Dear Editor,
Epigenetic regulation is crucial to the establishment and maintenance of the identity of a cell. Recent studies suggest that transcription is implemented amongst a mixture of various histone modifications1. It has also been recognized that to interrogate function of genetic information, comprehensively systematic profiling of the epigenome in multiple cell stages and types is required2. Chromatin immunoprecipitation (ChIP) has become one of the most critical assays to investigate the complex DNA-protein interactions3. Combined with profiling technologies such as microarrays (ChIP-on-chip) or high-throughput sequencing (ChIP-Seq), this assay becomes a great tool to study the epigenetic regulatory networks in cells4,5,6. However, the ChIP process produces limited amount of DNA due to the low yield of antibody pull-down, DNA damage during fragmentation and cleavage of DNA-protein complex, and complicated downstream analysis7. The conventional approaches have to consume a considerable amount of samples, typically 106 - 107 cells, to overcome this low-yield issue and obtain reliable results5. This limitation also restricts ChIP applications from precious primary tissue samples such as early embryonic cells or rare tumor stem cells.
ChIP-Seq, compared with ChIP-on-Chip, deeply sequences the target DNA fragments and generates highly comprehensive data with higher resolution, fewer artifacts, greater coverage and larger dynamic range6. Although recent application of automated microfluidic ChIP (AutoChIP) was successfully performed using 2 000 cells through locus-specific analysis by qPCR8, such assays do not achieve the comprehensiveness afforded by DNA sequencing approaches. Recently, several approaches have been developed to perform ChIP-Seq using as low as 10 000 or even only 5 000 cells7,9,10,11,14. However, all of these methods rely on ChIP reactions in tens of microliters and preamplification of ChIP product before sequencing library preparation, either through linear amplification (by in vitro transcription) or exponential amplification (by PCR), both of which potentially introduce significant bias. Adli et al.7 reported a modified protocol to realize the ChIP-Seq using 10 000 cells by revising the random primers used in amplification to reduce the primer self-annealing, with an optimized PCR condition to cover the GC-rich regions. Ng et al.14 developed another protocol to perform ChIP-Seq of H3K4me3 modification using 10 000 mouse primordial germ cells, requiring pre-amplification before the sequencing library preparation. Sachs et al.15 reported a chromatin immunoprecipitation study with low number of cells without pre-amplification, however, it needs at least 50 000 cells as starting material. Here we present a new method that implements a microfluidic device to facilitate the ChIP process, providing a technology to obtain the high-quality ChIP-Seq data from merely 1 000 mammalian cells, with no need of pre-amplification. The whole ChIP process has been greatly shortened to 8 h. Through this method, we have accomplished, for the first time, a rapid, semi-automated, and highly sensitive ChIP assay to investigate the genome-wide landscape of histone modification H3K4me3 using 1 000 mouse epiblast cells at E6.5, and found that the H3K4me3 landscape of post-implantation epiblast is more similar to that of the mEpiSCs than that of mESCs.
Microfluidic devices are the ideal reaction systems for handling small number of cells. We fabricated our PDMS-based device to treat four samples in parallel on a single chip (Figure 1A). Each reaction pipeline could accept no more than 1 200 cells. We first performed the whole ChIP assay on a microfluidic device, including trapping the magnetic beads that pre-incubated with antibody, binding of the sonicated chromatin fragments with H3K4me3 to the beads, and washing away the chromatin fragments without H3K4me3 modification. Alternatively, the micro-device can be used for fragmentation of chromatins through micrococcal nuclease (MNase) treatment on chip, after the cell permeabilization. After the enrichment we collected the DNA out of the device and finished the sequencing library construction in microcentrifuge tubes. We eliminated preamplification of the ChIP product. Instead, we combined the end-repair, adenylation, and ligation steps in a one-tube reaction and then used carrier DNA to facilitate high-efficiency purification of the ligated DNA product. Then we used PCR to amplify these ligated DNAs to get nanogram amount of DNA for sequencing. Conventionally, the experimental process is tedious, taking at least two days to complete the sample treatment3. However, our new protocol can greatly accelerate the whole process; the complete experimental procedure, including microfluidic-based cell permeabilization, chromatin fragmentation, antibody pull-down, and many washing steps, can be completed within 8 h.
Figure 1.
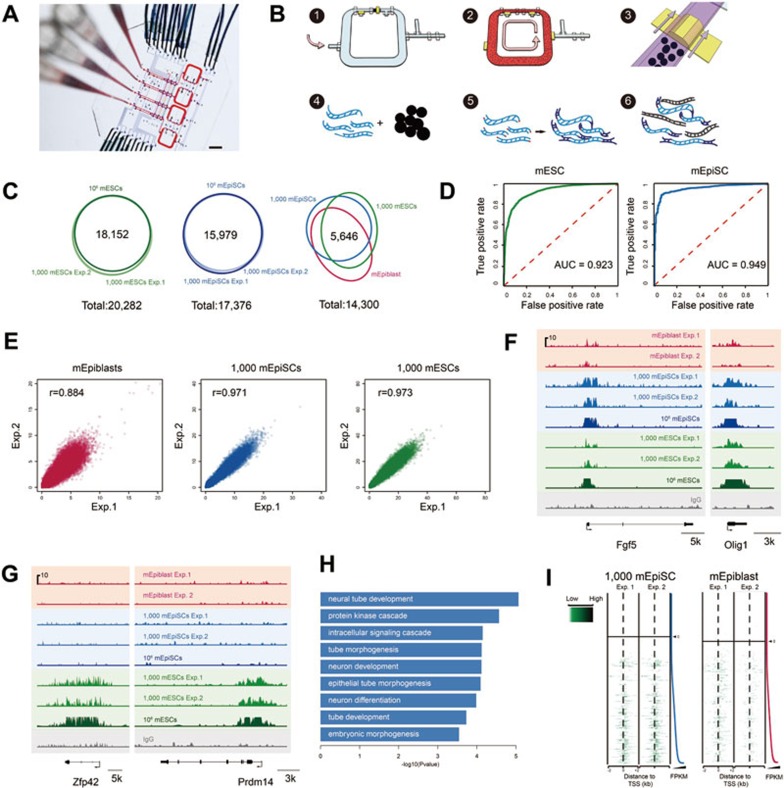
Microfluidic device and the ChIP-Seq from 1 000 mEpiSCs, 1 000 mESCs and epiblast cells of the E6.5 mouse embryos. (A) Optical micrograph of a 4-plex microfluidic device with control lines and sample inputs. Scale bar: 5 mm. (B) Key operational steps of a ChIP-Seq flow pipeline. Step 1: load the chromatin fragments to fill dead-end flow-channels; Step 2: mixing and immunoprecipitation; Step 3: trap the antibody-functionalized beads (Ab-beads) to form a column; Step 4: release the DNA from the chromatin-antibody-bead complex; Step 5: end-repair, adenylation, and ligation; Step 6: amplification for sequencing. (C) The Venn diagram of enriched TSS regions of mEpiSCs, mESCs, and E6.5 epiblast cells. (D) The receiver operating characteristic (ROC) curve representing the true positive and false positive rates for the 1 000-cell experiment of mEpiSCs and mESCs. The standards are the enriched TSS regions called from the ChIP-Seq experiments using one million cells. (E) The correlation of the enrichment of H3K4me3 markers around TSS regions of epiblast cells of E6.5 mouse embryos, mEpiSCs, and mESCs. Each point represents an individual gene. (F) Representative loci of the ChIP-Seq of the H3K4me3 markers for three cell types, E6.5 epiblast cells, mEpiSCs, and mESCs, showing the peaks shared by all these three types of pluripotent cells. (G) Representative loci of the ChIP-Seq of the H3K4me3 markers specifically enriched in mESCs. (H) Gene ontology terms enriched in epiblast cells of E6.5 mouse embryos and mEpiSCs, compared to mESCs. (I) Heatmap showing read distribution around TSS regions of different transcripts ranked by the FPKM.
We compared two approache for chromatin fragmentation, ultrasonic shearing and MNase digestion, and mainly used the former approach in our experiments since the result was more robust. The fragmentation step could be performed off-chip using the probe-free sonicator. 10 μl formaldehyde-crosslinked cell suspension was fragmented by the ultrasonic. Then the fragmented chromatin suspension was concentrated from 10 μl to 1 μl by evaporation.
Since the starting material is very limited and the reaction yield is intrinsically low, even losing a small fraction of the target DNA8 would potentially compromise sequencing efforts. We hence carefully designed the microfluidic channels to sequentially perform the necessary reactions without losing target DNA. We implement the dead-end filling method12 to transfer the chromatin fragments into a ring-shape chamber to react with antibody-coated beads (Figure 1B). Dead-end filling was practical because PDMS was gas permeable; the air in the ring-chambers was expelled and replaced by liquid within a few minutes. Integrated 3-valve peristaltic pumps circulate the liquid in the ring chambers, facilitating the immunoprecipitation. This step is the most challenging practice in traditional ChIP assays. The challenges come from the incomplete crosslinking, nonspecific adsorption, low-efficiency chromatin fragmentation, and low specificity of binding between the antibodies and histone. However, with reduced reaction volume and active mixing, the efficiency of the whole process has been greatly improved. The semi-automated microfluidic control improved the precision and synchronization among all reaction pipelines in the same chip, ensuring the high reproducibility of the assay. We found that the amount of the beads used in an individual assay was also a critical factor. Excess beads elevated the background signals while insufficient beads would decrease the enrichment efficiency.
The chromatin-antibody-bead complex was then flushed out of the chip and collected by microcentrifuge tubes. The complex was incubated with proteinase K at 68 °C for 2 h to release the DNA, which was then purified by phenol-chloroform-isoamyalcohol extraction. Precipitated DNA was re-suspended in 10 μl RNase-free H2O, and analyzed by qPCR using the specific primers (Supplemental methods) and by high-throughput sequencing.
To quantitatively assess the sensitivity and accuracy of this new protocol, we performed ChIP-Seq of H3K4me3 for mEpiSCs and mESCs from both a bulk amount (106) of cells and from 1 000 cells and compared the results. We found that for mEpiSCs, the ChIP-seq of the two 1 000-cell samples recovered 16 351 and 16 245 out of the 16 929 enriched transcription start site (TSS) regions with H3K4me3 peaks from the bulk sample, and for mESCs, the two 1 000-cell samples recovered 18 256 and 18 206 out of the 18 367 enriched TSS regions from the bulk sample, demonstrating the high sensitivity (on average 96.3% for mEpiSCs and 99.3% for mESCs) of our method. Moreover, 98.3% and 98.4% of the 16 636 and 16 516 enriched TSS regions from the 1 000 mEpiSCs overlapped with those from bulk sample, and 91.0% and 91.7% of the 19 917 and 19 733 enriched TSS regions from the 1 000 mESCs overlapped with those from bulk sample (Figure 1C and Supplementary information, Table S1), exhibiting the high accuracy of our method. The mean correlation coefficient is 0.94 when comparing the H3K4me3 profile of 1 000 mEpiSCs and 106 mEpiSCs, and 0.76 of 1 000 mESCs and 106 mESCs (Supplementary information, Figure S1J), proving the general agreement of the profile based on 1 000 cells of our method and that of standard ChIP-Seq. We plot the receiver operating characteristic (ROC) curves using the P-value ranked peaks from the 1 000-cell experiments against the positive TSS from the bulk samples (Figure 1D and Supplementary information, Figure S1G), and then calculated the area under curve (AUC) of mEpiSC and mESC as 0.949 and 0.923, indicating our method a good classifier to distinguish positive TSSs from negative ones. These results clearly prove that our method is able to recover a majority of H3K4me3 peaks from as low as 1 000 mammalian cells with a very low false positive rate.
Next we tested the robustness of our 1 000-cell ChIP-Seq method. Besides mEpiSCs and mESCs, we also performed ChIP-Seq on two biological replicates of the H3K4me3 marker from the 1 000 epiblast cells of E6.5 mouse embryos. We found that most of the TSS regions that have enrichment peaks are overlapped between these duplicates (Figure 1E). The correlation coefficients (r) of these replicates are 0.884 (epiblast cells of E6.5 mouse embryos), 0.971 (mEpiSCs), and 0.973 (mESCs), which are comparable to the previous report that required 10 000 cells7.
Since pluripotent mEpiSC cells were derived from epiblast cells of E6.5 mouse embryos, we asked whether the H3K4me3 landscape of mEpiSCs in vitro was similar to that of E6.5 epiblast in vivo. We found that, in general, the H3K4me3 pattern of mEpiSCs was very similar to that of epiblast from E6.5 embryos. Among the top 10 000 P-value ranked peaks around the TSS regions, 7 000 in mEpiSCs overlapped with those in E6.5 epiblast (Figure 1C). When we compared the mESCs and the E6.5 epiblast cells, the overlapped peak number was 6 680. These proved that the H3K4me3 epigenetic landscape of epiblast cells of E6.5 mouse embryos is more similar to that of mEpiSCs than to that of mESCs as expected.
Since H3K4me3 was an active marker for gene expression, we analyzed the transcriptome of mEpiSCs and E6.5 epiblast cells by RNA-Seq. We found that the gene expression profiles of mEpiSCs and E6.5 epiblast cells were very similar as well (r = 0.940) for transcripts with FPKM ≥ 0.1 in at least one of the samples. We compared, in-depth, the ChIP-Seq result at some important gene loci for early embryonic development and found high similarity among epiblast cells of E6.5 mouse embryos, mEpiSCs, and mESCs (Figure 1F). At the same time, we also found the specific gene loci only enriched in mESCs (Figure 1G). Gene ontology terms showed that, compared with mESCs, both epiblast cells of E6.5 mouse embryos and mEpiSCs enriched for the ectodermal differentiation-related characteristics such as neural tube development and neuronal differentiation (Figure 1H). Furthermore, the RNA expression level of the genes clearly correlated with the enrichment of H3K4me3 around their TSS regions both in mEpiSCs and E6.5 epiblast cells (Figure 1I and Supplementary information, Figure S1M). This correlation verifies the previous assumption that EpiSCs are a reliable in vitro model for post-implantation epiblast cells13.
In summary, we have developed a highly sensitive ChIP-Seq method by combining microfluidic chip-based chromatin immunoprecipitation with one-tube carrier sequencing library preparation. The integrated device is able to finish the major steps of ChIP, including concentration of the cells from tens of microliters to nanoliters, fixation and permeabilization of the cells, fragmentation of chromatins, binding of the target chromatin fragments onto the beads, as well as the elution of enriched chromatin fragments. Subsequently, without any preamplification, the purified DNA fragments were converted into a sequencing library by a one-tube reaction containing end-repair, adenylation, and ligation followed by carrier PCR. We have demonstrated that this microfluidic-assisted ChIP-Seq method works robustly for as low as 1 000 mammalian cells. We have shown that the quality of H3K4me3 profile acquired by our method from 1 000 cells is comparable to that of traditional approach using bulk materials. Moreover, our method is highly reproducible with the correlation coefficient of the two biological replicates of E6.5 epiblast cells, mEpiSCs and mESCs as high as 0.884, 0.971, and 0.973, respectively. Finally we have demonstrated that the H3K4me3 epigenetic landscape of mEpiSCs is very similar to that of epiblast cells from E6.5 mouse embryos, validating that mEpiSC is an appropriate in vitro model to study the epigenetic regulation of post-implantation epiblast cells in vivo. Our method will permit thorough analysis of the epigenomic landscape of early embryos or other situations in which only a very limited amount of materials are available.
Acknowledgments
We thank Prof Azim Surani for kindly giving us the mEpiSC cell line, and thank Tao Chen, Dr Yongfan Men, Zhilong Yu, Zitian Chen, Dr Liang Zhao, Haiwei Qiu, Lu Yang and Dr Yun Zhang for discussion and experimental help. This work was supported by the Ministry of Science and Technology of China (2011CB809106, 2012CB966704) and the National Natural Science Foundation of China (21222501, 21327808, 31271543, 91313302). AMS was supported by the Whitaker International Biomedical Engineering Fellowship.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Supplementary information Figures S1A–S1M, Methods and Material and Table S1
References
- 1Berger SL. Nature 2007; 447:407–412. [DOI] [PubMed]
- 2Shen Y, Yue F, McCleary DF, et al. Nature 2012; 488:116–120. [DOI] [PMC free article] [PubMed]
- 3O'Geen H, Nicolet CM, Blahnik K, et al. Biotechniques 2006; 41:577–580. [DOI] [PMC free article] [PubMed]
- 4Park PJ. Nat Rev Genet 2009; 10:669–680. [DOI] [PMC free article] [PubMed]
- 5Hawkins RD, Hon GC, Ren B. Nat Rev Genet 2010; 11:476–486. [DOI] [PMC free article] [PubMed]
- 6Furey TS. Nat Rev Genet 2012; 13:840–852. [DOI] [PMC free article] [PubMed]
- 7Adli M, Zhu J, Bernstein BE. Nat Methods 2010; 7:615–618. [DOI] [PMC free article] [PubMed]
- 8Wu AR, Hiatt JB, Lu R, et al. Lab Chip 2009; 9:1365–1370. [DOI] [PMC free article] [PubMed]
- 9Shankaranarayanan P, Mendoza-Parra M-A, Walia M, et al. Nat Methods 2011; 8:565–567. [DOI] [PubMed]
- 10Gilfillan GD, Hughes T, Sheng Y, et al. BMC Genomics 2012; 13:645. [DOI] [PMC free article] [PubMed]
- 11Shankaranarayanan P, Mendoza-Parra M-A, van Gool W, et al. Nat Prot 2012; 7:328–339. [DOI] [PubMed]
- 12Marcy Y, Ouverney C, Bik EM, et al. Proc Natl Acad Sci USA 2007; 104:11889–11894. [DOI] [PMC free article] [PubMed]
- 13Tesar PJ, Chenoweth JG, Brook FA, et al. Nature 2007; 448:196–199. [DOI] [PubMed]
- 14Sachs M, Onodera C, Blaschke K, et al. Cell Rep 2013; 3:1777–1784. [DOI] [PMC free article] [PubMed]
- 15Ng JH, Kumar V, Muratani M, et al. Dev Cell 2013; 24:324–333. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information Figures S1A–S1M, Methods and Material and Table S1