Abstract
A novel bacterial type III secretion effector, VopV, from the enteric pathogen Vibrio parahaemolyticus has been identified as a key factor in pathogenicity due to its interaction with cytoskeletal actin. One of the repeat units in the long repetitive region of VopV, named VopVrep1, functions as an actin-binding module. Despite its importance in pathogenesis, the manner in which the effector binds to actin and the subsequent effects on actin dynamics remain unclear. Here, we report the molecular basis of the VopVrep1/actin interaction. VopVrep1 exists as an unstructured protein in solution but potently and specifically binds filamentous actin (F-actin) and not globular actin (G-actin). The F-actin/VopVrep1 complex was directly visualized at 9.6-Å resolution using electron cryomicroscopy (cryoEM) and helical image reconstitution. The density map revealed the binding site of VopVrep1 at the interface between two actin strands, which is close to the binding site of the bicyclic heptapeptide toxin phalloidin. Consistent with this observation, VopVrep1 alone prevented the depolymerization of F-actin. Overall, VopVrep1 demonstrated unique characteristics in comparison to known actin-binding proteins, but was relatively similar to phalloidin. The phalloidin-like behavior, targeting the interstrand region of actin filaments to stabilize the filament structure, likely contributes to the pathogenicity of V. parahaemolyticus.
Vibrio parahaemolyticus is a food-borne pathogen that causes acute gastroenteritis in humans1. The type III secretion system (T3SS), which is the molecular machinery that delivers bacterial effectors into the cytoplasm of infected host cells, is essential for the pathogenicity of this bacterium2,3,4. Recent reports have shown that one of the type III secretion (T3S) effectors, VopV, plays an important role in the enterotoxicity of V. parahaemolyticus5,6. Among the three repeat units within the long repeat region in VopV, VopVrep1 has been identified as the key unit for F-actin binding and VopV enterotoxicity (Fig. 1(a); UniProt: Q87GF9)5,7.
Figure 1. VopVrep1 was an unstructured protein that demonstrated high affinity for F-actin.
(a) The repeating structure of VopV was composed of three patterns of repeat sequences. The VopVrep1 sequence used in this study is shown below, and the residues are color-coded by their hydrophobicity (charged: green; polar: cyan; glycine: yellow; and hydrophobic: gray) to emphasize the high proportion of hydrophilic residues. (b) Circular dichroism analysis of VopVrep1 revealed a typical random coil spectrum with a minimum peak at approximately 196 nm. (c), (d) The binding affinities of VopVrep1 and phalloidin with cytoskeletal actin were analyzed using isothermal titration calorimetry. The dissociation constant (Kd) and binding stoichiometry (N) obtained by curve fitting analysis are shown in the inset. The binding pattern of VopVrep1 (c, left) was similar to that of phalloidin (d, left). After the addition of VopVrep1, phalloidin did not bind to actin (c, right), as was observed for the addition of VopVrep1 to phalloidin-saturated actin (d, right).
The actin cytoskeleton is involved in various cellular processes, and its behavior is strictly controlled by numerous actin-binding proteins and their regulators8,9,10.To date, a lot of actin-binding proteins have been identified; these proteins affect cellular actin dynamics in various ways, such as sequestering actin monomers, severing or capping the filaments, forming networks or bundles of filaments, and supporting nucleation of filament formation10. These different interactions are mediated by individual actin-binding motifs and correspond to the functional diversity of the actin-protein interactions11,12. A number of pathogenic bacteria abuse the versatile ability of actin cytoskeleton to promote its infection13,14. Effectors take control of actin cytoskeleton by directly interacting with the actin molecule as actin-binding proteins or by indirectly interfering with the actin architecture via its regulation system14,15.
Despite our increasing knowledge of actin-binding proteins including bacterial effectors, VopVrep1 has no sequence homology with known actin-binding proteins5 and thus, researchers are not sure how VopVrep1 interacts with actin or how VopVrep1 affects actin dynamics. Here, we analyzed the molecular characteristics of VopVrep1 as a novel actin-binding protein. Direct visualization of the interaction between F-actin and VopVrep1 using electron cryomicroscopy (cryoEM) revealed the unique binding mechanism of VopVrep1 and demonstrated that VopVrep1 had a stabilizing effect on F-actin.
Results
VopVrep1 exists as an unstructured protein in solution, but potently and specifically binds F-actin
First, we overexpressed a construct containing VopVrep1 (residues 361–428) and purified the resulting recombinant protein. The circular dichroism spectrum of VopVrep1 was clearly indicative of a random coil pattern (Fig. 1(b)), demonstrating that the recombinant VopVrep1 did not form a stable fold in solution. Indeed, the high proportion of hydrophilic, polar, and glycine residues in VopVrep1 (~67.6%, Fig. 1(a)) was consistent with the typical characteristics reported for intrinsically unstructured proteins16. To assess whether the recombinant protein bound to actin, the binding affinity of VopVrep1 to cytoskeletal actin was determined using isothermal titration calorimetry (ITC). Without a stable fold in solution, VopVrep1 bound to actin with high affinity (Kd = 54.4 nM) at a binding stoichiometry of N = 1.06, which is comparable to the well-known F-actin-binding toxin phalloidin17,18, which has a binding affinity of Kd = 36.5 nM and a binding stoichiometry of N = 1.16 in our experiment (Fig. 1(c,d) and Supplementary Table S1). These values indicate that their stoichiometries are both 1.0. The affinity of VopVrep1 was as high as or higher than that of other well-studied actin-binding proteins/peptides, such as profilin (Kd = 0.1–5 μM)19, ADF/cofilin (Kd = 0.1–10 μM)19, Wiskott-Aldrich syndrome protein homology domain 2 peptides (Kd = 0.052–27 μM)20, and the nebulin repeat fragment (Kd = 220 μM)21, suggesting that VopVrep1 was well adapted for actin binding. Because actin exists in a dynamic equilibrium between F-actin and G-actin, we next examined the binding preference of VopVrep1 to each form of actin. An increase in the G-actin/F-actin ratio induced by the F-actin depolymerizing toxin latrunculin22 caused a decrease in the binding stoichiometry of VopVrep1 to N = 0.28 (Supplementary Fig. S1 and Table S1), indicating that VopVrep1 preferentially bound F-actin. Conversely, an increase in the F-actin/G-actin ratio using the F-actin-stabilizing toxin phalloidin18,23,24 unexpectedly resulted in complete inhibition of the actin/VopVrep1 interaction (Fig. 1(c)). One possible explanation for these observations is that the bindings of phalloidin and VopVrep1 are mutualy exclusive. The observation that VopVrep1 conversely prevented the binding of phalloidin to actin (Fig. 1(d)) supported this possibility.
Direct visualization of the VopVrep1/F-actin interaction by cryoEM
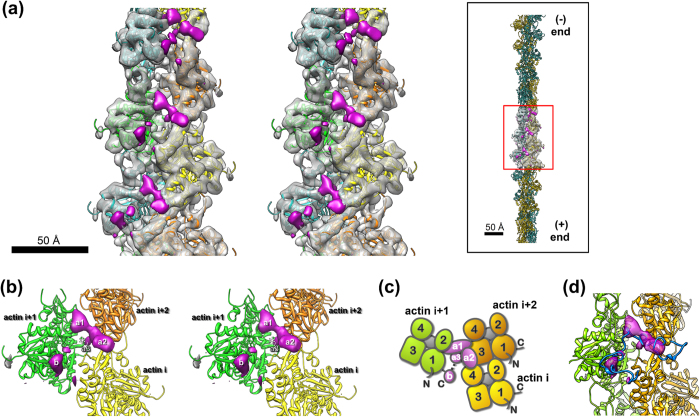
We then used cryoEM image analysis to elucidate the mode of VopVrep1 binding to cytoskeletal F-actin at the molecular level; the unstructured nature of VopVrep1 and the filamentous structure of actin made it nearly impossible to use other approaches, such as single-crystal X-ray crystallography or nuclear magnetic resonance (NMR) spectroscopy. Recent advances in the cryoEM method applied to F-actin25 have enabled the direct visualization of the human cytoskeletal actin/VopVrep1 complex at 9.6-Å resolution (Fig. 2(a)). The densities corresponding to the actin molecules were readily assigned to the atomic model of skeletal muscle F-actin without significant conformational change from the previously determined structure (Fig. 2(a) and Supplementary Fig. S2). One apparent exception was observed at the N-terminal region, which oriented in different direction from the model (Supplementary Fig. S2). One possible cause is the difference in sequence between cytoskeletal actin (β- and γ-actins, in this study) and skeletal muscle actin (α-actin, Fujii, et al. 2010)25. We should also note that the N-terminal structure of actin is also not observed in recent high resolution studies of F-actin composed of α-actin from striated muscle26,27. In addition to the density of the actin filament, densities corresponding to VopVrep1 were clearly observed along the interface of the two parallel actin strands in a repetitive manner (Fig. 2(a,b), colored magenta). One major density (sites a1-a2) was located at the cleft between the two actin strands (Fig. 2(a,b)), and an additional density (site b) was found immediately adjacent to the C-terminal region of one actin molecule (actin i + 1; Fig. 2(b,c)). Therefore, the VopVrep1 densities simultaneously contacted three actin molecules. The major elongated density of sites a1-a2 of VopVrep1 occupied the interface surrounded by the three actin subunits. There is a density extended from the connecting region between densities a1-a2 into the filament interior (named a3 in Fig. 2(b,c)), which is located deep inside the interstrand cleft. At the a1 site, the density was contacted domains 1 and 2 of an actin i + 1 molecule (Fig. 2(b,c)). The density extended toward the opposite actin strand, reaching domain 3 of actin i + 2 and further stretched to domain 4 of another actin i molecule running across the longitudinal interface (Fig. 2(b,c)). The additional density at site b was located near the C-terminal region at domain 1 of actin i + 1 around the extension of the interface of two actin strands (Fig. 2(b,c)). Because of the limited resolution of the map, we used the atomic model of actin (PDB ID: 3MFP)25 to predict the actin residues involved in the interaction with VopVrep1 (Supplementary Fig. S3 and Table S2). The density a3, which is located in the interior of actin filament, is close to the binding site of phalloidin18,24 (indicated by asterisk in Fig. 2(b)). Because the binding stoichiometry was determined by ITC analysis, the densities described above (i.e., sites a1, a2, a3, and b) are likely to correspond to one VopVrep1 molecule. The size of VopVrep1, 68 residues, was more than sufficient to explain the observed densities, suggesting that some regions connecting the densities were still unstructured and therefore not visible in the map. At a lower threshold, weak densities extending and connecting the major densities are observed (Supplementary Fig. S4). The volume ratio of the extra densities to the actin density is 7.7% and 13.0% at the high and low threshold, respectively. As a coarse estimation, the mass for VopVrep1 includes 29 to 48 residues (calculated from the residue number of actin), meaning that part of 68 residues of VopVrep1 is invisible in this reconstitution due to disorder. This concept was consistent with its natively unstructured properties, as demonstrated in a simulated model (Fig. 2(d)).
Figure 2. Structure of the F-actin/VopVrep1 complex visualized by cryoEM image analysis at 9.6-Å resolution.
(a) A magnified view of the F-actin/VopVrep1 complex in stereo. The long axis of the actin filament is vertical, as shown in the inset. The density map is represented as a transparent envelope and is filled by actin models that are alternately colored cyan/green and orange/yellow according to the two actin strands. The extra regions that likely correspond to VopVrep1 are colored magenta. For clarity, the extra density derived from the N-terminal region of actin was colored dark gray. Three repetitive clusters are observed in this view. (b) Detailed stereo view of the VopVrep1 densities on the F-actin model. Three distinguishable densities, a1, a2, and b, were in close contact with three actin units i, i + 1, and i + 2. (c) A schematic representation of the relative spatial arrangement of the VopVrep1 densities (magenta) and surrounding actin domains numbered 1-4 with ATP/ADP (gray). (d) A binding model of VopVrep1 (blue) in an elongated form, fitted to the observed densities. The chains outside the densities would be unstructured.
Bound VopVrep1 stabilizes the F-actin protecting from depolymerization.
In the density map, VopVrep1 occupied a key position surrounded by three actin monomers that reinforced both the lateral and longitudinal interactions within the filament, suggesting the ability of VopVrep1 to stabilize F-actin. In addition to the observed binding mode, the specific, high-affinity recognition of F-actin results in F-actin stabilization19. As expected, the F-actin-stabilizing activity of VopVrep1 was demonstrated by the observation that VopVrep1 strongly protected the actin filament from depolymerization in low-salt conditions (Fig. 3) to the same extent as phalloidin. These results suggested that VopVrep1 played a role in anchoring cytoskeletal actin and simultaneously interfered with actin dynamics by stabilizing F-actin.
Figure 3. F-actin protection from depolymerization by VopVrep1.
(a) F-actin stabilization was examined using a high-speed cosedimentation assay. A mixture of depolymerizing F-actin in the absence or presence of VopVrep1, latrunculin B, and phalloidin was ultracentrifuged after 0, 2, or 4 h incubation. The pellet fractions (F-actin) were separated using SDS-PAGE, and the gels were stained with Coomassie blue. (b) The stabilized F-actin shown in (a) was analyzed using densitometry. The relative F-actin ratio was calculated as the amount of F-actin pelleted at each time point divided by that of zero time point amount.
Discussion
In this study, a series of biophysical analyses of VopVrep1 revealed its peculiar characteristics as an actin-binding protein. VopVrep1 bound across two actin strands; therefore, the binding likely depended on the higher-order structure of F-actin. Such a binding mode may account for the specificity of F-actin observed in the ITC analysis. In particular, the binding site of VopVrep1 did not include the hydrophobic cleft between domains 1 and 3 of the actin molecule (Fig. 2(c)), which is recognized by the vast majority of actin-binding proteins12. The interstrand region of the actin filament is known to be used as a binding site of the muscle actin-stabilizing factor nebulin28 and the Salmonella-derived T3S effector SipA29. The actin-binding domain of SipA (SipA446–684, 238 residues) is significantly larger than VopVrep1, and the domain itself possesses a stable fold30, in contrast to the intrinsically unstructured VopVrep1. Electron microscope image analysis of the F-actin/SipA446-684 complex has revealed that the binding site of SipA446-684 involves a more extended region in addition to the filament interface29. Conversely, although the actin-binding fragment of nebulin is relatively small (~35 residues) and intrinsically unstructured in solution21, as is VopVrep1, the affinity of the nebulin fragment to F-actin is significantly weaker than that of VopVrep1 (nebulin, Kd = 200–500 μM vs. VopVrep1, Kd = 54.4 nM)21. The actin-binding motif of nebulin (SDxxYK)21 was not present in the VopVrep1 sequence. Therefore, our data demonstrated that VopVrep1 possessed novel characteristics as an actin-binding protein/peptide, targeting the interstrand interface of F-actin with significantly high affinity through its intrinsic unstructured nature.
F-actin stabilization by VopVrep1 appears to be at least partially responsible for the observed anomaly of cytoskeletal actin in V. parahaemolyticus-infected small intestinal epithelial cells31, supported by the observation that the actin cytoskeleton in microvilli, which are the target of V. parahaemolyticus infection, is in dynamic equilibrium32 and is thus susceptible to perturbation by extrinsic factors. The homeostasis of F- and G-actin is a well-known target for pathogenic effectors15. With regard to V. parahaemolyticus, VopL is known to possess F-actin nucleation activity33,34; thus, VopV and VopL likely facilitate the excessive formation of F-actin in a concerted manner.
VopVrep1 is a small, flexible unit that is a stand-alone module properly tailored for binding and stabilizing F-actin. Full-length VopV contains five homologous VopVrep1 units separated by two other repetitive units (Fig. 1(a))5, which simultaneously but individually act on F-actin, resulting in even higher affinity or leading to actin bundling, as observed in VopV-affected cells5. Its behavior as an actin-binding unit and the phalloidin-like potency of VopVrep1 suggest that this sequence may have versatile applications in cell biology studies, particular when considering the broad use of phalloidin as an imaging tool to monitor F-actin. In combination with genetic engineering techniques, VopVrep1 could be employed as an inducible factor to target F-actin and cause F-actin stabilization or actin bundling (using a tandem arrangement with an appropriate spacer). Such applications would be useful for the analysis of actin dynamics, F-actin structure35, and programmed cell death via F-actin abnormalities in living cells in tissues36.
Advanced techniques in cryoEM image analysis revealed the interaction of VopVrep1 with F-actin, which is the primary molecular step in the pathogenicity of V. parahaemolyticus. Unexpectedly, VopV was found to target a similar site as phalloidin and demonstrated a unique binding mode for F-actin that differed from that of known actin-binding proteins. The unstructured nature of VopVrep1 may contribute to its ability to enter the elongated binding site at the interstrand region of F-actin while maintaining its affinity and specificity. This novel and well-organized binding mode for VopVrep1 provides a glimpse into the unusual association between the pathogenic effector of V. parahaemolyticus and the fundamental human cytoskeleton.
Methods
For details of the MD calculation for making simulated model of VopVrep1 see Supplemental Methods.
Overexpression and purification of VopVrep1
The VopVrep1 sequence was subcloned into a pET-32b vector (Novagen) containing a modified multiple cloning site to express a fusion protein composed of thioredoxin, His6, and a thrombin protease site followed by VopVrep1. BL21(DE3) cells (Novagen) were transformed with this vector and cultivated in LB medium supplemented with 100 mg/mL ampicillin at 37 °C. Protein expression was induced with 0.2 mM IPTG, and cells were further incubated at 20 °C for 6 h. The cells were harvested by centrifugation, flash frozen in liquid N2, and thawed on ice. The cells were then resuspended in lysis buffer (20 mM Tris-HCl, pH 7.5, and 500 mM NaCl), treated with 0.2% Triton X-100 for 20 min on ice, and disrupted by ultrasonication. The cell debris was then removed by centrifugation. The supernatant was applied to a Ni-charged HiTrap Chelating column (GE Healthcare) equilibrated with lysis buffer. The target protein was eluted with lysis buffer containing 300 mM imidazole. The fusion tags were removed through thrombin cleavage (Sigma-Aldrich) for 18 h at 25 °C during dialysis in Tris-HCl (pH 7.5) and 5 mM NaCl. The VopVrep1 protein was further applied to a HiTrap SP column (GE Healthcare) and eluted using a 5–300 mM NaCl gradient. After concentration using an Amicon ultra concentrator (Millipore), VopVrep1 was further purified by size-exclusion chromatography using a HiLoad 26/60 Superdex 75 prep grade column (GE Healthcare). The purity was verified using SDS-PAGE analysis. Purified VopVrep1 was stored at –80 °C prior to use.
Circular dichroism (CD) spectroscopy
VopVrep1 was dialyzed in 20 mM Tris-HCl (pH 7.2). The CD spectrum of 125 μM VopVrep1 was acquired using a JASCO J-720 spectropolarimeter in a 1-mm quartz cell at 20 °C.
Isothermal titration calorimetry (ITC)
VopVrep1 and cytoskeletal actin were dialyzed in 30 mM Tris-HCl (pH 7.5) and 75 mM KCl. The titration experiment was conducted using a MicroCal iTC200 calorimeter. The measurement was performed in general actin buffer (20 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM MgCl2, and 0.2 mM ATP) at 25 °C. It was assumed that almost all the actin molecules formed F-actin in this solution. Phalloidin was dissolved in DMSO and used at a concentration of 110 μM in general actin buffer containing 1% DMSO. The sample cell was filled with 8.8 μM actin, and 110 μM VopVrep1 or phalloidin was loaded in the titration syringe. To account for the diluted DMSO in the case of its use as a drug, 1% DMSO was added to the actin solution. An inhibition assay was performed using a sequential titration experiment in which the actin solution that was titrated by the first binding sample, that is, VopVrep1 or phalloidin, was used for the second titration assay with phalloidin or VopVrep1, respectively. The obtained data were analyzed using Origin 7 Software (OriginLab Corporation) and fitted using a one-site binding model.
CryoEM
Image collection
The F-actin/VopVrep1 sample retrieved after ITC titration (12.5 μM VopVrep1, 7.8 μM actin) was used for cryoEM image data collection and analysis. The experimental procedures for cryoEM were identical to those of a previously published report25. Briefly, the sample solution was loaded on Quantifoil holey carbon molybdenum grid (R0.6/1.0, Quantifoil) and plunge-frozen into liquid ethane using Vitrobot (FEI). Image collection was performed at temperature of 50–60 K using a JEOL JEM3200FSC electron microscope equipped with a liquid helium-cooled specimen stage, an Ω-type energy filter, and a field-emission electron gun operated at 200 kV. In total, 445 images were recorded on a CCD camera (TemCam-F415MP, TVIPS) at magnification of around 170,100× and a defocus range of 1.0–2.4 μm. The image pixel size corresponded to 1.677 Å.
Image analysis
Image analysis using the iterative helical real-space reconstitution method (IHRSR)37 was performed using EMAN 1.9338 and SPIDER 15.0639. The defocus and astigmatism of each image were determined using CTFFIND40. In total, 39,910 image segments of F-actin were boxed, and ~121,000 actin molecules were included. Images were corrected for phase and amplitude contrast transfer function (CTF). The images were aligned and cross-correlated to a series of reference projection images, and a 3D image was reconstituted by back projection. The 3D images were improved by imposing helical symmetry parameters in an iterative manner and refined until convergence.
Model analysis
The rabbit α-skeletal actin structure (PDB ID: 3MFP) was fitted to the density, and symmetrical actin units were generated using UCSF Chimera software41. In the iterative process of image analysis, the helical symmetry and axial repeat distance were refined and converged to a subunit rotation of –166.8° and an axial repeat of 27.6 Å, which resulted in a helical symmetry of approximately 41 subunits/19 turns (~2.158). This value of the F-actin/VopVrep1 complex corresponded to a helical structure slightly overtwisted from that of native F-actin, which was reported in a study of rabbit skeletal muscle actin with approximately 67 subunits/31 turns (~2.161)25 and was relatively close to the phalloidin-bound form of F-actin with approximately 69 subunits/32 turns (~2.156)18.
F-actin stabilization assay
F-actin stabilization was performed using the Actin Binding Protein Biochem Kit (Cytoskeleton Inc.). Briefly, F-actin was pre-assembled from purified human platelet nonmuscle actin (1 mg/mL) at room temperature for 1 h in F-buffer containing 0.2 mM CaCl2, 50 mM KCl, 2 mM MgCl2, and 1 mM ATP in 5 mM Tris-HCl (pH 8.0). The assembled F-actin was then depolymerized by a 1:5 dilution with G-buffer (0.2 mM CaCl2 in 5 mM Tris-HCl, pH 8.0) in the absence or presence of VopVrep1, latrunculin, and phalloidin under the indicated final concentrations. After 0, 2, or 4 h of incubation on ice, the samples were ultracentrifuged (120,000 × g for 2 h) at 4 °C. The pellets were analyzed using SDS-PAGE, and the gels were stained with Coomassie blue. The density of the pelleted F-actin in individual fractions was determined using ImageJ software (National Institutes of Health).
Additional Information
How to cite this article: Nishimura, M. et al. A repeat unit of Vibrio diarrheal T3S effector subverts cytoskeletal actin homeostasis via binding to interstrand region of actin filaments. Sci. Rep. 5, 10870; doi: 10.1038/srep10870 (2015).
Supplementary Material
Acknowledgments
This work was supported by JSPS KAKENHI Grant Number 25711010 to T.F.; 21390013 to Y.K.; 21227006 and 25000013 to K.N.
Footnotes
Author Contributions M.N., T.F., H.H., S.N., K.N. and T.I. designed the experiments and wrote the paper. M.N., H.H., H.I., D.M. and T.K. performed all of the biochemical and biophysical experiments. T.F. and F.M. performed all of the cryoelectron microscopy experiments. T.K., T.O., Y.K., S.N., K.N. and T.I. planned and supervised the project.
References
- Park K. S. et al. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72, 6659–6665 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama T. et al. Identification and characterization of VopT, a novel ADP-ribosyltransferase effector protein secreted via the Vibrio parahaemolyticus type III secretion system 2. Cell Microbiol. 9, 2598–2609 (2007). [DOI] [PubMed] [Google Scholar]
- Burdette D. L., Seemann J. & Orth K. Vibrio VopQ induces PI3-kinase-independent autophagy and antagonizes phagocytosis. Mol. Microbiol. 73, 639–649 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyoshi H., Kodama T., Iida T. & Honda T. Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infect. Immun. 78, 1772–1780 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyoshi H. et al. VopV, an F-actin-binding type III secretion effector, is required for Vibrio parahaemolyticus-induced enterotoxicity. Cell Host Microbe 10, 401–409 (2011). [DOI] [PubMed] [Google Scholar]
- Zhou X. et al. Remodeling of the intestinal brush border underlies adhesion and virulence of an enteric pathogen. Mbio. 5, e01639–e01614 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino K. et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361, 743–749 (2003). [DOI] [PubMed] [Google Scholar]
- Wear M., Schafer D. A. & Cooper J. A. Actin dynamics: assembly and disassembly of actin networks. Curr. Biol. 10, R891–R895 (2000). [DOI] [PubMed] [Google Scholar]
- Pollard T. D. & Cooper J. A. Actin, a central player in cell shape and movement. Science 326, 1208–1212 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M. D. & Kwiatkowski A. V. Control of actin dynamics by allosteric regulation of actin binding proteins. Int. Rev. Cell Mol. Biol. 303, 1–25 (2013). [DOI] [PubMed] [Google Scholar]
- dos Remedios C. G. et al. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol. Rev. 83, 433–473 (2003). [DOI] [PubMed] [Google Scholar]
- Dominguez R. Actin-binding proteins--a unifying hypothesis. Trends Biochem. Sci. 29, 572–578 (2004). [DOI] [PubMed] [Google Scholar]
- Bhavsar A. P., Guttman J. A. & Finlay B. B. Manipulation of host-cell pathways by bacterial pathogens. Nature 449, 827–834 (2007). [DOI] [PubMed] [Google Scholar]
- Navarro-Garcia F., Serapio-Palacios A., Ugalde-Silva P., Tapia-Pastrana G. & Chavez-Duenas L. Actin cytoskeleton manipulation by effector proteins secreted by diarrheagenic Escherichia coli pathotypes. Biomed Res. Int. 2013, 374395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktories K., Lang A. E., Schwan C. & Mannherz H. G. Actin as target for modification by bacterial protein toxins. FEBS J. 278, 4526–4543 (2011). [DOI] [PubMed] [Google Scholar]
- Dyson H. J. & Wright P. E. Intrinsically unstructured proteins and their functions. Nat Rev. Mol. Cell Biol. 6, 197–208 (2005). [DOI] [PubMed] [Google Scholar]
- Dancker P., Low I., Hasselbach W. & Wieland T. Interaction of actin with phalloidin: polymerization and stabilization of F-actin. Biochim. Biophys. Acta 400, 407–414 (1975). [DOI] [PubMed] [Google Scholar]
- Oda T., Namba K. & Maeda Y. Position and orientation of phalloidin in F-actin determined by X-ray fiber diffraction analysis. Biophys. J. 88, 2727–2736 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A. Actin binding proteins that change extent and rate of actin monomer-polymer distribution by different mechanisms. Mol. Cell Biochem. 190, 67–74 (1999). [PubMed] [Google Scholar]
- Chereau D. et al. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. U S A 102, 16644–16649 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfuhl M., Winder S. J. & Pastore A. Nebulin, a helical actin binding protein. EMBO J. 13, 1782–1789 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector I., Shochet N. R., Blasberger D. & Kashman Y. Latrunculins--novel marine macrolides that disrupt microfilament organization and affect cell growth: I. Comparison with cytochalasin D. Cell Motil. Cytoskeleton 13, 127–144 (1989). [DOI] [PubMed] [Google Scholar]
- Wieland T. Modification of actins by phallotoxins. Naturwissenschaften 64, 303–309 (1977). [DOI] [PubMed] [Google Scholar]
- Steinmetz M. O. et al. Evaluating atomic models of F-actin with an undecagold-tagged phalloidin derivative. J. Mol. Biol. 276, 1–6 (1998). [DOI] [PubMed] [Google Scholar]
- Fujii T., Iwane A. H., Yanagida T. & Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature 467, 724–728 (2010). [DOI] [PubMed] [Google Scholar]
- von der Ecken J. et al. Structure of the F-actin-tropomyosin complex. Nature 519 114–117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin V. E., Orlova A., Vos M. R., Schröder G. F. & Egelman E. H. Near-atomic resolution for one state of F-actin. Structure 23 173–182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukoyanova N. et al. Each actin subunit has three nebulin binding sites: implications for steric blocking. Curr. Biol. 12, 383–388 (2002). [DOI] [PubMed] [Google Scholar]
- Galkin V. E. et al. The bacterial protein SipA polymerizes G-actin and mimics muscle nebulin. Nat Struct. Biol. 9, 518–521 (2002). [DOI] [PubMed] [Google Scholar]
- Mitra K., Zhou D. & Galan J. E. Biophysical characterization of SipA, an actin-binding protein from Salmonella enterica. FEBS Lett. 482, 81–84 (2000). [DOI] [PubMed] [Google Scholar]
- Ritchie J. M. et al. Inflammation and disintegration of intestinal villi in an experimental model for Vibrio parahaemolyticus-induced diarrhea. PLoS Pathog 8, e1002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waharte F., Brown C. M., Coscoy S., Coudrier E. & Amblard F. A two-photon FRAP analysis of the cytoskeleton dynamics in the microvilli of intestinal cells. Biophys. J. 88, 1467–1478 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namgoong S. et al. Mechanism of actin filament nucleation by Vibrio VopL and implications for tandem W domain nucleation. Nat Struct. Mol. Biol. 18, 1060–1067 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Cheng H. C., Brautigam C. A., Tomchick D. R. & Rosen M. K. Mechanism of actin filament nucleation by the bacterial effector VopL. Nat Struct. Mol. Biol. 18, 1068–1074 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small J., Rottner K., Hahne P. & Anderson K. I. Visualising the actin cytoskeleton. Microsc. Res. Tech. 47, 3–17 (1999). [DOI] [PubMed] [Google Scholar]
- Posey S. C. & Bierer B. E. Actin stabilization by jasplakinolide enhances apoptosis induced by cytokine deprivation. J. Biol. Chem. 274, 4259–4265 (1999). [DOI] [PubMed] [Google Scholar]
- Egelman E. H. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 (2000). [DOI] [PubMed] [Google Scholar]
- Ludtke S. J., Baldwin P. R. & Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999). [DOI] [PubMed] [Google Scholar]
- Frank J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996). [DOI] [PubMed] [Google Scholar]
- Mindell J. A. & Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003). [DOI] [PubMed] [Google Scholar]
- Pettersen E. F. et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.