Abstract
Abscisic acid (ABA) regulates various developmental processes and stress responses in plants. Protein phosphorylation/dephosphorylation is a central post-translational modification (PTM) in ABA signaling. However, the phosphoproteins regulated by ABA under osmotic stress remain unknown in maize. In this study, maize mutant vp5 (deficient in ABA biosynthesis) and wild-type Vp5 were used to identify leaf phosphoproteins regulated by ABA under osmotic stress. Up to 4052 phosphopeptides, corresponding to 3017 phosphoproteins, were identified by Multiplex run iTRAQ-based quantitative proteomic and LC-MS/MS methods. The 4052 phosphopeptides contained 5723 non-redundant phosphosites; 512 phosphopeptides (379 in Vp5, 133 in vp5) displayed at least a 1.5-fold change of phosphorylation level under osmotic stress, of which 40 shared common in both genotypes and were differentially regulated by ABA. Comparing the signaling pathways involved in vp5 response to osmotic stress and those that in Vp5, indicated that ABA played a vital role in regulating these pathways related to mRNA synthesis, protein synthesis and photosynthesis. Our results provide a comprehensive dataset of phosphopeptides and phosphorylation sites regulated by ABA in maize adaptation to osmotic stress. This will be helpful to elucidate the ABA-mediate mechanism of maize endurance to drought by triggering phosphorylation or dephosphorylation cascades.
Drought is one of globally environmental stress that greatly hampers crop production. The frequent occurrence of drought with rising temperature poses a serious challenge to sustainable crop production1. Maintaining crop yield stability in a changing climate is needed to guarantee a food supply for the increasing world population. Particularly, maize (Zea mays L.), one of the major food crops globally, is often exposed to drought stress. So, improving maize for increased drought tolerance is a priority in breeding programs2.
At the molecular level, understanding the mechanism of crops response to drought is useful to develop genotypes with improved drought tolerance3. Most known regulatory genes, e.g., transcription factors (TFs) and protein kinases, are characterized as important stress regulators based on their transcriptional induction by various stresses. However, many proteins are biologically active in vivo only after undergoing some kind of post translational modification (PTM), e.g., WRKY4 and ZmCPK45. For example, protein phosphorylation plays a critical role in regulating many biological functions including stress endurance through signal transduction6. Many regulatory proteins and enzymes can be switched on and off by phosphorylation and dephosphorylation to control a wide range of cellular processes or signal relays2. In maize response to drought stress, 138 phosphopeptides display highly significant changes and their corresponding proteins affect epigenetic control, gene expression, cell cycle-dependent processes and phytohormone-mediated responses6; in bread wheat response to drought stress, 31 phosphoproteins have significant change of phosphorylation level and are mainly involved in three biological processes: RNA transcription/processing, stress/detoxification/defense, and signal transduction7. Moreover, previous studies also indicate that there are different phosphoprotein changes in different crops response to drought stress. Thus, characterizing protein phosphorylation and its dynamics in cell response to stresses will contribute to understanding signaling pathways and stress endurance mechanisms in crops.
Plant hormone abscisic acid (ABA) is involved in regulating several major processes, such as seed dormancy, germination and seedling growth, and various stress responses. ABA can regulate different sets of stress-responsive genes to initiate the synthesis of various proteins, including TFs, enzymes, and molecular chaperones8. Protein phosphorylation belongs to a type of rapidly PTMs in the ABA-regulated signaling pathway7. ABA-regulated phosphoproteins have been analyzed in Arabidopsis9,10,11,12 and rice10,13,14. However, it remains unknown whether in vivo phosphosites of many drought stress-responsive protein kinases are involved in ABA-triggered maize response to drought stress. Recently, iTRAQ-based quantitative proteomic and LC-MS/MS methods demonstrate the power of quantitative analysis for protein phosphorylation. Using these methods, a total of 1625 unique phosphopeptides have been detected from 1126 phosphoproteins in soybean root hairs, of which 273 phosphopeptides corresponding to 240 phosphoproteins are significantly regulated in response to Bradyrhizobium japonicum15.
Maize viviparous-5 (vp5) is deficient in ABA biosynthesis16,17, with much reduced ABA content in seeds, roots and leaves compared to its wild-type Vp5. Thus, the mutant vp5 and wild-type Vp5 are useful for the studies of ABA-regulated phosphoproteins in maize. In this study, multiplex run iTRAQ-based quantitative phosphoproteomic analysis and LC-MS/MS methods were performed to identify and compare the differential phosphoproteins in maize under osmotic stress. As a result, up to 4052 unique phosphopeptides, corresponding to 3017 phosphoproteins, were identified, and their phosphorylation levels were analyzed as ABA-dependent or independent.
Results
Differentially accumulated phosphopeptides under osmotic stress
The ABA content in vp5 and Vp5 leaves was measured by ELISA. Under osmotic stress, the increased ABA content in Vp5 and vp5 leaves was 0.6863 and 0.0403 ng/g · dry weight, respectively; the increased ABA content in Vp5 leaves was about 17 times that in vp5 leaves (Fig. 1). This difference in ABA accumulation facilitates the study of the ABA-regulated signaling pathways in maize exposed to osmotic stress.
Figure 1. ABA content in maize ABA-deficient mutant vp5 and wild-type Vp5 leaves under normal conditions (control) or 8 h osmotic stress (OS).
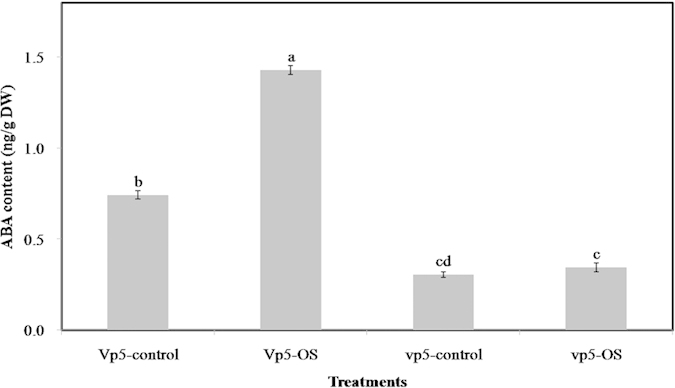
Values are means ± SE (n = 5).
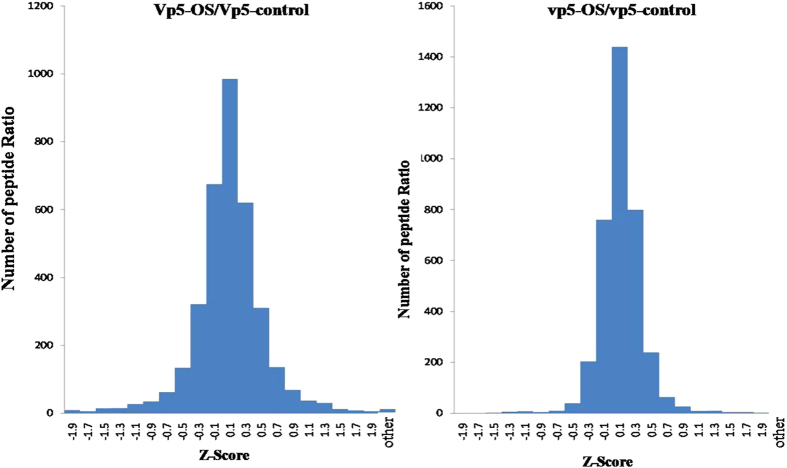
Total leaf proteins from vp5 and Vp5 seedlings exposed to osmotic stress were isolated and analyzed as shown in the work flowchart (Fig. 2). Simultaneously, osmotic stress and control iTRAQ ratios for each run were converted to z-scores to normalize the data (Fig. 3), resulting in the identification of 4052 unique phosphopeptides (correspond to 3017 proteins) at a false discovery rate (FDR) of 5%. Among the 4052 unique phosphopeptides, 53.84% contained only a single phosphoryl group, 37.81% contained two, 7.34% contained three, and 1.03% contained four and above. At a FDR of 1%, there were 3240 phosphorylated peptides and 153 non-phosphorylated peptides; the ratio of phosphoenrichment was 95.49%. At a FDR of 5%, there were 4052 phosphorylated peptides and 221 non-phosphorylated peptides; the ratio of phosphoenrichment was 94.84%.
Figure 2. iTRAQ 4-plex labeling and LC MS/MS workflow of identifying phosphorous proteins in leaves of maize ABA mutant vp5 and wild-type Vp5 seedlings under osmotic stress (OS).
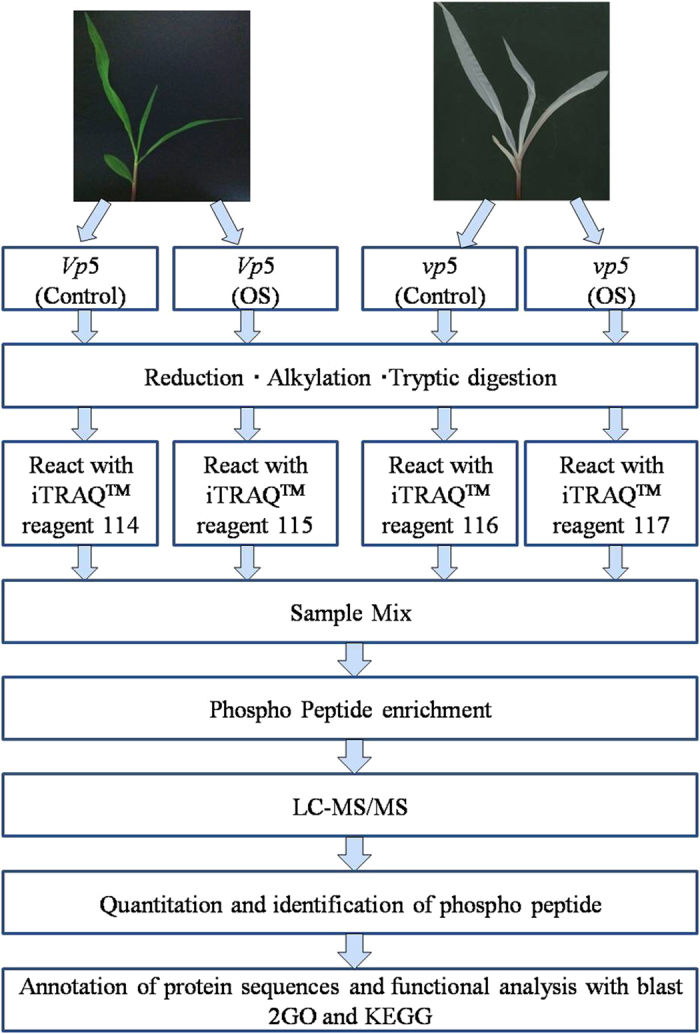
Figure 3. Z-scores frequency distribution of differential peptides in maize wild type Vp5 and mutant vp5 under osmotic stress.
iTRAQ ratios between osmotic stress (OS) and controls for each run were converted to z-scores to normalize the data. Positive z-score values represent proteins up-regulated by OS and negative values represent proteins down-regulated by OS. Z-scores between −0.9 and 0.9 indicates proteins not significantly altered, between ±0.9 and 1.96 moderately altered, and ≥1.96 and ≤−1.96 significantly altered ≥2-fold during osmotic stress (>95% confidence).
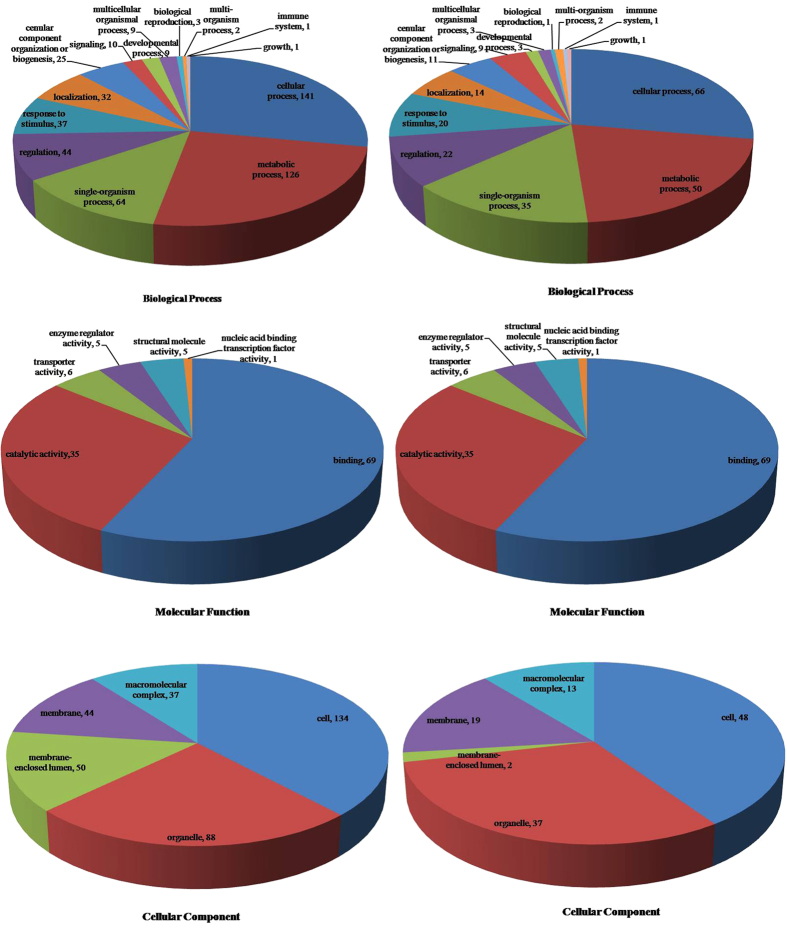
The proteins corresponding to the identified phosphoryled peptides in vp5 and Vp5 exposed to osmotic stress were annotated using Blast2GO according to the cell component and biological and molecular function (Fig. 4).
Figure 4. The distribution of differentially phosphorylated proteins in maize response to osmotic stress.
The 160 proteins identified were classified according to their known or predicted cellular component, molecular function, biological process, and signaling pathway. Left, Vp5; right, vp5.
Concerning cell component, 308 and 119 phosphoproteins were annotated in Vp5 and vp5, respectively, showing an unbiased distribution in different compartments. Thus, no protein enrichment procedure was introduced during protein extraction.
Concerning the biological process, phosphoproteins corresponding to the identified phosphopeptides in both genotypes were classified into 14 categories. The top categories with the highest number of phosphoproteins were cellular processes (28% in Vp5 and 27.72% in vp5), metabolic processes (25% in Vp5 and 21% in vp5) and single organism processes (12.70% in Vp5 and 4.96 in vp5), and these three functional categories were the most important in maize response to osmotic stress.
Concerning the molecular function, phosphoproteins corresponding to the identified phosphopeptides in both genotypes were classified into 9 categories. The top 3 categories with the highest number of phosphoproteins were transcription factor activity (57.57% in Vp5 and 57.24% in vp5), catalytic activity (28.95% in Vp5 and 28.92% in vp5) and transporter activity (4.28% in Vp5 and 5.95% in vp5).
Of the 4,052 phosphopeptides identified, there were 379 and 133 phosphopeptides with ≥1.5 folds (increased) or ≤0.6 folds (decreased) only in Vp5 and vp5, respectively, 40 in both genotypes (Fig. 5). This change was equivalent to a significant expression ratio according to the standard with p-value <0.05 (Table 1, Tables S1 and S2). These significant phosphopeptides corresponded to 472 phosphoproteins. In order to further test the significance of 512 phosphopeptides, FDR attained by Benjamini-Hochberg method at 5% level were used to adjust p-values (correction for multiple comparisons). As a result, 36 phosphopeptides were no significant difference, which corresponded to 36 phosphoproteins, including C0PLA9 and B8A0C6 (Table 1) and other 34 listed in Table S1. Among the 36 phosphoproteins, other phosphopeptides of B4F8Q3, B4FZY1, B6TDL6 and Q9ATM4, were significant (Table 1, Tables S1 and S2).
Figure 5. Venn diagram showing the number of proteins with significant changes of phosphorylation levels in maize vp5 and Vp5 leaves exposed to osmotic stress.
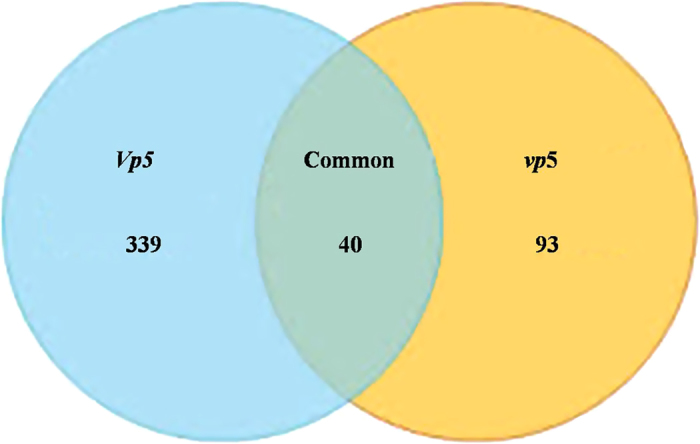
Table 1. Proteins with more than 1.5-folds phosphorylation level change in two maize genotypes response to ABA and osmotic stress.
| UniProt ID | >Protein name | >Sequence of phosphorylati-on peptides | >PhosphoRS-Site Probabilities (>75%) | >Ion score |
Vp5: OS/control |
vp5: OS/control |
Vp5: OS/control |
vp5: OS/control |
>T-test | >Regulation of ABA and osmotic stress for peptides phosphosites | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | Averagea | P-Value/FDR | Averagea | P-Value/FDR | |||||||
| B6TE49 | probable receptor-like protein kinase at1g33260-like | gGFsTVYLAsLSSSR | S(4):77.9 | 18 | 2.151 | 2.481 | 3.261 | 0.299 | 0.343 | 0.301 | 2.631 | 0.000/0.000 | 0.314 | 0.000/0.000 | 0.020 | Up-regulated by osmotic stress with ABA-dependent way |
| K7V8B2 | tata-binding protein-associated factor 172-like | sSAGtTPSk | S(1):100.0; T(5):99.9 | 15 | 4.345 | 3.219 | 2.340 | 0.455 | 0.221 | 0.599 | 3.301 | 0.000/0.000 | 0.425 | 0.000/0.000 | 0.044 | Up-regulated by osmotic stress with ABA-dependent way |
| K7TWA4 | regulatory-associated protein of tor 1-like | fRtPPVsPPQHDFLPGLR | T(3):100.0; S(7):100.0 | 13 | 0.232 | 0.362 | 0.490 | 0.232 | 0.622 | 0.455 | 0.361 | 0.000/0.000 | 0.436 | 0.000/0.000 | 0.505 | Down-regulated by osmotic stress with ABA-independent way |
| *C0PLA9 | nodulin-like protein | eEVTEDSENASSSTtALGGsNQDLSSGk | S(20):99.9 | 43 | 1.813 | 1.592 | 1.203 | 0.403 | 0.556 | 0.469 | 1.536 | 0.041/0.057 | 0.476 | 0.000/0.000 | 0.032 | Up-regulated by osmotic stress with ABA-dependent way |
| *B4FBC9 | patellin family protein | aAEADsEEEk | S(6):100.0 | 44 | 2.015 | 2.745 | 2.489 | 0.505 | 0.477 | 0.375 | 2.416 | 0.000/0.002 | 0.452 | 0.000/0.000 | 0.014 | Up-regulated by osmotic stress with ABA-dependent way |
| K7U7E1 | brefeldin a-inhibited guanine nucleotide-exchange protein 1-like | vLENVHQPsFLQk | S(9):100.0 | 17 | 0.409 | 0.378 | 0.312 | 0.789 | 0.400 | 0.543 | 0.366 | 0.000/0.000 | 0.577 | 0.000/0.001 | 0.179 | Down-regulated by osmotic stress with ABA-independent way |
| *K7TWZ6 | clustered mitochondria isoform x1 | qcDVLsPEEYsDEGWQAASmR | S(6):99.6 | 17 | 2.267 | 2.092 | 2.823 | 0.561 | 0.443 | 0.650 | 2.394 | 0.000/0.001 | 0.551 | 0.000/0.001 | 0.008 | Up-regulated by osmotic stress with ABA-dependent way |
| *K7V8M7 | mdr-like abc transporter | qIsINk | S(3):100.0 | 21 | 0.578 | 0.628 | 0.642 | 0.571 | 0.555 | 0.604 | 0.616 | 0.010/0.021 | 0.577 | 0.000/0.000 | 0.175 | Down-regulated by osmotic stress with ABA-independent way |
| *B8A0C6 | phosphatidate phosphatase lpin2-like | eLVPGGEDsGtGSDDEtVNEPEPPAR | S(9):75.0; T(11):75.0; S(13):75.0; T(17):75.0 | 21 | 2.012 | 1.812 | 2.166 | 0.676 | 0.488 | 0.595 | 1.997 | 0.001/0.005 | 0.586 | 0.000/0.001 | 0.003 | Up-regulated by osmotic stress with ABA-dependent way |
| *B4FWX5 | dihydroxy-acid mitochondrial-like | nAMVIVmALGGstNAVLHLIAIAR | S(12):100.0; T(13):100.0 | 16 | 1.803 | 1.503 | 2.071 | 0.602 | 0.613 | 0.593 | 1.792 | 0.008/0.017 | 0.603 | 0.000/0.001 | 0.020 | Up-regulated by osmotic stress with ABA-dependent way |
| *B4FS10 | TPA: hypothetical protein ZEAMMB73_767959 | aAGGDDSGsGGGFNLGGLGGLFAk | S(7):50.0; S(9):50.0 | 25 | 0.538 | 0.617 | 0.476 | 0.605 | 0.922 | 0.409 | 0.544 | 0.004/0.011 | 0.646 | 0.000/0.001 | 0.449 | Down-regulated by osmotic stress with ABA-independent way |
| *B8A0C6 | phosphatidate phosphatase lpin2-like | eLVPGGEDSGtGSDDETVNEPEPPAR | T(11):75.0 | 45 | 1.567 | 1.632 | 2.166 | 0.612 | 0.656 | 0.595 | 1.788 | 0.048/0.060 | 0.621 | 0.001/0.004 | 0.029 | Up-regulated by osmotic stress with ABA-dependent way |
| *K7V792 | splicing factor 3b subunit 1-like isoform x1 | mADADAtPAAGGAtPGATPSGAWDAtPk | T(7):99.7; T(26):100.0 | 26 | 1.786 | 1.565 | 2.019 | 0.666 | 0.612 | 0.671 | 1.790 | 0.000/0.001 | 0.650 | 0.003/0.011 | 0.010 | Up-regulated by osmotic stress with ABA-dependent way |
| *B6U1M6 | transposon protein | lALPLAGGHVtDNDGEGTAERPTk | T(11):93.7 | 11 | 0.513 | 0.491 | 0.578 | 1.612 | 1.499 | 1.480 | 0.527 | 0.005/0.011 | 1.530 | 0.005/0.017 | 0.003 | Up-regulated by osmotic stress, but down-regulated by ABA |
| *B6U0Y9 | atp binding protein | aVQVSPILDGNQtDADSNTAGEEVASR | T(13):96.4 | 52 | 1.512 | 1.492 | 1.557 | 1.647 | 1.590 | 1.558 | 1.520 | 0.004/0.011 | 1.598 | 0.000/0.001 | 0.190 | Up-regulated by osmotic stress with ABA-independent way |
| *B6SP06 | glycine-rich protein 2b | sLNDGDAVEYTVGsGNDGR | S(14):99.9 | 41 | 2.360 | 1.781 | 2.014 | 1.943 | 1.484 | 1.437 | 2.052 | 0.002/0.007 | 1.621 | 0.005/0.017 | 0.034 | Up-regulated by osmotic stress with ABA-dependent and independent way |
| *B6TB18 | lipid phosphate phosphatase 3 | eTLNDVESGsAR | S(10):100.0 | 69 | 0.175 | 0.143 | 0.307 | 1.570 | 1.439 | 1.234 | 0.208 | 0.000/0.000 | 1.415 | 0.005/0.016 | 0.014 | Up-regulated by osmotic stress, but down-regulated by ABA |
| K7U2M6 | heat shock protein sti | dVEPEPEAEPmDLtDEEkDR | T(14):100.0 | 11 | 0.353 | 0.212 | 0.495 | 1.523 | 1.533 | 1.484 | 0.353 | 0.005/0.012 | 1.513 | 0.005/0.016 | 0.007 | Up-regulated by osmotic stress, but down-regulated by ABA |
| K7USN0 | 2og-fe oxygenase family protein | aPVMMVAAAPARPmVmASSGTGGGNIsk | S(27):75.0 | 15 | 1.857 | 1.788 | 2.201 | 1.487 | 1.568 | 1.513 | 1.949 | 0.005/0.011 | 1.523 | 0.005/0.016 | 0.091 | Up-regulated by osmotic stress with ABA-independent way |
| *B4FKD1 | nucleoporin nup53-like | eGSPmDGVVQyQQQSPTTPSGQQSQQQk | Y(11):75.0 | 14 | 0.512 | 0.508 | 0.581 | 1.523 | 1.545 | 1.488 | 0.534 | 0.004/0.011 | 1.519 | 0.004/0.016 | 0.002 | Up-regulated by osmotic stress, but down-regulated by ABA |
| B6TCM5 | duf1664 domain family protein isoform 1 | hNMANAVssMTkHLEQVQssLAAAk | S(19):99.6; S(20):99.6 | 26 | 3.516 | 3.110 | 3.123 | 1.539 | 1.496 | 1.700 | 3.250 | 0.004/0.010 | 1.578 | 0.004/0.014 | 0.009 | Up-regulated by osmotic stress with ABA-dependent and independent way |
| Q3MQ01 | autophagy protein 5 | sQEAEQALAsPAEAGFAk | S(10):81.9 | 11 | 1.861 | 1.512 | 1.631 | 1.651 | 1.499 | 1.508 | 1.668 | 0.029/0.043 | 1.553 | 0.003/0.011 | 0.180 | Up-regulated by osmotic stress with ABA-independent way |
| *K7UKU6 | protein decapping 5-like | iGQLNDEPNGYEDDVIEDDEIsPR | S(22):100.0 | 54 | 2.432 | 2.821 | 2.066 | 1.523 | 1.613 | 1.528 | 2.440 | 0.000/0.001 | 1.555 | 0.002/0.009 | 0.045 | Up-regulated by osmotic stress with ABA-dependent and independent way |
| *P04711 | phosphoenolpyruvate carboxylase | hHsIDAQLR | S(3):100.0 | 35 | 5.006 | 3.801 | 4.766 | 1.974 | 1.447 | 1.377 | 4.524 | 0.000/0.000 | 1.599 | 0.001/0.006 | 0.011 | Up-regulated by osmotic stress with ABA-dependent and independent way |
| *O04014 | tpa:40s ribosomal protein s6 | dRRsEsLAk | S(4):100.0; S(6):100.0 | 14 | 10.27 | 2.875 | 2.829 | 1.467 | 1.641 | 1.828 | 5.324 | 0.000/0.000 | 1.645 | 0.001/0.005 | 0.288 | Up-regulated by osmotic stress with ABA-dependent and independent way |
| C0P2E1 | disease resistance protein rga2-like | aHFPVImLYSFtsTyDVk | Y(15):94.0 | 16 | 3.023 | 3.640 | 4.087 | 1.533 | 1.831 | 1.557 | 3.583 | 0.000/0.000 | 1.640 | 0.001/0.005 | 0.024 | Up-regulated by osmotic stress with ABA-dependent and independent way |
| B7ZYP2 | pentatricopeptide repeat-containing protein at4g22760-like | aGDIPAARAmFEAmPARDVVsWNSMVAGLAk | S(21):80.0 | 14 | 0.389 | 0.455 | 0.299 | 1.678 | 1.723 | 1.523 | 0.381 | 0.000/0.000 | 1.641 | 0.001/0.004 | 0.000 | Down-regulated by osmotic stress with ABA-dependent way |
| K7U3J5 | set domain protein sdg117 | dDTIVcsPVDLSDAcQSGmDR | S(7):92.7 | 12 | 3.544 | 2.809 | 3.211 | 1.812 | 1.578 | 1.593 | 3.188 | 0.000/0.000 | 1.661 | 0.001/0.003 | 0.010 | Up-regulated by osmotic stress with ABA-dependent and independent way |
| *K7UKZ7 | transcription elongation factor spt6-like | eScPtLLSFDSDEDNEDIESDAR | T(5):79.0 | 17 | 0.623 | 0.647 | 0.712 | 1.701 | 1.631 | 1.669 | 0.661 | 0.002/0.007 | 1.667 | 0.000/0.003 | 0.001 | Down-regulated by osmotic stress with ABA-dependent way |
| *B4FXH0 | act-domain containing protein kinase family protein | iEDMDSAyDsDASEEGDDDGDDLSVR | Y(8):84.9 | 18 | 0.347 | 0.251 | 0.196 | 1.701 | 1.723 | 1.651 | 0.265 | 0.000/0.000 | 1.692 | 0.000/0.002 | 0.001 | Down-regulated by osmotic stress with ABA-dependent way |
| *K7TW55 | translocase of chloroplast chloroplastic-like | gGNLGPTEAEAETDDGGEEPASGDGEtPASLAAPMPVVESk | T(27):83.3 | 63 | 1.822 | 1.529 | 1.532 | 2.120 | 1.593 | 1.681 | 1.628 | 0.004/0.011 | 1.798 | 0.000/0.001 | 0.130 | Up-regulated by osmotic stress with ABA-independent way |
| M1H548 | arginine serine-rich protein 45-like | rsPsPPPRR | S(2):100.0; S(4):100.0 | 10 | 0.345 | 0.234 | 0.123 | 2.072 | 1.503 | 1.638 | 0.234 | 0.000/0.000 | 1.738 | 0.000/0.001 | 0.008 | Down-regulated by osmotic stress with ABA-dependent way |
| *K7V1A7 | chromatin structure-remodeling complex protein syd-like isoform x4 | aAVVAELFGDATEGGSDQPLPsPR | S(22):94.8 | 26 | 2.084 | 1.824 | 1.508 | 1.562 | 1.624 | 2.127 | 1.805 | 0.011/0.022 | 1.771 | 0.000/0.001 | 0.929 | Up-regulated by osmotic stress with ABA-independent way |
| *B6UEN7 | ubiquitin ligase protein cop1 | aAsAsPQGPAEEGEGPADR | S(3):100.0; S(5):100.0 | 28 | 2.492 | 3.032 | 2.412 | 1.704 | 1.600 | 1.801 | 2.645 | 0.000/0.000 | 1.702 | 0.000/0.001 | 0.063 | Up-regulated by osmotic stress with ABA-dependent and independent way |
| *O04014 | tpa: 40s ribosomal protein s6 | sEsLAk | S(1):100.0; S(3):100.0 | 20 | 4.269 | 3.875 | 4.829 | 1.867 | 1.671 | 1.828 | 4.324 | 0.000/0.000 | 1.789 | 0.000/0.000 | 0.009 | Up-regulated by osmotic stress with ABA-dependent and independent way |
| *Q8W149 | cell division cycle 5-like | eSQtPLLGGDNPELHPSDFSGVtPR | T(4):99.9; T(23):99.9 | 47 | 1.578 | 2.420 | 1.953 | 2.199 | 1.503 | 1.763 | 1.984 | 0.003/0.009 | 1.822 | 0.000/0.000 | 0.750 | Up-regulated by osmotic stress with ABA-independent way |
| *B8A298 | histone-lysine n- h3 lysine-9 specific suvh1-like | dSESsPQPPIAAPAESGk | S(5):95.5 | 13 | 3.661 | 2.500 | 3.604 | 3.044 | 1.528 | 1.790 | 3.255 | 0.000/0.000 | 2.120 | 0.000/0.000 | 0.086 | Up-regulated by osmotic stress with ABA-dependent and independent way |
| C0P2E1 | disease resistance protein rga2-like | aHFPVImLYsFtstYDVk | S(10):78.0; T(12):78.0; S(13):78.0; T(14):78.0 | 17 | 0.653 | 0.281 | 0.483 | 2.101 | 2.105 | 1.976 | 0.472 | 0.001/0.003 | 2.061 | 0.000/0.000 | 0.006 | Down-regulated by osmotic stress with ABA-dependent way |
| *B6TXK5 | uncharacterized protein LOC100277637 | gPHAstDDEEEEDDDDEDAYEVER | S(5):99.9; T(6):99.9 | 18 | 1.547 | 1.588 | 1.778 | 2.020 | 2.122 | 2.501 | 1.638 | 0.003/0.009 | 2.214 | 0.000/0.000 | 0.017 | Up-regulated by osmotic stress, but down-regulated by ABA |
| *O04014 | tpa: 40s ribosomal protein s6 | skLsAAAk | S(1):100.0; S(4):100.0 | 16 | 9.494 | 10.39 | 11.99 | 2.572 | 2.373 | 2.786 | 10.63 | 0.000/0.000 | 2.577 | 0.000/0.000 | 0.007 | Up-regulated by osmotic stress with ABA-dependent and independent way |
| *P24993 | photosystem ii phosphoprotein | atQtVEDSSRPkPk | T(2):100.0; T(4):100.0 | 15 | 2.148 | 1.578 | 1.557 | 4.661 | 2.294 | 2.004 | 1.761 | 0.002/0.006 | 2.987 | 0.000/0.000 | 0.199 | Up-regulated by osmotic stress, but down-regulated by ABA |
aEach value represents the average of three biological replicas. The average is significant at a p < 0.05 and a false discovery rate (FDR) <0.05 level. Moreover, these peptides whose UniProt ID are signed with * are also significant under FDR <0.01. FDR values attained by Benjamini-Hochberg method were shown in column and were used to adjust p-values (correction for multiple comparisons). These phosphopeptides whose FDR values were signed with delete line ‘—’ were not significant. ‘-’, not measured. T-test is used to identify whether the difference is significant. A T-test value <0.05 is considered to be significant. OS = osmotic stress.
In order to prove that the observed changes in phosphopeptide abundances were due to the changes in phosphorylation state or the abundance change, protein abundance was also measured using the iTRAQ technique. As a result, among 472 phosphoproteins, 187 phosphoproteins changed in abundance but in no significant level; 10 phosphoproteins changed in abundance with a significant level only in Vp5 or vp5; no changes in abundance of the rest 275 phosphoproteins were detected (Tables S3–S5). Except C0P8S9 and K7U4E0, eight (B4G1E6, B6T0F0, B6STN4, B6T6R3, B6TPC9, K7VBH0, B6TM56 and K7UCK7) of the 10 differential abundance phosphoproteins resulted from a significant change in phosphorylation state. For example, B4G1E6 had significant phosphopeptide abundances but no significant protein abundances in Vp5, whereas it had significant protein abundances but no significant phosphopeptide abundances in vp5; K7UCK7 had significant folds of phosphopeptide only in vp5 which existed significant difference compared to protein abundances (Table S4).
Furthermore, for the 379 phosphopeptides in Vp5, the numbers of phosphoRS sites at S, T and Y residues were 585 (56.41%), 194 (33.16%) and 61 (10.42%), respectively. For the 133 phosphopeptides in vp5, the numbers of phosphoRS sites were 181 (51.93%), 66 (36.46%) and 21 (11.60%), respectively. For each peptide, the PhosphoRS site probabilities above 75% indicate that a site is truly phosphorylated (Table 1, Tables S1 and S2).
Our data showed that many phosphoproteins were differentially phosphorylated and involved in a series of DNA/RNA-related processes and protein synthesis/degradation (Table 2). This was consistent with the results attained by using Blast2GO software to analyze the biological function, cellular components and molecular function (Fig. 4).
Table 2. Most abundance phosphoproteins mediated ABA signaling pathways under osmotic stress.
| >Protein/peptide sequence | >UniProt ID | >Protein name | >PhosphoRS-Site Probabilities (>75%) | >Ion score |
Vp5: OS/control |
vp5: OS/control |
Vp5: OS/control |
vp5: OS/control |
t-test | Regulation of ABA and osmotic stress for peptides phosphosites | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | Averagea | P-Value | Averagea | P-Value | |||||||
| Transporter | ||||||||||||||||
| aLGSFRsNA | *Q9ATM4 | aquaporin pip2-7 | S(7):100.0 | 16 | 0.656 | 0.611 | 0.650 | 0.762 | 0.718 | 0.755 | 0.639 | 0.021 | 0.745 | 0.028 | 0.000 | Down-regulated by osmotic stress with ABA-dependent way |
| aLGsFR | Q9ATM4 | aquaporin pip2-7 | S(4):100.0 | 17 | 0.559 | 0.702 | 0.728 | 0.831 | 0.825 | 0.824 | 0.663 | 0.047 | 0.827 | 0.138 | 0.096 | Down-regulated by osmotic stress with ABA-independent way |
| aLGsFRsNA | Q9ATM4 | aquaporin pip2-7 | S(4):100.0; S(7):100.0 | 17 | 0.436 | 0.436 | 0.347 | 0.862 | 0.923 | 0.822 | 0.406 | 0.000 | 0.869 | 0.153 | 0.002 | Down-regulated by osmotic stress with ABA-dependent way |
| lGsSAsFSR | *Q9XF58 | aquaporin pip2-4-like | S(3):97.3; | 27 | 0.494 | 0.498 | 0.714 | 1.192 | 1.203 | 1.126 | 0.569 | 0.002 | 1.174 | 0.221 | 0.025 | Down-regulated by osmotic stress with ABA-dependent way |
| xtPLIAGLAVAAtALAGR | B6T195 | mitochondrial import inner membrane translocase subunit tim14 | T(2):100.0; T(13):100.0 | 13 | 2.210 | 2.004 | 2.451 | 0.989 | 1.024 | 0.719 | 2.222 | 0.018 | 0.911 | 0.163 | 0.027 | Up-regulated by osmotic stress with ABA-independent way |
| rPAsLR | B4FZY1 | Na+/H+ antiporter | S(4):100.0 | 14 | 0.641 | 0.621 | 0.553 | 1.032 | 0.988 | 1.099 | 0.605 | 0.048 | 1.040 | 0.880 | 0.016 | Down-regulated by osmotic stress with ABA-dependent way |
| gFVPFVPGsPTESsLPLLPGNEN | *B4FZY1 | Na+/H+ antiporter | S(9):91.3 | 28 | 2.946 | 2.823 | 2.969 | 1.139 | 0.936 | 0.789 | 2.913 | 0.000 | 0.954 | 0.396 | 0.003 | Up-regulated by osmotic stress with ABA-dependent way |
| gQsALGsALGLIsR | *B6U6U2 | hexose transporter | S(3):100.0; S(7):100.0; S(13):100.0 | 75 | 1.683 | 1.692 | 1.634 | 1.071 | 1.048 | 1.095 | 1.670 | 0.026 | 1.071 | 0.580 | 0.003 | Up-regulated by osmotic stress with ABA-dependent way |
| tQtGSSSNR | B6U937 | probable sugar phosphate/ phosphate translocator at3g17430-like | T(1):80.0 | 10 | 6.023 | 4.674 | 3.324 | 0.758 | 0.813 | 0.869 | 4.674 | 0.000 | 0.813 | 0.106 | 0.041 | Up-regulated by osmotic stress with ABA-dependent way |
| lSNsFLAITDsFR | C0PEW7 | vacuolar amino acid transporter 1-like | S(11):94.9 | 29 | 1.653 | 1.553 | 1.653 | 1.041 | 1.141 | 1.241 | 1.620 | 0.041 | 1.141 | 0.357 | 0.019 | Up-regulated by osmotic stress with ABA-dependent way |
| tPLGAAYEPPSAAAGGGGTtPVNIR | *C0PLZ2 | probable peptide nitrate transporter at5g13400-like | T(20):95.8 | 28 | 0.522 | 0.526 | 0.580 | 1.392 | 0.823 | 1.129 | 0.543 | 0.043 | 1.115 | 0.400 | 0.075 | Down-regulated by osmotic with ABA-dependent way |
| sAsTPR | K7U2V8 | zinc transporter | S(3):97.7 | 15 | 1.733 | 1.930 | 1.833 | 0.945 | 1.113 | 1.045 | 1.832 | 0.007 | 1.034 | 0.760 | 0.000 | Up-regulated by osmotic stress with ABA-dependent way |
| qSsLNAAGTssMAVLR | K7UMX4 | solute carrier family facilitated glucose transporter member 8 | S(10):97.8 | 55 | 1.728 | 1.821 | 1.691 | 0.912 | 1.121 | 1.054 | 1.747 | 0.018 | 1.029 | 0.701 | 0.005 | Up-regulated by osmotic stress with ABA-dependent way |
| nsVSsPIMTR | Q6UNK5 | abc transporter b family member 1-like | S(2):76.7 | 24 | 2.429 | 2.169 | 2.175 | 1.294 | 1.330 | 1.378 | 2.258 | 0.001 | 1.334 | 0.047 | 0.013 | Up-regulated by osmotic stress with ABA-dependent way |
| sSEGVFVGAFLSMSStAVVskFLVEk | B6SP24 | K+ efflux antiporter 5-like | T(16):87.3 | 17 | — | — | — | 0.454 | 0.436 | 0.424 | 0.438 | 0.000 | Up-regulated by ABA | |||
| kAssLQR | B6SV26 | vacuolar amino acid transporter 1-like | S(3):100.0; S(4):100.0 | 19 | 1.022 | 0.911 | 0.920 | 1.628 | 1.423 | 1.682 | 0.951 | 0.817 | 1.578 | 0.002 | 0.013 | Down-regulated by ABA |
| sTGTAAtGGsDAGLEEGk | B6UH65 | zinc transporter 2 precursor | S(1):78.4 | 10 | 0.686 | 0.645 | 0.665 | 2.999 | 3.120 | 2.973 | 0.665 | 0.069 | 3.031 | 0.000 | 0.001 | Down-regulated by ABA |
| nYLTPFFTsQTDDDNDDDFSQQPQNR | B4FS09 | sodium hydrogen exchanger 6-like | S(9):74.6 | 12 | — | — | — | 2.301 | 2.340 | 2.201 | 2.281 | 0.000 | Down-regulated by ABA | |||
| eGSPMDGVVQyQQQSPTTPSGQQSQQQk | B4FKD1 | nucleoporin nup53-like | Y(11):75.0 | 14 | 0.512 | 0.508 | 0.581 | 1.523 | 1.545 | 1.488 | 0.534 | 0.005 | 1.519 | 0.004 | 0.002 | Up-regulated by osmotic stress, but down-regulated by ABA |
| tsDADsEAGSGSGGGGR | C0P5C4 | abc1 family protein | S(2):95.0 | 16 | 0.701 | 0.774 | 0.787 | 1.660 | 1.594 | 1.612 | 0.754 | 0.197 | 1.622 | 0.001 | 0.003 | Down-regulated by ABA |
| Ubiquitin-conjugating enzyme family protein-like | ||||||||||||||||
| ntPSmPPAVSTSSAsR | B4FHK6 | ubiquitin-conjugating enzyme e2 22-like | T(2):76.1 | 11 | 0.456 | 0.511 | 0.405 | 0.782 | 0.790 | 0.757 | 0.457 | 0.000 | 0.776 | 0.053 | 0.004 | Down-regulated by osmotic stress with ABA-dependent way |
| eVNAGIASVsR | *B4FAG8 | e3ubiquitin-protein ligase rhf2a-like isoform x1 | S(10):100.0 | 30 | 0.605 | 0.589 | 0.646 | 0.912 | 0.800 | 0.845 | 0.613 | 0.028 | 0.852 | 0.196 | 0.020 | Down-regulated by osmotic stress with ABA-dependent way |
| rHsTGQstPDR | *B4FAG8 | e3ubiquitin-protein ligase rhf2a-like isoform x1 | S(3):97.0; T(8):97.0 | 28 | 2.211 | 1.890 | 2.017 | 1.401 | 1.201 | 1.303 | 2.039 | 0.002 | 1.302 | 0.057 | 0.002 | Up-regulated by osmotic stress with ABA-dependent way |
| aDsPSEGLTcGSQNLPAETcPk | *K7V4D9 | e3ubiquitin-protein ligase upl4-like | S(3):99.8 | 23 | 2.888 | 2.521 | 2.666 | 1.010 | 0.987 | 0.949 | 2.692 | 0.000 | 0.982 | 0.665 | 0.003 | Up-regulated by osmotic stress with ABA-dependent way |
| sAsPSTS | C0P3H1 | Ubiquitin carboxyl-terminal hydrolase isozyme l5-like | S(3):96.4; | 16 | 0.555 | 0.512 | 0.597 | 0.957 | 0.879 | 1.020 | 0.555 | 0.006 | 0.952 | 0.704 | 0.002 | Down-regulated by osmotic stress with ABA-dependent way |
| lGVDVNtmPAItDk | B4G0Z1 | e3ubiquitin-protein ligase ubr7-like | T(12):99.6 | 11 | 0.601 | 0.499 | 0.558 | 1.230 | 1.110 | 1.192 | 0.553 | 0.001 | 1.177 | 0.221 | 0.000 | Down-regulated by osmotic stress with ABA-dependent way |
| aAsAsPQGPAEEGEGPADR | *B6UEN7 | ubiquitin ligase protein cop1 | S(3):100.0; S(5):100.0 | 28 | 2.492 | 3.032 | 2.412 | 1.704 | 1.600 | 1.801 | 2.645 | 0.010 | 1.702 | 0.001 | 0.063 | Up-regulated by osmotic stress with ABA-dependent and independent way |
| dVsNAsELATEMQYER | *K7TFK8 | e3ubiquitin-protein ligase upl1-like | S(3):100.0; S(6):100.0 | 28 | 2.132 | 1.929 | 1.958 | 1.153 | 1.099 | 0.900 | 2.007 | 0.009 | 1.051 | 0.473 | 0.005 | Up-regulated by osmotic stress with ABA-dependent way |
| lRPGQPDAVQDAStSDmEDASTSSGGQR | *K7TFK8 | e3ubiquitin-protein ligase upl1-like | T(14):76.0 | 41 | 1.656 | 1.626 | 1.701 | 1.420 | 1.766 | 1.087 | 1.661 | 0.029 | 1.424 | 0.020 | 0.390 | Up-regulated by osmotic stress with ABA-independent way |
| eNEGSSSsAGESSSmDIDk | *B6T6V5 | ubiquitin carboxyl-terminal hydrolase 6-like | S(8):79.5 | 17 | 0.612 | 0.584 | 0.658 | 1.312 | 1.113 | 1.215 | 0.618 | 0.029 | 1.213 | 0.169 | 0.008 | Down-regulated by osmotic stress with ABA-dependent way |
| sALLsYSDTVR | B6T6V5 | ubiquitin carboxyl-terminal hydrolase 6-like | S(5):75.0 | 13 | 0.580 | 0.592 | 0.512 | 1.799 | 1.064 | 1.200 | 0.561 | 0.008 | 1.354 | 0.048 | 0.070 | Down-regulated by osmotic stress with ABA-dependent way |
| Zinc finger transcription factor | ||||||||||||||||
| dLVVDtDDGGNANR | *B6UB08 | zinc finger protein 652-a- partial | T(6):100.0 | 46 | 0.570 | 0.521 | 0.564 | 0.809 | 0.812 | 1.089 | 0.552 | 0.006 | 0.903 | 0.101 | 0.057 | Down-regulated by osmotic stress with ABA-independent way |
| dPSINQVAsPVAAPEPVGAILPk | *B4FX96 | Zinc finger ccch type domain containing protein zfn-like 1 | S(9):100.0 | 26 | 0.655 | 0.776 | 0.516 | 1.016 | 0.877 | 0.769 | 0.649 | 0.047 | 0.887 | 0.358 | 0.087 | Down-regulated by osmotic stress with ABA-independent way |
| eQGsIGITANDDPyNGNEmSPSDQR | K7UHH6 | zinc finger c-x8-c-x5-c-x3-h type family protein | S(4):100 | 40 | 0.245 | 0.360 | 0.495 | 0.808 | 0.904 | 1.007 | 0.367 | 0.000 | 0.906 | 0.443 | 0.001 | Down-regulated by osmotic stress with ABA-dependent way |
| dPAVGsSPAVsNNk | *B4FLK4 | zinc finger protein 207-like isoform x1 | S(6):97.3 | 41 | 0.431 | 0.423 | 0.454 | 1.277 | 1.292 | 1.311 | 0.436 | 0.000 | 1.293 | 0.086 | 0.000 | Down-regulated by osmotic stress with ABA-dependent way |
| dWNQNFEVsPTDYLPQDSR | *B6U194 | zinc finger c-x8-c-x5-c-x3-h type family protein | S(9):80.0 | 12 | 0.401 | 0.393 | 0.536 | 0.901 | 0.993 | 0.821 | 0.443 | 0.000 | 0.905 | 0.411 | 0.038 | Down-regulated by osmotic stress with ABA-dependent way |
| lQPADsIEGTVIDRDcDEVDDAAQDSGAR | *B4FY62 | tpa:c3hc zinc finger-like family protein | S(6):99.9 | 18 | 0.422 | 0.493 | 0.412 | 1.112 | 1.030 | 0.980 | 0.442 | 0.000 | 1.041 | 0.891 | 0.006 | Down-regulated by osmotic stress with ABA-dependent way |
| dcDEVDDAAQDsGAR | *B4FY62 | tpa:c3hc zinc finger-like family protein | S(12):100.0 | 35 | 2.981 | 1.879 | 2.006 | 1.355 | 1.439 | 1.402 | 2.288 | 0.001 | 1.399 | 0.017 | 0.139 | Up-regulated by osmotic stress with ABA-independent way |
| cMVsLsPPPPk | K7UE59 | ring zinc finger domain superfamily protein | S(4):100.0; S(6):100.0 | 21 | 0.498 | 0.556 | 0.587 | 1.332 | 1.376 | 1.385 | 0.547 | 0.002 | 1.364 | 0.020 | 0.000 | Down-regulated by osmotic stress with ABA-dependent way |
| gANEEVsSINVDEDPNVPYERsPNAAIAk | *K7UBL3 | zinc finger c-x8-c-x5-c-x3-h type family protein | S(7):95.1; S(22):100.0 | 35 | 0.409 | 0.627 | 0.570 | 0.925 | 1.328 | 1.624 | 0.535 | 0.002 | 1.293 | 0.131 | 0.041 | Down-regulated by osmotic stress with ABA-dependent way |
| dSSANPPPsPGTTYGPVGSISk | *B6SW01 | zinc finger ccch type domain-containing protein zfn-like 3 | S(9):83.6 | 16 | 0.534 | 0.424 | 0.624 | 0.980 | 0.883 | 1.039 | 0.527 | 0.005 | 0.967 | 0.698 | 0.001 | Down-regulated by osmotic stress with ABA-dependent way |
| lGGsDGNsEDDMDNDk | *B7ZXU2 | serrate-related c2h2 zinc-finger family protein | S(4):100.0; S(8):100.0 | 29 | 1.694 | 1.721 | 1.417 | 1.220 | 1.179 | 1.274 | 1.611 | 0.048 | 1.224 | 0.151 | 0.088 | Up-regulated by osmotic stress with ABA-independent way |
| ePGEGtSS | B6TD33 | zinc finger ccch domain-containing protein 11-like | T(6):83.3 | 17 | 1.594 | 1.623 | 1.856 | 1.226 | 1.112 | 1.345 | 1.691 | 0.027 | 1.228 | 0.149 | 0.010 | Up-regulated by osmotic stress with ABA-dependent way |
| nVDVDsDGER | *K7UZK2 | zinc finger ccch domain-containing protein 44-like | S(6):100.0 | 26 | 2.282 | 1.968 | 2.223 | 1.365 | 1.063 | 1.211 | 2.158 | 0.001 | 1.213 | 0.192 | 0.001 | Up-regulated by osmotic stress with ABA-dependent way |
| vEsSLVGSDDVLDSASDSPPsVk | C0P2B1 | phd zinc finger | S(21):74.8 | 15 | 2.334 | 2.410 | 2.309 | 1.350 | 1.597 | 1.441 | 2.351 | 0.000 | 1.463 | 0.008 | 0.003 | Up-regulated by osmotic stress with ABA-dependent way |
| nTHPPEPESIDGINDtGVQTPQQFR | *B4FZ17 | dhhc-type zinc finger domain-containing protein | T(16):49.0 | 13 | — | — | — | 3.911 | 3.890 | 3.589 | 3.797 | 0.000 | Down-regulated by ABA | |||
| eQGsIGItANEDPYNANEMSPSDQR | *B4FX77 | zinc finger c-x8-c-x5-c-x3-h type family protein | S(4):100.0; T(8):94.7 | 22 | 1.372 | 1.375 | 1.489 | 0.650 | 0.580 | 0.630 | 1.412 | 0.143 | 0.620 | 0.001 | 0.002 | Up-regulated by ABA |
| vPQDEEESGDDDEDEEADEHNNtLcGTcGTNDSk | B6TG72 | phd finger protein | T(27):32.2 | 10 | 1.320 | 1.344 | 1.462 | 0.431 | 0.660 | 0.778 | 1.375 | 0.162 | 0.623 | 0.001 | 0.008 | Up-regulated by ABA |
| sQPPDAAASPDASIssPSSLGGGGGDAADADAIEk | *K7UCK7 | zinc finger ccch type domain-containing protein zfn-like 6 | S(15):79.2 | 22 | 0.998 | 0.899 | 1.031 | 0.656 | 0.645 | 0.663 | 0.976 | 0.976 | 0.655 | 0.000 | 0.011 | Up-regulated by ABA |
| Ribosomal protein | ||||||||||||||||
| gQAAATAsk | *B4FCE7 | 60s ribosomal protein l2 | S(8):100.0 | 33 | 0.649 | 0.633 | 0.624 | 0.912 | 0,847 | 0.967 | 0.635 | 0.049 | 0.940 | 0.458 | 0.018 | Down-regulated by osmotic stress with ABA-dependent way |
| asAAtSA | *O04014 | tpa: 40s ribosomal protein s6 | S(2):100.0; T(5):100.0 | 29 | 2.017 | 1.865 | 1.947 | 0.992 | 1.254 | 0.913 | 1.943 | 0.005 | 1.053 | 0.920 | 0.024 | Up-regulated by osmotic stress with ABA-dependent way |
| vsEELR | *O04014 | tpa: 40s ribosomal protein s6 | S(2):100.0 | 16 | 1.773 | 1.541 | 1.476 | 0.981 | 0.989 | 1.121 | 1.597 | 0.044 | 1.030 | 0.358 | 0.046 | Up-regulated by osmotic stress with ABA-dependent way |
| fTADDVAAAAGGAAAtGAsLQEID | *B6TPG2 | 60s ribosomal protein l26-1 | T(16):100.0; S(19):100.0 | 20 | 0.456 | 0.387 | 0.523 | 1.301 | 1.299 | 1.286 | 0.455 | 0.000 | 1.295 | 0.075 | 0.003 | Down-regulated by osmotic stress with ABA-dependent way |
| eEsDDDMGFSLFD | *B6UE07 | 60s acidic ribosomal protein p2a | S(3):100.0 | 44 | 1.920 | 2.079 | 1.631 | 1.013 | 0.995 | 1.174 | 1.877 | 0.003 | 1.061 | 0.261 | 0.049 | Up-regulated by osmotic stress with ABA-dependent way |
| fASVPcGGGGVAVAAAsPAAGGAAPTAEAk | *B6UE07 | 60s acidic ribosomal protein p2a | S(17):80.0 | 27 | 0.498 | 0.487 | 0.536 | 1.371 | 1.104 | 1.240 | 0.507 | 0.002 | 1.238 | 0.127 | 0.010 | Down-regulated by osmotic stress with ABA-dependent way |
| kAsGGGGDDEEEE | B4FCK4 | 40s ribosomal protein s9 | S(3):100.0 | 19 | 0.987 | 1.082 | 1.020 | 1.560 | 1.487 | 1.550 | 1.030 | 0.643 | 1.532 | 0.004 | 0.010 | Down-regulated by ABA |
| eEEkAPEPAEEsDEEMGFSLFDD | *B4FWI0 | 60s acidic ribosomal protein p0 | S(12):100.0 | 12 | — | — | — | 1.752 | 1.687 | 1.681 | 1.706 | 0.000 | Down-regulated by ABA | |||
| WD-40 repeat protein | ||||||||||||||||
| vSNNDSEPDsPSGSPNR | *B6TM01 | transducin wd-40 repeat | S(10):100.0 | 49 | 1.608 | 1.264 | 1.921 | 1.070 | 0.804 | 1.020 | 1.598 | 0.022 | 0.965 | 0.801 | 0.043 | Up-regulated by osmotic stress with ABA-dependent way |
| gRSsPVVsGSPSQNSDGSmsSWR | B4FMI7 | wd repeat-containing protein 89 homolog | S(20):83.9 | 17 | 1.221 | 1.262 | 1.276 | 1.776 | 1.660 | 1.676 | 1.253 | 0.321 | 1.704 | 0.000 | 0.013 | Down-regulated by ABA |
| ssPVVsGSPSQNSDGSmSSWR | B4FMI7 | wd repeat-containing protein 89 homolog | S(2):96.0 | 12 | 3.083 | 3.880 | 3.912 | 1.240 | 1.022 | 1.012 | 3.625 | 0.000 | 1.091 | 0.701 | 0.018 | Up-regulated by osmotic stress with ABA-dependent way |
| Arginine serine-rich splicing factor | ||||||||||||||||
| gNNGDDEHRGsPRGsQsP | C0HIN5 | arginine serine-rich splicing factor rs2z37a transcript i | S(11):100.0; S(15):100.0; S(17):100.0 | 15 | 0.336 | 0.301 | 0.485 | 0.756 | 0.801 | 0.785 | 0.374 | 0.000 | 0.781 | 0.063 | 0.020 | Down-regulated by osmotic stress with ABA-dependent way |
| sEGSSSsSFGR | *B7ZYN1 | serine arginine repetitive matrix protein 2-like | S(7):79.6 | 27 | 0.601 | 0.552 | 0.705 | 0.850 | 0.798 | 0.844 | 0.620 | 0.038 | 0.831 | 0.190 | 0.028 | Down-regulated by osmotic stress with ABA-dependent way |
| eRsPGAR | B6SY05 | arginine serine-rich splicing factor rsp41 | S(3):100.0 | 17 | 1.680 | 1.781 | 2.356 | 0.878 | 0.880 | 0.825 | 1.939 | 0.008 | 0.861 | 0.218 | 0.042 | Up-regulated by osmotic stress with ABA-dependent way |
| gGtPPR | K7V1I2 | arginine serine-rich splicing factor sr45_2 transcript i | T(3):100.0 | 18 | 0.310 | 0.308 | 0.387 | 0.859 | 0.949 | 0.827 | 0.335 | 0.000 | 0.878 | 0.234 | 0.011 | Down-regulated by osmotic stress with ABA-dependent way |
| qYRsPsADR | K7U6X8; B4FD63 | serine arginine repetitive matrix protein 2-like isoform x2 | S(4):99.9; S(6):100.0 | 11 | 0.626 | 0.504 | 0.613 | 0.866 | 0.945 | 0.941 | 0.581 | 0.017 | 0.917 | 0.616 | 0.029 | Down-regulated by osmotic stress with ABA-dependent way |
| aAcsGsP | *M1GS93 | splicing arginine serine-rich 2 | S(4):100.0; S(6):100.0 | 26 | 0.555 | 0.564 | 0.560 | 0.849 | 0.835 | 0.916 | 0.559 | 0.011 | 0.867 | 0.199 | 0.007 | Down-regulated by osmotic stress with ABA-dependent way |
| sYTPDDINDR | *B4FQ73 | serine arginine-rich splicing factor 33-like | S(1):100.0 | 11 | 1.789 | 1.954 | 1.864 | 0.977 | 1.010 | 0.945 | 1.869 | 0.007 | 0.977 | 0.654 | 0.002 | Up-regulated by osmotic stress with ABA-dependent way |
| Heterogeneous nuclear ribonucleoprotein | ||||||||||||||||
| ssQGGGGYR | C0P8S9 | heterogeneous nuclear ribonucleoprotein a2 | S(2):100.0 | 12 | 0.597 | 0.567 | 0.685 | 0.812 | 0.845 | 0.815 | 0.616 | 0.029 | 0.824 | 0.114 | 0.040 | Down-regulated by osmotic stress with ABA-dependent way |
| sPAGGQNYAmSR | *B8A134 | heterogeneous nuclear ribonucleoprotein 1-like | S(1):100.0 | 12 | 0.587 | 0.543 | 0.618 | 0.897 | 0.834 | 0.847 | 0.583 | 0.015 | 0.859 | 0.202 | 0.008 | Down-regulated by osmotic stress with ABA-dependent way |
| lGsPIGYVGLNDDSGSILSSMSR | *B8A134 | heterogeneous nuclear ribonucleoprotein 1-like | S(3):95.1 | 13 | 2.221 | 2.340 | 2.179 | 0.987 | 1.021 | 1.163 | 2.247 | 0.001 | 1.057 | 0.289 | 0.006 | Up-regulated by osmotic stress with ABA-dependent way |
| qPsEEPEEQVDLEGDDDGmDDDDAGYR | *K7UBY5 | heterogeneous nuclear ribonucleoprotein r-like | S(3):100.0 | 74 | 1.789 | 1.801 | 1.894 | 0.867 | 0.887 | 1.080 | 1.828 | 0.010 | 0.945 | 0.845 | 0.002 | Up-regulated by osmotic stress with ABA-dependent way |
| rGsRDDsEEPEEDDDNDER | *K7UBY5 | heterogeneous nuclear ribonucleoprotein r-like | S(3):100.0; S(7):100.0 | 29 | 1.923 | 2.088 | 2.265 | 1.031 | 1.176 | 0.963 | 2.092 | 0.002 | 1.057 | 0.880 | 0.016 | Up-regulated by osmotic stress with ABA-dependent way |
| dDsEEPEEDDDNDER | *K7UBY5 | heterogeneous nuclear ribonucleoprotein r-like | S(3):100.0 | 55 | 1.577 | 1.779 | 2.056 | 1.208 | 1.302 | 1.436 | 1.804 | 0.002 | 1.315 | 0.880 | 0.021 | Up-regulated by osmotic stress with ABA-dependent way |
| eANPGGsGGGR | B8A305 | heterogeneous nuclear ribonucleoprotein 1-like isoform x1 | S(7):100.0 | 11 | 0.565 | 0.612 | 0.671 | 1.210 | 0.988 | 1.102 | 0.616 | 0.031 | 1.100 | 0.508 | 0.028 | Down-regulated by osmotic stress with ABA-dependent way |
OS = osmotic stress.
aEach value represents the average of three biological replicas. A p value <0.05 in regard of FDR <0.05 is considered to be significant. Nevertheless, these peptides whose UniProt ID are signed with * are also significant under FDR <0.01. ‘-’, not measured. T-test is used to identify whether the difference is significant. A T-test value <0.05 is considered to be significant.
Phosphorylation motifs in phosphopeptides
To determine whether the phosphorylated versions of the identified phosphopeptides had different phosphorylation site motifs in both genotypes and whether ABA affected the motifs, Motif-X online software was used to predict the motif specificity of the phosphopeptides. The motifs SP and TP were common in both genotypes response to osmotic stress; 12 motifs were only predicted in Vp5; 2 motifs were only predicated in vp5 (Table 3). These results indicated a high sensitivity and specificity of phosphorylation sites in maize response to ABA under osmotic stress.
Table 3. Phosphorylation motif of proteins with significant phosphorylation sites in Vp5 and vp5 under osmotic stress.
| Genotype | # | Motif | Motif Score | Foreground Matches | Foreground size | Background Matches | Background size | Fold Increase |
|---|---|---|---|---|---|---|---|---|
| 1 | . . . . . . . . . . S P . . . . . p . . . | 21.42 | 20 | 369 | 3885 | 1013205 | 14.14 | |
| 2 | . . . . . . P . . . S P . . . . . . . . . | 20.07 | 15 | 349 | 3609 | 1009320 | 12.02 | |
| 3 | . . . . . . . . . . S P . . . . . . . . . | 13.92 | 52 | 334 | 45568 | 1005711 | 3.44 | |
| 4 | . . . . . . . . . R S . . . . . . . . P . | 16.77 | 11 | 282 | 2404 | 960143 | 15.58 | |
| 5 | . . . . . . . . . R S . . . . . . . . . . | 7.15 | 36 | 271 | 46916 | 957739 | 2.71 | |
| 6 | . . . . . . . . . G S . . . . . . . . . . | 5.78 | 37 | 235 | 61833 | 910823 | 2.32 | |
| W | 7 | . . . . . . . A . D S . . . . . . . . . . | 10.34 | 9 | 198 | 2666 | 848990 | 14.48 |
| 8 | . . . . . . . R . . S . . . . . . . . . . | 5.42 | 27 | 189 | 45461 | 846324 | 2.66 | |
| 9 | . . . . . . . . . . S S . . . . . A . . . | 8.25 | 11 | 162 | 6216 | 800863 | 8.75 | |
| 10 | . . . . . . . . . AT P . . . . . . . . . | 18.63 | 9 | 101 | 1982 | 574595 | 25.83 | |
| 11 | . . . . . . . . . . T P . . . . . . . . . | 8.70 | 21 | 92 | 27413 | 572613 | 4.77 | |
| 12 | P . . . . . . . . . T P . . . . . . . . . | 5.06 | 14 | 71 | 26983 | 545200 | 3.98 | |
| 13 | . . . . . A . . . . T . . . . . . . . . . | 4.46 | 13 | 57 | 31931 | 518217 | 3.70 | |
| 14 | . . . . . . . . . . T . S . . . . . . . . | 3.95 | 13 | 44 | 44728 | 486286 | 3.21 | |
| M | 1 | . . . . . . . . . . S P . . . . . . . . . | 11.70 | 35 | 167 | 53062 | 1013205 | 4.00 |
| 2 | . . . . . . . . . . S . . D . . . . . . . | 7.76 | 26 | 132 | 53429 | 960143 | 3.54 | |
| 3 | . . . . . . . . . . S . . . . . . G . . . | 4.09 | 18 | 106 | 55755 | 906714 | 2.76 | |
| 4 | . . . . . . . . . . T P . . . . . . . . . | 7.50 | 15 | 52 | 29395 | 574595 | 5.64 |
In the present study, 34 phosphoproteins (Table 4) were found to contain several phosphopeptides. Notably, these peptides had specific phosphorylation characteristics in response to ABA and osmotic stress. Particularly, the phosphorylation level of two different peptides in 13 phosphorylation proteins was up-regulated or down-regulated in Vp5, whereas there was no change in vp5 under drought stress. These results indicated that ABA regulated the phosphorylation of different peptides of one protein with contrasting influence in maize response to osmotic stress. In contrast, B4F808 and C0HF00 were up-regulated or down-regulated in vp5, whereas no changes were detected in Vp5 under osmotic stress. The different phosphopeptides of the other 19 phosphoproteins had similar response to ABA under osmotic stress. Overall, this result showed the diversity of the phosphorylation sites and their specificity in maize response to ABA and stress treatments.
Table 4. Comparison of different phosphopeptides belonging to one protein in response to ABA and osmotic stress.
| Protein accession | Protein name | Sequence | Vp5: OS/control | vp5: OS/control |
|---|---|---|---|---|
| B4F8Q3 | btb poz domain-containing protein at5g66560-like | dVADEGNEEEGsEAEtPGR | 4.768 | 0.882 |
| aIAQTIMANEGGAAGsGEEGGEsDGGGTWR | 0.635 | 1.307 | ||
| B4FAG8 | e3 ubiquitin-protein ligase rhf2a-like isoform x1 | eVNAGIASVsR | 0.613 | 0.852 |
| rHSTGQstPDR | 2.039 | 1.302 | ||
| B4FK28 | tpa:rna-binding protein | sNTSsIGSPGPGR | 0.597 | 0.866 |
| sPAGVGQNYAMNR | 0.588 | 1.031 | ||
| B4FWC4 | rna-binding protein 39-like isoform x1 | aVEPAPPQANGSGsGSGEkDR | 2.213 | 1.009 |
| nLVQSNATsGGAASGGAR | 0.645 | 1.036 | ||
| B4FZY1 | Na+/H+ antiporter | rPAsLR | 0.605 | 1.040 |
| gFVPFVPGsPTESsLPLLPGNEN | 2.913 | 0,954 | ||
| B6SS20 | tpa:phototropin family protein kinase | dALPAEVEAPAPAPAPAPPEsTTEk | 2.021 | 1.046 |
| sEGEQEPVEPAPPVMAsPLVAPGtPSGGASLk | 1.763 | 1.271 | ||
| B6T245 | zn- - containing protein | gsPmPVSsPWSGGALAENTDNIASR | 1.714 | 1.045 |
| gsPMPVsSPWSGGALAENTDNIASR | 0.509 | 1.224 | ||
| B6T6V5 | ubiquitin carboxyl-terminal hydrolase 6-like | eNEGSSSsAGESSSmDIDk | 0.618 | 1.213 |
| sALLsYSDTVR | 0.561 | 1.354 | ||
| B6TDL6 | uncharacterized membrane protein at1g16860-like | lSGPQsSGVNPmAR | 0.595 | 0.991 |
| rLsGPQSsGVNPmAR | 1.830 | 1.100 | ||
| B6TI42 | at-hook protein 1 | qQQQQQLAPSPAPLNLAPTGVAAGPSsPPSR | 0.575 | 0.923 |
| ePFGLPktPAtPPSSGGTQGLR | 0.339 | 1.177 | ||
| B6UBN4 | j domain-containing protein required for chloro-plast accumulation response 1-like isoform x2 | nDDGTSYAYsVPTsPNASMNNYLAQGAAR | 0.454 | 1.006 |
| gMDSSmPtsPSQQMSNR | 0.617 | 1.117 | ||
| B6UE07 | 60s acidic ribosomal protein p2a | eEsDDDMGFSLFD | 1.877 | 1.061 |
| fASVPcGGGGVAVAAAsPAAGGAAPTAEAk | 0.507 | 1.238 | ||
| B8A134 | heterogeneous nuclear ribonucleoprotein 1-like | sPAGGQNYAmSR | 0.583 | 0.859 |
| lGsPIGYVGLNDDSGSILSSMSR | 2.247 | 1.057 | ||
| B8A307 | transmembrane expressed | dQEGGQPTGPEVVADDEVTsHR | 0.513 | 0.956 |
| sNsVSTtGNENLR | 1.725 | 1.092 | ||
| C0HIM6 | integrin-linked protein kinase family protein | qLsSGAAR | 0.579 | 0.926 |
| gGPDGSsAHQQLAVPENLDATmR | 0.312 | 1.050 | ||
| C0HIQ2 | something about silencing protein 10-like isoform x4 | qIAGGDDsmDEQEDETQENVWGR | 2.497 | 0.909 |
| qIAGGDDsMDEQEDETQENVWGR | 0.617 | 1.212 | ||
| C0P9I0 | unknown | eTGDGEEGEEEDASAAtGDEVVk | 1.979 | 1.139 |
| eTGDGEEGEEEDAsAAtGDEVVk | 1.973 | 1.023 | ||
| C0PJF1 | basic proline-rich | sPSQQPPR | 1.645 | 0.754 |
| rPPsPPAPAPPAAEELTEAGTEER | 1.729 | 0.977 | ||
| C0PM56 | chloroplast post-illumination chlorophyll fluorescence increase protein | lDIVSGcTDPSSDmFDPLATVDDGScPLEsDSEE | 1.677 | 1.051 |
| lDIVSGcTDPSSDMFDPLATVDDGScPLESDsEE | 1.762 | 0.988 | ||
| C4J2P1 | protein kinase superfamily protein | asPEPGEVSGGR | 1.729 | 0.992 |
| sVsPADSSVPGQWk | 0.483 | 1.104 | ||
| K7TFK8 | e3 ubiquitin-protein ligase upl1-like | dVsNAsELATEMQYER | 2.007 | 1.051 |
| lRPGQPDAVQDAStSDmEDASTSSGGQR | 1.661 | 1.424 | ||
| K7U2M6 | heat shock protein sti | dVEPEPEAEPmDLtDEEk | 0.541 | 1.084 |
| dVEPEPEAEPMDLtDEEk | 0.242 | 0.988 | ||
| K7U4E0 | protein furry homolog isoform x1 | asEmDAVGLVFLsSADVQIR | 0.536 | 1.005 |
| sGQLLPALItmSGPLSGVR | 0.581 | 1.376 | ||
| K7UBY5 | heterogeneous nuclear ribonucleoprotein r-like | qPsEEPEEQVDLEGDDDGmDDDDAGYR | 1.828 | 0.945 |
| rGsRDDsEEPEEDDDNDER | 2.091 | 1.057 | ||
| dDsEEPEEDDDNDER | 1.804 | 1.515 | ||
| K7UT89 | jumonji-like transcription factor family protein | dTVAEDSAHATEEsGEENLQEk | 2.399 | 1.208 |
| dTVAEDsAHATEEsGEENLQEk | 2.924 | 1.240 | ||
| K7V792 | splicing factor 3b subunit 1-like isoform x1 | mADADAtPAAGGATPGATPSGAWDAtPk | 1.776 | 0.802 |
| lLAtPTPLGtPLYAIPEENR | 0.537 | 0.942 | ||
| mADADAtPAAGGAtPGAtPSGAWDAtPk | 0.319 | 1.127 | ||
| K7VBC2 | vacuolar proton atpase a1-like | fLGTSEmDPDSEPDsAR | 1.861 | 0.982 |
| fLGTSEMDPDSEPDsAR | 0.447 | 1.054 | ||
| O48547 | nonphototropic hypocotyl protein expressed | vsEELR | 1.597 | 1.030 |
| ssETGsR | 1.905 | 1.008 | ||
| eDPLLDsDDERPDsFDDDFR | 1.774 | 1.109 | ||
| Q6JN48 | ethylene-insensitive protein 2-like | sIVDSTPYVSDDGPPsLTFSR | 0.569 | 0.874 |
| sYYDPsSVDGNENAGSPAYSk | 0.427 | 1.458 | ||
| Q8W149 | cell division cycle 5-like | eIQtPNPMAtPLAsPGPGItPR | 1.752 | 1.030 |
| eIQtPNPMATPLAsPGPGITPR | 2.139 | 0.932 | ||
| Q9ATM4 | aquaporin pip2-7 | aLGSFRsNA | 0.639 | 0.745 |
| aLGsFR | 0.663 | 0.827 | ||
| aLGsFRsNA | 0.406 | 0.869 | ||
| B4F808 | nucleic acid binding protein | eLALLNstLREDsPHPGsVsPFsNGGmkR | 0.410 | |
| eLALLNstLREDSPHPGsVsPFsNGGmkR | 1.593 | |||
| B6SVF2 | gtp binding protein | asAEPLRFtVTPGDAFGDGPPVGmsEAAk | 1.247 | 0.364 |
| asAEPLRFtVTPGDAFGDGPPVGMsEAAk | 1.261 | 0.646 | ||
| C0HF00 | vacuolar protein sorting-associated protein 41 homolog | sNSGQDsDGGMDDEDGSPSGQSR | 1.032 | 0.624 |
| sNSGQDsDGGmDDEDGSPSGQSR | 1.808 |
Effect of ABA on peptide phosphosites regulated by osmotic stress
The mechanisms of plant response to stress include both ABA-dependent and ABA-independent processes18. In this study, a total 472 phosphorylation peptides were changed with an l.5-fold increase, including 40 in two genotypes, 339 only in Vp5 and 93 only in vp5. Specially, among 40 phosphopeptides identified in both genotypes (Table 1), the phosphorylation level of some phosphopeptides (corresponding to protein ID: B6TE49 and K7V8B2) increased in Vp5 but decreased in vp5, indicating that these phosphopeptides were up-regulated by osmotic stress in an ABA-dependent way; the phosphorylation level of some phosphopeptides (corresponding to protein ID: K7TWA4 and Q3MQ01), was not obviously different in two genotypes, indicating that they were regulated by osmotic stress in an ABA-independent way; The phosphorylation level of some phosphopeptides (corresponding to protein ID: B6SP06, K7USN0 and B6TCM5) (Table 1), increased in both genotypes, but the increase was more significant in Vp5 under osmotic stress, indicating that they were regulated by osmotic stress in an ABA-dependent or ABA-independent way. Overall, the phosphorylation levels of 27 proteins were up-regulated by osmotic stress (9 in an ABA-dependent way, 12 in an ABA-dependent or ABA-independent way and 4 in an ABA-independent way), and 2 down-regulated by ABA; the phosphorylation levels of 13 were down-regulated by osmotic stress: 10 in an ABA-dependent way and 3 in an ABA-independent way.
Among the 339 phosphopeptides whose phosphoryltion level were identified with fold change >1.5 only in Vp5 (Table S1), 183 were down-regulated by osmotic stress, of which 156 had significant increase folds compared to vp5, indicating a down-regulation in an ABA-dependent way; 27 had no significant increase folds compared to vp5, indicating a down-regulation in an ABA-independent way; 156 were up-regulated by osmotic stress, of which 136 had significant increase fold compared to vp5, indicating an up-regulation in an ABA-dependent way; 20 had no significant increase folds compared to vp5, indicating an up-regulation in an ABA-independent way (Table S1).
Among the 93 phosphopeptides whose phosphorylation level were identified with fold change >1.5 only in vp5 under osmotic stress (Table S2), 34 peptides with >1.5 fold increase had significant difference in vp5 compared to Vp5, indicating a down-regulation by ABA; 19 peptides with >1.5 fold decrease had significant difference in vp5 compared to Vp5, indicating an up-regulation by ABA; three were down-regulated both in vp5 and Vp5 but without significant difference between them, indicating a down-regulation by osmotic stress in an ABA-independent way; two were up-regulated in vp5 and Vp5 but without significant difference between them, indicating an up-regulated by osmotic stress in an ABA-independent way; one was up-regulated more in vp5 than in Vp5, indicating up-regulated by osmotic stress but down-regulated by ABA. Particularly, among the 93 phosphopeptides, the phosphorylation levels of 34 peptides were detected in vp5 but not in Vp5, of which 22 were significantly up-regulated and 12 significantly down-regulated under osmotic stress (Table S2).
Phosphorylation of ubiquitin and transporters
Ubiquitin is a highly conserved protein found in all eukaryotic species. This small protein is involved in the destruction of endogenous target proteins via the ubiquitin 26S proteasome system, which is the primary proteolysis mechanism in eukaryotic cells19. In the present study, 11 phosphopeptides corresponding to 8 ubiquitin proteins were identified during osmotic stress (Table 2). The phosphorylation level of ubiquitin-conjugating enzyme e2 22-like (B4FHK6), e3ubiquitin-protein ligase rhf2a-like x1 (B4FAG8: eVNAGIASVsR), ubiquitin carboxyl-terminal hydrolase isozyme l5-like (C0P3H1), e3ubiquitin-protein ligase ubr7-like (B4G0Z1) and ubiquitin carboxyl-terminal hydrolase 6-like (B6T6V5) was decreased by osmotic stress in Vp5, whereas no obvious change occurred in vp5. In contrast, the phosphorylation level of e3ubiquitin-protein ligase rhf2a-like isoform x1 (B4FAG8: rHSTGQstPDR), e3ubiquitin-protein ligase upl4-like (K7V4D9), ubiquitin ligase protein cop1 (B6UEN7) and e3ubiquitin-protein ligase upl1-like (K7TFK8) was increased by osmotic stress in Vp5, whereas no obvious change occurred in vp5. These results indicated that ubiquitination played an important role in ABA regulating maize response to osmotic stress.
All types of transporters are important for turgor pressure and water-potential regulation, which is crucial to the growth and survival of plants under water stress. In the present study, the phosphorylation level of 16 transporters related to the cell ion/water-potential regulation was significantly changed (Table 2). Particularly, the phosphorylation level of aquaporin PIP2–5 (Q9ATM7) and aquaporin pip2–4-like (Q9XF58) was decreased by osmotic stress in Vp5, whereas no difference occurred in vp5; the phosphorylation level of probable sugar phosphate/phosphate translocator at3g17430-like (B6U937), vacuolar amino acid transporter 1-like (C0PEW7), hexose transporter (B6U6U2), zinc transporter (K7U2V8), solute carrier family facilitated glucose transporter member 8 (K7UMX4) and abc transporter b family member 1-like (Q6UNK5) was significantly up-regulated by osmotic stress in Vp5, where there was no difference in vp5; Na+/H+ antiporter (B4FZY1) had two different phosphopeptides whose phosphorylation level was up-regulated or down-regulated by osmotic stress in Vp5, whereas no difference occurred in vp5; the phosphorylation level changes of K+ efflux antiporter 5-like (B6SP24) and sodium hydrogen exchanger 6-like (B4FS09) was only detected in vp5. These results indicated that ABA might regulate the phosphorylation states of transporter proteins to maintain cell solute and ion homeostasis under osmotic stress.
Phosphorylation of chloroplast proteins
vp5 seedlings have light green leaves under dim light conditions. Nevertheless, vp5 seedlings have white leaves under high light conditions due to photooxidation of chlorophyll20,21,22. Vp5 seedlings had green leaves. This difference of morphology is helpful to identify the chloroplast-related phosphoproteins. In the present study, there were 20 chloroplast proteins corresponding to 23 phosphopeptides whose phosphorylation level was significantly changed by osmotic stress (Table 5). The phosphorylation level of 5 phosphoproteins (B6UBN4, B4FQ59, B6SVI8, B6T9S5 and C4JAR6) was significantly increased (B6SVI8: decreased) by osmotic stress in vp5, whereas had no significant change in Vp5 response to osmotic stress; the phosphorylation level of 8 phosphoproteins (Protein ID: B4FAW3, B4FSE2, B4FVB8, B4FZ38, B6SS20, C0PM56, K7U926 and P22275), was significantly increased by osmotic stress in Vp5, whereas there was no differences in vp5 response to osmotic stress; the phosphorylation level of the rest 8 phosphoproteins had an opposite response under osmotic stress.
Table 5. Phosphorylated chloroplast proteins in maize leaves regulated by ABA under osmotic stress.
| Protein accession | Protein name | Sequence | Phosphorylation level/protein abundance | Vp5: OS/control | vp5: OS/control | Regulation of ABA and osmotic stress for peptides phosphosites | |
|---|---|---|---|---|---|---|---|
| B4FAW3 | photosystem i reaction center subunit ii | gFVAPQLDPSTPSPIFGGStGGLLR | Phosphorylation level | 2.281 | 1.295 | Up-regulated by osmotic stress with ABA-independent way | |
| Protein abundance | 0.775 | 0.968 | |||||
| B4FSE2 | protochlorophyllide reductase b | aQAAAVSSPSVTPAsPSGk | Phosphorylation level | 1.701 | 1.048 | Up-regulated by osmotic stress with ABA-dependent way | |
| Protein abundance | 0.935 | 0.935 | |||||
| B4FVB8 | serine threonine-protein kinase chloroplastic-like | tIkEsMDELNSQR | Phosphorylation level | 1.509 | 0.943 | Up-regulated by osmotic stress with ABA-dependent way | |
| Protein abundance | 1.102 | 0.923 | |||||
| B4FZ38 | fructose–bisphosphatase | dGsPPR | Phosphorylation level | 1.815 | 0.668 | Up-regulated by osmotic stress with ABA-dependent way | |
| Protein abundance | — | — | |||||
| B4G1V3 | ribonucleoprotein chloroplastic-like | gGGGGGGGGsFVDSGNk | Phosphorylation level | 0.522 | 0.871 | Down-regulated by osmotic stress with ABA-dependent way | |
| Protein abundance | 0.990 | 0.893 | |||||
| B6SS20 | tpa:phototropin family protein kinase | dALPAEVEAPAPAPAPAPPEsTTEk | Phosphorylation level | 2.021 | 1.046 | Up-regulated by osmotic stress with ABA-dependent way | |
| sEGEQEPVEPAPPVMAsPLVAPGtPSGGASLk | 1.763 | 1.271 | Up-regulated by osmotic stress with ABA-independent way | ||||
| Protein abundance | 1.087 | 1.015 | |||||
| B6STN4 | chlorophyll a-b binding protein 2 | vGsFGEGR | Phosphorylation level | 0.561 | 1.054 | Down-regulated by osmotic stress with ABA-dependent way | |
| Protein abundance | 1.621 | 0.863 | |||||
| B6TM56 | chloroplast outer envelope 24 kd protein | nSADGAGAADAEsR | Phosphorylation level | 0.270 | 1.345 | Down-regulated by osmotic stress with ABA-dependent way | |
| Protein abundance | 0.597 | 1.537 | |||||
| B6TS38 | ribose-5-phosphate isomerase | gsAAAsPPPSGk | Phosphorylation level | 0.618 | 1.031 | Down-regulated by osmotic stress with ABA-independent way | |
| Protein abundance | 1.037 | 1.011 | |||||
| B6UBN4 | j domain-containing protein required for chloroplast accumulation response 1-like isoform x2 | nDDGTSYAYsVPTsPNASMNNYLAQGAAR | Phosphorylation level | 0.454 | 1.006 | Down-regulated by osmotic stress with ABA-dependent way | |
| gMDSSmPtsPSQQMSNR | 0.617 | 1.117 | Down-regulated by osmotic stress with ABA-dependent way | ||||
| Protein abundance | 1.060 | 0.985 | |||||
| C0PM56 | chloroplast post-illumination chlorophyll fluorescence increase protein | lDIVSGcTDPSSDmFDPLATVDDGScPLEsDSEE | Phosphorylation level | 1.677 | 1.051 | Up-regulated by osmotic stress with ABA-dependent way | |
| lDIVSGcTDPSSDMFDPLATVDDGScPLESDsEE | 1.762 | 0.988 | Up-regulated by osmotic stress with ABA-dependent way | ||||
| Protein abundance | — | — | |||||
| C0PNN7 | atp synthase gamma chain chloroplast (h(+)-transporting two-sector atpase f -atpase atpc1) | nLsIAYNR | Phosphorylation level | 0.618 | 0.936 | Down-regulated by osmotic stress with ABA-dependent way | |
| Protein abundance | — | — | |||||
| K7U926 | stress enhanced protein chloroplastic-like isoform x2 | sLsIIR | Phosphorylation level | 1.824 | 1.034 | Up-regulated by osmotic stress with ABA-dependent way | |
| Protein abundance | — | — | |||||
| K7VLY6 | blue-light photoreceptor phr2-like | lNsAtYSVISPLPSSTPGLSR | Phosphorylation level | 0.615 | 1.054 | Down-regulated by osmotic stress with ABA-dependent way | |
| Protein abundance | — | — | |||||
| P22275 | phosphoenolpyruvate carboxylase | Phosphorylation level | Phosphorylation level | 2.248 | 1.090 | Up-regulated by osmotic stress with ABA-dependent way | |
| Protein abundance | Protein abundance | 0.983 | 0.987 | ||||
| P31927 | sucrose-phosphate synthase | gAGGGGGGGDPRsPTk | Phosphorylation level | 0.603 | 1.074 | Down-regulated by osmotic stress with ABA-dependent way | |
| Protein abundance | 1.002 | 0.985 | |||||
| B4FQ59 | phosphoribulokinase precursor | lTsVFGGAAEPPk | Phosphorylation level | 0.980 | 1.577 | Down-regulated by ABA | |
| Protein abundance | 1.113 | 0.932 | |||||
| B6SVI8 | protein lutein deficient chloroplastic-like | aATTPAmPAtGLssAGASPFR | Phosphorylation level | — | 0.371 | Up-regulated by ABA | |
| Protein abundance | — | — | |||||
| B6T9S5 | ferredoxin--nadp leaf isozyme | mAAVTAAAIsLsSSSASSxAAAAk | Phosphorylation level | 0.889 | 1.591 | Down-regulated by ABA | |
| Protein abundance | — | — | |||||
| B6UBN4 | j domain-containing protein required for chloroplast accumulation response 1-like isoform x2 | nDDGTSYAYsVPtSPNASmNNYLAQGAAR | Phosphorylation level | — | 1.952 | Down-regulated by ABA | |
| Protein abundance | 1.060 | 0.985 | |||||
| C4JAR6 | rubisco subunit binding-protein beta subunit | sSEGTGSFPsPAAsPQPSR | Phosphorylation level | 1.058 | 1.563 | Down-regulated by ABA | |
| Protein abundance | — | — |
Responses of kinases and phosphatases to osmotic stress
The responses of enzymes, including protein kinases and phosphatases, are notable. In this study, 34 protein kinases/phosphatases were found to be involved in the ABA regulating maize response to osmotic stress (Table 6). The phosphorylation levels of the top 30 protein kinases/phosphatases (except B7ZYR5: atsEEERSGGtPPAAPtP) was significantly increased or decreased by osmotic stress in Vp5, whereas had no significant change in vp5 response to osmotic stress; by contrast, the phosphorylation levels of cyclin-dependent kinase family protein (K7VGC6), calcium-dependent protein kinase (Q41790) and tpa: leucine-rich repeat receptor-like protein kinase family protein (B7ZYR5) was significantly increased or decreased in vp5 response to osmotic stress, whereas had not obvious change in Vp5 response to stress. These results showed that ABA was involved in the phosphorylation and dephosphorylation of the 34 protein kinases/phosphatases. Particularly, some phosphopeptide belonged to the same protein kinases/phosphatases but had a different response to osmotic stress. For example, the phosphorylation levels of two different peptides, vAFNDTPTTVFWtDyVATR and qLsSGAAR of the map kinase family protein isoform 1 (B8A0M9) were up-regulated and down-regulated in Vp5 response to osmotic stress, respectively, but with no significant change in vp5 response to osmotic stress.
Table 6. Kinases and phosphatases regulated by ABA under osmotic stress.
| Protein accession | Protein name | Sequence | Vp5: OS/control | vp5: OS/control | Regulation of ABA and osmotic stress for peptides phosphosites |
|---|---|---|---|---|---|
| B4FAE7 | tpa: protein kinase superfamily protein | dAGFQsAEEGGsGTFR | 0.590 | 1.036 | Down-regulated by osmotic stress with ABA-dependent way |
| B4FGQ3 | probable receptor-like protein kinase at5g56460-like | aEsPkIQsPSER | 0.614 | 1.231 | Down-regulated by osmotic stress with ABA-dependent way |
| B4FVB8 | serine threonine-protein kinase chloroplastic-like | tIkEsMDELNSQR | 1.509 | 0.943 | Up-regulated by osmotic stress with ABA-dependent way |
| B4FXH0 | act-domain containing protein kinase family protein | iEDmDSAYDsDAsEEGDDDGDDLSVR | 2.322 | 0.819 | Up-regulated by osmotic stress with ABA-dependent way |
| B4FY41 | protein kinase chloroplastic-like | rLSGsAsPLPAPAtGSPLPGSSR | 1.900 | 0.925 | Up-regulated by osmotic stress with ABA-dependent way |
| B4FZ38 | fructose–bisphosphatase | dGsPPR | 1.815 | 0.668 | Up-regulated by osmotic stress with ABA-dependent way |
| B6SS20 | tpa: phototropin family protein kinase | dALPAEVEAPAPAPAPAPPEsTTEk | 2.021 | 1.046 | Up-regulated by osmotic stress with ABA-dependent way |
| sEGEQEPVEPAPPVMAsPLVAPGtPSGGASLk | 1.763 | 1.271 | Up-regulated by osmotic stress with ABA-independent way | ||
| B6SVR9 | protein kinase | hsQPDLsGPPPPk | 0.617 | 1.073 | Down-regulated by osmotic stress with ABA-dependent way |
| B6SWV6 | nad kinase 1 | sLSPAPIPIPAsPGIR | 3.890 | 1.049 | Up-regulated by osmotic stress with ABA-dependent way |
| B6SXI8 | tpa: protein kinase superfamily protein | sGPGPsFANR | 0.516 | 0.986 | Down-regulated by osmotic stress with ABA-independent way |
| B6SYP7 | cdpk-related protein kinase | aDHDADPSGAGSVAPPsPLPANGAPLPAtPR | 1.774 | 0.984 | Up-regulated by osmotic stress with ABA-independent way |
| B7ZXP0 | tpa: snrk sapk family protein kinase | sTVGTPAYIAPEVLLk | 2.182 | 0.920 | Up-regulated by osmotic stress with ABA-dependent way |
| B7ZYP6 | pyruvate orthophosphate dikinase | sDsGAGR | 0.589 | 0.999 | Down-regulated by osmotic stress with ABA-dependent way |
| B7ZYR5 | tpa: leucine-rich repeat receptor-like protein kinase family protein | aATSSAAAAAGsGATR | 0.385 | 1.130 | Down-regulated by osmotic stress with ABA-dependent way |
| atsEEERSGGtPPAAPtP | 1.000 | 1.574 | Down-regulated by ABA | ||
| B8A0M9 | tpa: map kinase family protein isoform 1 | vAFNDTPTTVFWtDyVATR | 2.941 | 1.211 | Up-regulated by osmotic stress with ABA-dependent way |
| C0HIM6 | integrin-linked protein kinase family protein | qLsSGAAR | 0.579 | 0.926 | Down-regulated by osmotic stress with ABA-independent way |
| gGPDGSsAHQQLAVPENLDATmR | 0.312 | 1.050 | Down-regulated by osmotic stress with ABA-dependent way | ||
| C0P5V5 | tpa: act-domain containing protein kinase family protein | gAsPPPPPSAGGAAGR | 0.520 | 1.083 | Down-regulated by osmotic stress with ABA-dependent way |
| C0P8J5 | tpa: act-domain containing protein kinase family protein | sVQVSPILDGNQtDsDSNTAGEEVASR | 2.738 | 1.136 | Up-regulated by osmotic stress with ABA-dependent way |
| C0PKH3 | serine threonine-protein kinase afc3-like | gGAsPPWR | 0.627 | 1.080 | Down-regulated by osmotic stress with ABA-dependent way |
| C0PKN2 | phosphoglycerate dehydrogenase | gLVEPVsSTFVNLVNADYtAk | 2.589 | 1.044 | Up-regulated by osmotic stress with ABA-dependent way |
| C4IYD7 | c-type lectin receptor-like tyrosine-protein kinase at1g52310-like isoform x1 | sGtsTSATsPmLPLEVRtPR | 0.416 | 0.955 | Down-regulated by osmotic stress with ABA-dependent way |
| C4J2P1 | protein kinase superfamily protein | asPEPGEVSGGR | 1.729 | 0.992 | Up-regulated by osmotic stress with ABA-dependent way |
| sVsPADSSVPGQWk | 0.483 | 1.104 | Down-regulated by osmotic stress with ABA-dependent way | ||
| K7TWL5 | casein kinase i | mATEsDsDSDAR | 0.443 | 1.106 | Down-regulated by osmotic stress with ABA-dependent way |
| K7UAY1 | serine threonine-protein kinase ctr1 | tNVDPSIsIPGFVsSQIDNPTTTk | 0.365 | 1.180 | Down-regulated by osmotic stress with ABA-dependent way |
| K7UJC3 | serine threonine-protein kinase at5g01020-like | fmDPGLEAQysPRAAEAAAk | 0.283 | 1.100 | Down-regulated by osmotic stress with ABA-dependent way |
| K7UNQ6 | proline-rich receptor-like protein kinase perk2-like | aSsSSTSAADPNPNk | 0.370 | 1.050 | Down-regulated by osmotic stress with ABA-dependent way |
| K7UW53 | proline-rich receptor-like protein kinase perk1-like | fFGSYSSSDyDSGQYNEDmk | 2.064 | 1.088 | Up-regulated by osmotic stress with ABA-dependent way |
| K7VGC6 | cyclin-dependent kinase family protein | iPDLNLQDGPmVLsPPR | 1.597 | 1.047 | Up-regulated by osmotic stress with ABA-dependent way |
| K7VN66 | leucine-rich repeat receptor-like protein kinase family protein | gLTASGGDFTSsSk | 0.534 | 1.005 | Down-regulated by osmotic stress with ABA-dependent way |
| A0MBZ8 | gck-like kinase mik | fSSYEDMSNSGTVVQTQNEDPEtPR | 0.408 | Up-regulated by ABA | |
| B4FQ59 | phosphoribulokinase precursor | lTsVFGGAAEPPk | 0.980 | 1.577 | Down-regulated by ABA |
| C0PHB9 | probable receptor-like protein kinase at5g56460-like | vSSTAkPEsPPkVQsPSEVDR | 1.227 | 1.580 | Down-regulated by ABA |
| K7VGC6 | cyclin-dependent kinase family protein | iPDLNLQDGPMVLsPPR | 1.230 | 0.649 | Up-regulated by ABA |
| Q41790 | calcium-dependent protein kinase | aPAPDsGR | 1.133 | 1.575 | Down-regulated by ABA |
Signaling pathways regulated by ABA under osmotic stress
According to the KEGG results, signal pathways related to phosphoproteins with significant changes of phosphorylation level in Vp5 (Table S6) and vp5 (Table S7) response to osmotic stress were classified into 47 and 35 categories under osmotic conditions, respectively. For Vp5, the top 3 categories with the highest number of phosphoproteins were spliceosome (13), carbon metabolism (9) and biosynthesis of amino acids (7), RNA transport (7), and the mRNA surveillance pathway (7). For vp5, the top 3 categories with the highest number of phosphoproteins were spliceosome (5), the PI3K-Akt signaling pathway (4) and photosynthesis, ribosome, RNA transport, the mRNA surveillance pathway, cell cycle, and protein processing in endoplasmic reticulum (3). Particularly, 15 signal pathways, including glycolysis/gluconeogenesis, pentose phosphate pathway, glycine, serine and threonine metabolism, plant hormone signal transduction, fructose and mannose metabolism, circadian rhythm–plant, photosynthesis-antenna proteins, cysteine and methionine metabolism, glyoxylate and dicarboxylate metabolism, one carbon pool by folate, nicotinate and nicotinamide metabolism, porphyrin and chlorophyll metabolism, sulfur metabolismvaline, proteasome, protein export, lysosome and peroxisome, were only found in Vp5; however, meiosis–yeast, galactose metabolism, and cell cycle-yeast were only found in vp5 (Table S7).
The top five signaling pathways in both genotypes all included spliceosome, RNA transport and the mRNA surveillance pathway. The three pathways are involved in mRNA synthesis and processing. In the present study, 27 identified proteins (K7V792, C0HIN5, B6U3A0, M1GS93, B4FUX9, K7TTT8, C0P8S9, B4FX58, Q8W149, C0PMQ0, B6SY05, B4FQ73, K7VZN2, B6T2W8, K7VKP3, P11143, M1H548, K7V1I2, B8A134, B4FK28, B8A305, C4J0D7, B4FX58, B6SGQ1, M1H548 K7V1I2 B4FKD1 C0PL59 B4FX58 B6T7C2 K7V0H) belonged to the three pathways, of which 26 proteins except K7VKP3 were regulated by ABA under osmotic stress. This was consistent with the signaling pathways related to protein synthesis, such as the biosynthesis of amino acids and ribosome, which had the second greatest number of proteins (B8A367, C0HHU2, B6TS38, C0HIV2, B4FRM3, B4FWX5, C0PKN2, B6TPG2, B4FCE7, O04014, B4FCK4, B4FWI0), and only B4FWI0 was not regulated by ABA under osmotic stress. Moreover, the signaling pathways related to photosynthesis, such as carbon fixation in photosynthetic organisms and photosynthesis, had the third greatest number of proteins (B7ZYP6, B6TS38, B4FRM3, B4FZ38, P04711, B4FQ59, C0PNN7, P24993, B4FAW3, B6T9S5, P05022), and all were regulated by ABA under osmotic stress. These results indicated that the three pathways related to mRNA synthesis, protein synthesis and photosynthesis played a vital role in ABA enhancing maize endurance to osmotic stress.
Discussion
ABA governs many aspects of plant physiology, and the induced reversible phosphorylation of proteins is an important regulator of ABA signaling23. The degree of specificity and redundancy among these factors is hotly debated. Previously, there had been no comprehensive survey of phosphorylation sites regulated by ABA in maize exposed to osmotic stress. We have performed a comparative, global analysis of ABA effects on maize protein phosphorylation under osmotic stress using the ABA mutant vp5 and wild-type Vp5 and identified known associations with ABA pathways and proteins that contain strongly induced phosphorylation sites.
ABA regulation of phosphorylation at transcriptional and post-translational levels
The interaction between specific transcription factors and their cis-elements causes the expression of stress inducible genes. Abiotic stress regulation also occurs at post-transcriptional and post-translational levels. The former involves pre-mRNA processing, which starts with intron splicing and exon joining24. In Arabidopsis, the phosphorylation state of the ABA-responsive element binding protein 3, the bZIP family transcription factor, GsZFP1, an ABA-responsive C2H2-type zinc finger protein, and the Topless transcription repressor was regulated by exogenous ABA treatment9,25. TFs which involved in ABA-mediated gene expression are increasingly recognized as promising candidates to create useful transgenic crops that can tolerate drought stress26. Our data showed that many ABA-regulated phosphoproteins were involved in a series of DNA/RNA-related processes and protein syntheses/degradation under osmotic stress (Table 2, Tables S1 and S2). ABA triggered the phosphorylation or dephosphorylation of 17 zinc finger protein transcription factors and other transcription factors, such as the gata transcription factor (B6TFI9), and 6 ribosomal proteins under osmotic stress. These results imply that phosphoproteins participating in gene transcription and translation may be major targets for regulatory phosphorylation during osmotic stress and that ABA-mediated transcriptional regulation plays a crucial role in many cellular processes of plants response to stress.
Ubiquitination and transporter–mediated ABA signaling under osmotic stress
Ubiquitination is a major modifier of signaling in all eukaryotes that causes the conjugation of ubiquitin to the lysine residues of acceptor proteins. The targeted protein is then subjected to degradation by the 26S proteasome, which is the major protein degradation system in eukaryotes and greatly influences plant growth and development by modulating the activity, localization, and stability of proteins under stress19. Many signaling details of ABA responses to abiotic stresses, such as salt and dehydration stress have been well elucidated in large studies using ABA mutants27,28. In salt and/or drought stress signaling, many E3 ligases mediate the stress response in ABA-dependent and ABA-independent pathways19. In this study, by using the maize ABA-deficient mutant vp5 and wild-type Vp5, the phosphorylation level of 8 phosphoproteins related to the ubiquitin/26S proteasome system was regulated by osmotic stress in an ABA-dependent way. These results indicate that the changes in expression abundance or modification state of the ubiquitin/26S proteasome complex protein subunits directly reflected the related-protein degradation, or not, during some biological processes and was necessary for many processes involved in plant responses to abiotic stresses.
Plasma membrane intrinsic proteins have been shown to be primary channels mediating water uptake in plant cells and their regulation via phosphorylation events29. In Arabidopsis, the phosphorylation level of plasma membrane intrinsic protein 2-A/B (PIP2-A/B), intrinsic protein 3, intrinsic protein 2–8 and intrinsic protein 2–4 was found to significantly decrease after ABA treatment up to 30 min7. Na+/H+ exchangers in the plasma membrane or vacuole have been recognized as one of the key regulatory mechanisms mediating cellular signaling by maintaining ion homeostasis. Previous studies indicated Na+/H+ exchangers can be up-regulated by salt, drought and heat stress30 and ABA treatment31. In the present study, the phosphorylation level of two aquaporins, one mitochondrial import inner membrane translocase subunit tim14 and one Na+/H+ antiporter involved in the signaling of ABA- regulated maize response to osmotic stress. Moreover, the phosphorylation states of other important transporters, such as probable sugar phosphate/phosphate translocator at3g17430-like (B6U937), vacuolar amino acid transporter 1-like (C0PEW7), hexose transporter (B6U6U2), zinc transporter (K7U2V8), solute carrier family facilitated glucose transporter member 8 (K7UMX4), abc transporter b family member 1-like (Q6UNK5), Na+/H+ antiporter (B4FZY1), K+ efflux antiporter 5-like (B6SP24) and sodium hydrogen exchanger 6-like (B4FS09) were regulated by ABA under osmotic stress. In summary, these results show that the phosphorylation and dephosphorylation of transporters might help the cell to maintain solute and ion stability, which might play an active role in ABA-regulated plant adaptation to osmotic stress.
Phosphorylation states of chloroplast proteins regulated by osmotic stress in an ABA-dependent way
Photosynthesis is a key process affected by environmental stress. The expression patterns of most photosynthesis-related proteins are complex under drought stress32. ABA signal transduction has been extensively studied, and numerous signaling components have been identified, including the chloroplast envelope-localized ABA receptor33, which provides stronger evidence that ABA plays an active role in regulating chloroplast response to stress. Previous reports have shown that PLASTID MOVEMENT IMPAIRED1 involved in blue-light-induced chloroplast movement, functions in ABA-response pathways and participates in the regulation of ABA accumulation during periods of water deficit at the seedling stage34. Other reports have also shown that some chloroplast proteins, such as the light-harvesting chlorophyll a/b binding proteins, ATP synthase, 2-cys peroxiredoxin BAS1, elongation factor 1a, phosphoglycerate kinase, protochlorophyllide reductase A, rubisco large chain, fructokinase-2, β-glucosidase, glyceraldehyde-3-phosphate dehydrogenase A, and phosphoribulokinase are involved in ABA signal transduction and play a positive role in maize response to ABA and drought stress32. In the present study, the phosphorylation level of 21 chloroplast proteins displayed significant differences between Vp5 and vp5 under osmotic stress. However, taking into account the fact that phosphorylation changes of chloroplasts proteins in white leaves of vp5 might be due to a carotenoid side-effect rather than a direct effect of ABA, so we supposed that the phosphorylation of the chloroplast proteins might by regulated by osmotic stress in an ABA-dependent or –independent way.
Phosphorylated protein kinases and phosphatases that are associated with signal perception and transduction
Protein phosphorylation, which plays a key role in most cellular activities, is a reversible process mediated by protein kinase and phosphatases. The interplay between phosphatases and kinases strictly controls biological processes, such as metabolism, transcription, cell cycle progression, differentiation, cytoskeletal arrangement and cell movement, apoptosis, intercellular communication, and immunological functions35,36. Recent studies have established a simple ABA signaling model consisting of three core components: PYR/PYL/RCAR receptors, 2C-type protein phosphatases, and SnRK2 protein kinases. This model highlights the importance of protein phosphorylation mediated by SnRK2. Other protein kinases, e.g., Ca2+ dependent protein kinase (CDPK) and mitogen-activated protein kinase (MAPK), have been identified as ABA signaling factors37,38. In fact, Arabidopsis snrk2.2/2.3/2.6 triple-mutant plants are nearly completely insensitive to ABA; most of the phosphoproteins regulated by ABA are triggered by SnRK2s-mediated phosphorylation. These proteins are involved in flowering time regulation, RNA and DNA binding, miRNA and epigenetic regulation, signal transduction, chloroplast function, and many other cellular processes39. Moreover, in maize, research results show that ZmPYL3 and ZmPP2C16 proteins are the most likely members of the receptors and the second components of the ABA signaling pathway, respectively4. In this study, 33 kinases and 1 phosphatase were identified under osmotic stress (Table 5). The phosphorylation level of CDPK-related protein kinase (B6SYP7), SNRK SAPK family protein kinase (B7ZXP0), and map kinase family protein isoform 1 (B8A0M9) was up-regulated by osmotic in an ABA-dependent way. Thus, our results did not only prove this model but also highlighted the importance of protein phosphorylation that is mediated by these kinases in maize responses to osmotic stress and ABA signaling.
Overall, protein phosphorylation/dephosphorylation is a central PTM in plant hormone signaling, which usually results in a functional change of the target protein by changing enzyme activity, cellular location, or association with other proteins. In this study, we have identified up to 3,484 unique phosphopeptides, corresponding to 2,863 phosphoproteins using Multiplex run iTRAQ-based quantitative proteomic and LCMS/MS methods. Differential phosphorylation and expression patterns of individual protein isoforms were detected in maize response to osmotic stress and ABA. Our results provide a comprehensive dataset of phosphopeptides and phosphorylation sites regulated by ABA in maize adaptation to osmotic stress. This will be helpful to elucidate the ABA-mediate mechanism of maize endurance to drought by triggering phosphorylation or dephosphorylation cascades.
Methods
Plant material and treatments
Maize mutant vp5 and wild-type Vp5 seedlings were used in this study. The vp5 mutant is deficient in ABA biosynthesis and has decreased amounts of ABA16. Homozygous recessive kernels (vp5/vp5) lack carotenoids, resulting in white endosperm and embryos, which is easily distinguishable from the yellow, wild type kernels (Vp5/-). Because the recessive mutation is lethal in the homozygous state, it is maintained as a heterozygote. Seeds of vp5 and Vp5 plants were obtained by selfing plants grown from heterozygous seeds (Maize Genetics Stock Center, Urbana, IL, USA).
Vp5 and vp5 seeds were germinated on moistened filter paper after being surface-sterilized for 10 min in 2% hypochlorite and then rinsed in distilled water. After germination for 2 d, both vp5 and Vp5 seedlings were cultured in Hogland’s nutrient solution in a light chamber (day 28 °C/night 22 °C, relative humidity 75%) under 400 μmol m−2 s−1 photosynthetically active radiations with a 14/10 h (day/night) cycle. After 2 weeks, the seedlings were subjected to osmotic stress by placing them in a −0.7 MPa PEG6000 solution for 8 h at 28 °C under relative humidity 40%. Control seedlings were maintained at 28 °C under relative humidity 75%. Subsequently, leaves of treated and untreated seedlings were sampled, immediately frozen in liquid N2 and stored at −80 °C until analysis. Three or five replicates were performed for each treatment.
Protein Extraction
Total proteins from the second new expand leaf of the maize seedlings were extracted according to the following procedure. Approximately 0.5 g of fresh leaves from each biological replicate were ground into a fine power in liquid N2 in a mortal and further ground in a 4 ml SDS buffer (30% sucrose, 2% SDS, 100 mM Tris-HCl, pH 8.0, 50 mM EDTA-Na2, 20 mM DTT) and 4 ml phenol (Tris-buffered, pH 8.0) in a 10 ml tube, followed by the addition of 1 mM phenylmethanesulfonyl fluoride (PMSF) and PhosSTOP Phosphatase Inhibitor Cocktail (one tablet/10 ml; Roche, Basel, Switzerland) to inhibit protease and phosphatase activity. The mixture was thoroughly vortexed for 30 s and the phenol phase was separated by centrifugation at 14,000 × g and 4 °C for 15 min. The upper phenol phase was pipetted into fresh 10 mL tubes and four fold volumes of cold methanol plus 100 mM ammonium acetate were added. After centrifugation at 14,000 × g and 4 °C for 15 min, the supernatant was carefully discarded and the precipitated proteins were washed twice with cold acetone. Finally, the protein mixtures were harvested by centrifugation. Using a 2-D Quant Kit (Amersham Bioscience, America) containing bovine serum albumin (BSA) (2 mg/mL) as the standard, we carried out the measurement of protein content. To enhance the quantitative accuracy, extracted proteins from every biological replicate were adjusted to the same concentration for the subsequent analysis7,39.
Protein digestion and iTRAQ labeling
Protein digestion was performed according to the FASP procedure40,38, and the resulting peptide mixture was labeled using the 4-plex iTRAQ reagent according to the manufacturer’s instructions (Applied Biosystems). Briefly, 200 μg of proteins for each sample were incorporated into 30 μl of STD buffer (4% SDS, 100 mM DTT, 150 mM Tris-HCl pH 8.0). The detergent DTT and other low-molecular-weight components were removed using UA buffer (8 M Urea, 150 mM Tris-HCl pH 8.0) by repeated ultrafiltration (Microcon units, 30 kD). Then, 100 μl of 0.05 M iodoacetamide in UA buffer was added to block reduced cysteine residues, and the samples were incubated for 20 min in darkness. The filters were washed with 100 μl of UA buffer three times and then washed twice with 100 μl of DS buffer (50 mM trimethylammonium bicarbonate at pH 8.5). Finally, the protein suspensions were digested with 2 μg of trypsin (Promega) in 40 μl of DS buffer overnight at 37 °C, and the resulting peptides were collected as a filtrate. The peptide content was estimated by UV light spectral density at 280 nm using an extinction coefficient of 1.1 of 0.1% solution that was calculated on the basis of the frequency of tryptophan and tyrosine in vertebrate proteins.
For labeling, each iTRAQ reagent was dissolved in 70 μl of ethanol and added to the respective peptide mixture. The samples, Vp5-control, Vp5-OS (osmotic stress), vp5-control, and vp5-OS, were multiplexed and vacuum dried. Three independent biological experiments were performed.
Peptide fractionation with strong cation exchange (SCX) chromatography for proteomic analysis
iTRAQ labeled peptides were fractionated by SCX chromatography using the AKTA Purifier system (GE Healthcare). The dried peptide mixture was reconstituted and acidified with 2 ml buffer A (10 mM KH2PO4 in 25% of ACN, pH 2.7) and loaded onto a PolySULFOETHYL 4.6 × 100 mm column (5 μm, 200 Å, PolyLC Inc, Maryland, USA.). The peptides were eluted at a flow rate of 1 ml/min with a gradient of 0–10% buffer B (500 mM KCl, 10 mM KH2PO4 in 25% of ACN, pH 2.7) for 2 min, 10–20% buffer B for 25 min, 20–45% buffer B for 5 min, and 50–100% buffer B for 5 min. The elution was monitored by absorbance at 214 nm, and fractions were collected every 1 min. The collected fractions (about 30 fractions) were finally combined into 10 pools and desalted on C18 Cartridges (Empore™ SPE Cartridges C18 (standard density), bed I.D. 7 mm, volume 3 ml, Sigma). Each fraction was concentrated by vacuum centrifugation and reconstituted in 40 μl of 0.1% (v/v) trifluoroacetic acid. All samples were stored at −80 °C until LC-MS/MS analysis.
Liquid chromatography (LC)—electrospray ionization (ESI) tandem MS (MS/MS) analysis by Q Exactive for proteomic analysis
Experiments were performed on a Q Exactive mass spectrometer that was coupled to Easy nLC (Proxeon Biosystems, now Thermo Fisher Scientific). 10 μl of each fraction was injected for nanoLC-MS/MS analysis. The peptide mixture (5 μg) was loaded onto a the C18-reversed phase column (Thermo Scientific Easy Column, 10 cm long, 75 μm inner diameter, 3μm resin) in buffer A (0.1% Formic acid) and separated with a linear gradient of buffer B (80% acetonitrile and 0.1% Formic acid) at a flow rate of 250 nl/min controlled by IntelliFlow technology over 140 min. MS data was acquired using a data-dependent top10 method dynamically choosing the most abundant precursor ions from the survey scan (300–1800 m/z) for HCD fragmentation. Determination of the target value is based on predictive automatic gain control (pAGC). Dynamic exclusion duration was 60 s. Survey scans were acquired at a resolution of 70,000 at m/z 200 and resolution for HCD spectra was set to 17,500 at m/z 200. Normalized collision energy was 30 eV and the underfill ratio, which specifies the minimum percentage of the target value likely to be reached at maximum fill time, was defined as 0.1%. The instrument was run with peptide recognition mode enabled.
Phosphopeptide enrichment by TiO2 beads
The labeled peptides were mixed, concentrated by a vacuum concentrator and resuspended in 500 μL of loading buffer (2% glutamic acid/65% ACN/ 2% TFA). Then, TiO2 beads were added and then agitated for 40 min. The centrifugation was performed for 1 min at 5000 g, resulting in the first beads. The supernatant from the first centrifugation was mixed with additional TiO2 beads, resulting in the second beads that were collected as before. Both bead groups were combined and washed three times with 50 μL of washing buffer I (30% ACN/3%TFA) and then washed three times with 50 μL of washing buffer II (80% ACN/0.3% TFA) to remove the remaining non-adsorbed material. Finally, the phosphopeptides were eluted with 50 μL of elution buffer (40% ACN/15% NH4OH)41, followed by lyophilization and MS analysis.
MS/MS for phosphoproteomics analysis
Five μl of the phosphopeptide solution mixed with 15 μl of 0.1% (v/v) trifluoroacetic acid and then 10 μl of the solution mixture was injected into a Q Exactive MS (Thermo Scientific) equipped with Easy nLC (Proxeon Biosystems, now Thermo Scientific) for nanoLC-MS/MS analysis. The peptide mixture was loaded onto a C18-reversed phase column (15 cm long, 75 μm inner diameter, RP-C18 3 μm, packed in-house) in buffer A (0.1% Formic acid) and separated with a linear gradient of buffer B (80% acetonitrile and 0.1% Formic acid) at a flow rate of 250 nL/min controlled by IntelliFlow technology over 240 min. The peptides were eluted with a gradient of 0%–60% buffer B from 0 min to 200 min, 60% to 100% buffer B from 200 min to 216 min, 100% buffer B from 216 min to 240 min.
For MS analysis, peptides were analyzed in positive ion mode. MS spectra were acquired using a data-dependent top10 method dynamically choosing the most abundant precursor ions from the survey scan (300–1800 m/z) for HCD fragmentation. Determination of the target value is based on predictive Automatic Gain Control (pAGC). Dynamic exclusion duration was 40 s. Survey scans were acquired at a resolution of 70,000 at m/z 200, and the resolution for the HCD spectra was set to 17,500 at m/z 200. Normalized collision energy was 27 eV, and the under fill ratio, which specifies the minimum percentage of the target value likely to be reached at maximum fill time, was defined as 0.1%. The instrument was run with peptide recognition mode enabled.
Data analysis
MS/MS spectra were searched using Mascot 2.2 (Matrix Science) embedded in Proteome Discoverer 1.4 against the uniprot_Zea_mays_87227_20150504.fasta (87227 sequences, download May 4th, 2015) and the decoy database. The parameters used in Mascot searches for normal peptides were as follows: Peptide mass tolerance: 20 ppm, MS/MS tolerance: 0.1 Da, Enzyme: Trypsin, max missed cleavage: 2, Fixed modification: Carbamidomethyl (C), iTRAQ4plex(K), iTRAQ4plex(N-term), Variable modification: Oxidation (M), FDR ≤0.01. The protein and peptide probabilities were set at 50 and 60%, respectively. Only proteins with at least two unique peptides with a Mascot score of at least 25 and detected in at least two replicates were further used. For peptides after phosphopeptide enrichment, the following options were used. Peptide mass tolerance: 20 ppm, MS/MS tolerance: 0.1 Da, enzyme: trypsin, max missed cleavage: 2, fixed modification: Carbamidomethyl (C), iTRAQ4plex (K), iTRAQ4plex (N-term), variable modification: Oxidation (M), phosphorylation (S/T/Y). The score threshold for peptide identification was set at a 5% or 1% false discovery rate (FDR), and the PhosphoRS site probabilities estimate the probability (0–100%) of each phosphorylation site. The PhosphoRS site probabilities above 75 percent indicate that a site is truly phosphorylated42.
For each replicate of both proteomics and phosphoproteomics, iTRAQ ratios between osmotic stress (OS) and controls for each run were converted to z-scores to normalize the data.
Bioinformatics
The molecular functions of the identified proteins were classified according to their gene ontology annotations and their biological functions. The subcellular localization of the unique proteins identified in this study was predicted using the publicly available program WolfPsort ( http://wolfpsort.org). Protein-protein interaction networks were analyzed using the publicly available program STRING ( http://string-db.org/). STRING is a database of known and predicted protein-protein interactions. The interactions include direct (physical) and indirect (functional) associations, and they are derived from four sources: the genomic context, high-throughput experiments, coexpression and previous knowledge. STRING quantitatively integrates the interaction data from these sources for a large number of organisms and, where applicable, transfers information between these organisms.
Motif-X online software ( http://motif-x.med.harvard.edu/motif-x.html) was used to find phosphorylation site motifs in the identified maize proteins and to predict the specificity of these motifs based on the identified phosphopeptide sequences. The parameters were set to peptide length = 21, occurrence = 5, and statistical significance for p-values of less than 0.000001.
NABA assay
Maize leaves (0.5–1.0 g) were ground in liquid N2 with a mortar, extracted with 2 ml of ice-cold 80% methanol containing 1 mM butylated hydroxytoluene to prevent oxidation, and then stored overnight at 4 °C. The extracts were centrifuged at 12000 g for 15 min at 4 °C. The pellets were extracted once and stored at 4 °C for 1 h. The two resulting supernatants were combined and passed through a C18 Sep-Pak cartridge (Waters, Milford, MA, USA). The efflux was collected and dried in N2. The residues were then dissolved in 10 mM phosphate buffer solution (pH 7.4) and concentrations of ABA were determined in enzyme-linked immunosorbent assay (ELISA)32. Statistical analyses of the physiological measurements were conducted using independent Student’s t-tests with SPSS statistics software (version 17.0).
Statistical analysis
The phosphoproteins, phosphopeptides and ABA assays were the mean of three replicates. The means were compared by a one-way analysis of variance and Duncan’s multiple range test at a 5% level of significance. FDR attained by Benjamini-Hochberg method were used to adjust p-values (correction for multiple comparisons). The significance of difference between Vp5 and vp5 were compared by T-Test analysis at a 5% level.
Additional Information
How to cite this article: Hu, X. et al. Quantitative iTRAQ-based proteomic analysis of phosphoproteins and ABA-regulated phosphoproteins in maize leaves under osmotic stress. Sci. Rep. 5, 15626; doi: 10.1038/srep15626 (2015).
Supplementary Material
Acknowledgments
This research was supported by the Program for Science & Technology Innovation Talents (13HASTIT001 to XLH), the National Natural Science Foundation of China (Grant 31171470 to XLH) and Scientific Innovation Talent for Henan Province (Grant X to XLH), the Program for Innovative Research Team (in Science and Technology) (15IRTSTHN015 to WW) in University of Henan Province, and the Plan for Scientific Innovation Talent of Henan Province (144200510012 to WW) for financial support.
Footnotes
Author Contributions X.L.H. and W.W. conceived the study and participated in its design. N.N.L. and L.J.W. carried out the experiments. C.Q.L., T.X.L., C.H.L. and L.Z. contributed samples. X.L.H. and W.W. analyzed the data and drafted the manuscript.
References
- Harrison M. T., Tardieu F., Dong Z., Messina C. D. & Hammer G. L. Characterizing drought stress and trait influence on maize yield under current and future conditions. Global Change Biol. 20, 867–878 (2014). [DOI] [PubMed] [Google Scholar]
- Yang S., Vanderbeld B., Wan J. & Huang Y. Narrowing down the targets: towards successful genetic engineering of drought-tolerant crops. Mol. Plant. 3, 469–490 (2010). [DOI] [PubMed] [Google Scholar]
- Ribaut J. M., Betran J., Monneveux P. & Setter T. Drought tolerance in maize. In Bennetzen J. & Hake Sarah C. (Eds.), Handbook of Maize: Its Biology books,311–344 (Springer, 2009). [Google Scholar]
- Wang Y. G. et al. Interaction between abscisic acid receptor PYL3 and protein phosphatase type 2C in response to ABA signaling in maize. Gene 549, 179–185 (2014). [DOI] [PubMed] [Google Scholar]
- Jiang S. et al. A maize calcium-dependent protein kinase gene, ZmCPK4, positively regulated abscisic acid signaling and enhanced drought stress tolerance in transgenic Arabidopsis. Plant Physiol. Bioch. 71, 112–120 (2013). [DOI] [PubMed] [Google Scholar]
- Bonhomme L., Valot B., Tardieu F. & Zivy M. Phosphoproteome dynamics upon changes in plant water status reveal early events associated with rapid growth adjustment in maize leaves. Mol. Cell. Proteomics 11, 957–72 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. et al. Phosphoproteome analysis reveals new drought response and defense mechanisms of seedling leaves in bread wheat (Triticum aestivum L.). J. Proteomics 109, 290–308 (2014). [DOI] [PubMed] [Google Scholar]
- Shinozaki K. & Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223 (2000). [PubMed] [Google Scholar]
- Kline K. G., Barrett-Wilt G. A. & Sussman M. R. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. USA 107, 15986–15991 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Hoehenwarter W. & Weckwerth W. Comparative analysis of phytohormone-responsive phosphoproteins in Arabidopsis thaliana using TiO2-phosphopeptide enrichment and mass accuracy precursor alignment. Plant J. 63, 1–17 (2010). [DOI] [PubMed] [Google Scholar]
- Isner J. C., Nuhse T. & Maathuis F. J. The cyclic nucleotide cGMP is involved in plant hormone signalling and alters phosphorylation of Arabidopsis thaliana root proteins. J. Exp. Bot. 63, 3199–3205 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T. et al. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6, rs8 (2013). [DOI] [PubMed] [Google Scholar]
- He H. & Li J. Proteomic analysis of phosphoproteins regulated by abscisic acid in rice leaves. Biochem. Bioph. Res. Co. 371, 883–888 (2008). [DOI] [PubMed] [Google Scholar]
- Zhang Z. et al. Proteomic and phosphoproteomic determination of ABA's effects on grain-filling of Oryza sativa L. inferior spikelets. Plant Sci. 185–186, 259–273 (2012). [DOI] [PubMed] [Google Scholar]
- Nguyen T. H. et al. Quantitative phosphoproteomic analysis of soybean root hairs inoculated with Bradyrhizobium japonicum. Mol. Cell. Proteomics 11, 1140–1155 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud C. S., Wang J. & Sussex I. M. Control of in vitro growth of viviparous embryo mutants of maize by abscisic acid. Dev. Genet. 1, 325–330 (1980). [Google Scholar]
- Hable W. E., Oishi K. K. & Schumaker K. S. Viviparous-5 encodes phytoene desaturase, an enzyme essential for abscisic acid (ABA) accumulation and seed development in maize. Mol. Gen. Genet. 257, 167–176 (1998). [DOI] [PubMed] [Google Scholar]
- Tuteja N. Abscisic acid and abiotic stress signaling. Plant Sign. Behav. 2, 135–138 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. Y., Li J. H., Liu H. H., Chong K. & Xu Y. Y. Roles of ubiquitination-mediated protein degradation in plantresponses to abiotic stresses. Environ. Exp. Bot. 114, 92–103 (2015). [Google Scholar]
- Neill S. J., Horgan R., & Parry A. D. The carotenoid and abscisic acid content of viviparous kernels and seedlings of Zea mays L. Planta 169, 87–96 (1986). [DOI] [PubMed] [Google Scholar]
- Maluf M. P., Saab I. N., Wurtzel E. T. & Sachs M. M. The viviparous12 maize mutant is deficient in abscisic acid, carotenoids, and chlorophyll synthesis. J. Exp. Bot. 48, 1259–1268 (1997). [Google Scholar]
- Anderson I. C. & Robertson D. S. Role of carotenoids in protecting chlorophyll from photodestruction. Plant Physiol. 35, 531–534 (1960). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T. et al. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 51, 1821–1839 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra R. et al. Abscisic acid and abiotic stress tolerance–different tiers of regulation. J. Plant Physiol. 171, 486–496 (2014). [DOI] [PubMed] [Google Scholar]
- Luo X. et al. Over-expression of GsZFP1, an ABA-responsive C2H2-type zinc finger protein lacking a QALGGH motif, reduces ABA sensitivity and decreases stomata size. J. Plant Physiol. 169, 1192–1202 (2012). [DOI] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Shinozaki K. & Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–25 (2011). [DOI] [PubMed] [Google Scholar]
- Kim S. Y. Recent advances in ABA signaling. J. Plant Biol. 50, 117–121 (2007). [Google Scholar]
- Osakabe Y., Yamaguchi-Shinozaki K., Shinozaki K. & Tran L. S. ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 202, 35–49 (2014). [DOI] [PubMed] [Google Scholar]
- Horie T. et al. Mechanisms of water transport mediated by PIP aquaporins and their regulation via phosphorylation events under salinity stress in barley roots. Plant Cell Physiol. 52, 663–675 (2011). [DOI] [PubMed] [Google Scholar]
- Baltierra F., Castillo M., Gamboa M. C., Rothhammer M. & Krauskopf E. Molecular characterization of a novel Na+/H+ antiporter cDNA from Eucalyptus globulus. Biochem. Biophys. Res. Commun. 430, 535–40 (2013). [DOI] [PubMed] [Google Scholar]
- Song S. Y., Chen Y., Chen J., Dai X. Y. & Zhang W. H. Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234, 331–45 (2011). [DOI] [PubMed] [Google Scholar]
- Hu X. et al. Abscisic acid refines the synthesis of chloroplast proteins in maize (Zea mays) in response to drought and light. PloS one 7, e49500 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y. Y. et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443, 823–826 (2006). [DOI] [PubMed] [Google Scholar]
- Rojas-Pierce M., Whippo C. W., Davis P. A., Hangarter R. A. & Springer P. S. PLASTID MOVEMENT IMPAIRED1 mediates ABA sensitivity during germination and implicates ABA in light-mediated Chloroplast movements. Plant Physiol. Bioch. 83, 185–193 (2014). [DOI] [PubMed] [Google Scholar]
- Johnson L. N. The regulation of protein phosphorylation. Biochem. Soc. Trans. 37, 627–641 (2009). [DOI] [PubMed] [Google Scholar]
- Pjechová M., Hernychova L., Tomašec P., Wilkinson G. W. & Vojtěšek B. Ananlysis of phosphoproteins and signalling pathways by quantitative proteomics. Klin Onkol. 27, S116–120 (2014). [DOI] [PubMed] [Google Scholar]
- Danquah A., Zelicourt A. D., Colcombet J. & Hirt H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 32, 40–52 (2014). [DOI] [PubMed] [Google Scholar]
- Umezaw T., Takahashi F. & Shinozaki K. Chapter two–phosphorylation networks in the abscisic acid signaling pathway. Enzymes 35, 27–56 (2014). [DOI] [PubMed] [Google Scholar]
- Wang P. C. et al. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 110, 11205–11210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. L., Xiong E. H., Wang W., Scali M. & Cresti M. Universal sample preparation method integrating trichloroacetic acid/acetone precipitation with phenol extraction for crop proteomic analysis. Nat. Protoc. 9, 362–374 (2014). [DOI] [PubMed] [Google Scholar]
- Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P. & Jorgensen T. J. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873–886 (2005). [DOI] [PubMed] [Google Scholar]
- Taus T. et al. Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.