Abstract
Ovules are essential for plant reproduction and develop into seeds after fertilization. SPOROCYTELESS/NOZZLE (SPL/NZZ) has been known for more than 15 years as an essential factor for ovule development in Arabidopsis, but the biochemical nature of SPL function has remained unsolved. Here, we demonstrate that SPL functions as an adaptor-like transcriptional repressor. We show that SPL recruits TOPLESS/TOPLESS-RELATED (TPL/TPR) co-repressors to inhibit the CINCINNATA (CIN)-like TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors. We reveal that SPL uses its EAR motif at the C-terminal end to recruit TPL/TPRs and its N-terminal part to bind and inhibit the TCPs. We demonstrate that either disruption of TPL/TPRs or overexpression of TCPs partially phenocopies the defects of megasporogenesis in spl. Moreover, disruption of TCPs causes phenotypes that resemble spl-D gain-of-function mutants. These results define the action mechanism for SPL, which along with TPL/TPRs controls ovule development by repressing the activities of key transcription factors. Our findings suggest that a similar gene repression strategy is employed by both plants and fungi to control sporogenesis.
Keywords: SPOROCYTELESS/NOZZLE, transcriptional repressor, TPL/TPR co-repressors, TCP transcription factors, sporogenesis
Introduction
In flowering plant female reproduction, an ovule consists of the haploid female gametophyte and the diploid integuments and develops into a seed after fertilization. The first step of ovule development is the formation of a protrusion from the internal wall of the carpel. The protrusion then elongates and forms a finger-like nucellus. At the tip of the nucellus, only one hypodermal cell differentiates into an archespore, which further elongates and differentiates into a megasporocyte. A megasporocyte goes through meiosis to produce four haploid megaspores. In Arabidopsis, three of the megaspores near the distal end of the ovule are degenerated and the remaining cell develops into the haploid embryo sac by undergoing three rounds of mitosis. In the mature ovule, the embryo sac contains one egg cell, one central cell, two synergids and three antipodal cells. Extensive genetic studies in Arabidopsis have identified several components that play key roles in ovule development (see reviews1,2,3,4). Among them, SPOROCYTELESS/NOZZLE (SPL/NZZ) is a key regulator responsible for promoting the formation of megasporocyte and integuments during ovule development5,6,7,8.
The spl/nzz mutants were initially identified as sterile mutants5,6, which displayed a complete failure of male and female gametophyte formation. Detailed analysis showed that SPL/NZZ controlled sporogenesis and played pivotal roles in the differentiation of megasporocytes. In the embryo sac of spl/nzz mutants, an archesprorial cell is formed but it fails to differentiate into a megasporocyte. SPL/NZZ also promotes the growth of ovule integuments8, the differentiation of microsporocytes9, the identity of stamen10, and other lateral organ development11. It was reported that SPL/NZZ physically interacted with the YABBY protein INNER NO OUTER (INO) in the regulation of ovule development12. Some important downstream genes of SPL/NZZ have also been identified. During ovule development, SPL/NZZ represses some important ovule developmental genes including BELL1 (BEL1), AINTEGUMENTA (ANT) and INO12, and the auxin biosynthesis gene YUCCA211,13. It has been shown that AGAMOUS activates the expression of SPL/NZZ by directly binding to the CArG box at the 3′-region of the gene during microsporogenesis9. Recently, the plant hormone cytokinin has been reported to promote the expression of SPL/NZZ, and SPL/NZZ in turn activates the auxin transport gene PIN1 during ovule development14. However, the exact action mode of SPL/NZZ is still not understood.
We previously isolated a dominant spl-D mutant from an Arabidopsis activation tagged mutant collection and found that SPL controls lateral organ morphogenesis by regulating auxin homeostasis11. Here, we demonstrate that SPL/NZZ is actually a transcriptional repressor. The C-terminal end of SPL/NZZ contains a typical EAR motif (ERF-associated amphiphilic repression), which is a well-characterized repression domain with the consensus sequence LXLXL15,16. We show that SPL/NZZ uses its EAR motif to recruit the known transcriptional co-repressor TOPLESS/TOPLESS-RELATED (TPL/TPRs). We demonstrate that TPL and SPL have overlapping expression patterns and that the homozygous mutants tpl-1 and spl displayed similar defects in ovule development. Furthermore, we discover that SPL/NZZ, through its N-terminal part without the EAR motif, directly interacts with the CINCINNATA (CIN)-like TCP transcription factors. Disruption of CIN-like TCPs leads to distorted ovule arrangement in ovaries similar to those observed in the gain-of-function mutant spl-D. Overexpression of the CIN-like TCPs resulted in aborted ovules similar to those observed in the spl loss-of-function mutant. Our results indicate that SPL serves as an adaptor-like transcriptional repressor to control ovule development by recruiting TPL/TPR co-repressors to suppress the activities of CIN-like TCP transcription factors in Arabidopsis. Interestingly, both plants and fungi adopt a similar gene repression strategy to control sporogenesis, suggesting that this strategy may be an evolutionarily conserved mechanism.
Results
SPL/NZZ is an EAR motif-containing transcriptional repressor
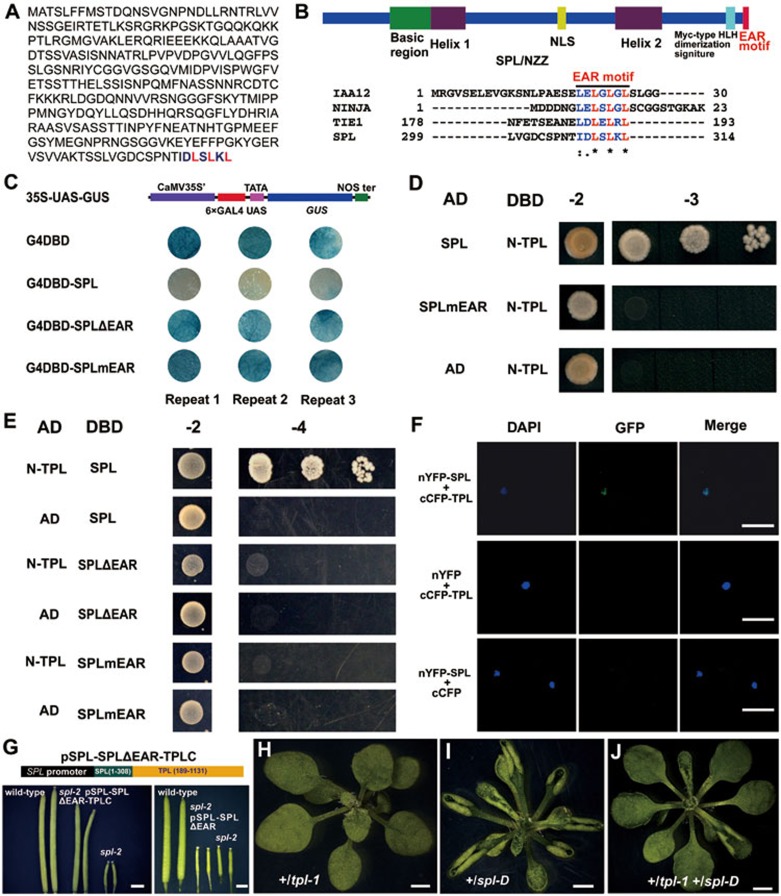
SPL/NZZ was reported as a nuclear protein with several structural features including a basic region in the N-terminus and two putative helixes5. We re-analyzed the protein sequence of SPL and found that the last five amino acid residues at the C-terminal end, i.e., LSLKL, resembled the EAR motif (LXLXL; Figure 1A). Because many known repressors including IAA12, NINJIA, and TIE1 all contain the EAR motif17,18,19 (Figure 1B), SPL may also have transcriptional repression activity. To test this hypothesis, we co-transformed GAL4 DNA binding domain (G4DBD) or G4DBD-SPL with a reporter construct 35S-UAS-GUS19 into tobacco leaves. The results showed that GUS activities were highly repressed in the combination of 35S-UAS-GUS and G4DBD-SPL, suggesting that SPL has transcriptional repression activities (Figure 1C). To further determine whether the EAR motif in SPL is required for the repression activity, we generated two other fusion constructs, G4DBD-SPLΔEAR in which the EAR motif was deleted and G4DBD-SPLmEAR in which the conserved leucine residues in the EAR motif were mutated to alanine residues. We found that the GUS activities were not repressed in the tobacco leaves in the combinations of the reporter and either G4DBD-SPLΔEAR or G4DBD-SPLmEAR (Figure 1C), indicating that the EAR motif is required for the repression activity of SPL.
Figure 1.
SPL/NZZ is a transcriptional repressor interacting with the TPL co-repressor. (A, B) SPL contains a typical EAR motif. The full-length protein sequence of SPL. The EAR motif at the C-terminal end is indicated by color letters (A). The domain structures of SPL are represented by different color blocks. The EAR motif is shared by other known repressors (B). (C) SPL shows transcriptional repression activity dependent on the C-terminal EAR motif. The transcription activity of SPL was investigated in tobacco leaves. The small circles were tobacco leaf discs co-transformed with different construct combinations. Every combination was repeated for three times. (D, E) Yeast two-hybrid assays indicated that SPL interacted with TPL protein by the EAR motif. Transformed yeast cells were spotted on control medium (2), or selective medium SD-Leu-Trp-His (−3), or SD-Ade-Leu-Trp-His (−4) in 10-fold, 100-fold and 1 000-fold dilutions. The empty vectors were used as the controls. (F) BiFC analysis verified the interaction of SPL and TPL. From left to right, DAPI staining of the nucleus, GFP fluorescence, and merge of DAPI and GFP. Scale bar, 30 μm. (G) pSPL-SPLΔEAR-TPLC fusion protein complemented the male and female sterility of spl-2. Top, schematic representation of pSPL-SPLΔEAR-TPLC. Bottom left, the siliques from wild-type, spl-2 transformed with pSPL-SPLΔEAR-TPLC and spl-2. Bottom right, pSPL-SPLΔEAR could not complement the male and female sterility of spl-2. (H-J) The genetic interaction of spl-D and tpl-1. The up-curled leaf phenotype of heterozygous spl-D was released in the background of tpl-1 mutant. 21-day-old heterozygous tpl-1 mutant (G). 21-day-old heterozygous spl-D mutant (H). 21-day-old heterozygous tpl-1 spl-D double mutant (I). Scale bar, 1 mm.
SPL/NZZ interacts with TPL/TPR co-repressors
EAR motif-containing repressors often interact with TPL/TPR co-repressors17,18,19. To test whether SPL could interact with TPL/TPRs, we fused GAL4 activation domain (AD) to SPL or SPLmEAR, and DNA binding domain (DBD) to the N-terminal region of TPL (N-TPL), a region sufficient for interaction of TPL with EAR motif-containing proteins17,20, to conduct the yeast two-hybrid (YTH) assays. The results showed that SPL interacted with TPL, whereas SPLmEAR did not (Figure 1D). We further fused DBD to either SPL, SPLmEAR, or SPLΔEAR, and AD to the N-TPL, and the YTH results confirmed that SPL interacted with TPL and that the EAR motif was required for the interaction (Figure 1E). Bimolecular fluorescence complementation (BiFC) assays also confirmed the interaction between SPL and TPL in vivo (Figure 1F). Furthermore, SPL also interacted with the N-terminus of the other four TPRs (Supplementary information, Figure S1).
To further test the hypothesis that SPL regulates plant development by recruiting TPL through the EAR motif, we first identified a 4.4-kb-long fragment of the SPL promoter, pSPL, which reproduced the expression pattern of SPL (Supplementary information, Figure S2A and S2B)9. We used the SPL promoter to drive SPLΔEAR-TPLC in which SPLΔEAR fused with the C-terminus of TPL (TPLC) to generate pSPL-SPLΔEAR-TPLC (Figure 1G). We then identified SAIL_519_H07 T-DNA insertion mutant as spl-2. The spl-2 contained a T-DNA insertion located 206 bp upstream to the start codon ATG of SPL (Supplementary information, Figure S2C) and displayed male and female sterility (Supplementary information, Figure S2D and S2E, Figure 1G). We transformed pSPL-SPLΔEAR-TPLC or the control pSPL-SPLΔEAR into spl-2. It was clear that pSPL-SPLΔEAR-TPLC complemented the male and female sterility of spl-2 (Figure 1G), whereas pSPL-SPLΔEAR did not (Figure 1G), suggesting that the SPLΔEAR-TPLC can functionally substitute SPL. To further demonstrate the essential roles of EAR motif for SPL function, we generated 35S-SPLmEAR plants in which SPLmEAR was driven by a CaMV 35S promoter (Supplementary information, Figure S2F). We hypothesized that SPLmEAR could compete with endogenous SPL, thus causing a dominant-negative effect. Indeed, overexpression of SPLmEAR led to male and female sterility, resembling spl-2 (Supplementary information, Figure S2D-S2H; Figure 1G), indicating that the EAR motif was crucial for SPL functions. We also observed epinastic leaves in the 35S-SPLmEAR plants, a phenotype opposite to the hyponastic leaves in spl-D mutants, suggesting that SPLmEAR may also compete with and disrupt the functions of some other unknown proteins redundant to SPL in leaves (Supplementary information, Figure S2I-S2L). When we crossed spl-D with a dominant-negative mutant tpl-117, we found that the hyponastic leaf phenotype of +/spl-D was partially rescued in the +/spl-D +/tpl-1, suggesting that TPL is required for the SPL function11 (Figure 1H-1J). These results demonstrate that SPL represses transcription by interacting with TPL/TPRs through the EAR motif.
TPL is expressed during ovule development
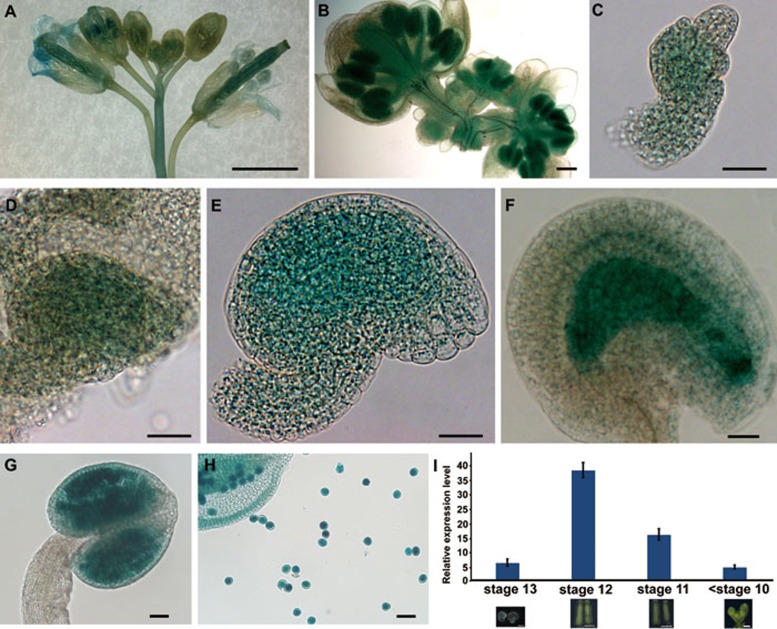
To test whether TPL participates in ovule development, we first analyzed the expression pattern of TPL in the ovules using the TPLP-GUS plants19. TPL was expressed ubiquitously in the pistils, stamens and pollens (Figure 2A, 2B, 2G, and 2H), and expressed dynamically during ovule development. GUS signal was first relatively strong in the proximal region and weak in the nucellus in the young ovules (Figure 2C). Then GUS signal became stronger and more ubiquitously distributed in both proximal region and nucellus (Figure 2D and 2E). In the mature ovules, GUS staining was mainly observed in the nucellus (Figure 2F). This expression pattern of TPL is consistent with that revealed by in situ hybridizations21. Quantitative real-time PCR (qRT-PCR) analysis confirmed that the expression of SPL and TPL were overlapped (Figure 2I). These results suggest that TPL may be involved in ovule development.
Figure 2.
The expression pattern of TPL. (A, B) GUS staining of inflorescences. (C-E) TPL was expressed in young ovules. (F) TPL was expressed strongly in the nucellus of a mature ovule. (G, H) TPL was highly expressed in an anther and pollens. (I) The overlapping expression of TPL and SPL revealed by qRT-PCR. The relative expression levels of TPL in the mature ovules from flower stage 13, the pistils from flower stage 12 or flower stage 11, and the flower buds before stage 10. The expression level of SPL was set to 1.0 in the different corresponding tissues. The error bars represent the SD of three biological replicates. Scale bars, 1 mm (A), 100 μm (B), 20 μm (C-F), and 50 μm (G, H).
TPL/TPR co-repressors are required for ovule development
To reveal the possible roles of TPL/TPR co-repressors during ovule development, we quantitatively and statistically analyze the phenotypes of tpl-1, a dominant-negative mutant of TPL/TPRs22. We grew and identified 1 638 homozygous tpl-1 and found 1 210 plants with cup-shaped cotyledons, 372 with one cotyledon, 5 with no cotyledon, and 51 plants with two cotyledons which were similar to wild-type (WT) plants. Only 184 plants grew to adult stage and flowered. We dissected the siliques of 116 tpl-1 individual plants and found variable aborted ovules (Figure 3I and 3J). We took one silique from each of 61 tpl-1 plants and 10 WT plants, and observed the ovules. In WT siliques only 3.1% of the ovules (17/550) were aborted, whereas 43.8% (550/1 254) of ovules in tpl-1 siliques were aborted, suggesting that TPL/TPRs are required for ovule development.
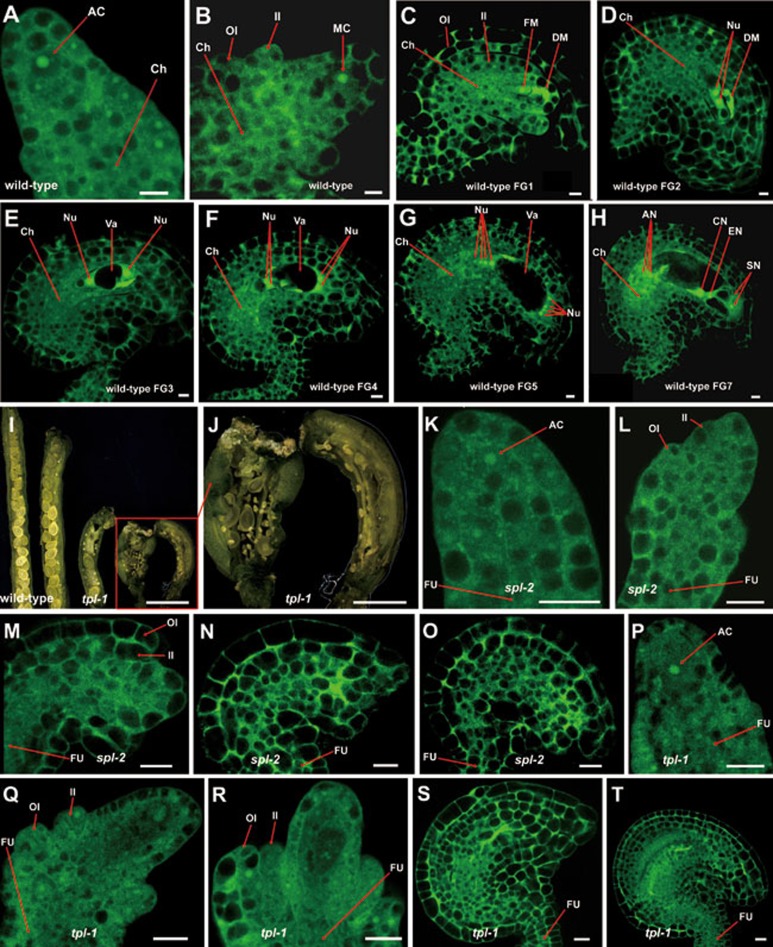
Figure 3.
The developmental ovules from wild type, spl-2 and tpl-1 observed by confocal laser scanning microscopy (CLSM). (A-H) The developmental ovules from wild type observed by CLSM. An archesporial cell was formed in the primordium (A). A megasporocyte could be found (B). Functional and degenerating megaspores were observed (C). Vacuole formation, nuclear division, nuclear migration and fusion, and cellularization could be found (D-H). Scale bar, 5 μm. (I, J) The dissected siliques from wild-type and four tpl-1 individual plants. Aborted ovules were prevalent in tpl-1 siliques. From left to right, silique from wild-type plant, four siliques from tpl-1 plants (I). The close-up view of aborted ovules in tpl-1 siliques (J). Scale bars, 1 mm (I) and 500 μm (J). (K-O) The ovules from spl-2 mutant. An archesporial cell was formed in the ovule of spl-2 (K). No megasporocyte could be found (L). No events, such as vacuole formation, nuclear division, and cellularization could be found (M-O). (P-T) The ovules from tpl-1 plants. An archesporial cell was also observed (P). No megasporocytes or functional megaspores could be found in the ovules (Q, R). And no nuclei, vacuoles, egg cell, central cell or synergid cells could be found in the ovules (S, T). Scale bar, 10 μm (K-T). AC, Archesporial cell; MC, Megasporocyte; OI, Outer integument; II, Inner integument; FM, Functional megaspore; DM, Degenerating megaspore; Nu, Nucleus; Va, Vacuole; AN, Antipodal cell nucleus; CN, Central cell nucleus; EN, Egg cell nucleus; SN, Synergid cell nucleus; FU, Funiculus.
We further adopted confocal laser scanning microscopy (CLSM) to analyze the ovules of spl-2 and tpl-1 mutants. In the WT ovules, an archesporial cell was formed in the primordium (Figure 3A) and then differentiated into a megasporocyte (Figure 3B). The megasporocyte underwent meiosis, and one of the meiotic products developed into a functional megaspore (Figure 3C). Then the megaspore produced a two-nucleated embryo sac (Figure 3D and 3E). After a series of events, i.e., vacuole formation, two other rounds of nuclear division, nuclear migration and fusion, and cellularization (Figure 3E, 3F, 3G, and 3H), a seven-celled embryo sac containing one egg cell, one central cell, two synergid cells and three antipodal cells was formed (Figure 3H). However, in the spl-2 and tpl-1 mutants showing low fertility, although the archesporial cells were formed in nearly all the observed ovules (Figure 3K and 3P), no megasporocytes were found in 41.8% (69/165) of tpl-1 ovules, a phenotype similar to that of spl-2 (Figure 3L and 3Q). In the ovules of spl-2 and tpl-1 without megasporocytes, vacuole formation, nuclear division and cellularization were not observed (Figure 3M, 3N, 3R, and 3S). Both spl-2 and tpl-1 produced ovules without egg cells, central cells, synergid cells and antipodal cells (Figure 3O and 3T). The development of tpl-1 ovules with megasporocytes was frequently arrested at different stages, indicating that TPL/TPRs may also be essential for megagametogenesis (Supplementary information, Figure S3A-S3F). The tpl-1 mutants also produced defective pollens (Supplementary information, Figure S3G and S3H). The observation that spl-2 and tpl-1 displayed similar ovule defects further supports our hypothesis that both SPL and TPL are involved in ovule development.
SPL interacted with CIN-like TCP transcription factors
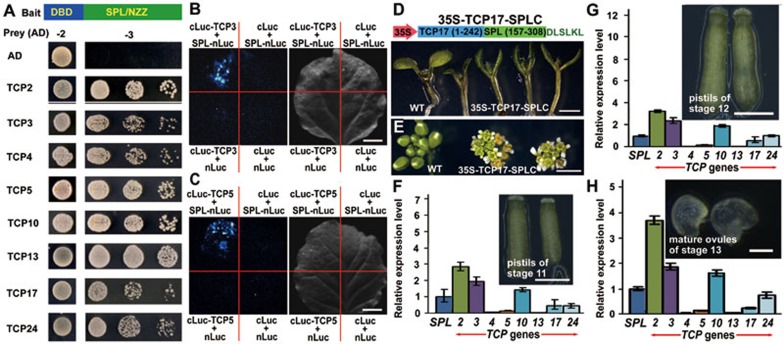
We conducted a yeast two-hybrid screen using SPL as a bait to identify proteins that interact with SPL23. Several classes of transcription factors were identified from the screen, including the CIN-like TCPs (Supplementary information, Figure S4A) and the known SPL-interacting protein INO12. We further verified that all eight CIN-like TCPs interact with SPL in yeast cells (Figure 4A). Moreover, we demonstrated that CIN-like TCPs interacted with the N-terminal part of SPL without the EAR motif (Supplementary information, Figure S4B). Firefly luciferase complementation imaging assays also confirmed that SPL interacted with TCP2, TCP3, TCP5, TCP10, TCP17, or TCP24 in vivo (Figure 4B and 4C; Supplementary information, Figure S4C-S4F). The fact that SPL interacts with TPL/TPR co-repressors with its C-terminal EAR motif and interacts with TCPs with its N-terminal part raises the possibility that SPL serves as a bridge to link TPL/TPRs to TCPs. To test the hypothesis, we first conducted qRT-PCR analysis and found that the TCP direct target genes were downregulated in the SPL gain-of-function mutant spl-D and in jaw-5D mutant in which the microRNA miR319 was overexpressed to inhibit five CIN-like TCP genes19,24, suggesting that overproduction of SPL could repress TCP activities (Supplementary information, Figure S5A)11,25,26,27. The repression of the TCP target genes was largely relieved by the disruption of TPL/TPRs (Supplementary information, Figure S5A). We then used a CaMV 35S promoter to drive TCP17 which was fused with the C-terminal half of SPL (Figure 4D). All of the 10 independent 35S-TCP17-SPLC transgenic plants, but not the controls, displayed up-curled cotyledons and produced unfurled flower buds that were also observed in SPL overexpression plants (Figure 4D and 4E; Supplementary information, Figure S5B, S5C and S5D)10,11, confirming the association between SPL and TCPs. We then crossed spl-D with jaw-5D. The jaw-5D +/spl-D double mutants produced more highly serrated and more deeply lobed leaves than jaw-5D or +/spl-D (Supplementary information, Figure S5E-S5G), suggesting that spl-D synergistically interacted with jaw-5D. These results suggest that SPL connects CIN-like TCP transcription factors with TPL/TPR co-repressors.
Figure 4.
SPL interacts with CIN-like TCPs and most of them are expressed during ovule development. (A) SPL interacted with CIN-like TCP transcription factors in yeast cells. Transformed yeast cells were spotted on control medium (−2) or selective medium (−3) in 10-fold, 100-fold and 1 000-fold dilutions. The empty vector was used as the control. (B, C) SPL was confirmed to interact with TCP3 (B) or TCP5 (C) in vivo by firefly luciferase complementation imaging assays. (D, E) Overexpression of TCP17-SPLC fusion protein led to the up-curled cotyledons (D) and unfurled flower buds (E) similar to those of SPL overexpression lines. Top of D, schematic representation of 35S-TCP17-SPLC construct. (F-H) The relative expression levels of CIN-like TCPs in the pistils from stage 11 (F) and stage 12 (G), and the mature ovules from stage 13 (H). The expression level of SPL was set to 1.0. The error bars represent the SD of three biological replicates. Scale bars, 1 cm (B, C), 1 mm (D, E), 200 μm (F, G), 50 μm (H).
Most of CIN-like TCPs are expressed during ovule development
To test whether CIN-like TCPs play roles in ovule development, we first analyzed the expression of CIN-like TCP genes during ovule development. Ovules start initiation at flower stage 8. The megasporocytes are differentiated at stage 11 and the megagametogenesis begins at stage 12. Ovules are mature at stage 13 (Supplementary information, Figure S6A)28. We tested the expression of SPL in the flowers of different stages, i.e., flower buds before stage 10, the flowers of stage 11, stage 12 and stage 13 (Supplementary information, Figure S6B). The dynamic expression of SPL was consistent with the results of SPL-GUS staining9. We then tested the expression of the CIN-like TCPs and found that the CIN-like TCP genes were all expressed in these flowers (Supplementary information, Figure S6C-S6F). We next isolated the pistils from flowers at stage 11, stage 12, and the mature ovules from flower stage 13 for more precise analysis. qRT-PCR analysis showed that six TCP genes were expressed in these tissues, with high expression from TCP2, TCP3 and TCP10, and low expression from TCP5, TCP17 and TCP24 (Figure 4F-4H), suggesting that CIN-like TCPs may participate in ovule development.
The activities of CIN-like TCPs are essential for ovule development
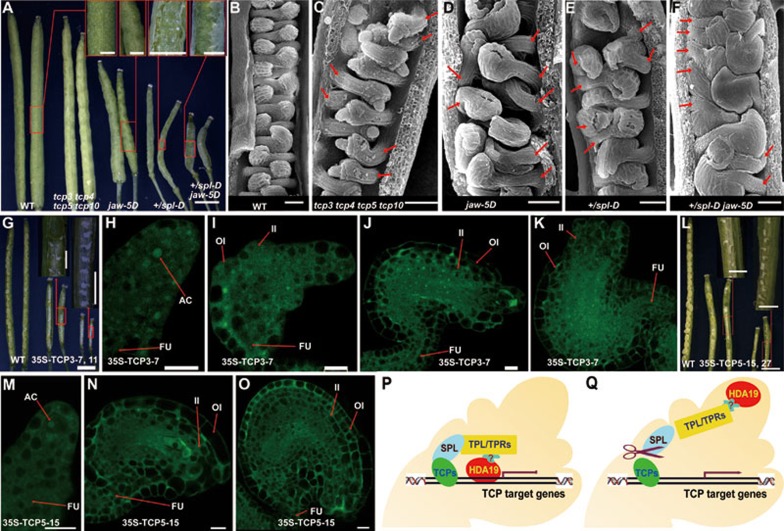
To investigate the possible roles of CIN-like TCPs during ovule development, we examined the ovule development of the four mutants, i.e., tcp3 tcp4 tcp5 tcp10 quadruple mutant, jaw-5D in which TCP2, TCP3, TCP4, TCP10 and TCP24 were downregulated, spl-D, and jaw-5D +/spl-D double mutant. Interestingly, all these four mutants produced seeds that were protruded from siliques and the double mutant jaw-5D +/spl-D displayed enhanced phenotypes (Figure 5A). Scanning electron microscope analysis showed that the arrangement of ovules, which are normally interdigitated in WT ovaries (Figure 5B), were frequently disrupted in tcp3 tcp4 tcp5 tcp10, jaw-5D and +/spl-D mutants: two ovules arose consecutively from the same side of the locule (Figure 5C-5E). In jaw-5D +/spl-D, more ovules were initiated consecutively and crowded at one side of the locule (Figure 5F), indicating that disruption of the CIN-like TCPs caused the abnormal ovule initiation in the ovaries.
Figure 5.
The CIN-like TCPs are implicated in ovule development. (A) The seeds are protruded from siliques from the CIN-like TCP disruption mutants and spl-D. Scale bars, 1 mm and 250 μm in the insets. (B-F) The interdigitated array of ovules was disrupted in the CIN-like TCP disruption mutants and spl-D. The interdigitated array of ovules in young pistil of wild type (B). The abnormal arrays of ovules in tcp3 tcp4 tcp5 tcp10 (C), jaw-5D (D), spl-D (E) and jaw-5D +/spl-D(F). The red arrows indicate the consecutive ovules. Scale bar, 50 μm. (G-K) The TCP3overexpression lines produced aborted ovules. The dissected siliques from wild-type and two 35S-TCP3 plants (G). The ovule of 35S-TCP3 plant produced an archesporial cell (H). No megasporocyte was formed in the ovule (I). No egg cells, central cells, synergid cells and antipodal cells were formed in the ovules (J, K). The integuments of the ovules displayed retarded growth. Scale bars, 1 mm (G) and 250 μm in insets of G, and 10 μm (H-K). (L-O) Overexpression of TCP5 caused aborted ovules similar to those in spl mutants. The dissected siliques showed high ratios of aborted ovules (L). The archesporial cell in the tip of the primordium contained a big nucleus (M). No megasporocytes or megaspores were observed in the ovule (N). The nucellus contained no egg cells, central cells, synergid cells and antipodal cells (O). Scale bars, 1 mm (L) and 250 μm in the insets of L and 10 μm (M-O). AC, Archesporial cell; FU, Funiculus; OI, Outer integument; II, Inner integument. (P, Q) The working model of SPL. (P) In the context of SPL expression, the N-terminus of SPL interacts with the CIN-like TCPs, while the EAR motif at the C-terminus interacts with TPL/TPR co-repressors. The activities of TCPs are modified by the connection with TPL/TPRs. The TCP downstream genes are possibly regulated so that the differentiation of megasporocytes is promoted. (Q) When SPL is inactivated, TPL/TPRs are dissociated from CIN-like TCPs. The excessive activities of TCPs alter the expression of TCP downstream genes and cause abnormal ovule development.
We further used a CaMV 35S promoter to overexpress TCP3 and TCP5, two representative members from the two main clades of CIN-like TCPs (Supplementary information, Figure S4A). All of the six independent 35S-TCP3 lines and nine of the 35S-TCP5 lines produced aborted ovules (Figure 5G and 5L). CLSM analysis showed that archesporial cells with a big and obvious nucleus were formed in the primordia (Figure 5H and 5M). However, in 46.7% (28/60) ovules from 35S-TCP3 lines and 42.2% (65/154) ovules from 35S-TCP5 lines, no megasporocytes or megaspores were differentiated (Figure 5I and 5N). In the ovules without megasporocytes, no egg cells, central cells, synergid cells and antipodal cells were formed (Figure 5J, 5K and 5O), resembling the spl ovules. The integuments of ovules from 35S-TCP3 plants also exhibited retarded growth (Figure 5J and 5K), a phenotype also observed in spl/nzz mutants6. Our results suggest that the activities of CIN-like TCPs are essential for normal ovule development and that SPL is an important negative regulator that modulates the activities of TCPs.
Taken together, our results indicate that SPL recruits TPL/TPR co-repressors to suppress the activities of CIN-like TCPs and possibly controls the expression of the TCP downstream genes27, thus promoting cell differentiation of megasporocytes during ovule development (Figure 5P). When SPL is inactivated, TPL/TPR co-repressors are disassociated from CIN-like TCPs. The excessive activities of TCPs alter the expression of TCP downstream genes and cause the failure of megasporogenesis and abnormal ovule development (Figure 5Q).
Discussion
In this study, we demonstrate that SPL/NZZ controls ovule development by recruiting the TPL/TPR transcription co-repressors to suppress CIN-like TCP transcription factors. First, we show that the five amino acid residues at the C-terminal end of SPL is a typical EAR motif that is required for transcriptional repression activity. Second, SPL physically interacts with TPL/TPR co-repressors through the EAR motif. Third, disruption of TPL/TPRs phenocopies the ovule defects observed in spl mutants5,6. Fourth, we show that six CIN-like TCPs are expressed in ovules and that they interact with SPL. Fifth, disruption of CIN-like TCPs leads to abnormal ovule arrangement, a phenotype also observed in spl-D gain-of-function mutants. Finally, either overexpression of the CIN-like TCPs or inactivation of SPL results in aborted ovules. These findings reveal the SPL action mechanism: SPL functions as an adaptor-like transcriptional repressor linking TPL/TPR co-repressors to the key transcription factors in the megasporogenesis during ovule development.
SPL is also known to regulate pollen development in Arabidopsis5,9. It is not clear whether SPL regulates pollen development by a similar mechanism, i.e., by recruiting TPL/TPR co-repressors to certain transcription factors. However, we did find that TPL was highly expressed in stamens and pollens, and that tpl-1 mutants also produced defective pollens. The fact that pSPL-SPLΔEAR-TPLC complemented not only the female sterility but also the male sterility of spl-2 strongly suggests that SPL-TPL/TPRs may also function in pollen development. Recently, TPL/TPR co-repressors were reported to directly interact with EAR motif-containing transcription repressors DAZ1 and DAZ2 in microgametogenesis and male germ cell division29, supporting that TPL/TPRs-mediated gene repression plays pivotal roles not only in ovule development but also in pollen development.
The action mechanism of SPL discovered in this study is analogous to those employed in auxin signaling17, jasmonic acid (JA) signaling18, circadian clock regulation30, meristem maintenance31,32, floral organ development21 and leaf development19. The common feature of these pathways is that the TPL/TPR co-repressors are recruited, through an adaptor-like repressor or not, to repress the activities of certain transcription factors. For example, TPL/TPR co-repressors are directly recruited to transcription factors that contain the EAR motifs, i.e., PRRs, WUSCHEL (WUS), RAMOSA1 (RA1) and APETALA2 (AP2), to regulate circadian clock, meristem maintenance, and floral organ development, respectively21,30,32,33. In auxin signaling, JA signaling and leaf development, however, the adaptor-like repressors (i.e., AUXs/IAAs, NINJA, and TIEs, respectively), through their EAR motifs, recruit TPL/TPR co-repressors to repress AUXIN RESPONSIVE FACTORs, MYCs, and TCP transcription factors, respectively17,18,19. In this study, we found that, in ovule development, the adaptor-like repressor SPL recruited TPL/TPR co-repressors through its C-terminal EAR motif and interacted with CIN-like TCP transcription factors through its N-terminal part without the EAR motif, suggesting that similar mechanism has been adopted to regulate ovule development as in auxin and JA signaling, and leaf development regulation. The ovules are the main organs for plant reproduction, whereas leaves are the main organs for vegetative growth. Interestingly, both SPL and TIEs recruit TPL/TPR co-repressors and regulate CIN-like TCP activities in ovules and leaves. This supports the hypothesis that similar regulatory mechanisms are adopted in ovules and leaves during plant evolution19,34. However, unlike TIEs-TPL/TPR complexes, which delay leaf cell differentiation19, SPL-TPL/TPR complexes actually promote the differentiation of the archesporial cell into the megasporocytes and promote the growth of ovule integuments.
An ovule can be distinguished as the proximal funiculus which is the stalk linking an ovule to the ovary wall, the central integuments and the distal nucellus7. SPL participates in the formation of all the three parts7,8,12. SPL controls the cell number of the proximal funiculus by repressing the expression of ANT and INO7. In the central region, SPL and BEL1 coordinate to determine the identity of ovule chalaza where the integuments and nucellus are joined, since the chalaza of nzz bel1 double mutant was partially substituted by the tissue of the funiculus7. SPL also interacts with INO and represses the expression of INO and ANT to control outer integument development and thus the formation of ovule adaxial-abaxial axis8,12. In the distal region, SPL represses the expression of ANT, BEL1, and INO in the formation of the nucellus and/or the megasporocyte7. These findings suggest that SPL acts as a repressor in these ovule developmental processes; but how SPL suppress these genes at the molecular level has long been unclear. Based on the working model of SPL from this study, it is possible that SPL connects CIN-like TCPs and/or other transcriptional factors with the TPL/TPR co-repressors and thus represses the expression of ANT, BEL1, and INO. However, it seems that the roles of SPL are rather complicated during ovule development. In addition to repressing the transcription of ANT, BEL1, and INO, SPL also promotes the expression of PIN1 and WUS in the ovule35. The possible explanation might be that SPL represses the activities of some unknown suppressors and releases the transcription of PIN1 and/or WUS. The finding that SPL interacts with TPL/TPRs and TCPs during ovule development adds a new layer of the complexity of the SPL-mediated regulation network. Whether ANT, BELL1, and INO are direct target genes of CIN-like TCPs and whether TCPs and INO bind to similar regions of SPL need to be investigated in the future.
TCP transcription factors are grouped into two classes, Class I TCPs and Class II TCPs, based on the sequence similarities of the TCP domains36. CINCINNATA (CIN)-like TCPs belong to the Class II TCPs and promote leaf cell differentiation36. CIN-like TCPs have long been reported to play pivotal roles in leaf development, because disruption of CIN-like TCPs causes excessive growth at the leaf margins in Antirrhinum and Arabidopsis24,37. In addition to the CIN-like TCP genes, most of the Class I TCP genes are also expressed in leaves26,38,39, but Class I TCPs are functionally redundant, because even the pentuple mutants of the Class I TCP genes (i.e., tcp8 tcp15 tcp21 tcp22 tcp23) have almost no obvious phenotypes38. Because the Class I TCPs and Class II TCPs bind to different optimal cis-regulatory elements but with overlapping consensus sequences, it is possible that two classes of TCPs compete with the same binding sites36. It is clear that CIN-like TCPs play essential roles in ovule development, but whether Class I TCPs are also involved in ovule development still needs verification.
SPL controls multiple layers of plant development by affecting cell fate determination, cell differentiation, and organ identity5,6,7,8,9,10,11. This work not only demonstrates the essential roles of CIN-like TCP transcription factors and TPL/TPR co-repressors in ovule development, but also clarifies the mechanism of how these two groups of proteins are involved in the SPL-mediated regulation of ovule development. The temporal and spatial TCP activities are essential for organ development26,40. The recruitment of TPL/TPR co-repressors by SPL to the TCPs revealed an accurate and flexible mechanism in control of the CIN-like TCP activities in the megasporogenesis during ovule development. Interestingly, disruption of Tup1, the TPL ortholog in S. cerevisiae, or disruption of TPL orthologs in fungal pathogens including Penicillium marneffei, Candida albicans, Neurospora crassa and Ustilago maydis, led to defective spore production41,42,43,44,45,46, suggesting that Tup1 co-repressors are essential for sporogenesis in fungi, a process similar to plant sporogenesis. Notably, Tup1 needs to be recruited to specific transcription factors by adaptor protein Suppressor of snf1 6 (Ssn6) and the homozygous ssn6 diploid S. cerevisiae fails to sporulate47. Our findings suggest that TPL/Tup1 co-repressor-involved mechanism probably represents an evolutionarily conserved strategy to control sporogenesis by both plants and fungi.
Materials and Methods
Plant materials and growth conditions
The Arabidopsis thaliana Columbia-0 (Col-0) ecotype and Landsberg erect (Ler) ecotype for topless-1 (tpl-1) were used. The spl-D was a gain-of-function mutant isolated from our T-DNA insertion mutant collection11. The spl-2 (SAIL_519_H07) was obtained from Syngenta Arabidopsis Insertion Library (SAIL) collection. The seeds of the dominant-negative allele tpl-1 were provided by Dr Jeff A Long (University of California, Los Angeles). Dr Tomotsugu Koyama (Kyoto University) kindly provided the seeds of tcp3 tcp4 tcp5 tcp10 multiple knock-out mutants27. The Arabidopsis seeds were grown on half-strength Murashige and Skoog (1/2 MS) medium. The herbicide DL-phosphinothricin (20 μg/ml), kanamycin (50 g/ml) or hygromycin (50 μg/ml) was supplemented in 1/2 MS medium for screening the mutants or transgenic plants. The Arabidopsis seeds were synchronized in 4 °C refrigerator for 3 days and then were grown in the growth chamber with the temperature of 22 ± 2 °C and long day condition (16-h light and 8-h dark). After 7 days, the green seedlings were transferred to soil and grown in the green house under the same conditions as described above. The Nicotiana benthamiana was grown in the same green house for SPL/NZZ transcriptional activity assays, the BiFC analysis and firefly luciferase complementation imaging assays. The primers used in this work are provided in Supplementary information, Table S1.
PCR analysis and gene expression assays
The primers of LB3, spl-F, spl-R were used for genotyping spl-2 mutant. The genotyping analysis of spl-D and jaw-5D was carried out as described previously11,19. PCR were performed using the cycle condition: 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 1 min.
For quantitative RT-PCR, total RNAs of the seedlings, flowers, ovules or pistils from WT, spl-D, tpl-1, tpl-1/spl-D or jaw-5D were extracted using TRIzol reagent (Invitrogen). Total RNAs were reverse-transcribed and quantitative RT-PCR were performed as described previously48,49. Briefly, three biological repeats were executed using a SYBR Green real-time PCR Master Mix (TOYOBO) using ABI 7500 Fast Real-Time PCR System (Applied Biosystems). The 2−ΔΔCT method was used to calculate the relative expression level of each gene50. ACT8 was used as an internal control.
Generation of binary constructs and transformation
To generate pSPL-SPLΔEAR-TPLC, the 4.4-kb-long SPL promoter was amplified from genomic DNA of WT Arabidopsis using the primer pair of pATSP-attB4 and pATSP-attBr1 or that of pSPL-F and pSPL-R. The fragment was cloned into pDONRP4P1R (Invitrogen) to generate pEN-L4-4.4k-R1 by the BP reaction or into pENTR/D-TOPO to get pENTRY-4.4k by TOPO cloning. pSPL-GUS was generated by LR reaction between pENTRY-4.4k and pKGWFS7. SPL-F and SPL-TPLC-EAR-R primer pair was used for amplifying the fragment containing SPLΔEAR with a linker of a short 5′-end of TPLC. TPLC-SPL-EAR-F and TPL-R primer pair was used for amplifying the fragment containing a linker of 3′-end of SPLΔEAR with TPLC. The two fragments were denatured and annealed to be template for amplifying the SPLΔEAR-TPLC fusion with the primers SPL-F and TPL-R. The SPLΔEAR-TPLC fusion fragment was cloned into pENTR/D-TOPO to generate pENTRY-SPLΔEAR-TPLC. pSPL-SPLΔEAR-TPLC was generated by LR reaction with the plasmids of pEN-L4-4.4k-R1, pH7m24GW3 and pENTRY-SPLΔEAR-TPLC. To generate pSPL-SPLΔEAR, SPLΔEAR were amplified from cDNA of spl-D with the primer pairs of SPL-F and SPL-ΔEAR-R. The fragment was cloned into pENTR/D-TOPO to generate pENTRY-SPLEAR. pSPL-SPLΔEAR was generated by LR reaction with the plasmids of pEN-L4-4.4k-R1, pH7m24GW3 and pENTRY-SPLΔEAR.
To generate 35S-SPLmEAR construct, SPLmEAR which encodes the SPL protein containing a mutated EAR motif was amplified from cDNA of spl-D with the primer pair of SPL-F and SPL-mEAR-R. The DNA fragment was cloned into pENTR/D-TOPO to generate pENTRY-SPLmEAR. 35S-SPLmEAR was generated by LR reaction with the plasmids of pENTRY-SPLmEAR and pK2GW7.
To generate 35S-TCP17-SPLC, the sequence encoding C-terminus of SPL (from residue 157 to 314) was amplified using primers SPLC-F and SPLC-R and cloned into pDONRP2rP3 (Invitrogen) to generate pEN-R2-SPLC-L3. The pENTRY-TCP17N and pEN-L4-35S-R1 were generated as described previously. 35S-TCP17-SPLC was generated by LR reactions using the plasmids of pK7m34GW, pEN-L4-35S-R1, pENTRY-TCP17N and pEN-R2-SPLC-L3. To generate 35S-TCP17 and 35S-SPLC, the coding region of TCP17 or the C-terminus of SPL was amplified using primers TCP17-1 and TCP17-2 or SPLC-1 and SPLC-2. The fragments were cloned into pENTR/D-TOPO to generate pENTRY-TCP17 or pENTRY-SPLC. 35S-TCP17 or 35S-SPLC was generated by LR reaction between the plasmids pK2GW7 and pENTRY-TCP17 or pENTRY-SPLC.
To generate TCP overexpression constructs, the coding region of TCP3 or TCP5 was amplified from genomic DNA from Arabidopsis by primer pairs of TCP3-1 and TCP3-2 or TCP5-1 and TCP5-2. The fragments were cloned into pENTR/D-TOPO to generate pENTRY-TCP3 or pENTRY-TCP5. 35S-TCP3 or 35S-TCP5 was generated by LR reaction between the plasmids pK2GW7 and pENTRY-TCP3 or pENTRY-TCP5.
Constructs were transformed into Agrobacterium tumefaciens GV3101/pMP90 and then into Arabidopsis as described previously51.
Staining and microscopy
For GUS staining, flowers from pSPL-GUS or TPLP-GUS transgenic lines were soaked in 90% acetone solution for 20 min on the ice. The tissues were washed twice in the phosphate buffer and then were put into GUS staining buffer containing 0.5 mg/ml 5-bromo-4-chloro-3-indolyl glucuronide. The samples were vacuumed for 10 min and were stained overnight in 37 °C incubator. The staining buffer was then changed to 70% ethanol for clearing chlorophyll. The ovules were dissected and observed using microscope (OLYMPUS SPE microscope). The Alexander's staining of pollens was performed as described previously49. The DAPI (4, 6-diamidino-2-phenylindole) staining and the scanning electron microscopy was performed as described previously19.
The observation of ovules using CLSM was described previously52. Briefly, the inflorescences with open flowers removed was fixed in 4% glutaraldehyde (in 12.5 mM cacodylate, pH 6.9), and vacuumed until the tissues were all sunk in the bottom of the container. The inflorescences were fixed overnight at room temperature and then were gradually dehydrated using 15%, 30%, 50%, 70%, 80%, 90%, 95% and 100% ethanol. The tissues were cleared in the solution (2 volume of benzyl benzoate : 1 volume benzyl alcohol) for at least 2 h. The pistils were dissected and the ovules were observed using a confocal laser scanning microscope (Leica TCS SPE confocal microscope).
Yeast two hybrid assays
To test the interaction of SPL and TPL/TPRs, SPL was amplified from cDNA of spl-D with the primer pairs of SPL-F and SPL-R. The fragment was cloned into pENTR/D-TOPO to generate pENTRY-SPL. The prey constructs of AD-SPL or AD-SPLmEAR were generated by LR reactions between pDEST22 (Invitrogen) and pENTRY-SPL or pENTRY-SPLmEAR. The bait construct of pDEST32-NTPL (DBD-NSPL) was generated as described previously19. To confirm the interaction of SPL and TPL/TPRs, we also generated the constructs of DBD-SPL, DBD-SPLmEAR and DBD-SPLΔEAR using LR reactions between pDEST32 (Invitrogen) and pENTRY-SPL, pENTRY-SPLmEAR or pENTRY-SPLΔEAR. The pENTRY-NTPL and pENTRY-NTPRs were generated as described previously19. The AD-NTPL and AD-NTPRs were generated by LR reactions between pDEST22 and pENTRY-NTPL or pENTRY-NTPRs. The bait and prey plasmids or the blank pDEST22 were co-transformed into AH109 (Clontech) yeast strain.
To investigate the interactions between CIN-like TCPs and SPL or SPLΔEAR, TCP genes were cloned into pDEST22 as preys19. Bait plasmids of DBD-SPL or DBD-SPLEAR and prey plasmids of pDEST22-TCPs or the blank pDEST22 were co-transformed into AH109 yeast cells.
The yeast cells were selected on the medium supplemented with 2.5 mM 3-amino-1,2,4 triazole and SD-Leu-Trp-His or SD-Ade-Leu-Trp-His.
Transient expression, BiFC and firefly luciferase complementation imaging assays
The G4BD-SPL, G4BD-SPLmEAR or G4DBD-SPLΔEAR was amplified from DBD-SPL, DBD-SPLmEAR or DBD-SPLΔEAR using primers of G4BD-1 and SPL-R, SPLmEAR-R or SPLΔEAR-R. The PCR products were cloned into pENTR/D-TOPO to generate pENTRY-G4BD-SPL, pENTRY-G4BD-SPLmEAR or pENTRY-G4BD-SPLΔEAR. The effector constructs were obtained by LR reactions between pK2GW7 and pENTRY-G4BD-SPL, pENTRY-G4BD-SPLmEAR or pENTRY-G4BD-SPLΔEAR. The plasmids of effectors were co-transformed into the leaves of N. benthamiana with the reporter 35S-UAS-GUS and pCam-P19 using the method mediated by Agrobacterium GV3101/pMP90. GUS staining was described previously19.
To test the interaction between SPL and TPL, the construct cCFP-TPL was generated by LR reaction between pcCFPxGW and pENTRY-TPL19. LR reaction was performed to generate nYFP-SPL using the plasmids of pnYFPxGW and pENTRY-SPL. BiFC analysis was performed as described previously with slight modifications. These plasmids were transformed into Agrobacterium GV3101/pMP90. The nYFP-SPL was co-transformed with pCam-P19 and cCFP-TPL into the leaves of N. benthamiana. The empty vectors of pcCFPxGW and pnYFPxGW were co-transformed with nYFP-SPL and cCFP-TPL as controls. The plants were incubated in dark for 24 h and then in light for 72 h. The leaves were observed using the microscope (Leica TCS SPE confocal microscope).
To test the interactions of SPL and TCPs in vivo, SPL was amplified from pENTRY-SPL using SPL-KPN1-F and SPL-SAL1-R. The PCR fragment was digested by Kpn I and Sal I and cloned into the vector pCAMBIA-nLuc to generate SPL-nLuc53. The TCP genes were amplified using corresponding primer pairs and cloned into pCAMBIA-cLuc to generate cLuc-TCPx53. These plasmids were transformed into Agrobacterium GV3101/pMP90. Firefly luciferase complementation imaging assays were carried out as described previously53. Briefly, SPL-nLuc and Luc-TCPx, and the control combinations were co-transformed with pCam-P19 into the leaves of N. benthamiana, respectively. Every combination was repeated more than three times in different leaves. The plants were incubated in dark for 24 h and then in light for 48 h. The leaves were sprayed with 1 mM luciferin and observed under a low-light cooled CCD imaging apparatus (lumazone 1300B, Bio-One Scientific Instrument).
Acknowledgments
We thank Dr Yunde Zhao (University of California at San Diego, USA) for valuable suggestions. We thank Dr Jeff Long (University of California, Los Angeles, USA) and Dr Tomotsugu Koyama (Kyoto University, Japan) for kindly providing the seeds of tpl-1 and tcp3 tcp4 tcp5 tcp10. This work was supported by the National Key Basic Research Program of China (973-2012CB944801), the National Natural Science Foundation of China (31270321), and National Transformation Science and Technology Program (2014ZX08009003-003-003). This work was also partially supported by the 111 Project (B06001).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
SPL interacts with other members of TPL family proteins using the C-terminal EAR motif.
Identification of 4.4-kb-long promoter of SPL and spl-2.
The other phenotypes of tpl-1 plants.
SPL interacts with CIN-like TCPs.
SPL was associated with CIN-like TCPs.
The CIN-like TCP genes were all expressed in flowers.
The primer list used in this work.
References
- 1Shi DQ, Yang WC. Ovule development in Arabidopsis: progress and challenge. Curr Opin Plant Biol 2011; 14:74–80. [DOI] [PubMed] [Google Scholar]
- 2Yang WC, Shi DQ, Chen YH. Female gametophyte development in flowering plants. Annu Rev Plant Biol 2010; 61:89–108. [DOI] [PubMed] [Google Scholar]
- 3Sundaresan V, Alandete-Saez M. Pattern formation in miniature: the female gametophyte of flowering plants. Development 2010; 137:179–189. [DOI] [PubMed] [Google Scholar]
- 4Liu J, Qu LJ. Meiotic and mitotic cell cycle mutants involved in gametophyte development in Arabidopsis. Mol Plant 2008; 1:564–574. [DOI] [PubMed] [Google Scholar]
- 5Yang WC, Ye D, Xu J, Sundaresan V. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 1999; 13:2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K. Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acad Sci USA 1999; 96:11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Balasubramanian S, Schneitz K. NOZZLE regulates proximal-distal pattern formation, cell proliferation and early sporogenesis during ovule development in Arabidopsis thaliana. Development 2000; 127:4227–4238. [DOI] [PubMed] [Google Scholar]
- 8Balasubramanian S, Schneitz K. NOZZLE links proximal-distal and adaxial-abaxial pattern formation during ovule development in Arabidopsis thaliana. Development 2002; 129:4291–4300. [DOI] [PubMed] [Google Scholar]
- 9Ito T, Wellmer F, Yu H, et al. The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 2004; 430:356–360. [DOI] [PubMed] [Google Scholar]
- 10Liu X, Huang J, Parameswaran S, et al. The SPOROCYTELESS/NOZZLE gene is involved in controlling stamen identity in Arabidopsis. Plant Physiol 2009; 151:1401–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Li LC, Qin GJ, Tsuge T, et al. SPOROCYTELESS modulates YUCCA expression to regulate the development of lateral organs in Arabidopsis. New Phytol 2008; 179:751–764. [DOI] [PubMed] [Google Scholar]
- 12Sieber P, Petrascheck M, Barberis A, Schneitz K. Organ polarity in Arabidopsis. NOZZLE physically interacts with members of the YABBY family. Plant Physiol 2004; 135:2172–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 2006; 20:1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Bencivenga S, Simonini S, Benkova E, Colombo L. The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. Plant Cell 2012; 24:2886–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 2001; 13:1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Hiratsu K, Mitsuda N, Matsui K, Ohme-Takagi M. Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem Biophys Res Commun 2004; 321:172–178. [DOI] [PubMed] [Google Scholar]
- 17Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 2008; 319:1384–1386. [DOI] [PubMed] [Google Scholar]
- 18Pauwels L, Barbero GF, Geerinck J, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010; 464:788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Tao Q, Guo D, Wei B, et al. The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 2013; 25:421–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Causier B, Ashworth M, Guo W, Davies B. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol 2012; 158:423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Krogan NT, Hogan K, Long JA. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 2012; 139:4180–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Long JA, Woody S, Poethig S, Meyerowitz EM, Barton MK. Transformation of shoots into roots in Arabidopsis embryos mutant at the TOPLESS locus. Development 2002; 129:2797–2806. [DOI] [PubMed] [Google Scholar]
- 23Ou B, Yin KQ, Liu SN, et al. A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol Plant 2011; 4:546–555. [DOI] [PubMed] [Google Scholar]
- 24Palatnik JF, Allen E, Wu X, et al. Control of leaf morphogenesis by microRNAs. Nature 2003; 425:257–263. [DOI] [PubMed] [Google Scholar]
- 25Schommer C, Palatnik JF, Aggarwal P, et al. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 2008; 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Koyama T, Furutani M, Tasaka M, Ohme-Takagi M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 2007; 19:473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 2010; 22:3574–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Alvarez-Buylla ER, Benitez M, Corvera-Poire A, et al. Flower development. Arabidopsis Book 2010; 8: e0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Borg M, Rutley N, Kagale S, et al. An EAR-dependent regulatory module promotes male germ cell division and sperm fertility in Arabidopsis. Plant Cell 2014; 26:2098–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Wang L, Kim J, Somers DE. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci USA 2013; 110:761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Kieffer M, Stern Y, Cook H, et al. Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 2006; 18:560–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Gallavotti A, Long JA, Stanfield S, et al. The control of axillary meristem fate in the maize ramosa pathway. Development 2010; 137:2849–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Ikeda M, Mitsuda N, Ohme-Takagi M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 2009; 21:3493–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Kelley DR, Gasser CS. Ovule development: genetic trends and evolutionary considerations. Sex Plant Reprod 2009; 22:229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Sieber P, Gheyselinck J, Gross-Hardt R, Laux T, Grossniklaus U, Schneitz K. Pattern formation during early ovule development in Arabidopsis thaliana. Dev Biol 2004; 273:321–334. [DOI] [PubMed] [Google Scholar]
- 36Martin-Trillo M, Cubas P. TCP genes: a family snapshot ten years later. Trends Plant Sci 2010; 15:31–39. [DOI] [PubMed] [Google Scholar]
- 37Nath U, Crawford BC, Carpenter R, Coen E. Genetic control of surface curvature. Science 2003; 299:1404–1407. [DOI] [PubMed] [Google Scholar]
- 38Aguilar-Martinez JA, Sinha N. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front Plant Sci 2013; 4:406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Herve C, Dabos P, Bardet C, et al. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol 2009; 149:1462–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Efroni I, Blum E, Goldshmidt A, Eshed Y. A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 2008; 20:2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41Elias-Villalobos A, Fernandez-Alvarez A, Ibeas JI. The general transcriptional repressor Tup1 is required for dimorphism and virulence in a fungal plant pathogen. PLoS Pathog 2011; 7: e1002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42Friesen H, Hepworth SR, Segall J. An Ssn6-Tup1-dependent negative regulatory element controls sporulation-specific expression of DIT1 and DIT2 in Saccharomyces cerevisiae. Mol Cell Biol 1997; 17:123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43Todd RB, Greenhalgh JR, Hynes MJ, Andrianopoulos A. TupA, the Penicillium marneffei Tup1p homologue, represses both yeast and spore development. Mol Microbiol 2003; 48:85–94. [DOI] [PubMed] [Google Scholar]
- 44Braun BR, Johnson AD. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 1997; 277:105–109. [DOI] [PubMed] [Google Scholar]
- 45Yamashiro CT, Ebbole DJ, Lee BU, et al. Characterization of rco-1 of Neurospora crassa, a pleiotropic gene affecting growth and development that encodes a homolog of Tup1 of Saccharomyces cerevisiae. Mol Cell Biol 1996; 16:6218–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Mizuno T, Nakazawa N, Remgsamrarn P, Kunoh T, Oshima Y, Harashima S. The Tup1-Ssn6 general repressor is involved in repression of IME1 encoding a transcriptional activator of meiosis in Saccharomyces cerevisiae. Curr Genet 1998; 33:239–247. [DOI] [PubMed] [Google Scholar]
- 47Schultz J, Carlson M. Molecular analysis of SSN6, a gene functionally related to the SNF1 protein kinase of Saccharomyces cerevisiae. Mol Cell Biol 1987; 7:3637–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Zhao Y, Wei T, Yin KQ, et al. Arabidopsis RAP2.2 plays an important role in plant resistance to Botrytis cinerea and ethylene responses. New Phytol 2012; 195:450–460. [DOI] [PubMed] [Google Scholar]
- 49Wang WY, Zhang L, Xing S, et al. Arabidopsis AtVPS15 plays essential roles in pollen germination possibly by interacting with AtVPS34. J Genet Genom 2012; 39:81–92. [DOI] [PubMed] [Google Scholar]
- 50Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 51Qin G, Gu H, Zhao Y, et al. An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 2005; 17:2693–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52Shi DQ, Liu J, Xiang YH, Ye D, Sundaresan V, Yang WC. SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 2005; 17:2340–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53Chen H, Zou Y, Shang Y, et al. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 2008; 146:368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SPL interacts with other members of TPL family proteins using the C-terminal EAR motif.
Identification of 4.4-kb-long promoter of SPL and spl-2.
The other phenotypes of tpl-1 plants.
SPL interacts with CIN-like TCPs.
SPL was associated with CIN-like TCPs.
The CIN-like TCP genes were all expressed in flowers.
The primer list used in this work.