Abstract
In this study, we evaluated genetic variants of the androgen metabolism genes CYP17A1, CYP3A4, and CYP3A43 to determine whether they play a role in the development of prostate cancer (PCa) in Korean men. The study population included 240 pathologically diagnosed cases of PCa and 223 age-matched controls. Among the 789 single-nucleotide polymorphism (SNP) database variants detected, 129 were reported in two Asian groups (Han Chinese and Japanese) in the HapMap database. Only 21 polymorphisms of CYP17A1, CYP3A4, and CYP3A43 were selected based on linkage disequilibrium in Asians (r2 = 1), locations (SNPs in exons were preferred), and amino acid changes and were assessed. In addition, we performed haplotype analysis for the 21 SNPs in CYP17A1, CYP3A4, and CYP3A43 genes. To determine the association between genotype and haplotype distributions of patients and controls, logistic analyses were carried out, controlling for age. Twelve sequence variants and five major haplotypes were identified in CYP17A1. Five sequence variants and two major haplotypes were identified in CYP3A4. Four sequence variants and four major haplotypes were observed in CYP3A43. CYP17A1 haplotype-2 (Ht-2) (odds ratio [OR], 1.51; 95% confidence interval [CI], 1.04–2.18) was associated with PCa susceptibility. CYP3A4 Ht-2 (OR: 1.87; 95% CI: 1.02–3.43) was associated with PCa metastatic potential according to tumor stage. rs17115149 (OR: 1.96; 95% CI: 1.04–3.68) and CYP17A1 Ht-4 (OR: 2.01; 95% CI: 1.07–4.11) showed a significant association with histologic aggressiveness according to Gleason score. Genetic variants of CYP17A1 and CYP3A4 may play a role in the development of PCa in Korean men.
Keywords: androgens, CYP17A1, CYP3A4, CYP3A43, genetics, prostatic neoplasms
INTRODUCTION
Androgens likely play a key role in prostate growth and prostate cancer (PCa) development, and variants of genes involved in androgen metabolism may be related to increased risk of prostate disease.1,2 Because of the known hormone dependence of PCa, genetic alterations in the androgen pathway are among the most natural candidates for conferring genetic susceptibility to PCa.
Cytochrome P450c17α (CYP17A1), which is located in the 10q24.32 chromosomal region and encodes cytochrome P450c17α, is an enzyme with 17α-hydroxylase and 17,20-lyase activities at branch points in estrogen and testosterone biosynthesis.3 Previous studies of CYP17 and PCa risk have focused on a single-nucleotide polymorphism (SNP) in the 50-untranslated (50-UTR) promoter region (rs743572). The results of these studies have been inconclusive, with some studies finding lower risk in carriers of the wild-type allele,4,5,6 while others report that the variant allele is associated with reduced risk.7,8,9,10 A meta-analysis involving 2404 patients with PCa and 2755 controls has concluded that the rs743572 CYP17 polymorphism is unlikely to substantially alter the risk of PCa occurrence.11
The CYP3A subfamily is a group of enzymes that are key deactivators of testosterone. The CYP3A locus consists of four genes, CYP3A5, CYP3A7, CYP3A4, and CYP3A43, located on chromosome 7q21.1, with each gene containing 13 exons.
CYP3A4, which is a subgroup of the cytochrome P450 supergene family, has an important role in the metabolic transformation and elimination of many drugs, possibly more so than any other cytochrome. Its expression varies by as much as 40-fold in the liver and small intestine, with genetic variation contributing to this heterogeneity.12 CYP3A4 is involved in the oxidative deactivation of testosterone to biologically less active metabolites.13,14,15 Inhibition of this transformation results in increased bioavailability of testosterone. Changes in CYP3A4, which is also involved in the oxidative metabolism of finasteride,16 can impact the effectiveness of this drug as a chemopreventative agent for PCa. A germ-line genetic variant in the 5’-regulatory region of CYP3A4 (A to G transition, rs2740574) on chromosome 7 has been reported. Alternate names include CYP3A4*1B, −392A>G, and CYP3A4-V.17 The CYP3A4 variant G allele (referred to as the CYP3A4 variant) is more common among African-American men than Caucasian, Hispanic, or Asian men.18,19 Several previous studies have found that the CYP3A4 G variant is associated with higher PCa clinical grade and stage, especially among men who were diagnosed at an older age.18,19 In other research, the variant was shown to be inversely associated with risk among men with less aggressive PCa.20,21
CYP3A43 is expressed predominantly in the prostate and less significantly in the testis, kidney, and pancreas.22 The CYP3A43 enzyme is inactive, but splicing of the CYP3A43 exon 1 to CYP3A4 and CYP3A5 exons produces hybrid mRNA products, of which the longest CYP3A43/CYP3A4 chimeric isoform can hydroxylate testosterone.22 An association has been demonstrated between the G allele of rs680055 (60084G>C [Pro340Ala]) and the probability of developing PCa in Caucasian American men with a positive family history of the disease,23 while another association has been observed between the G allele and the probability of PCa in African-American men, after adjusting for age and pack-years of cigarette smoking.24 A significant association has been demonstrated between the rs680055 G allele and the probability of developing PCa in Caucasian American men with a history of benign prostatic hyperplasia (BPH).25
In this study, genetic variants of the androgen metabolism genes CYP17A1, CYP3A4, and CYP3A43 were evaluated to determine whether they play a role in the development of PCa in Korean men.
MATERIALS AND METHODS
Study population and blood samples
Blood samples used in this study were obtained from the Korea Prostate Bank (Seoul, Korea). Both PCa and BPH groups originated from a population of older men treated for urological problems at St. Mary's Hospital (Seoul, Korea) and were predominantly of Korean ancestry. Peripheral blood leukocyte samples were obtained for genotyping from 463 men (PCa: 240; BPH: 223) and stored at − 80°C. Subjects with BPH (n = 223) had no signs of prostate malignancy based on prostate-specific antigen (PSA) blood tests and digital rectal examination of the prostate at the time the samples were taken. The mean age of the BPH cohort was 68.2 years, while for the PCa cohort the mean age was 69.0 years.
Benign prostatic hyperplasia samples were used as the control group for several reasons. First, most males have evidence of BPH by the age of 70 or 80 years, and hence the presence of some degree of BPH is normal for the mean age at diagnosis in our PCa cohort (69.0 years). Truly normal samples would thus only be obtained in a much younger control cohort, which could introduce bias. Second, blood sample collection requires a hospital visit and a PCa screening procedure of patients, which would only be undertaken in subjects with evidence of symptoms of prostate enlargement.
Written informed consent was obtained from all the study participants. The study was approved by the Institutional Review Board of Chung-Ang University Hospital and the Catholic University Hospital (IRB no. C2008035, and CUMC09U092, respectively). Blood samples were collected in tubes containing sodium ethylenediaminetetraacetic acid from St. Mary's Hospital in Korea. The QIAamp blood extraction kit (Qiagen, Seoul, Korea) was used for DNA extraction.
Gleason scores were classified as low (Gleason score, 2–6), intermediate (3 + 4 or 4 + 3), or high (8–10) grade. The clinical or pathological regional stages were categorized into localized (T1 or T2N0M0), locally advanced (T3 or T4N0M0), or metastatic (Tx, N+ or M+) based on pathological or radiological reports. Clinical characteristics of studied cases are shown in Table 1.
Table 1.
Clinical characteristics of the study subjects
Single-nucleotide polymorphism selection and genotyping
Among the 789 SNP database variants detected, 129 have been reported in two Asian groups (Han Chinese and Japanese) in the HapMap database (release #27). Only 21 polymorphisms of CYP17A1, CYP3A4, and CYP3A43 were selected based on linkage disequilibrium (LD) in Asians (only one SNP if there were absolute LDs [r2 = 1]), locations (SNPs in exons were preferred), and amino acid changes (nonsynonymous SNPs were preferred). SNPs were genotyped using the TaqMan® assay.26 Genotyping quality control was performed in 10% of the samples by duplicate checking (rate of concordance in duplicates >99.9%).
Statistical analyses
Single-nucleotide polymorphism genotype frequencies were examined for Hardy–Weinberg equilibrium (HWE) using the χ2 statistic and all were found to be consistent (P > 0.05) with HWE among Korean controls. Data were analyzed using unconditional logistic repression to calculate odds ratio (OR) as an estimate of relative risk of PCa associated with SNP genotypes.27
To determine the association between genotype and haplotype distributions of patients and controls, logistic analysis was carried out, controlling for age (continuous value) as covariates in order to eliminate or reduce any confounding variables that might influence the findings. Significant associations are shown in bold font (P ≤ 0.05). To address the problem of multiple comparisons, Bonferroni correction was used. Lewontin's D’ (|D”|) and the LD coefficient r2 were examined to measure LD between all pairs of biallelic loci.28 Using PHASE algorithm version 2.0,29 the haplotypes were inferred from the successfully genotyped SNPs, and association analysis was performed using SAS version 9.1 (SAS Inc., Cary, NC, USA). To achieve optimal correction for multiple testing of markers, representing SNPs in LD with each other, the effective number of independent marker loci was calculated using SNP spectral decomposition (SNPSpD) software (http://genepi.qimr.edu.au/general/daleN/SNPSpD/), a program that is based on the SpD of matrices of pair-wise LD among markers.30
Genotypes of major homozygotes (A/A), heterozygotes (A/B), and minor homozygotes (B/B) were given codes of 0, 1, and 2; 0, 1, and 1; and 0, 0, and 1 in the co-dominant, dominant, and recessive models, respectively. Genetic effects of inferred haplotypes were analyzed in the same way as SNPs. For example, zero copies of haplotype-1 (Ht-1) (−/−, major homozygote), one copy of Ht-1 (−/Ht-1, heterozygote), and two copies of Ht-1 (Ht-1/Ht-1, minor homozygote) were coded as 0, 1, and 2 in the co-dominant model.
RESULTS
The clinical characteristics of the study subjects are summarized in Table 1. There was no significant difference in mean age between the two groups. Average prostate volume was 36.4 ± 15.2 cm3 in the PCa group and 49.0 ± 27.0 cm3 in the control group (P < 0.01). Average PSA levels in the PCa group were 25.2 ± 87.2 ng ml−1 and 6.1 ± 6.7 ng ml−1 in the control group (P < 0.01). Figure 1 and Figure 2 display the positions of the polymorphisms, haplotypes, and pairwise LD values among the SNPs for CYP17A1, CYP3A4, and CYP3A43, respectively.
Figure 1.
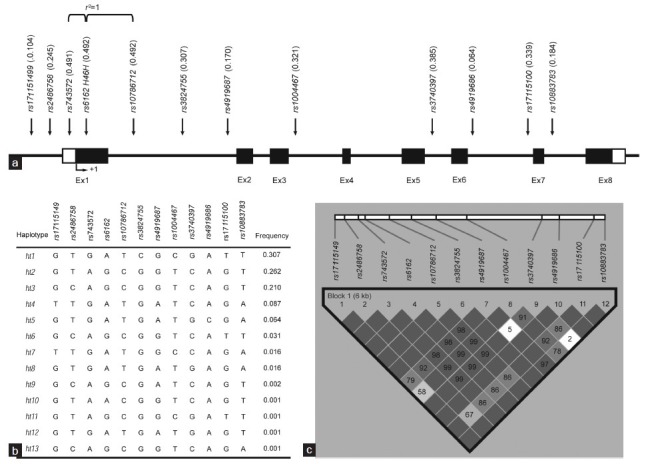
Gene map, haplotypes, and linkage disequilibrium (LD) coefficients of the CYP17A1 polymorphisms (a) Coding exons (Ex) are marked by black blocks, and 50- and 30-UTRs are marked by white blocks. (b) Haplotypes of CYP17A1. (c) LD coefficients (|D”| and r2) among the CYP17A1 polymorphisms.
Figure 2.
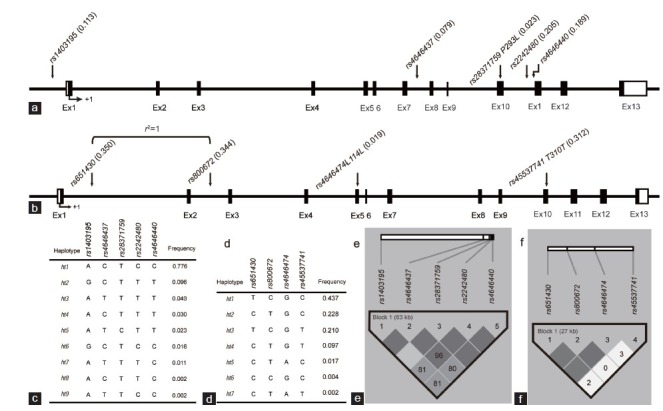
Gene map, haplotypes, and linkage disequilibrium (LD) coefficients of the CYP3A4 and CYP3A43 polymorphisms. (a) Coding exons are marked by black blocks, and 50- and 30-UTRs are marked by white blocks in map of CYP3A4. (b) Coding exons are marked by black blocks, and 50- and 30-UTRs are marked by white blocks in map of CYP3A43. (c) Haplotypes of CYP3A4. (d) Haplotypes of CYP3A43. (e) LD coefficients (|D”| and r2) among the CYP3A4 polymorphisms. (f) LD coefficients (|D”| and r2) among the CYP3A43 polymorphisms.
Twelve sequence variants were identified in CYP17A1: eight SNPs in the intron regions, two in the promoter region, one in the coding regions of the exon, and one in the 5’-UTR region of the exon (Figure 1a). Pairwise comparisons among all 12 polymorphisms revealed two sets of markers in absolute LD with each other (|D”| =1 and r2 = 1; Figure 1c). No significant deviations from HWE were observed in the polymorphisms (P > 0.05; data not shown). Five major haplotypes showed frequencies over 0.05 and accounted for 93% of the distribution (Figure 1b).
Five sequence variants were identified in CYP3A4: three SNPs in the introns, one in the promoter region, and one in the exon (Figure 2a). Pairwise comparisons among all five polymorphisms revealed two sets of markers in absolute LD with each other (|D”| =1 and r2 = 1; Figure 2e). No significant deviations from HWE were observed in the polymorphisms (P > 0.05; data not shown). Two major haplotypes showed frequencies over 0.05 and accounted for 87.2% of the distribution (Figure 2c).
Four sequence variants were observed in CYP3A43: two SNPs in the introns and two in the exons (Figure 2b). Pairwise comparisons among all four polymorphisms revealed two sets of markers in absolute LD with each other (|D”| =1 and r2 = 1; Figure 2f). No significant deviations from HWE were observed in the polymorphisms (P > 0.05; data not shown). Four major haplotypes showed frequencies over 0.05 and accounted for 97.6% of the distribution (Figure 2d).
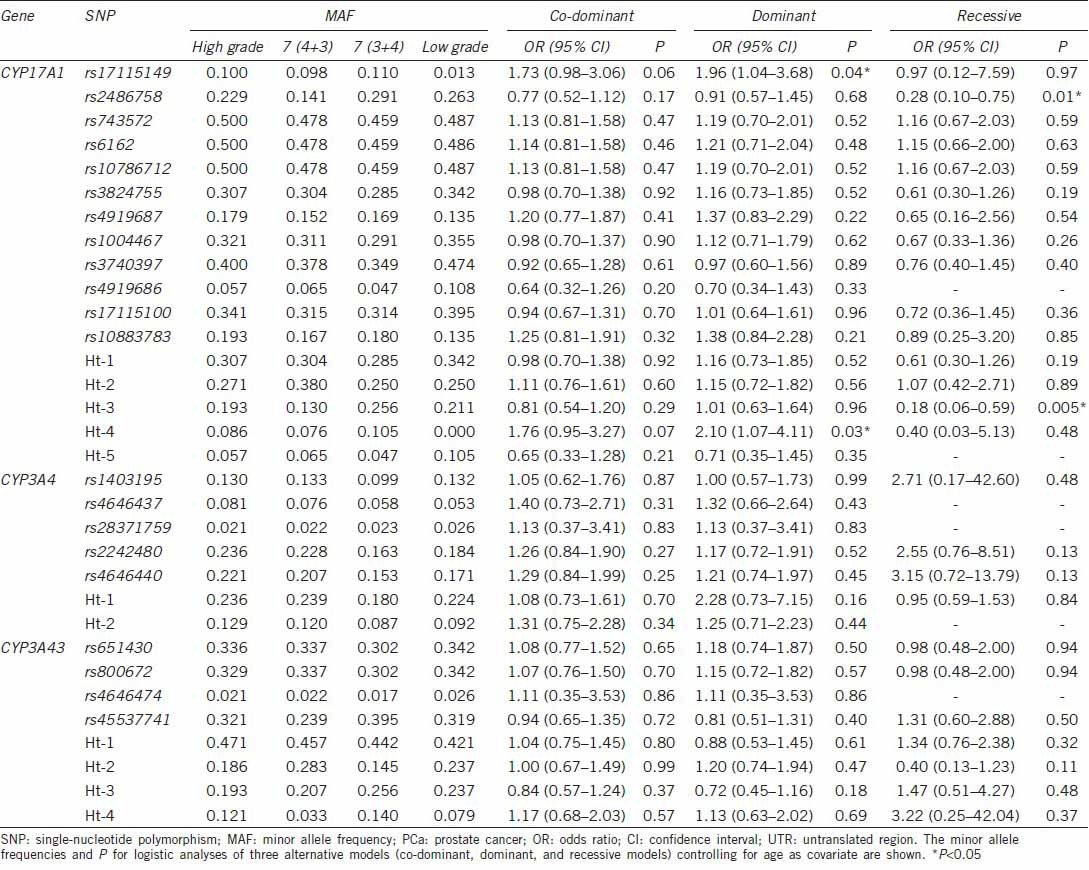
In logistic regression analysis, Ht-2 of CYP17A1 (OR: 1.51; 95% confidence interval [CI]: 1.04–2.18) was associated with PCa susceptibility in a dominant model, while Ht–2 of CYP3A43 (OR: 0.61; 95% CI: 0.42–0.88) showed a protective effect on PCa susceptibility in a dominant model (Table 2). Ht-2 of CYP3A4 (OR: 1.87; 95% CI: 1.02–3.43) was associated with the metastatic potential of PCa as determined by tumor stage in a co-dominant model (Table 3). rs17115149 (OR: 1.96; 95% CI: 1.04–3.68) and Ht-4 (OR: 2.01; 95% CI: 1.07–4.11) of CYP17A1 showed an association with the histologic aggressiveness determined using Gleason score in a dominant model, while rs2486758 (OR: 0.28; 95% CI: 0.10–0.75) and Ht-3 (OR: 0.18; 95% CI: 0.06–0.59) of CYP17A1 had a protective effect on histologic aggressiveness susceptibility in a recessive model (Table 4).
Table 2.
Logistic regression analysis of CYP17A1, CYP3A4, and CYP3A43 SNPs with the risk of PCa
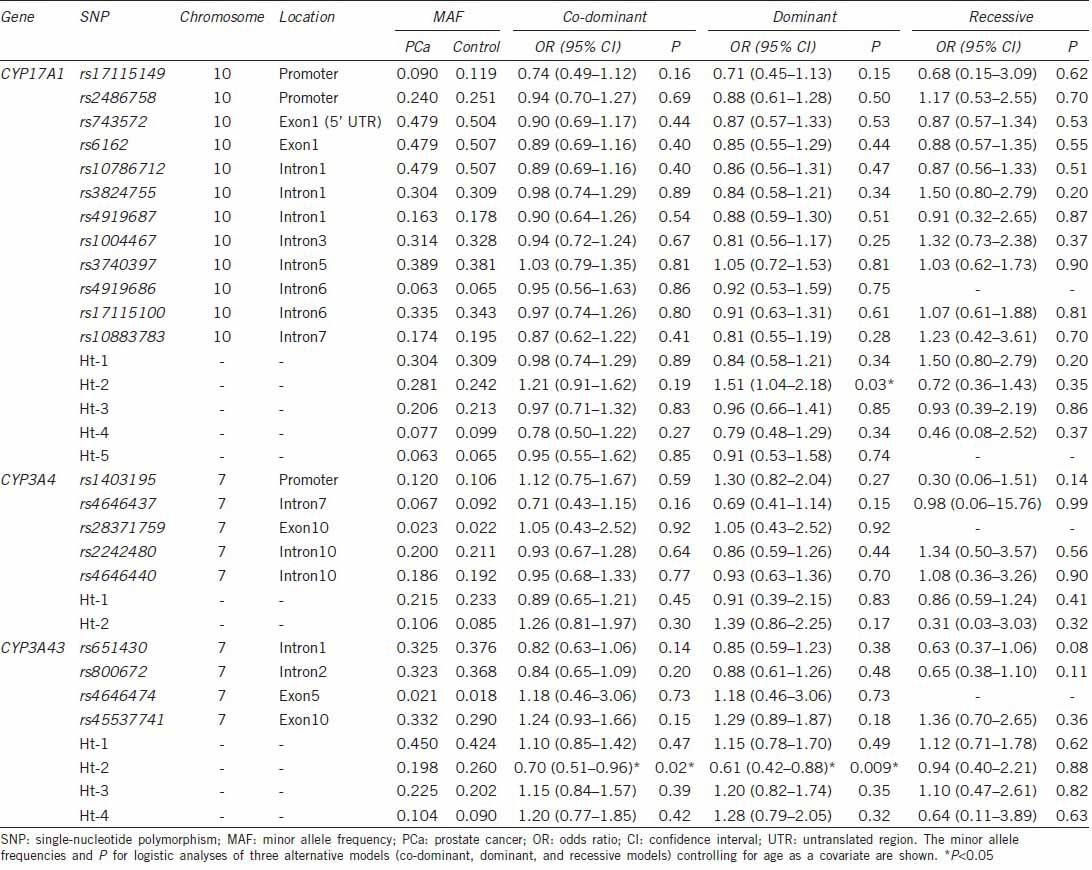
Table 3.
Logistic regression analysis of CYP17A1, CYP3A4, and CYP3A43 SNPs with the metastatic potential of PCa
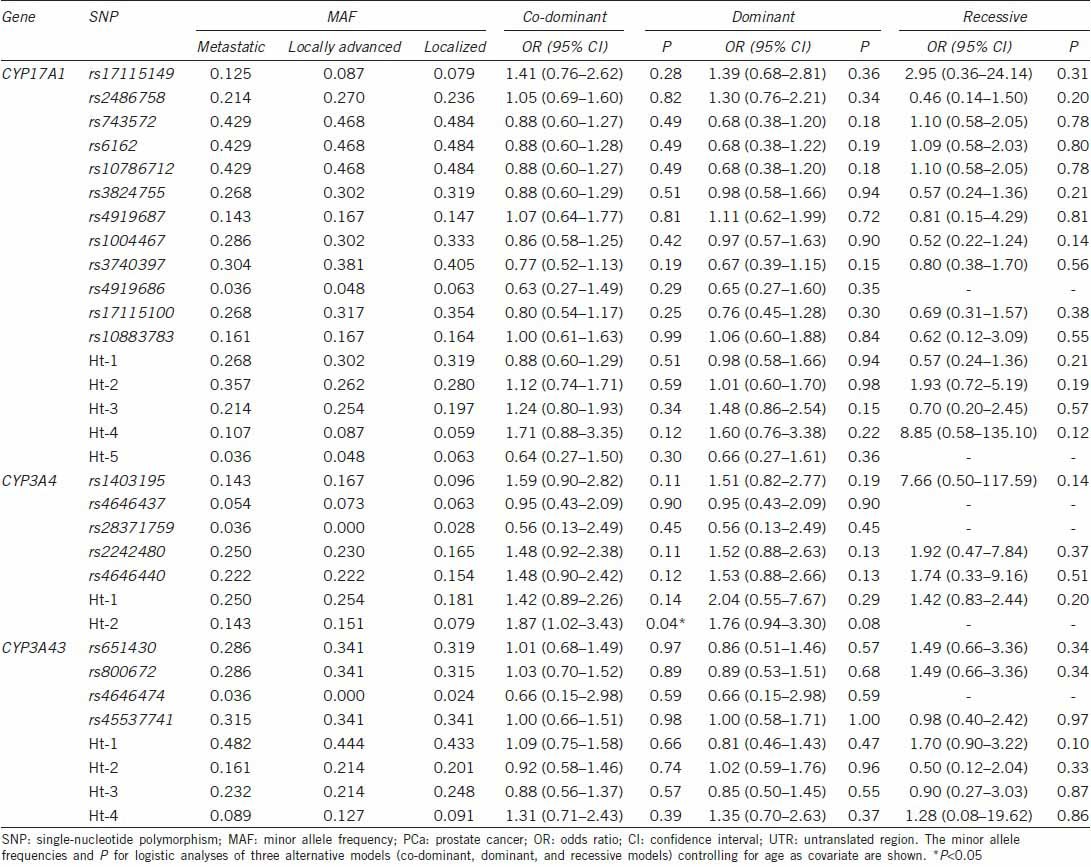
Table 4.
Logistic regression analysis of CYP17A1, CYP3A4, and CYP3A43 SNPs with the aggressiveness of PCa
DISCUSSION
Endogenous hormones, especially androgens, are required for essential prostate function, and they affect the proliferation and differentiation status of the luminal epithelium. Many studies of genes in the androgen pathway have been characterized by conflicting results, which can be attributed to population differences and biological, statistical, or technical factors. The underlying polymorphic genes of many human traits show independent patterns that are related to racial, ethnic, and geographic variation. As endogenous factors affecting the functional genome may differ, it is important to define the polymorphic spectrum of genes that are implicated in cancer causation in different populations.
Previous studies on CYP17 and PCa risk have focused on a SNP in the 50-UTR promoter region (rs743572). A single base-pair change, -34T > C, in this gene has been hypothesized to create an additional binding site (CCACT to CCACC) for the transcription factor Sp-1, which may lead to the increased transcription of the enzyme and enhanced steroid hormone production.31 The results have been contradictory, however, with some studies finding a lower risk in carriers of the wild-type allele,4,5,6 while others report that the variant allele is associated with reduced risk.7,8,9,10 A meta-analysis involving 2404 patients with PCa and 2755 controls concluded that the rs743572 CYP17 polymorphism was unlikely to substantially alter the risk of PCa occurrence.11 Yamada et al.32 recently reported that rs743572, rs6162, rs6163, and rs1004467, in CYP17A1 were significantly associated with a risk of progression to castration resistance PCa (P < 0.05). In our study, rs743572 was not associated with PCa susceptibility, metastatic potential or histologic aggressiveness. However, Ht-2 of CYP17A1 was significantly associated with PCa susceptibility, while rs17115149 and Ht-4 of CYP17A1 showed a significant association with histologic aggressiveness associated with Gleason scores.
The observation that the CYP3A4 genotype was associated with higher clinical stages and grades is consistent with the hypothesis that CYP3A4 may be associated with androgen-mediated increases in prostate cell proliferation or growth. Siemes et al.33 investigated the role of polymorphic CYP3A expression in the regulation of steroid hormone serum levels in males and the association between CYP3A variation and PCa incidence and mortality. They concluded that a common variant allele of CYP3A7 (*1C) is associated with decreased levels of estrone, estrone sulfate, dehydroepiandrosterone, and androstenedione, but no indications for an association with PCa incidence were observed. Rebbeck et al.18 in their study, they found that the CYP3A4 variant is associated with a higher clinical grade and stage, especially among men whose PCa was diagnosed at age 64 or older. Of Caucasian men with no family history of PCa, 46% with stage T3/T4 tumors and only 5% with T1 tumors had the CYP3A4 variant (OR: 9.45, 95% CI: 2.6–35; P < 0.001). Paris et al.19 found that, among 176 African-American men with PCa, those homozygous for the CYP3A4 variant presented with higher grades and stages, especially if they were 65 years of age or older at the time of diagnosis (OR: 2.4, 95% CI: 1.1–5.4). Plummer et al.21 have reported a case-control study of siblings that included 440 cases and 480 controls, more than 90% of which were Caucasian men. Of the Caucasian men, they found that the CYP3A4 variant was associated with the risk of PCa that was clinically aggressive at diagnosis (OR: 1.91, 95% CI: 1.02–3.57; P = 0.04) and inversely associated with the risk of less aggressive disease (OR: 0.08; 95% CI: 0.01–0.49; P = 0.006). The inverse association with less aggressive disease among Caucasian men was also found for several other CYP3A4 polymorphisms and in a CYP3A4 haplotype analysis.20 In this study, Ht-2 of CYP3A4 was significantly associated with PCa metastatic potential as determined by tumor stage.
Associations have been reported between the specific allele CYP3A43 coding SNP rs680055 and PCa, only after stratifying for a positive family history of PCa,23 a history of cigarette smoking,24 and a history of BPH.25 However, in this study, we did not find a significant association with PCa risk in the enrolled SNPs and haplotypes of CYP3A43. Only Ht-2 of CYP3A43 showed a significant protective effect on PCa susceptibility, but it was of no clinical importance.
The limitations of our study include the small sample size; the sample size limited our ability to test for associations with disease aggressiveness or disease extent. Future studies with larger sample sizes and collection of all relevant covariates should overcome these limitations.
The overall consistent findings of alterations in risk associated with CYP17A1 and CYP3A4 variants indicate that they may play a role in the development of PCa in Korean men and also be a determinant of risk for aggressive PCa.
The findings of this study support the role of genetic variants in genes that regulate androgen metabolism in the development of PCa in Korean men. The definition and clinical characteristics of PCa may have changed or shifted with time compared with PCa cases that were detected before the PSA era. A more detailed understanding of the importance of polymorphisms in androgen pathway genes requires more study.
AUTHOR CONTRIBUTIONS
JHH, HJK and SYL carried out the genetic studies. YSL performed the statistical analysis and helped to draft the manuscript. SCM conceived of the study, SCM and JHH participated in its design and coordination and JHH drafted the manuscript. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declare no competing interests.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Research Foundation of Korea (NRF-2013R1A1A2011824).
REFERENCES
- 1.Hsing AW. Hormones and prostate cancer: what's next? Epidemiol Rev. 2001;23:42–58. doi: 10.1093/oxfordjournals.epirev.a000795. [DOI] [PubMed] [Google Scholar]
- 2.Ross RK, Pike MC, Coetzee GA, Reichardt JK, Yu MC, et al. Androgen metabolism and prostate cancer: establishing a model of genetic susceptibility. Cancer Res. 1998;58:4497–504. [PubMed] [Google Scholar]
- 3.Sharp L, Cardy AH, Cotton SC, Little J. CYP17 gene polymorphisms: prevalence and associations with hormone levels and related factors. a HuGE review. Am J Epidemiol. 2004;160:729–40. doi: 10.1093/aje/kwh287. [DOI] [PubMed] [Google Scholar]
- 4.Yamada Y, Watanabe M, Murata M, Yamanaka M, Kubota Y, et al. Impact of genetic polymorphisms of 17-hydroxylase cytochrome P-450 (CYP17) and steroid 5alpha-reductase type II (SRD5A2) genes on prostate-cancer risk among the Japanese population. Int J Cancer. 2001;92:683–6. doi: 10.1002/1097-0215(20010601)92:5<683::aid-ijc1255>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 5.Kittles RA, Panguluri RK, Chen W, Massac A, Ahaghotu C, et al. Cyp17 promoter variant associated with prostate cancer aggressiveness in African Americans. Cancer Epidemiol Biomarkers Prev. 2001;10:943–7. [PubMed] [Google Scholar]
- 6.Gsur A, Haidinger G, Hinteregger S, Bernhofer G, Schatzl G, et al. Polymorphisms of glutathione-S-transferase genes (GSTP1, GSTM1 and GSTT1) and prostate-cancer risk. Int J Cancer. 2001;95:152–5. doi: 10.1002/1097-0215(20010520)95:3<152::aid-ijc1026>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 7.Stanford JL, Noonan EA, Iwasaki L, Kolb S, Chadwick RB, et al. A polymorphism in the CYP17 gene and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2002;11:243–7. [PubMed] [Google Scholar]
- 8.Wadelius M, Andersson AO, Johansson JE, Wadelius C, Rane E. Prostate cancer associated with CYP17 genotype. Pharmacogenetics. 1999;9:635–9. [PubMed] [Google Scholar]
- 9.Habuchi T, Liqing Z, Suzuki T, Sasaki R, Tsuchiya N, et al. Increased risk of prostate cancer and benign prostatic hyperplasia associated with a CYP17 gene polymorphism with a gene dosage effect. Cancer Res. 2000;60:5710–3. [PubMed] [Google Scholar]
- 10.Antognelli C, Mearini L, Talesa VN, Giannantoni A, Mearini E. Association of CYP17, GSTP1, and PON1 polymorphisms with the risk of prostate cancer. Prostate. 2005;63:240–51. doi: 10.1002/pros.20184. [DOI] [PubMed] [Google Scholar]
- 11.Ntais C, Polycarpou A, Ioannidis JP. Association of the CYP17 gene polymorphism with the risk of prostate cancer: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2003;12:120–6. [PubMed] [Google Scholar]
- 12.Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54:1271–94. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- 13.Waxman DJ, Attisano C, Guengerich FP, Lapenson DP. Human liver microsomal steroid metabolism: identification of the major microsomal steroid hormone 6 beta-hydroxylase cytochrome P-450 enzyme. Arch Biochem Biophys. 1988;263:424–36. doi: 10.1016/0003-9861(88)90655-8. [DOI] [PubMed] [Google Scholar]
- 14.Waxman DJ, Lapenson DP, Aoyama T, Gelboin HV, Gonzalez FJ, et al. Steroid hormone hydroxylase specificities of eleven cDNA-expressed human cytochrome P450s. Arch Biochem Biophys. 1991;290:160–6. doi: 10.1016/0003-9861(91)90602-f. [DOI] [PubMed] [Google Scholar]
- 15.Ozdemir V, Kalow W, Tang BK, Paterson AD, Walker SE, et al. Evaluation of the genetic component of variability in CYP3A4 activity: a repeated drug administration method. Pharmacogenetics. 2000;10:373–88. doi: 10.1097/00008571-200007000-00001. [DOI] [PubMed] [Google Scholar]
- 16.Huskey SW, Dean DC, Miller RR, Rasmusson GH, Chiu SH. Identification of human cytochrome P450 isozymes responsible for the in vitro oxidative metabolism of finasteride. Drug Metab Dispos. 1995;23:1126–35. [PubMed] [Google Scholar]
- 17.Home Page of the Human Cytochrome P450 (CYP) Allele Nomenclature Committee. [Last accessed on 2011 Jun 26]. Available from: http://www.cypalleles.ki.se/cyp3a4.htm .
- 18.Rebbeck TR, Jaffe JM, Walker AH, Wein AJ, Malkowicz SB. Modification of clinical presentation of prostate tumors by a novel genetic variant in CYP3A4. J Natl Cancer Inst. 1998;90:1225–9. doi: 10.1093/jnci/90.16.1225. [DOI] [PubMed] [Google Scholar]
- 19.Paris PL, Kupelian PA, Hall JM, Williams TL, Levin H, et al. Association between a CYP3A4 genetic variant and clinical presentation in African-American prostate cancer patients. Cancer Epidemiol Biomarkers Prev. 1999;8:901–5. [PubMed] [Google Scholar]
- 20.Loukola A, Chadha M, Penn SG, Rank D, Conti DV, et al. Comprehensive evaluation of the association between prostate cancer and genotypes/haplotypes in CYP17A1, CYP3A4, and SRD5A2. Eur J Hum Genet. 2004;12:321–32. doi: 10.1038/sj.ejhg.5201101. [DOI] [PubMed] [Google Scholar]
- 21.Plummer SJ, Conti DV, Paris PL, Curran AP, Casey G, et al. CYP3A4 and CYP3A5 genotypes, haplotypes, and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:928–32. [PubMed] [Google Scholar]
- 22.Finta C, Zaphiropoulos PG. Intergenic mRNA molecules resulting from trans-splicing. J Biol Chem. 2002;277:5882–90. doi: 10.1074/jbc.M109175200. [DOI] [PubMed] [Google Scholar]
- 23.Zeigler-Johnson C, Friebel T, Walker AH, Wang Y, Spangler E, et al. CYP3A4, CYP3A5, and CYP3A43 genotypes and haplotypes in the etiology and severity of prostate cancer. Cancer Res. 2004;64:8461–7. doi: 10.1158/0008-5472.CAN-04-1651. [DOI] [PubMed] [Google Scholar]
- 24.Stone A, Ratnasinghe LD, Emerson GL, Modali R, Lehman T, et al. CYP3A43 Pro(340) Ala polymorphism and prostate cancer risk in African Americans and Caucasians. Cancer Epidemiol Biomarkers Prev. 2005;14:1257–61. doi: 10.1158/1055-9965.EPI-04-0534. [DOI] [PubMed] [Google Scholar]
- 25.Rebbeck TR, Rennert H, Walker AH, Panossian S, Tran T, et al. Joint effects of inflammation and androgen metabolism on prostate cancer severity. Int J Cancer. 2008;123:1385–9. doi: 10.1002/ijc.23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ. Allelic discrimination using fluorogenic probes and the 5’ nuclease assay. Genet Anal. 1999;14:143–9. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 27.Morris JA, Gardner MJ. Calculating confidence intervals for relative risks (odds ratios) and standardised ratios and rates. Br Med J (Clin Res Ed) 1988;296:1313–6. doi: 10.1136/bmj.296.6632.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hedrick PW. Gametic disequilibrium measures: proceed with caution. Genetics. 1987;117:331–41. doi: 10.1093/genetics/117.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–9. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mononen N, Schleutker J. Polymorphisms in genes involved in androgen pathways as risk factors for prostate cancer. J Urol. 2009;181:1541–9. doi: 10.1016/j.juro.2008.11.076. [DOI] [PubMed] [Google Scholar]
- 32.Yamada T, Nakayama M, Shimizu T, Nonen S, Nakai Y, et al. Genetic polymorphisms of CYP17A1 in steroidogenesis pathway are associated with risk of progression to castration-resistant prostate cancer in Japanese men receiving androgen deprivation therapy. Int J Clin Oncol. 2013;18:711–7. doi: 10.1007/s10147-012-0430-8. [DOI] [PubMed] [Google Scholar]
- 33.Siemes C, Visser LE, de Jong FH, Coebergh JW, Uitterlinden AG, et al. Cytochrome P450 3A gene variation, steroid hormone serum levels and prostate cancer - The Rotterdam Study. Steroids. 2010;75:1024–32. doi: 10.1016/j.steroids.2010.07.001. [DOI] [PubMed] [Google Scholar]


