CCAP 19/4, 9/7A, 19/7B, 19/7C, 19/8, 19/14, 19/22 and 19/24) were almost identical, each sharing seven of the nine major peaks but missing peaks at m/z 7588 and 18101. However, CCAP 19/8 was missing a peak at m/z 6473 which was present in the other isolates (data not shown). Both rDNA and MALDI results suggest these strains belong to the same Dunaliella species yet the current CCAP nomenclature places CCAP 19/7A, 19/7B, and 19/7C as D. polymorpha, CCAP 19/4 as D. bioculata, CCAP 19/8 as D. quartolecta, CCAP 19/24 as D. tertiolecta and CCAP 19/14 and 19/22 as Dunaliella sp. CCAP 19/23 (Dunaliella sp.) and 19/31 (D. salina) also had identical spectra sharing seven major peaks but missing the peaks at m/z 5803 and 18101. The remaining eight isolates were each distinct from all other members of the clade.
Table 2. Presence (+) or absence (−) of nine major protein peaks (m/z up to ± 10 units) as identified by MALDI-TOF-MS from the eighteen Dunaliella isolates assigned to clade 2 in the ITS rDNA dendrogram (Fig. 1).
| m/z | 3045 | 4627 | 5803 | 7588 | 8711 | 9254 | 10157 | 13019 | 18101 |
|---|---|---|---|---|---|---|---|---|---|
| CCAP 19/4 | + | + | + | − | + | + | + | + | − |
| CCAP 19/5 | + | 4327 | + | + | − | − | + | + | − |
| CCAP 19/6B | 3091 | + | 5881 | − | + | − | + | + | − |
| CCAP 19/7A | + | + | + | − | + | + | + | + | − |
| CCAP 19/7B | + | + | + | − | + | + | + | + | − |
| CCAP 19/7C | + | + | + | − | + | + | + | + | − |
| CCAP 19/8 | + | + | + | − | + | + | + | + | − |
| CCAP 19/9 | + | + | − | + | + | + | + | + | − |
| CCAP 19/12 | + | + | + | + | + | − | + | + | − |
| CCAP 19/14 | + | + | + | − | + | + | + | + | − |
| CCAP 19/22 | + | + | + | − | + | + | + | + | − |
| CCAP 19/23 | + | + | − | + | + | + | + | + | − |
| CCAP 19/24 | + | + | + | − | + | + | + | + | − |
| CCAP 19/27 | + | + | − | − | + | + | + | + | − |
| CCAP 19/31 | + | + | − | + | + | + | + | + | − |
| CCAP 19/32 | − | + | − | − | − | 9278 | + | + | − |
| CCAP 19/33 | + | + | − | − | − | 9266 | + | − | − |
| CCAP 19/34 | + | + | + | − | + | + | + | + | + |
In cases where a similar peak with a m/z of more than 10 mass unit differences was observed the exact m/z values are indicated in the columns. These changes in peak mass would suggest potential oxidation and amino acid substitution respectively ( www.Unimod.org/). Some of the observed differences in the mass to charge ratios may be due to structural modifications such as amino acid oxidation, substitution or protein glycosylation; or uneven cell distribution due to the topology of the analyte/matrix mixture on the MALDI plate.
Clade 3 of the ITS phylogram includes CCAP 19/1 (D. parva), 19/2 (D. peircei), 19/3 (D. salina), and 19/20 (D. salina) with very similar ITS rDNA, also CCAP 19/19 (Dunaliella sp.) and 19/39 (D. salina). Mass spectra of these isolates are compared in Fig. 4. Although CCAP 19/1 did not generate a high quality sectrum, the MALDI fingerprint has marked similarities to the other strains in this clade and was included in the analysis. The CCAP 19/20 mass spectrum contained a polymeric pattern that did not interfere with clustering and resolution of the mass spectra, with peaks gap of m/z 142. Peaks at m/z 4357, 5380, 8714, 9134, 10735 were observed in most isolates.
Figure 4.
MALDI-TOF mass spectra of five of the six Dunaliella isolates assigned to clade 3 in the ITS rDNA dendrogram (Fig. 1): A) CCAP 19/1; B) CCAP 19/2; C) CCAP 19/3; D) CCAP 19/19; and E) CCAP 19/20. The spectra for CCAP 19/20 contained a polymeric pattern similar to that of CCAP 19/18 (Fig. 5) but did not interfere with spectral analysis. The spectra for CCAP 19/39 was of lower presentational quality and is omitted.
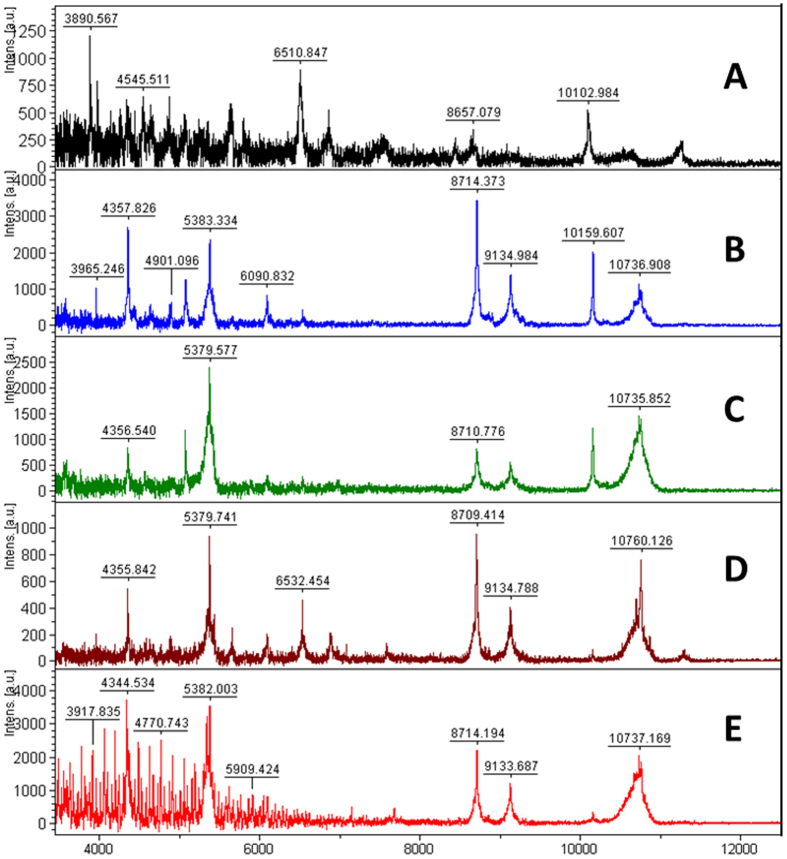
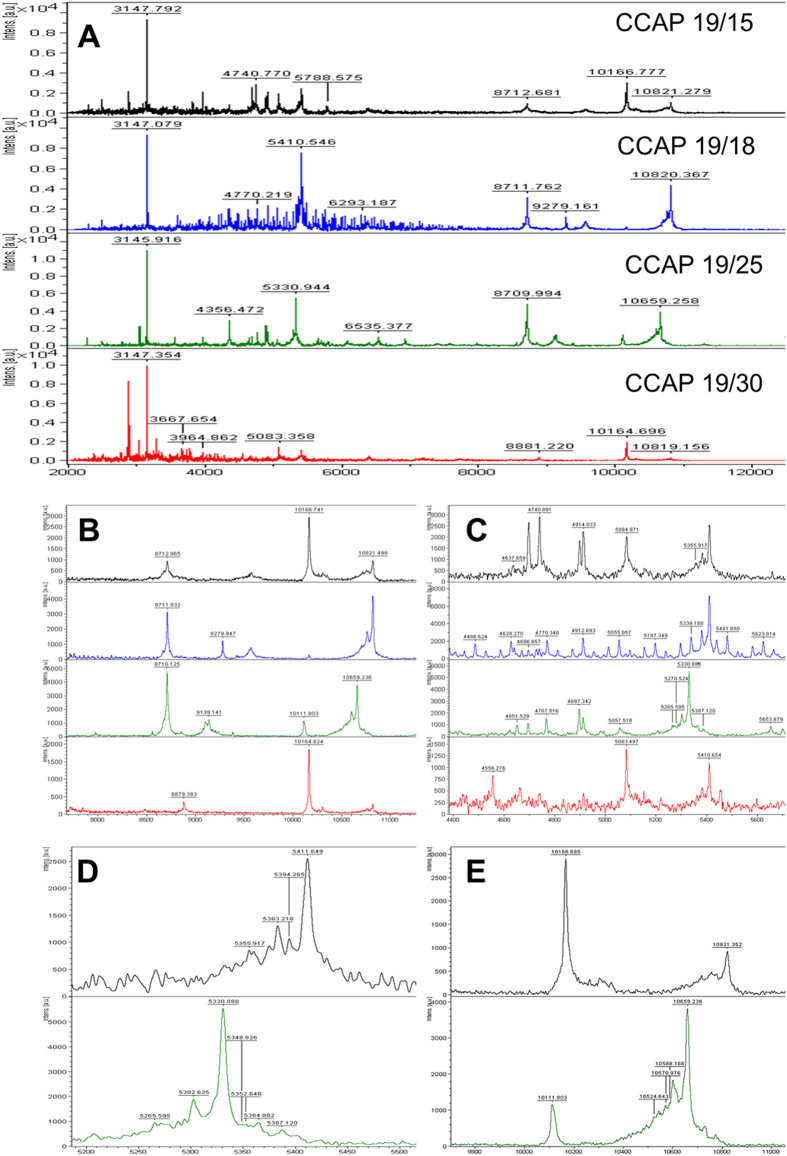
Clade 4 of the ITS phylogram contains three D. salina strains (CCAP 19/18, 19/25 and 19/30) and one strain (CCAP 19/15) not previously designated to species level. CCAP 19/15 and 19/30 have identical ITS sequences and their mass spectra are also very similar (Fig. 5), strongly indicating that both isolates are the same species. All the isolates in this clade have peaks at approximately m/z 3146, 3965, 5411, and 8711 (Fig. 5A,B). On this basis CCAP 19/15 is very likely D. salina. In this clade CCAP 19/25 possessed the most distinct fingerprint. The other three isolates had peaks at m/z 10166 ± 1 but CCAP 19/25 lacked this peak, instead it had unique peaks at m/z 9139, 10659 and 13489 (Fig. 5B-E). CCAP 19/18 yielded a polymer-type pattern similar to that observed from CCAP 19/20; again, the polymer did not interfere with spectral resolution.
Figure 5.
MALDI-TOF mass spectra of the four Dunaliella isolates assigned to clade 4 in the ITS rDNA dendrogram (Fig. 1). Most peaks had very similar mass to charge ratios. Panel A shows the mass spectra between m/z 2400-12200, the region of the spectra where most peaks appeared. Panels B (m/z 8000-12000) and C (m/z 3500-6000) present more detailed examples of similar and distinctive peaks between the four isolates. Panels D (m/z 5200-5700) and E (m/z 9000-11400) show examples of peak differences between CCAP19/15 and CCAP 19/25.
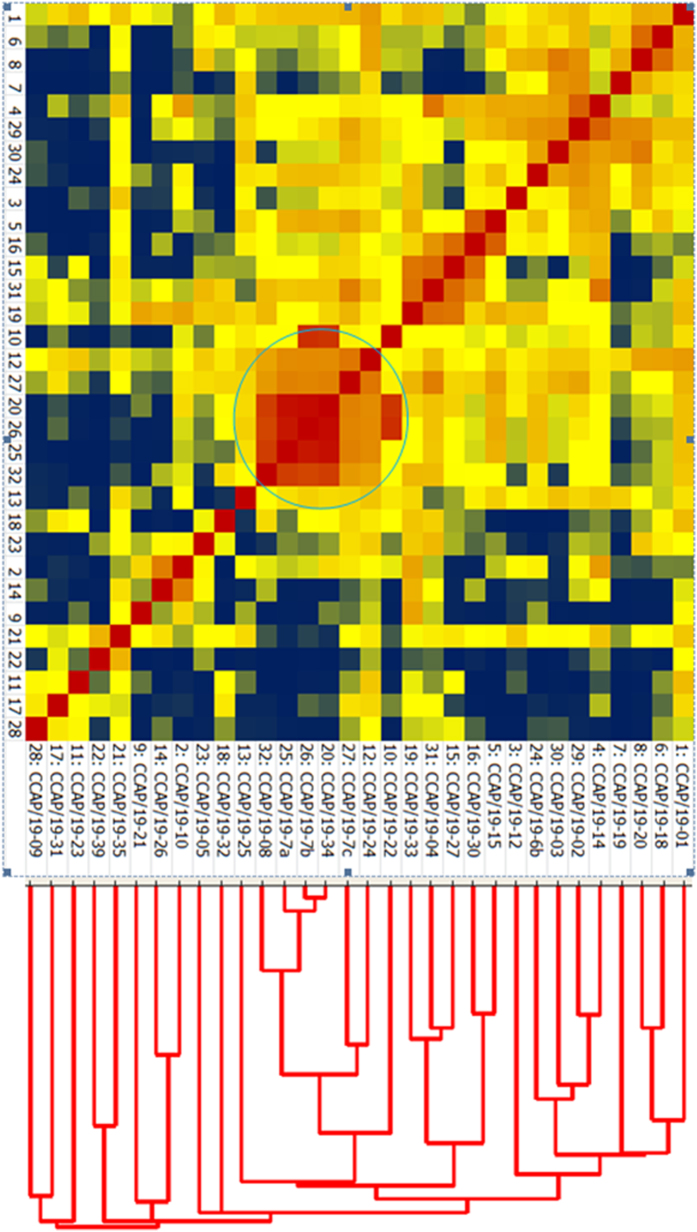
An overall view of the relatedness of the CCAP Dunaliella isolates based on MALDI mass spectral patterns is presented in Fig. 6 in both dendrogram and heat map formats. The most closely related isolates are indicated by red squares and the least in blue. The isolates CCAP 19/8, 19/24, 19/34, 19/7A, 19/7B, and 19/7C are at the core of the heat map due to the high degree of similarity across their spectra. Heat map mass spectral clustering of the isolates CCAP 19/10, 19/21 and 19/26 concurs with the ITS sequencing confirming that these isolates are closely related (Figs. 1 and 6).
Figure 6.
Composite correlation index based heat map and corresponding dendrogram generated from MALDI-TOF analysis for all thirty two CCAP Dunaliella isolates. Red colouration represents the highest degree of similarity, yellow corresponds to the 50th percentile of similarity, and blue represents the lowest similarity. Isolates with the most similar spectra are circled. Numbers on the bottom of the heat map correspond to those on the side. It is clear that biotyping by MALDI-TOF-MS has a greater strain resolving power than ITS rDNA sequencing (Fig. 1) alone.
Discussion
The use of ITS sequences to resolve microalgae taxonomy to species level is widely accepted although the tufA plastid gene sequence would appear to provide greater resolving power for the green algae18. Based on crossing experiments, it has been proposed that even a single Compensatory Base Change (CBC) in helices 2 and 3 of the ITS2 indicates sexual incompatibility and thus separate biological species32. Furthermore, secondary structure analysis can improve the phylogenetic resolution obtained from the primary sequence46.
Dunaliella species share very high ITS sequence similarity and similar phenotypic traits15,17,27,33,34,35,36,37; therefore, taxonomic definition of Dunaliella species requires a thorough understanding of the molecular and biochemical characteristics of the isolates. The current taxonomic designation of the thirty two CCAP isolates used in this study (Table 1) follows the nomenclature of the original depositor and was based primarily on a morphological approach. DNA barcoding has not previously been employed to re-evaluate these strains. The ITS phylogeny assigned thirty one of the thirty two strains to one of four main clades, the exception being CCAP19/35 D. acidophila which was clearly distinct from all other strains. This is supported by the requirement for this strain to be held in highly acidic conditions to allow growth whereas the other strains have pH optima closer to that of ambient seawater. Clade 2 was particularly interesting as all eighteen isolates had identical ITS sequences yet twelve of these strains have been assigned to eight separate species based on morphological traits, with the remaining six isolates as yet not classified to species level. Clearly there is a discrepancy between classical taxonomic approaches and ITS barcoding. Due to the high level of DNA sequence similarity the value of ITS2 structure analysis is limited for the differentiation of morphologically distinct isolates (Supplementary Fig. 1). If the barcoding approach is accurate then this serves to highlight the high degree of morphological plasticity exhibited by this genus.
Although the most reliable standard in algal taxonomy remains DNA-DNA hybridisation among species, it is not always feasible to apply this method to all isolates of interest18,47. Furthermore, as algal biotechnology continues to develop there is already a growing ground swell within the community aimed towards deploying mutant and transgenic strains; with the concomitant absolute requirement for methods capable of identifying biochemically distinct environmental isolates. Alarmingly, standard molecular barcoding methods are not capable of this level of resolution. There are suggestions that whole genome sequence data may eventually replace DNA-DNA hybridisation approaches48 and this would certainly provide the level of definition required to differentiate algae at species or even strain level. However, cost implications, particularly with respect to data assembly, interpretation and quality assurance still make this impracticable in the short to medium term.
In this study we have aimed to explore the utility of MALDI-TOF-MS biotyping as a method to bridge this gap between ITS based DNA barcoding and whole genome sequencing for the identification and differentiation of Dunaliella isolates. Chemotaxonomic techniques such as MALDI-TOF-MS biotyping are now commonly used in many microbiology laboratories worldwide and are capable of detecting very minor differences in the expression of major ribosomal and housekeeping genes. The method has been expanded to the identification and characterisation of eukaryotes40,42,43,44; yet it remains an underutilised technique in the study of microalgae, particularly from a biotechnology perspective49. Figure 3 clearly shows that the approach can easily distinguish between genera, yet the real perceived value of the approach lies with its potential to differentiate between isolates within the same genus and ideally within the same species. The target of the current study was to assess the feasibility of biotyping by MALDI-TOF-MS to discriminate between strains of Dunaliella held in the publically accessible CCAP collection ( http://www.ccap.ac.uk/). However, the authors support the assertion that sexual compatibility remains the gold standard for the phylogeny of this genus.
In clade 1 of the ITS phylogram CCAP 19/10 and 19/26 have identical ITS sequences and they share MALDI peaks at m/z 5074 ± 2 and 10144 ± 1; however, clustering based on MALDI data (Fig. 6) suggests that the strains are distinct. The third strain in clade 1, CCAP 19/21 has a mass difference in the two main peaks. The MALDI data therefore confirms that all three strains cluster together but also highlights differences between the strains. Interestingly, the MALDI analysis for D. acidophila CCAP 19/35 suggests it may not be entirely distinct from all other strains. There appeared to be some commonality with D. salina CCAP 19/39; however, this is not a strong indicator of relatedness. For clade 1 the ITS and MALDI approaches are generally consistent.
For clade 3 again there is broad agreement between the two approaches. Clade 4 tells a similar story with two strains, CCAP 19/30 and 19/15 in very close alignment across both techniques, yet MALDI suggests that CCAP 19/18 and 19/25 do not cluster with each other or either of the two clade 4 strains.
The most interesting differences are highlighted for the strains within clade 2, all of which have identical ITS sequences. Biotyping by MALDI-TOF-MS would suggest that four separate clusters are identifiable within this clade; 1) CCAP 19/14, 19/6B and 19/12; 2) CCAP 19/32 and 19/5; 3) CCAP 19/23, 19/31 and 19/9; and finally 4) CCAP 19/27, 19/4, 19/33, 19/22, 19/24, 19/7C, 19/34, 19/7B, 19/7A and 19/8. Clustering is based on the presence and absence of indicator peaks and also changes in peak mass to charge ratio (Table 2).
One concern of the MALDI approach is the presence and potentially confounding influence of bacteria or other microbial contaminants within the algae. As most algae cultures are non-axenic and in some cases there is an absolute requirement for bacteria-algae interactions to ensure normal healthy growth50, this may restrict the applicability of the method. If biotyping by MALDI-TOF-MS was found to only be applicable to axenic cultures, this would necessitate the addition of antibiotics to the cultures. None of the cultures used in the study were axenic; therefore, there is a risk that contaminants may potentially interfere with the acquisition of a pure algae mass spectra. Indeed, further investigation revealed that the polymer-like patterns observed from CCAP 19/18 and 19/20 were due to the presence of culturable bacteria (data not shown); although the bacterial MALDI-TOF spectrum did not share any common peaks with those of the respective Dunaliella isolates. Similarly, the spectra obtained from numerous bacteria isolated from two cultures of D. polymorpha (supplementary Figure 4; manuscript in preparation) did not overlap with the microalgae peaks. Indeed, the bacterial biomass relative to the algae biomass within the D. polymorpha cultures was too low to make any real impact on the alga’s spectral fingerprint. Antibiotic treatment of cultures supported this conclusion (data not shown).
There are contradictory reports as to the effect of culture media and culture age on bacterial protein mass-spectral patterns. Pennanec et al.51 and Ruelle et al.52 found no significant change in mass spectra fingerprint which may be explained by the dominance of peaks resulting from ribosomal and house-keeping proteins which generally remain unchanged under different growth conditions and throughout different growth stages53,54,55. However, Valentine et al.56 observed significant differences in the mass spectra of bacteria when grown on different media. Similarly, Salaün and coinvestigators57 noted that culture media did affect the spectral signature in bacteria but culture age had no effect. To explore whether culture age will alter Dunaliella mass spectra, a time series experiment was conducted using CCAP 19/27 and 19/7C obtaining mass spectra from a range of growth phases spanning exponential growth through to early senescence. The spectral pattern did not change for either isolate; the only difference was peak intensity (supplementary Fig. 2). Therefore, in the case of Dunaliella it would appear that the reproducibility of the spectra is independent of culture age. However, it should be noted that we did not determine whether growth media composition affected the spectra.
Biotyping by MALDI-TOF-MS has provided compelling evidence of much greater strain diversity than previously recognised within the CCAP Dunaliella collection, particularly where the shortcomings of DNA barcoding are evident. Taken together with DNA barcoding, biotyping by MALDI-TOF-MS should be considered as a part of a polyphasic approach to characterise microalgae and cyanobacteria. This information is not only of considerable value to the culture collection but also for those researchers that access and will eventually seek to exploit Dunaliella biodiversity for biotechnology goals.
Methods
Microalgae and cyanobacteria strains and culture conditions
Thirty two Dunaliella isolates (Table 1) and single strains of Isochrysis galbana (CCAP 927/1) and Chlorella vulgaris (CCAP 211/63) were obtained from the Culture Collection of Algae and Protozoa (CCAP; www.ccap.ac.uk). Single strains of Tetraselmis suecica and Arthrospira platensis (CCMP 1295) were obtained from Seasalter Shellfish Ltd (Kent, UK) and the Provasoli-Guillard National Centre for Marine Algae and Microbiota (NCMA; https://ncma.bigelow.org/) (formerly the Centre for Culture of Marine Phytoplankton).
All Dunaliella (except D. acidophila, CCAP 19/35) and T. suecica strains were batch cultured in 50 mL Erlenmeyer flasks with foam bungs using either sterile f/2 or double strength f/2 media58 following guidance on the CCAP website. Media were made with 1 μm filtered, UV sterilised and autoclaved natural seawater. Dunaliella acidophila was grown in AJS medium that was acidified to pH 1.5 using hydrochloric acid. Isochrysis galbana was cultured using f/2 supplemented with 1.06 × 10−4 M silicate, A. platensis was cultured using f/2 supplemented with 29.4 mM NaNO3 and adjusted to pH 8. Chlorella vulgaris was cultured in 250 mL Erlenmeyer flasks using Jaworski’s medium made with sterilised deionised water. Cultures were maintained within a temperature range of 18 ± 1 °C with a 16L:8D photoperiod. Lighting was provided by a combination of warm and cool fluorescent tubes giving a mean illuminance of 3000 lux. Culture optical densities (OD) were measured at 690 nm from 300 μL triplicate subsamples in a microplate using a FLUOstar OPTIMA fluorescence plate reader (BMG LABTECH). Prior to analysis the cultures were diluted into the OD range of 0.2-0.3 for consistency.
Genomic DNA preparation and ITS gene sequencing
Genomic DNA extraction was carried out using the GenEluteTM Bacterial DNA Kit (Sigma Aldrich, UK) with the following modifications. A 1.8 mL aliquot of each Dunaliella culture was centrifuged at 14,000 g. The pellets were washed and resuspended in Lysis Buffer P (Stratec) and Proteinase K (Invisorb) and incubated at 65 °C for 30 minutes. The lysate was centrifuged to remove cell debris and then isolation continued as described in the manufacturer’s protocol. PCR reactions were conducted using the ITS4 (5’-TCCTCCGCTTATTGATATGC-3’) and ITS5 (5’-GGAAGTAAAAGTCGTAACAAG-3) primer pair (Sigma Aldrich, UK)59. Each reaction was in a total volume of 20 μL which contained 1x HF buffer, 0.5 μM of each primer, 200 μM of each dNTP, 15 μg BSA (Promega), 0.4 U Phusion DNA polymerase (ThermoScientific) and 1 μL of genomic DNA. The cycling conditions were 98 °C for 5 minutes followed by 30 cycles of 98 °C for 30 s, 42 °C for 30 s, 72 °C for 1 minute followed by a final extension step at 72 °C for 10 minutes. Pure PCR products (~782 bp) were cleaned up using the GenEluteTM PCR Clean-Up Kit (Sigma Aldrich, UK) according to the manufacturer’s instructions. PCR reactions that contained multiple PCR fragments were cleaned using QIAEX II Gel Extraction Kit (QIAGEN) according to the manufacturer’s instructions. Bidirectional DNA sequencing was performed by Geneius Laboratories Ltd (Cramlington, UK) using the same primers as for PCR. Chromatograms were checked using Chromas Lite (v2.1.1) and consensus sequences were constructed using the CAP3 sequence assembly programme60. The sequences were deposited in GenBank® with the respective accession numbers listed in Table 1.
Sample preparation for MALDI-TOF analysis using whole cells
Twenty milligrams of the sample matrix (α-cyano-4-hydroxy-cinnamic acid; HCCA, Brucker Daltonics) was prepared by mixing 1 mL of 50% acetonitrile: 2.5% trifluoro-acetic acid (Sigma-Aldrich, UK). The matrix was vortexed and saturated by 30 minutes incubation at 25 °C in an ultrasonic water bath at 100% power (Grant Instruments, Cambridge) with a second vortex at 15 minutes. The matrix was then centrifuged at 14,000 g for 2 minutes (Sigma 1-15K microcentrifuge) and 50 μL aliquots prepared fresh for use. Preliminary work revealed that crude solvent based extracts of the algae isolates did not generate consistent high quality mass spectra (data not shown). In contrast, analysis using whole cells in water proved effective. One millilitre of each algae culture containing whole cells was centrifuged at 14,000 g for 5 minutes and the pellets washed twice with deionised water. Each pellet was then re-suspended in 50 μL of deionised water. Samples were mixed 1:1 with HCCA matrix and four 2 μL technical replicates were spotted on to a MTP 384 ground steel MALDI target plate (Bruker Daltonics) and air dried at room temperature for 20 minutes.
MALDI-TOF parameters and data acquisition
Mass spectrometry was done using an UltraFlex II MALDI-TOF TOF (Bruker Daltonics GmbH, Leipzig, Germany) mass spectrometer with fuzzy control of laser intensity. Ion source 1 was set at 25 kV and ion source 2 was set at 23.5 kV with a laser frequency of 50.0 Hz, a detector gain of 1,650 V, and a gating maximum of 1,500 Da. Spectra were recorded in the positive linear mode for the mass range of 2,000 to 20,000 m/z. Each spectrum was obtained by averaging 600 laser shots acquired in the automatic mode. For data acquisition and validation measurements were performed in Auto Execute mode. The spectra were externally calibrated using the Bacterial Test Standard (Bruker Daltonics). The standard consisted of seven ribosomal proteins from Escherichia coli with added RNase A and myoglobin to cover a range of ca. 3637 to 16957 m/z (Da).
Four independent samples of each microalgae and cyanobacteria isolate were placed on four separate spots on a ground steel MALDI target plate (see supplementary Figure 3). Each sample spot was read twice thereby producing eight spectra per isolate. The quality and mass accuracy of the peaks were examined using the FlexAnalysis software (Bruker Daltonics). The eight spectra were overlaid and a consensus spectrum generated which was added to the database for each isolate.
Time course growth study
To determine whether isolate spectra change with culture age the isolates CCAP 19/27 and CCAP 19/7C were batch cultured for 20 days using sterile f/2 medium in 500 mL Erlenmeyer flasks with foam bungs as described above. Cultures were aerated using compressed sterile-filtered air at 1.6 vessel volumes per minute. Cell counts were made using an improved Neubauer haemocytometer with an Olympus BH-2 brightfield microscope using Lugol’s solution as an immobilising agent. Samples that were collected on culture days 2, 9, 10, 12, 14, 18 and 20 were prepared for MALDI analysis as described above and the pellets stored at −80 °C. Prior to spotting, samples were thawed on ice and resuspended in 50 μL deionised water.
Bacterial interference experiment
Two strains of D. polymorpha (CCAP 19/7A and 19/7C) were selected to investigate if bacterial mass spectra could potentially confound the spectra produced from the Dunaliella cultures. Two separate 150 mL cultures per isolate were grown in sterilized f/2 medium with one culture treated with the antibiotics penicillin G and dihydrostreptomycin sulphate in a ratio of 200 μg mL−1: 50 μg mL−1. One millilitre samples from each culture were inoculated onto triplicate sterile marine agar plates after four days of growth. The plates were incubated for eight days at 18 oC and 16L:8D photoperiod. Whole bacterial colonies were spotted on to MALDI target plates and analysed as per the microalgae. Spectra from antibiotic and non-antibiotic treated D. polymorpha cultures were obtained for comparison.
Data analysis
A phylogenetic tree was constructed for ITS data using Clustal Omega’s neighbour joining clustering method with a bootstrap value of 100 ( http://www.ebi.ac.uk/Tools/msa/clustalo/). Further ITS2 analysis was performed using the default settings of the University of Wuerzberg ITS2 workbench ( http://its2.bioapps.biozentrum.uni-wuerzburg.de/) and ProfDist for phylogenetic analysis using both sequence and structure information. For MALDI data a composite correlation index (CCI) distance matrix was constructed using the Biotyper software. From the distance matrix a heat map was generated in Microsoft Excel (Microsoft Corporation). The dendrogram was constructed by clustering of observations using Minitab (Minitab, Inc.) with the average linkage method.
Additional Information
How to cite this article: Emami, K. et al. Proteomic-based biotyping reveals hidden diversity within a microalgae culture collection: An example using Dunaliella. Sci. Rep. 5, 10036; doi: 10.1038/srep10036 (2015).
Supplementary Material
Acknowledgments
This work has been co-funded by the North Sea Region Programme under the ERDF of the European Union, the Engineering and Physical Sciences Research Council (EPSRC) (EP/G037620/1) as part of the Biopharmaceutical Bioprocessing Technology Centre, and by Newcastle University.
Footnotes
Author Contributions K.E. designed the research, conducted the mass spectrometry, analysed data and wrote the manuscript; E.H. contributed to designing the research and data analysis; A.N. conducted the ITS studies and data analysis; C.M.B. conducted the algae time course studies and contributed to mass spectrometry data collection and analysis; F.M.L. contributed to the bacterial spectrometry; E.M. contributed funding for the study; J.G.D. provided algae cultures and contributed to data analysis; G.S.C. supervised the research, contributed funding, analysed data and wrote the manuscript. All authors contributed to discussions, writing and reviewing of the manuscript.
References
- Das S. Biotechnological exploitation of marine animals. In Animal Biotechnology: Models in Discovery and Translation, (ed Verma A. S., Singh A.) 541–562 Academic Press 2013). [Google Scholar]
- Mascarelli A. L. Gold rush for algae. Nature 461, 460–461 (2009). [DOI] [PubMed] [Google Scholar]
- Schubert C. Making fuels for the future. Nature 474, 531–532 (2011). [DOI] [PubMed] [Google Scholar]
- Worden A. Z. et al. Rethinking the marine carbon cycle: Factoring in the multifarious lifestyles of microbes. Science 347, 1257594DOI: 10.1126/science.1257594 (2015). [DOI] [PubMed] [Google Scholar]
- Ben-Amotz A. Industrial production of microalgal cell-mass and secondary products - Major industrial species. In Handbook of Microalgal Culture: Biotechnology and Applied Phycology (ed Richmond A.) 273–280 Blackwell Science Ltd 2004). [Google Scholar]
- Scott S. A. et al. Biodiesel from algae: Challenges and prospects. Curr. Opin. Biotech. 21, 277–286 (2010). [DOI] [PubMed] [Google Scholar]
- Barclay W. & Apt K. Strategies for Biopropecting Microalgae for Potential Commercial Application. In Handbook of Microalgal Culture: Applied Phycology and Biotechnology (ed Richmond A, Hu Q ) 69–79 (Wiley, 2013). [Google Scholar]
- Elliott L. G. et al. Establishment of a bioenergy-focused microalgal culture collection. Algal Res. 1, 102–113 (2012). [Google Scholar]
- Gachon C. M. M. et al. The CCAP knowledgebase: Linking protistan and cyanobacterial biological resources with taxonomic and molecular data. Syst. Biodivers 11, 407–413 (2013). [Google Scholar]
- Johnson T. S. et al. Genetic improvement of Biofuel plants: Recent progress and patents. Recent Pat DNA Gene Sequences 7, 2–12 (2013). [DOI] [PubMed] [Google Scholar]
- Tabernero A., Martín Del Valle E. A. & Galan M. A. Microalgae technology: A patent survey. Int. J. Chem. React. Eng. 11, 733–763 (2013). [Google Scholar]
- Cameotra S. S. Can microbes be patented? Biochem Biophys. Res. Comms. 430, 448 (2013). [DOI] [PubMed] [Google Scholar]
- Ryan Georgianna D. & Mayfield S. P. Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 488, 329–335 (2012). [DOI] [PubMed] [Google Scholar]
- Garfinkel M. S., Endy D., Epstein G. L. & Friedman R. M. Synthetic genomics: Options for governance. Ind. Biotech. 3, 333–365 (2007). [DOI] [PubMed] [Google Scholar]
- Gonzalez M. A., Gomez P. I. & Montoya R. Comparison of PCR-RFLP analysis of the ITS region with morphological criteria of various strains of Dunaliella. J. Appl. Phycol. 10, 573–580, doi: Doi 10.1023/A:1008035422784 (1998). [DOI] [Google Scholar]
- Olmos J., Paniagua J. & Contreras R. Molecular identification of Dunaliella sp. utilizing the 18S rDNA gene. Lett. Appl. Microbiol. 30, 80–84 (2000). [DOI] [PubMed] [Google Scholar]
- Borowitzka M. A. & Siva C. J. The taxonomy of the genus Dunaliella (Chlorophyta, Dunaliellales) with emphasis on the marine and halophilic species. J. Appl. Phycol. 19, 567–590, doi: DOI 10.1007/s10811-007-9171-x (2007). [DOI] [Google Scholar]
- Leliaert F. et al. DNA-based species delimitation in algae. Eur. J. Phycol. 49, 179–196 (2014). [Google Scholar]
- Kim K. M., Park J. H., Bhattacharya D. & Yoon H. S. Applications of next-generation sequencing to unravelling the evolutionary history of algae. Int. J. Syst. Evol. Microbiol. 64, 333–345 (2014). [DOI] [PubMed] [Google Scholar]
- Norton T. A., Melkonian M. & Andersen R. A. Algal biodiversity. Phycologia 35, 308–326, DOI:DOI 10.2216/i0031-8884-35-4-308.1 (1996). [DOI] [Google Scholar]
- Markovits A., Gianelli M. P., Conejeros R. & Erazo S. Strain selection for β-carotene production by Dunaliella. World J. Microb. Biot. 9, 534–537, DOI:Doi 10.1007/Bf00386289 (1993). [DOI] [PubMed] [Google Scholar]
- Harari A. et al. Supplementation with 9-cis β-carotene-rich alga Dunaliella improves hyperglycemia and adipose tissue inflammation in diabetic mice. J. Appl. Phycol. 25, 687–693, doi: DOI 10.1007/s10811-012-9903-4 (2013). [DOI] [Google Scholar]
- Herrero M., Jaime L., Martin-Alvarez P. J., Cifuentes A. & Ibanez E. Optimization of the extraction of antioxidants from Dunaliella salina microalga by pressurized liquids. J. Agr Food Chem. 54, 5597–5603, doi: Doi 10.1021/Jf060546q (2006). [DOI] [PubMed] [Google Scholar]
- Tang H. et al. Potential of microalgae oil from Dunaliella tertiolecta as a feedstock for biodiesel. Appl. Energy 88, 3324–3330 (2011). [Google Scholar]
- Hadi M. R., Shariati M. & Afsharzadeh S. Microalgal biotechnology: Carotenoid and glycerol production by the green algae Dunaliella isolated from the Gave-Khooni salt marsh, Iran. Biotech. Bioproc. Eng. 13, 540–544 (2008). [Google Scholar]
- Teodoresco E. C. Organisation et développement du Dunaliella, nouveau genre de Volvocacée - Polyblepharidée. Beihefte zum Botanischen Centralblatt. 18, 215–232 (1905). [Google Scholar]
- Assuncao P. et al. Molecular taxonomy of Dunaliella (Chlorophyceae), with a special focus on D. salina: ITS2 sequences revisited with an extensive geographical sampling. Aquat. Biosyst. 8, 2, DOI: 10.1186/2046-9063-8-2 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai J. C. & Coleman A. W. The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants. J. Mol. Evol. 44, 258–271, Doi 10.1007/Pl00006143 (1997). [DOI] [PubMed] [Google Scholar]
- Kvhier A., Coleman A. W. & Fabry S. Intraspecies relationships in algae: High-resolution taxonomy and short-term phylogeny using a combination of classical and novel molecular methods. Phycologia 36, 57–57 (1997). [Google Scholar]
- Coleman A. W., Suarez A. & Goff L. J. Molecular delineation of species and syngens in Volvocacean green-algae (Chlorophyta). J. Phycol. 30, 80–90, DOI 10.1111/j.0022-3646.1994.00080.x (1994). [DOI] [Google Scholar]
- Vynne N. G., Mansson M., Nielsen K. F. & Gram L. Bioactivity, chemical profiling, and 16S rRNA-based phylogeny of Pseudoalteromonas strains collected on a global research cruise. Mar. Biotechnol. (NY) 13, 1062–1073, DOI: 10.1007/s10126-011-9369-4 (2011). [DOI] [PubMed] [Google Scholar]
- Coleman A. W., Jaenicke J. & Starr R. C. Genetics and sexual behavior of the pheromone producer Chlamydomonas allensworthii (Chlorophyceae). J. Phycology 37, 345–349, DOI 10.1046/j.1529-8817.2001.037002345.x (2001). [DOI] [Google Scholar]
- Gomez P. I. & Gonzalez M. A. Genetic variation among seven strains of Dunaliella salina (Chlorophyta) with industrial potential, based on RAPD banding patterns and on nuclear ITS rDNA sequences. Aquaculture 233, 149–162, DOI 10.1016/j.aquaculture.2003.11.005 (2004). [DOI] [Google Scholar]
- Gonzalez M. A., Coleman A. W., Gomez P. I. & Montoya R. Phylogenetic relationship among various strains of Dunaliella (chlorophyceae) based on nuclear its rDNA sequences. J. Phycol. 37, 604–611, DOI 10.1046/j.1529-8817.2001.037004604.x (2001). [DOI] [Google Scholar]
- Assuncao P. et al. Phylogenetic position of Dunaliella acidophila (Chlorophyceae) based on ITS and rbcL sequences. J. Appl. Phycol. 24, 635–639, DOI 10.1007/s10811-011-9676-1 (2012). [DOI] [Google Scholar]
- Gomez P. I. & Gonzalez M. A. Genetic polymorphism in eight Chilean strains of the carotenogenic microalga Dunaliella salina Teodoresco (Chlorophyta). Biol. Res. 34, 23–30 (2001). [DOI] [PubMed] [Google Scholar]
- Muller T., Philippi N., Dandekar T., Schultz J. & Wolf M. Distinguishing species. RNA 13, 1469–1472, Doi 10.1261/Rna.617107 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caisova L., Marin B. & Melkonian M. A close-up view on ITS2 evolution and speciation - a case study in the Ulvophyceae (Chlorophyta, Viridiplantae). BMC Evol. Biol. 11, 262, DOI: 10.1186/1471-2148-11-262 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anhalt J. P. & Catherine F. Identification of bacteria using mass spectrometry. Anal. Chem. 47, 219–225, DOI: 10.1021/ac60352a007 (1975). [DOI] [Google Scholar]
- Kaufmann C. et al. Evaluation of matrix-assisted laser desorption/ionization time of flight mass spectrometry for characterization of Culicoides nubeculosus biting midges. Med. Vet. Entomol 25, 32–38, DOI: 10.1111/j.1365-2915.2010.00927.x (2011). [DOI] [PubMed] [Google Scholar]
- Stephan R. et al. Rapid and reliable species identification of scallops by MALDI-TOF mass spectrometry. Food Control 46, 6–9 (2014). [Google Scholar]
- Volta P., Riccardi N., Lauceri R. & Tonolla M. Discrimination of freshwater fish species by matrix-assisted laser Desorption/Ionization-Time Of Flight Mass Spectrometry (MALDI-TOF MS): a pilot study. J. Limnol. 71, 164–169, DOI 10.4081/jlimnol.2012.e17 (2012). [DOI] [Google Scholar]
- Wirth H. et al. MALDI-typing of infectious algae of the genus Prototheca using SOM portraits. J. Microbiol. Method 88, 83–97, DOI: 10.1016/j.mimet.2011.10.013 (2012). [DOI] [PubMed] [Google Scholar]
- Murugaiyan J., Ahrholdt J., Kowbel V. & Roesler U. Establishment of a matrix-assisted laser desorption ionization time-of-flight mass spectrometry database for rapid identification of infectious achlorophyllous green micro-algae of the genus Prototheca. Clin. Microbiol. Infect. 18, 461–467, DOI: 10.1111/j.1469-0691.2011.03593.x (2012). [DOI] [PubMed] [Google Scholar]
- McKenna T. et al. Analysis of a cyanobacterial bloom by matrix-assisted laser desorption/ionisation time of flight microscopy. In Proceedings of the 50th American Society for Mass Spectrometry: Conference on Mass Spectrometry and Allied Topics. Orlando, USA 621–622 (2012).
- Keller A. et al. ITS2 data corroborate a monophyletic chlorophycean DO-group (Sphaeropleales). BMC Evol. Biol. 8, 218, DOI: 10.1186/1471-2148-8-218 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Doss C. G. P., Patra B. C. & Bandyopadhyay S. DNA barcoding to map the microbial communities: Current advances and future directions. Appl. Microbiol. Biotech. 98, 3425–3436 (2014). [DOI] [PubMed] [Google Scholar]
- Konstantinidis K. T. & Tiedje J. M. Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187, 6258–6264, DOI: 10.1128/JB.187.18.6258-6264.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E. M. et al. Proteomics meets blue biotechnology: A wealth of novelties and opportunities. Mar Genomics 17, 35–42 (2014). [DOI] [PubMed] [Google Scholar]
- Lorenz M., Friedl T. & Day J. G. Perpetual maintenance of actively metabolizing microalgae cultures. In Algal Culturing Techniques (ed Andersen R.A. ) 145–155 Academic Press 2005). [Google Scholar]
- Pennanec X., Dufour A., Haras D. & Rehe K. A quick and easy method to identify bacteria by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom 24, 384–392, DOI: 10.1002/rcm.4404 (2010). [DOI] [PubMed] [Google Scholar]
- Ruelle V., El Moualij B., Zorzi W., Ledent P. & Pauw E. D. Rapid identification of environmental bacterial strains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom 18, 2013–2019, DOI: 10.1002/rcm.1584 (2004). [DOI] [PubMed] [Google Scholar]
- Teramoto K. et al. Phylogenetic classification of pseudomonas putida strains by MALDI-MS using ribosomal subunit proteins as biomarkers. Anal. Chem. 79, 8712–8719, DOI: 10.1021/ac701905r (2007). [DOI] [PubMed] [Google Scholar]
- Ryzhov V. & Fenselau C. Characterization of the protein subset desorbed by MALDI from whole bacterial cells. Anal. Chem. 73, 746–750 (2001). [DOI] [PubMed] [Google Scholar]
- Pineda F. J. et al. Microorganism identification by matrix-assisted laser/desorption ionization mass spectrometry and model-derived ribosomal protein biomarkers. Anal. Chem. 75, 3817–3822, DOI: 10.1021/ac034069b (2003). [DOI] [PubMed] [Google Scholar]
- Valentine N., Wunschel S., Wunschel D., Petersen C. & Wahl K. Effect of culture conditions on microorganism identification by matrix-assisted laser desorption ionization mass spectrometry. Appl. Environ. Microbiol. 71, 58–64, DOI: 10.1128/aem.71.1.58-64.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaun S. et al. Whole-cell spectroscopy is a convenient tool to assist molecular identification of cultivatable marine bacteria and to investigate their adaptive metabolism. Talanta 80, 1758–1770, DOI:DOI 10.1016/j.talanta.2009.10.020 (2010). [DOI] [PubMed] [Google Scholar]
- Guillard R. R. & Ryther J. H. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can. J. Microbiol. 8, 229–239 (1962). [DOI] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S. & Taylor J. W. in PCR Protocols: A Guide to Methods and Applications (eds Innis M. A., Gelfand D. H., Sninsky J. J., & White T. J. ) 315–322 Academic Press, Inc. 1990). [Google Scholar]
- Huang X. Q. & Madan A. CAP3: A DNA sequence assembly program. Genome Res. 9, 868–877, DOI:Doi 10.1101/Gr.9.9.868 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.