Abstract
Background
The genotypic diversity and cariogenicity of C. albicans from the dental plaque of children are poorly understood. This study aimed to explore the genotypic diversity and cariogenicity of C. albicans from children with early childhood caries and caries-free children.
Methods
Dental plaque samples from 238 children with early childhood caries and from 125 caries-free children were collected for C. albicans isolation. A PCR method based on 25S rDNA was used to analyze C. albicans genotypes, and the strains with different genotypes were tested with regard to acidogenicity and aciduricity.
Results
Among 129 C. albicans isolates, 79 (61.2 %) belonged to genotype A. The distribution frequency of genotypes A and C or genotypes B and C showed no significant difference between children with early childhood caries and caries-free children (p = 0.178 and 0.148), whereas genotypes A and B exhibited significantly different distributions (p = 0.010). No significant differences in aciduricity were found among the three genotypes, but the acidogenicity of genotypes B and C differed significantly from that of genotype A at pH 4.0.
Conclusions
The genotypic distribution of C. albicans is associated with the caries experience of children, and the genotype may be related to its acidogenicity at pH 4.0.
Keywords: Candida albicans, Cariogenicity, Early childhood caries, Genotype
Background
Early childhood caries (ECC) is defined as the presence of 1 or more decayed teeth (noncavitated or cavitated lesions), missing teeth (due to caries), or filled tooth surfaces in any primary tooth in a child 71 months of age or younger [1]. Due to the high prevalence in primary school children, ECC is a public health problem of great concern. Because caries are caused by microorganisms in dental plaque, knowledge about the relationship between the microorganisms in children’s dental plaque and ECC is important for ECC prevention.
C. albicans is often detected in children with dental caries, while this yeast is typically absent in caries-free children [2, 3]. These findings provide indirect evidence for the association of C. albicans with dental caries. In our previous study, the frequency of C. albicans isolated in dental plaque from children with ECC was confirmed to be higher than that in caries-free (CF) children (44.1 vs. 19.2 %, χ2 = 22.213, p < 0.001), which indicated that C. albicans is related to ECC [4].
Some reports have analyzed the genotypic distribution of C. albicans in the dental biofilm of primary school children with different caries statuses and found that a specific genotype was dominant in the dental biofilm of children with severe early childhood caries (S-ECC) [5, 6]. However, the correlations between different C. albicans genotypes and the cariogenicity of this yeast remain unknown.
Researchers have found that the virulence factors of C. albicans, such as invasiveness and drug resistance, are linked to the genotype [7–9] and that acidogenicity and aciduricity are important C. albicans virulence factors for dental caries. In our previous study, the C. albicans strains isolated from the dental plaque of ECC children were found to be more aciduric than those from CF children, indicating that the aciduricity of clinical C. albicans strains is associated with a child’s dental status. In contrast, no differences were found with regard to acidogenicity between the two groups [10]. Regardless, the relationships of acidogenicity and aciduricity with genotype remain unclear.
In this study, we hypothesized that the genotypic distribution of C. albicans from ECC and CF children differs and that different C. albicans genotypes exhibit variable acidogenicity and aciduricity. To test this hypothesis, C. albicans from the dental plaque of ECC and CF children was detected by 25S rDNA-based PCR, and the acidogenicity and aciduricity of different C. albicans genotypes were evaluated.
Methods
Subjects
In total, 363 children aged 3–5 years (mean ± SD, 3.2 ± 1.9) were randomly recruited from four kindergartens in Guangzhou, China, from Feb. to Apr. 2009 (200 boys and 163 girls). The children were healthy and had no history of antibiotic use for at least 1 month before the study, and none of their permanent teeth had yet erupted. The children were assigned to one of two groups according to their caries status: the ECC group (n = 238) or the CF group (n = 125). The diagnostic criteria for ECC used in this study were proposed by Drury et al. [1]. The parents of children were fully informed in writing and written informed consent for participation was obtained from the parents who decided to take participation in the study.
Approval was obtained from the Research Ethics Committee of Sun Yat-sen University before the study began.
Sample collection
Sample collection was performed in the morning, and the children were asked not to eat for 2 h before sample collection. Sterilized dental explorers were used for dental plaque collection. For the ECC children, dental plaque was obtained from caries lesions in the anterior teeth and/or molar teeth and then pooled. For the CF children, pooled dental plaque was obtained from the sound labial surfaces of upper anterior teeth and from the sound buccal surfaces of the upper and lower first primary molars. All samples were kept individually in sterile tubes containing brain heart infusion broth (HKM Co., Guangdong, China) and stored on ice; the samples were transported to the laboratory within 2 h after collection.
C. albicans isolation and identification
All samples were dispersed by vortexing for 30 s to disperse any yeast aggregates. A 20-ml aliquot of each sample was inoculated onto a plate containing selective medium (CHROMagar Candida; CAC, CHROMagar Co., Paris, France) and then cultured for 48 h at 37 °C [11]. To obtain as many representative genotypes of C. albicans in one sample as practically possible, we selected 5 colony-forming units (CFUs) per sample (approximately 30–300 CFUs per child) from each CAC plate using a method described previously [12] to minimize selection bias and to maximize randomness. All the colonies in each sample were transferred to Sabouraud dextrose broth (SDB) (HKM Co., Guangdong, China) containing glycerol (30 % v⁄v) and preserved at −80 °C.
The methods of subject recruitment, sample collection, and C. albicans isolation and identification were described in our previously published study [4]. The isolates of C. albicans were the stored isolates from the previous study [4].
DNA extraction
A boiling method was used for DNA extraction. Frozen suspensions of C. albicans strains were blended, and a 200-μl aliquot of each sample suspension was added to an Eppendorf tube and centrifuged for 8 min at 12,000 rpm. After the precipitate was ground in liquid nitrogen, it was allowed to stand for 5 min after the liquid nitrogen had evaporated; this procedure was repeated in triplicate. The precipitate was suspended in 30 μl of DNA extraction solution, boiled at 100 °C for 10 min, and centrifuged at 12,000 rpm for 3 min. The supernatant was stored at −20 °C until PCR amplification.
PCR amplification
The primers CA-INT- L (5′-ATAAGGGAAGTCGGCAAAATAGATCCGTAA-3′) and CA-INT-R (5′-CCTTGGCTGTGGTTTCGCTAGATAGTAGAT-3′) were designed as previously described [13]. The amplification mixture (total volume, 50 μl) contained 10 μl of PCR buffer; 1 μl each of dNTPs, primers, and Taq polymerase (Promega, Madison, WI, USA); 34 μl of ddH2O; and 2 μl of the template DNA. The amplification conditions were as follows: initial denaturation at 93 °C for 5 min, 40 cycles of denaturation at 93 °C for 30 s, primer annealing at 55 °C for 45 s, and extension at 72 °C for 45 s, with a final extension at 72 °C for 7 min. For identification, the PCR products were loaded onto 2 % agarose gels (Bio-Rad, Hercules, CA, USA) at 100 V for 30 min and then stained with an ethidium bromide solution. The C. albicans sequences were classified into five genotypes according to the band pattern: genotype A (450 bp), genotype B (840 bp), genotype C (450 and 840 bp), genotype D (1080 bp), and genotype E (1400 bp) [14, 15].
Suspension preparation
Twenty strains of each C. albicans genotype were examined for acid production and acid tolerance. Frozen stocks of the strains were inoculated in SDB and grown at 37 °C with shaking (150 rpm) under aerobic conditions. After the yeast cells were cultured for 17–24 h, they were harvested in the late exponential growth phase [16] and washed twice with phosphate-buffered saline (PBS, containing 0.05 mM Na2HPO4/KH2PO4, pH 6.8). The cells were suspended in PBS and adjusted to an optical density (OD) of 1.0 at 540 nm (equivalent to 1 × 108 cells/ml) for acid production and acid tolerance tests. The OD value was detected using an ultraviolet spectrophotometer (752S, Lengguang Tech. Co., Shanghai, China).
Acid production and acid tolerance assays
Sterile SDB (containing 100 mM glucose) was used for the acid production and acid tolerance assays; the pH of the medium was adjusted to different initial pH values of 7.0, 6.0, 5.5, 5.0, 4.5, and 4.0 (pH S-25, Shanghai Precision & Scientific Instrument Co., China) using sterile 2 mM HCl or 4 mM NaOH [17]. Briefly, 50-μl aliquots of suspensions of each of the different genotypes at an OD540 of 1.0 were added separately to 5 ml of SDB, cultured for 48 h at 37 °C and then centrifuged at 4500 rpm for 8 min at 4 °C. The pH of the final supernatant was detected using a standard pH meter to determine acid formation. The ∆pH (equal to the initial pH value minus the final pH value) was used to assess acid production.
To evaluate growth under acidic conditions, the precipitate was washed 3 times in PBS and then diluted with 2 ml of PBS. Then, the turbidity was measured at 540 nm (OD540) using a spectrophotometer.
These experiments were performed in triplicate. The mean ∆pH and OD540 were each calculated from three replicate samples. A greater ∆pH value indicated a greater ability to produce acid, and a greater OD540 value indicated better growth and higher aciduricity.
Statistical analysis
Data analysis was performed using SPSS 16.0 (SPSS, Chicago, IL, USA). The relationship between the genotypic diversity of C. albicans and different caries experiences was analyzed using chi-square tests. To avoid type I errors when comparing multiple groups via chi-square tests, a critical p-value-required correction was applied, and the p-value threshold was set to 0.017 in three-way comparisons. Differences in ∆pH and OD540 values among the three genotypes of C. albicans were analyzed by ANOVA. When a significant difference was found among the three genotypes, the LSD-t method was used to analyze the difference between two groups, and the level of significance for the statistical tests was set at 0.05.
Results
C. albicans genotypic distribution
C. albicans strains were isolated from 129 subjects (35.5 % of 363 children): 105 children with ECC and 24 CF children. Each child from whom C. albicans was isolated presented only one genotype, with genotypes A, B, and C being the three detected genotypes. Genotypes D and E were not detected. Among the isolates, 79 (61.2 %) belonged to genotype A; 20 (15.5 %), to genotype B; and 30 (23.3 %), to genotype C. Genotype A was the major component in both groups of children (56.2 % in the ECC group, 83.3 % in the CF group); the frequencies of genotype B were 19.0 % and 0, and the frequencies of genotype C were 24.8 and 16.7 %, respectively.
The frequencies of different genotypic distributions in the ECC and CF groups were significantly different (p = 0.042). Genotypes A and B were distributed in a significantly different manner in the two groups (p = 0.010): genotype A presented a higher frequency in the CF group, and genotype B presented a higher frequency in the ECC group. The distribution frequencies of genotypes A and C or genotypes B and C showed no significant difference between the two groups, and the p-values were 0.178 and 0.148, respectively (Table 1 and Fig. 1).
Table 1.
Genotypic distribution of C. albicans from dental plaque of children with different caries experiences, n (%)
| Group | Total number of isolates | Genotype A | Genotype Ba | Genotype Cb、c | p-value |
|---|---|---|---|---|---|
| ECC | 105 | 59 (56.2) | 20 (19.0) | 26 (24.8) | 0.042 |
| CF | 24 | 20 (83.3) | 0 | 4 (16.7) | |
| total | 129 | 79 (61.2) | 20 (15.5) | 30 (23.3) |
aComparison of the genotypic frequencies of A and B between ECC and CF groups (p = 0.010)
bComparison of the genotypic frequencies of A and C between ECC and CF groups (p = 0.178)
cComparison of the genotypic frequencies of B and C between ECC and CF groups (p = 0.148)
Fig. 1.
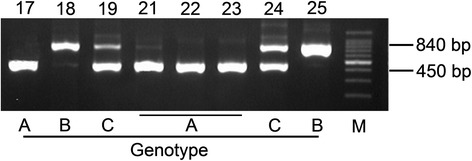
Different genotypic subgroups of C. albicans as determined by PCR. Lanes 17, 21, 22, and 23 show genotype A (the PCR amplification product is approximately 450 bp). Lanes 18 and 25 show genotype B (the PCR amplification product is approximately 840 bp). Lanes 19 and 24 show genotype C (two PCR amplification products: one band is approximately 450 bp, and the other band is approximately 840 bp)
Acidogenicity and aciduricity
The growth of C. albicans was gradually inhibited as the initial pH value of the medium decreased; however, C. albicans was able to grow at pH 4.0. As the initial pH of the cultures decreased, the OD540 values also decreased. The OD540 values for the C. albicans isolates of the three genotypes were not significantly different when cultured in media with different pH values (p > 0.05) (Table 2).
Table 2.
OD values of different genotypes of C. albicans isolates (mean ± SD)
| Genotype | OD540 value | ||||||
|---|---|---|---|---|---|---|---|
| 4.0 | 4.5 | 5.0 | 5.5 | 6.0 | 6.5 | 7.0 | |
| A (n = 20) | 1.41 ± 0.11 | 1.49 ± 0.10 | 1.52 ± 0.13 | 1.58 ± 0.14 | 1.58 ± 0.11 | 1.62 ± 0.14 | 1.63 ± 0.14 |
| B (n = 20) | 1.49 ± 0.13 | 1.53 ± 0.14 | 1.55 ± 0.14 | 1.57 ± 0.08 | 1.63 ± 0.10 | 1.65 ± 0.13 | 1.64 ± 0.16 |
| C (n = 20) | 1.49 ± 0.10 | 1.50 ± 0.07 | 1.54 ± 0.24 | 1.60 ± 0.15 | 1.61 ± 0.14 | 1.64 ± 0.14 | 1.64 ± 0.14 |
| p-value | 0.187 | 0.256 | 0.282 | 0.496 | 0.145 | 0.231 | 0.244 |
As the initial pH of the culture decreased, acid production by all C. albicans isolates was reduced. The final pH of all cultures ranged from 3.16 to 3.76. All strains exhibited the highest ∆pH values at pH 7.0. Among the three genotypes, no significant differences were found for ∆pH when cultured in media with pH values of 7.0, 6.5, 6.0, 5.5, 5.0, and 4.5 (p > 0.05). However, significant differences were found for ∆pH when the isolates were cultured in media at pH 4.0 (p = 0.029). After pairwise comparisons, no significant difference in ∆pH was observed between genotypes B and C (p = 0.836) when cultured in media at pH 4.0. In contrast, significant differences were found between genotypes A and B (p = 0.048) and between genotypes A and C (p = 0.019) (Table 3).
Table 3.
Acid formation abilities of different genotypes of C. albicans isolates (mean ± SD)
| Genotype | △pH value | ||||||
|---|---|---|---|---|---|---|---|
| 4.0 | 4.5 | 5.0 | 5.5 | 6.0 | 6.5 | 7.0 | |
| A (n = 20) | 0.84 ± 0.12 | 1.11 ± 0.11 | 1.46 ± 0.22 | 1.86 ± 0.19 | 2.37 ± 0.27 | 2.83 ± 0.20 | 3.27 ± 0.21 |
| B (n = 20) | 0.91 ± 0.09a, b | 1.21 ± 0.20 | 1.55 ± 0.12 | 1.82 ± 0.39 | 2.49 ± 0.24 | 2.87 ± 0.40 | 3.39 ± 0.23 |
| C (n = 20) | 0.93 ± 0.03c | 1.21 ± 0.08 | 1.64 ± 0.05 | 1.93 ± 0.26 | 2.57 ± 0.11 | 2.98 ± 0.15 | 3.38 ± 0.15 |
| p-value | 0.029 | 0.093 | 0.059 | 0.169 | 0.150 | 0.192 | 0.197 |
aComparison of acid formation abilities of C. albicans isolates between genotype A and B cultured at pH 4.0 (p = 0.048)
bComparison of acid formation abilities of C. albicans isolates between genotype B and C cultured at pH 4.0 (p = 0.836)
cComparison of acid formation abilities of C. albicans isolates between genotype A and C cultured at pH 4.0 (p = 0.019)
Discussion
C. albicans is detected more frequently in children with dental caries but is typically absent in caries-free children. This yeast can readily adhere to dental hard tissues in humans and form biofilms. This yeast also produces acid by metabolizing dietary sugars and carbohydrates, causing the dissolution of hydroxyapatite crystals in the enamel and dentin [16, 18]. C. albicans also produces acid at a pH value lower than 4.0 and shows strong acid tolerance [17].
To assess the genotypic identity of C. albicans strains, a PCR method based on 25S rDNA was employed for classification. To date, five genotypes of C. albicans have been identified, designated genotypes A, B, C, D, and E. Genotypes A, B, and C can be detected in the oral mucosa [15] and dental plaque [5, 6] of children. Genotype D is found in the periodontal pocket of systemically healthy patients with periodontitis [19], whereas genotype E is rarely present in the oral cavity. The results of this study revealed three genotypes of C. albicans strains in the examined children, genotypes A, B, and C, with the dominant strain being genotype A. These results are similar to a previous report showing that C. albicans genotype A is the dominant strain in the dental plaque of children [5, 6].
The genotypes of C. albicans from the oral cavities of different patients vary significantly possibly because the patients’ local oral conditions are unique [15]. In the present study, genotypes B and C were found more frequently in the ECC group than in the CF group. In particular, genotype B was not isolated from the CF group, whereas genotype A was more frequent in this group. Similar results were reported in previous studies, with genotypes B and C only detected in the carious site, not in the sound site, of S-ECC children [5] and with genotype B only detected in S-ECC children [6]. Such differences in the genotypic distribution of C. albicans may be related to differences in the sampling sites [5]. For the ECC children, the samples were obtained from caries lesions, whereas the samples for the CF children were obtained from sound surfaces. In the caries lesions, bad oral hygiene and high sugar concentrations in the cavities would have a greater influence on C. albicans colonization. Our findings suggested that different environments could facilitate the colonization of different genotypes of C. albicans and indicated that the distribution of C. albicans genotypes differed in children with different caries experiences.
In this study, the growth of C. albicans strains was gradually inhibited with a decreasing initial pH of the medium, and all C. albicans strains were able to grow at pH 4.0 and produce acid. However, the three genotypes did not significantly differ with regard to growth at any pH value, indicating that the three genotypes had similar acid tolerance and were all able to survive in a more acidic environment while continuing to produce acid. In this study, the acid-forming abilities of C. albicans genotypes B and C were significantly higher than the acid-forming ability of genotype A at pH 4.0 but not at pH 4.5–7.0. This result indicated that the genotype of C. albicans was related to its acidogenicity at pH 4.0. Notably, the pH value of the caries lesion is usually lower than that of the sound surface. The isolates of genotypes B and C were obtained from caries lesions, while some isolates of genotype A were obtained from sound surfaces. Therefore, the isolates of genotypes B and C would be able to survive in a more acid environment.
Moreover, C. albicans isolates of genotype A were detected more frequently in ECC children, and genotype A was less acidogenic at pH 4.0 than genotypes B and C. One reason for this finding is that C. albicans isolates were obtained from children with different caries status. C. albicans isolates from ECC children were found to be more acidogenic than those from CF children when cultured at pH 4.0 in our previous study [10]. In the present study, genotype A isolates were obtained from both ECC children and CF children, while all the genotype B isolates and most of the genotype C isolates were obtained from CF children.
Whether such genotypic diversity is related to the virulence of C. albicans remains uncertain. In patients with deep-seated infections, genotype A has been shown to be the most invasive isolate, whereas most non-invasive isolates belong to genotypes B and C [7]. Another study reported that of various clinical specimens, genotype C is the most invasive isolate, whereas most noninvasive isolates belong to genotype A [8]. However, in patients with bloodborne candidiasis, the genotypic distribution of C. albicans was not related to invasiveness [20]. In the present study, genotypes B and C were detected more frequently in the ECC group than in the CF group, and genotypes B and C showed significantly higher acid-forming abilities than genotype A when cultured at pH 4.0. In contrast, no relationship was found between the genotypic distribution of C. albicans and acid tolerance. Therefore, additional studies should be conducted to examine the relationship between cariogenicity and the genotype of C. albicans.
Conclusions
In this study, genotypes B and C were found more frequently in the ECC group than in the CF group, indicating that the distribution of C. albicans genotypes differs among children with different caries experiences. The acid-forming abilities of C. albicans genotypes B and C were significantly higher than that of genotype A when cultured at pH 4.0, and this result indicated that the genotype of C. albicans is related to acidogenicity at pH 4.0.
Acknowledgements
The authors express their gratitude to the principals and teachers of the kindergartens and to the children and their parents for their cooperation. This study was partially supported by the Chinese National Natural Science Fund (No. 81272554), the Guangdong Science and Technology Program Fund of China (No. 2010B050700004), and the Guangdong Natural Science Fund of China (No. 8151008901000076).
Abbreviations
- C. albicans
Candida albicans
- CF
Caries-free
- CFUs
Colony-forming units
- ECC
Early childhood caries
- OD
Optical density
- SDB
Sabouraud dextrose broth
- S-ECC
Severe early childhood caries
Footnotes
Rongmin Qiu and Wenqing Li contributed equally to this work.
Competing interest
None of the authors has competing interests.
Authors’ contributions
QRM, LWQ, and ZW designed the study, conducted the experiments, analyzed the data, and wrote the manuscript. LWQ and LY collected dental plaque samples, analyzed the data and interpreted the results. YDS and ZW designed and supervised the study and provided critical revision of the manuscript for important intellectual content. QRM and LWQ contributed equally to this work. All authors read and approved the final manuscript.
Contributor Information
Rongmin Qiu, Email: rongminqiu@hotmail.com.
Wenqing Li, Email: wendysums@hotmail.com.
Yan Lin, Email: linyanjm@gmail.com.
Dongsheng Yu, Phone: +86-20-8386-83844247, Email: yudsh@mail.sysu.edu.cn.
Wei Zhao, Phone: +86-20-8386-2553, Email: zhaowei3@mail.sysu.edu.cn.
References
- 1.Dury TF, Horowitz AM, Ismail AI, Maertens MP, Rozier RG, Selwitz RH. Diagnosing and reporting early childhood caries for research purposes. A report of a workshop sponsored by the National Institute of Dental and Craniofacial Research, the Health Resources and Services Administration, and the Health Care Financing Administration. J Public Health Dent. 1999;59:192–197. doi: 10.1111/j.1752-7325.1999.tb03268.x. [DOI] [PubMed] [Google Scholar]
- 2.De Carvalho FG, Silva DS, Hebling J, Spolidorio LC, Spolidorio DMP. Presence of mutans streptococci and Candida spp. in dental plaque/dentine of carious teeth and early childhood caries. Arch Oral Biol. 2006;51:1024–1028. doi: 10.1016/j.archoralbio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Raja M, Hannan A, Ali K. Association of oral candidal carriage with dental caries in children. Caries Res. 2010;44:272–276. doi: 10.1159/000314675. [DOI] [PubMed] [Google Scholar]
- 4.Qiu RM, Zhao W, Lin Y, Gao S, Luo W, Lu JX. Relationship between Candida albicans and early childhood caries [in Chinese] Chin J Stomatol Res: Electron ED. 2010;4:284–287. [Google Scholar]
- 5.Yang XQ, Zhang Q, Lu LY, Yang R, Liu Y, Zou J. Genotypic distribution of Candida albicans in dental biofilm of Chinese children associated with severe early childhood caries. Arch Oral Biol. 2012;57:1048–1053. doi: 10.1016/j.archoralbio.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 6.Li WQ, Yu DS, Gao S, Lin JC, Chen ZY, Zhao W. Role of Candida albicans -secreted aspartyl proteinases (Saps) in severe early childhood caries. Int J Mol Sci. 2014;15:10766–10779. doi: 10.3390/ijms150610766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karahan ZC, Güriz H, Ağirbaşli H, Balaban N, Göçmen JS, Aysev D, Akar N. Genotype distribution of Candida albicans isolates by 25S intron analysis with regard to invasiveness. Mycoses. 2004;47:465–469. doi: 10.1111/j.1439-0507.2004.01022.x. [DOI] [PubMed] [Google Scholar]
- 8.Gurbuz M, Kaleli I. Molecular analysis of Candida albicans isolates from clinical specimens. Mycopathologia. 2010;169:261–267. doi: 10.1007/s11046-009-9263-z. [DOI] [PubMed] [Google Scholar]
- 9.Liu XP, Fan SR, Bai FY, Li J, Liao QP. Antifungal susceptibility and genotypes of Candida albicans strains from patients with vulvovaginal candidiasis. Mycoses. 2009;52:24–28. doi: 10.1111/j.1439-0507.2008.01539.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhao W, Qiu RM, Lin Y, Lu JX, Yu DS. Cariogenicity of Candida albicans isolated from children with early childhood caries and caries free [in Chinese] Chin J Obstet Gynecol Pediatrics: Electron ED. 2011;7:295–298. [Google Scholar]
- 11.Odds FC, Bernaerts R. CHROMagar Candida–a new differential isolation medium for presumptive identification of clinically important Candida species. J Clin Microbiol. 1994;32:1923–1929. doi: 10.1128/jcm.32.8.1923-1929.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munson MA, Banerjee A, Watson TF, Wade WG. Molecular analysis of the microflora associated with dental caries. J Clin Microbiol. 2004;42:3023–3029. doi: 10.1128/JCM.42.7.3023-3029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCullogh MJ, Clemons KV, Stevens DA. Molecular and phenotypic characterization of genotypic C. albicans subgroups and comparison with Candida dubliniensis and Candida stellatoidea. J Clin Microbiol. 1999;37:417–421. doi: 10.1128/jcm.37.2.417-421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura M, Watanabe K, Mikami Y, Yazawa K, Nishimura K. Molecular characterization of new clinical isolates of Candida albicans and C. dubliniensis in Japan: analysis reveals a new genotype of C. albicans with group I intron. J Clin Microbiol. 2002;39:4309–4315. doi: 10.1128/JCM.39.12.4309-4315.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi QG, Hu T, Zou XD. Frequency, species and molecular characterization of oral Candida in hosts of different age in China. J Oral Pathol Med. 2005;34:352–356. doi: 10.1111/j.1600-0714.2005.00326.x. [DOI] [PubMed] [Google Scholar]
- 16.Nikawa H, Yamashiro H, Makihira S. In vitro cariogenic potential of Candida albicans. Mycoses. 2003;46:471–478. doi: 10.1046/j.0933-7407.2003.00888.x. [DOI] [PubMed] [Google Scholar]
- 17.Klinke T, Kneist S, de Soet JJ, Mauersberger S, Forster A, Klimm W. Acid production by oral strains of Candida albicans and lactobacilli. Caries Res. 2009;43:83–91. doi: 10.1159/000204911. [DOI] [PubMed] [Google Scholar]
- 18.Sen BH, Safavi KE, Spångberg LS. Colonization of C. albicans on cleaned human dental hard tissues. Arch Oral Biol. 1997;42:513–520. doi: 10.1016/S0003-9969(97)00026-5. [DOI] [PubMed] [Google Scholar]
- 19.Barros LM, Boriollo MF, Alves AC, Klein MI, Gonçalves RB, Höfling JF. Genetic diversity and exoenzyme activities of Candida albicans and Candida dubliniensis isolated from the oral cavity of Brazilian periodontal patients. Arch Oral Biol. 2008;53:1172–1178. doi: 10.1016/j.archoralbio.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Millar BC, Moore JE, Xu J, Walker MJ, Hedderwick S, McMullan R. Genotypic subgrouping of clinical isolates of Candida albicans and Candida dubliniensis by 25S intron analysis. Lett Appl Microbiol. 2002;35:102–106. doi: 10.1046/j.1472-765X.2002.01135.x. [DOI] [PubMed] [Google Scholar]


