Abstract
The lignite biodegradation procedure to produce water-soluble humic materials (WSHM) with a Penicillium stain was established by previous studies in our laboratory. This study researched the effects of WSHM on the growth of Bradyrhizobium liaoningense CCBAU05525 and its nodulation on soybean. Results showed that WSHM enhanced the cell density of CCBAU05525 in culture, and increased the nodule number, nodule fresh weight and nitrogenase activity of the inoculated soybean plants. Then the chemical compounds of WSHM were analyzed and flavonoid analogues were identified in WSHM through tetramethyl ammonium hydroxide (TMAH)-py-GC/MS analysis. Protein expression profiles and nod gene expression of CCBAU05525 in response to WSHM or genistein were compared to illustrate the working mechanism of WSHM. The differently expressed proteins in response to WSHM were involved in nitrogen and carbon metabolism, nucleic acid metabolism, signaling, energy production and some transmembrane transports. WSHM was found more effective than genistein in inducing the nod gene expression. These results demonstrated that WSHM stimulated cell metabolism and nutrient transport, which resulted in increased cell density of CCBAU05525 and prepared the bacteria for better bacteroid development. Furthermore, WSHM had similar but superior functions to flavone in inducing nod gene and nitrogen fixation related proteins expression in CCBAU05525.
Soybean is an important legume that forms symbiotic root nodules with Bradyrhizobium liaoningense, bacteria that reduce atmospheric nitrogen to ammonium, which is a form of nitrogen available for plant uptake. To initiate this symbiosis process, flavonoids secreted by soybean would activate the transcription of nod genes of rhizobia, which encode lipochitooligosaccharide signal molecules, also termed Nod factors. After being recognized by the epidermal cells of the host, these Nod factors induce root hair curling and cell division, and result in root nodule formation. Rhizobia entrapped in the curled root hair could enter the roots through infection threads1.
Agricultural systems could acquire approximately 40 million tonnes of nitrogen each year through symbiotic nitrogen fixation between legumes and rhizobia2. Symbiotic nitrogen fixation is a gift from the nature which is beneficial to increasing crop yields and nutrient-use efficiency meanwhile reducing N fertilizer application. Thus, practices to stimulate nodulation, such as inoculation of soybean with rhizobia, are of great significance to sustainable agriculture; and great efforts have been made to improve the efficiency of symbiotic nitrogen fixation. Previously, the application of whey (40 tonne ha−1) and inoculation with rhizobia enhanced the nodule numbers, crop growth and yield of chickpea under field conditions, which was speculated that these might be resulted from the positive effects of organic and inorganic nutrients in the whey on the plant development and the stimulation effects of whey on rhizobia activity3. Supplement of pea seeds with Nod factors secreted by Rhizobium leguminosarum bv. viciae strain GR09 before planting resulted in a significant increase in nodule numbers, nitrogenase activity and pea yield under greenhouse and field conditions4,5. Treatment with flavonoids such as luteolin or increased flavonoids production in the plant through genetic engineering enhanced both the number of nodules and N2 fixation of alfalfa6. Flavonoids were the most potent compounds for inducing nod gene expression, while other non-flavonoid compounds such as betaines and xanthones were also able to induce nod genes in rhizobia. However, these molecules could not move in the soil freely because of their positive charges7,8. Furthermore, the functioning mechanism of whey was unclear, and it was economically infeasible to apply these compounds in the field to increase legume yield. Thus, it is of value to search for other compounds that could enhance the symbiotic nitrogen fixation efficiency and be applicable under field conditions to improve legume yield.
Humic substances are polyelectrolytic macromolecular compounds originated from the chemical and biological degradation of plants and animal residues and microbial cells9, and they play an important role in the global carbon and nitrogen cycling. Humic materials were reported to have some positive effects on different organisms, specifically through accelerating seed germination10, improving rhizome growth by stimulating ATPase activity or functioning as auxin-like phytohormone to promote the plant growth11,12, and promoting the efficient utilization of nutrients by plants13,14. However, the effects of humic materials on symbiotic nitrogen fixation had been rarely investigated except in the reports of Tan and Tantiwiramanond15 that soil humic and fulvic acids extracted by 0.1 M NaOH could increase the plant dry weight and nodule mass but not the nodule number of soybeans and peanuts under greenhouse conditions. In addition, Til’ba and Sinegovskaya16 proved in a field experiment that seed coating with sodium humate, rhizobia, ammonium molybdate and leaf spraying with sodium humate increased the nodule number and nitrogen-fixing efficiency of soybean, and the yield hence increased by 22% compared with the control. Even though, the exact role of humic materials in symbiotic nitrogen fixation and its mechanism were still unclear. Recently, the production of water-soluble humic acids (WSHM) by the biodegradation of lignite using a fungal strain Penicillium sp. P6 or an alkali-producing bacterial community was reported17,18. Furthermore, it was found previously that these humic acids could protect against the disruption of bacterial diversity that ensued from the application of urea to the soil19, and the soybean productivity was increased by 12.6% ~ 26.3% when this extract of humic acids was applied in fields (our unpublished data). In order to investigate the effects of WSHM on the growth and nodulation of rhizobia, we performed this study using Bradyrhizobium liaoningense CCBAU05525 as a model, and the functioning mechanisms of WSHM were estimated through proteomics for the first time.
Results
The effects of WSHM on nodulation of soybean under greenhouse conditions
The symbiotic features of soybean inoculated with B. liaoningense CCBAU05525 in responding to the supplement of WSHM under greenhouse conditions were shown in Table 1. The number of nodules per plant increased by 19.4% and 30.5% respectively in the 300 and 500 mg L−1 WSHM treatments, but no significant increase was observed in the 1000 mg L−1 WSHM treatment (Table 1). Moreover, the nodule fresh weight in the 500 mg L−1 WSHM treatment increased by 36.0% compared with control. Furthermore, the nitrogenase activity per plant significantly increased (from 15.1% to 30.2%) in the 300, 500 and 1000 mg L−1 WSHM treatments compared with control. Based on these observations, WSHM was concluded to have positive effects on the nodulation of soybean inoculated with B. liaoningense CCBAU05525.
Table 1. Effects of water-soluble humic material (WSHM) treatments on the nodulation of soybean in greenhousea.
| WSHM concentration (mg L−1) |
||||
|---|---|---|---|---|
| Nodulation | 0 (control) | 300 | 500 | 1000 |
| Number of nodules | 36.0±1.00b | 43.0±4.35a | 47.0±3.00a | 38.0±5.00b |
| Fresh weight of nodules (g) | 0.436±0.0610b | 0.547±0.0330ab | 0.593±0.113a | 0.554±0.0950ab |
| Nitrogenase activity (nmol C2H4 h−1) | 22.0±2.93c | 28.6±1.30a | 25.3±0.320b | 26.9±0.810ab |
aValues are given as mean ± standard deviation of triplicate samples for nodule analysis; values with different alphabets in the same line are significantly (P < 0.05) different from each other, according to LSD test.
The effects of WSHM on the Growth of B. liaoningense CCBAU05525
As shown in Fig. 1, the cell density of B. liaoningense CCBAU05525 in YM broth was significantly increased by WSHM treatments at the concentrations of 100, 200, 500, 1000 and 1500 mg L−1. The highest cell density (7.98 ± 0.56 × 109 cfu mL−1) on the fourth day of incubation observed at the WSHM concentrations of 500 mg L−1 was almost 8 times greater than that of control. The growth curves of B. liaoningense CCBAU05525 (available as Supplementary Fig. S1) showed that CCBAU05525 in the 500 mg L−1 WSHM solutions increased by 9.9 × 107 mL−1. Whereas, the cell density of B. liaoningense CCBAU05525 in YM broth culture supplied with WSHM increased to 8 × 1010 mL−1, which was 8 times greater than that of B. liaoningense CCBAU05525 in YM broth culture without WSHM.
Figure 1. Effects of WSHM on the growth of Bradyrhizobium liaoningense CCBAU05525 in YM medium.
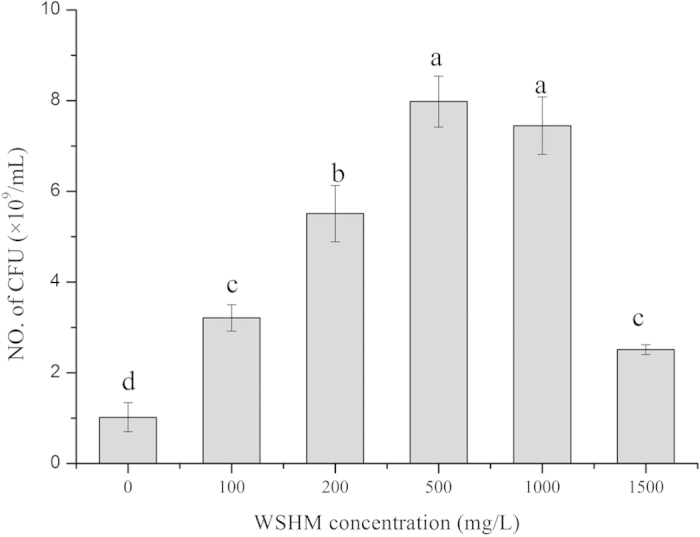
The data were obtained on the fourth day of incubation with shaking 140 rpm at 28 °C. Values given are mean ± standard deviation of triplicate samples; Bars with different letters are significantly different (P < 0.05) from each other, according to LSD test.
Identification of the chemical compounds in WSHM
By TMAH-py-GC/MS analysis, 68 compounds were identified in WSHM which fell into 4 classes: aromatics, aliphatic, nitrogen compounds and other compounds (Table 2). The aromatics that included methoxy and hydroxy benzenes, aromatic ketones and phenolic acids were the main components of the WSHM. Fatty acid methyl esters and dicarboxylic acid dimethyl esters were detected in WSHM. Sterol isomers and diterpene resin acids contents, originated mainly from lipids were also one of the important components in WSHM. Besides, N-containing compounds such as pyrroles, imidazoles, pyrimidines, piperidines and indoles were identified. The indole-like compounds identified in WSHM, indicated its potential in promoting plant growth. In addition, it was noteworthy that the 3-Hydroxy-7-methoxy-3-phenyl-4-chromanone in the WSHM had similar structure of flavonoids (C6-C3-C6). Thus, WSHM might act as flavonoid in the symbiosis between soybean and rhizobia.
Table 2. Chemical structures in water-soluble humic material detected by TMAH-py-GC/MS.
| Peak No. | Retention time (min) | Compound | Area% | Formula | Mr | CAS |
|---|---|---|---|---|---|---|
| 1 | 2.300 | 2-methylfuran | 0.69 | C5H6O | 82 | 534-22-5 |
| 2 | 2.395 | Acrylic acid methyl ester | 2.44 | C4H6O2 | 86 | 96-33-3 |
| 3 | 2.535 | Methyl propionate | 1.87 | C4H8O2 | 88 | 554-12-1 |
| 4 | 3.600 | Methacrylic acid methyl ester | 0.87 | C5H8O2 | 100 | 80-62-6 |
| 5 | 4.210 | 2-chloroethyl methyl ether | 2.25 | C3H7ClO | 94 | 627-42-9 |
| 6 | 4.415 | (Dimethylamino) acetonitrile | 4.43 | C4H8N2 | 84 | 926-64-7 |
| 7 | 4.475 | Pyrrole | 2.45 | C4H5N | 67 | 109-97-7 |
| 8 | 4.710 | Toluene | 4.06 | C7H8 | 92 | 108-88-3 |
| 9 | 5.145 | 2-Hydroxy-2-methyl-6-hepten-3-one | 0.54 | C8H14O2 | 142 | 996-61-2 |
| 10 | 5.575 | 3-(Octylamino) propanenitrile | 0.58 | C11H22N2 | 182 | 29504-89-0 |
| 11 | 5.690 | Hexamethylcyclotrisiloxane | 0.15 | C6H8O3Si3 | 222 | 541-5-9 |
| 12 | 5.845 | N,N-dimethyl-2-phosphinoethanamine | 1.07 | C4H12NP | 105 | 161944-90-7 |
| 13 | 6.175 | 2-cyclopenten-1-one | 0.82 | C5H6O | 82 | 930-30-3 |
| 14 | 6.755 | Ethylbenzene | 0.52 | C8H10 | 106 | 100-41-4 |
| 15 | 6.815 | 2,5-Dimethylpyrrole | 0.38 | C6H9N | 95 | 625-84-3 |
| 16 | 6.960 | Dimethylbenzene | 1.17 | C8H10 | 106 | 95-47-6 |
| 17 | 7.435 | 1,3,5,7-Cyclooctatetraene | 1.09 | C8H8 | 108 | 629-20-9 |
| 18 | 7.960 | Methoxybenzene | 0.68 | C7H8O | 108 | 100-66-3 |
| 19 | 8.985 | 6-(Hydroxy-phenyl-methyl)-2,2-dimethyl-cyclohexanone | 1.23 | C15H20O2 | 232 | CID557652 (NCBI no.) |
| 20 | 9.205 | Oktamethylcyklotetrasiloxan | 0.39 | C8H24O4Si4 | 296 | 556-67-2 |
| 21 | 9.555 | Phenol | 3.13 | C6H6O | 94 | 108-95-2 |
| 22 | 9.655 | 3-(Octylamino)Propanenitrile | 1.03 | C11H22N2 | 182 | 29504-89-0 |
| 23 | 10.095 | (+)-dimethyl 2,3-O-benzylidene-D-tartrate | 2.27 | C13H14O6 | 266 | 38270-70-1 |
| 24 | 10.275 | Butanedioic acid, dimethyl ester | 2.86 | C6H10O4 | 146 | 106-65-0 |
| 25 | 10.860 | 2-Methylphenol | 0.77 | C7H8O | 108 | 95-48-7 |
| 26 | 11.010 | 2-Propenoic acid, 2-benzoylamino-3-phenyl,ethyl ester | 0.57 | C18H17NO3 | 295 | 32089-78-4 |
| 27 | 11.080 | 3-Ethyl-2-hydroxy-2-cyclopenten-1-one | 0.35 | C7H10O2 | 126 | 21835-1-8 |
| 28 | 11.350 | 3,4,4-Trimethyl-2-cyclopenten-1-one | 3.25 | C8H12O | 124 | 30434-65-2 |
| 29 | 11.455 | N-methylsuccinimide | 4.4 | C5H7NO2 | 113 | 1121-7-9 |
| 30 | 12.300 | o-Dimethoxybenzene | 0.94 | C8H10O2 | 138 | 91-16-7 |
| 31 | 13.550 | 2,4-Imidazolidinedione,3,5,5-trimethyl- | 0.8 | C6H10N2O2 | 142 | 6345-19-3 |
| 32 | 13.600 | S-(2,5-Dihydroxyphenyl) cyclohexylthiocarbamate | 1.1 | C13H17NO3S | 267 | 345259-5-4 |
| 33 | 14.465 | N-methylindolea | 0.65 | C9H9N | 131 | 603-76-9 |
| 34 | 14.575 | Dimethyl(2E,4E)-2,4-hexadienedioate | 0.52 | C8H10O4 | 170 | 1733-37-5 |
| 35 | 14.940 | Benzeneacetonitrile | 1.09 | C8H7N | 117 | 140-29-4 |
| 36 | 15.430 | Piperidine, 1,1-methylenebis- | 3.44 | C11H22N2 | 182 | 880-9-1 |
| 37 | 15.855 | 1,2,4-Trimethoxybenzene | 0.33 | C9H12O3 | 168 | 135-77-3 |
| 38 | 15.930 | 1,3-Dimethylindolea | 0.35 | C10H11N | 145 | 875-30-9 |
| 39 | 16.030 | Benzoic acid, 4-methoxy-, methyl ester | 0.5 | C9H10O3 | 166 | 121-98-2 |
| 40 | 16.160 | Methyl 5-oxo-2-pyrrolidinecarboxylate | 2.55 | C6H9NO3 | 143 | 54571-66-3 |
| 41 | 16.790 | N-methylphthalimide | 0.73 | C9H7NO2 | 161 | 550-44-7 |
| 42 | 17.060 | 2,4(1H,3H)-Pyrimidinedione | 2.01 | C7H10N2O2 | 154 | 4401-71-2 |
| 43 | 17.440 | 3,4,5-Trimethoxybenzylamine | 0.56 | C10H15NO3 | 197 | 18638-99-8 |
| 44 | 17.955 | Dodecanoic acid, methyl ester | 1.370 | C13H26O2 | 214 | 111-82-0 |
| 45 | 18.485 | (3Z)-1-Methyl-1H-indole-2,3-dione 3-hydrazonea | 0.490 | C9H9N3O | 175 | 3265-23-4 |
| 46 | 18.755 | Methyl 3,5-dimethoxybenzoate | 3.140 | C10H12O4 | 196 | 2150-37-0 |
| 47 | 18.870 | Benzoic acid, 3,4-dimethoxy-,methyl ester | 0.920 | C10H12O4 | 196 | 2150-38-1 |
| 48 | 20.425 | 3,4,5-Trimethoxybenzoic acid, methyl ester | 1.950 | C11H14O5 | 226 | 1916-7-0 |
| 49 | 20.480 | Methyl tetradecanoate | 0.910 | C15H30O2 | 242 | 124-10-7 |
| 50 | 21.320 | Pentadecanoic acid, methyl eater | 0.510 | C16H32O2 | 256 | 7132-64-1 |
| 51 | 22.115 | 1H-purine-2,6-dione, 3,7-dihydro-1,3,7-trimethyl | 0.530 | C8H10N4O2 | 194 | 58-8-2 |
| 52 | 22.350 | Hexadecanoic acid, methyl ester | 0.660 | C17H34O2 | 270 | 112-39-0 |
| 53 | 22.760 | Hexadecanoic acid, methyl ester | 3.430 | C17H34O2 | 270 | 112-39-0 |
| 54 | 23.510 | 4-[2-(4-Hydroxy-phenyl)-viny]-benzoic acid | 0.850 | C15H12O3 | 240 | 152027-61-7 |
| 55 | 23.680 | Cyclopropaneoctanoic acid, 2-hexyl-,methyl ester | 0.450 | C18H34O2 | 282 | 10152-61-1 |
| 56 | 23.745 | 3-Hydroxy-7-methoxy-3-phenyl-4-chromanoneb | 1.140 | C16H14O4 | 270 | 18380-57-9 |
| 57 | 24.590 | 9-Octadecenoic acid (Z)-,methyl ester | 1.240 | C19H36O2 | 296 | 112-62-9 |
| 58 | 24.645 | 9-Octadecenoic acid (Z)-,methyl ester | 0.710 | C19H36O2 | 296 | 112-62-9 |
| 59 | 24.830 | Octadecanoic acid, methyl ester | 1.340 | C19H38O2 | 298 | 112-61-8 |
| 60 | 25.690 | 10-Nonadecenoic acid, methyl ester | 0.460 | C20H38O2 | 310 | 56599-83-8 |
| 61 | 26.085 | 2-Phenanthrenamine, 9,10-dihydro-7-nitro- | 8.500 | C14H12N2O2 | 240 | 18264-82-9 |
| 62 | 26.760 | 1-Phenanthrenecarboxylic acid, 7-ethenyl-1,2,3,4,4a,4b,5,6,7,8,10,10a -dodecahydro-1,4a,7-trimethyl-,methyl ester, (1R,4aR,4bS,7S,10aR)- | 1.930 | C21H32O2 | 316 | 1686-62-0 |
| 63 | 26.860 | 6-Methyl-4-phenyl-3-cyanopyridine-2(1H)-thione | 1.980 | C13H10N2S | 226 | 78564-23-5 |
| 64 | 26.920 | Methyl 5-[2-(3-furyl)ethyl]-1,4a-dimethyl-6-methylidene-decalin-1-carboxylate | 2.070 | C21H30O3 | 330 | 10267-15-9 |
| 65 | 27.060 | Methyl dehydroabietyate | 2.010 | C21H30O2 | 314 | 1235-74-1 |
| 66 | 27.540 | Methyl abietate | 0.490 | C21H32O2 | 316 | 127-25-3 |
| 67 | 29.145 | 7-Oxodehydroabietic acid, methyl ester | 0.600 | C21H28O3 | 328 | 110936-78-2 |
| 68 | 30.175 | Tetracosanoic acid, methyl ester | 0.440 | C25H50O2 | 382 | 2442-49-1 |
aIndole-like Compounds.
bFlavonoid-like compound.
Proteome analysis of B. liaoningense in response to WSHM and genistein
In the assay of proteomics, a total of 1,900 protein spots were identified by 2-D electrophoresis across all samples, 15 of which were up-regulated and 15 down-regulated by WSHM treatment. 7 and 9 proteins were up- and down-regulated respectively by genistein , all of which were among the 30 WSHM-affected proteins (Fig. 2). Among these 30 proteins, 27 were identified based on their similarity with reported sequences, which were divided into three groups: 1) protein expression was increased or decreased by both WSHM and genistein treatments; 2) protein expression was increased by genistein but decreased or not significantly altered by WSHM; 3) protein expression was increased by WSHM but decreased or not significantly altered by genistein (Table 3).
Figure 2. 2-D Electrophoresis of the proteins of B. liaoningense CCBAU05525 following different treatments.
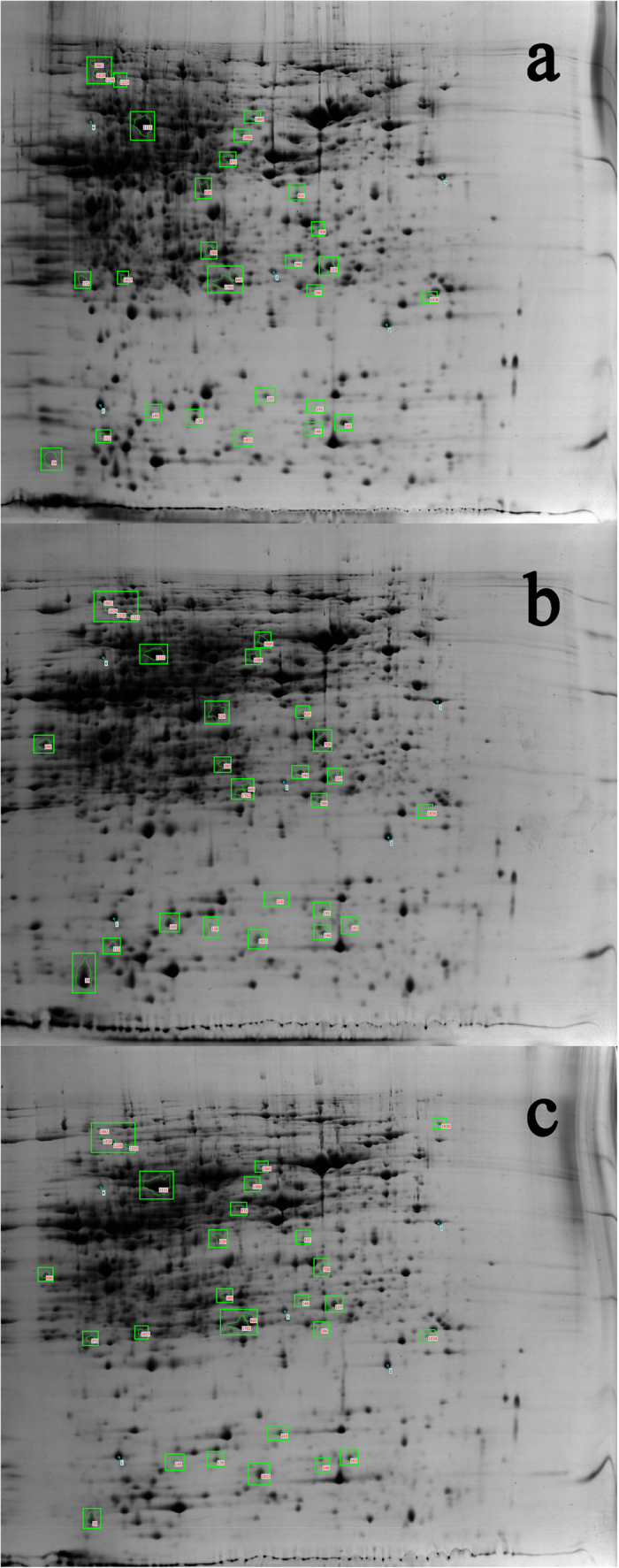
a: control; b: treatment with water-soluble humic materials; c: treatment with genistein.
Table 3. Proteins identified in B. liaoningense CCBAU 05525 treated by genistein and water-soluble humic acid.
| Category | Spot No. | Protein description* | Protein source | NCBI Access NO. | Participation in (function) | WSHM vs. control | Genistein vs. control |
|---|---|---|---|---|---|---|---|
| 1 | 39 | nitrogen regulatory protein PII | Bradyrhizobium japonicum USDA 110 | gi|27375723 | regulate glutamine synthetase | 5.58 | 1.41 |
| 168 | iron-regulated outer membrane protein | Mannheimia haemolytica BOVINE | gi|261491674 | adapt to the iron-restricted environment inside the host | 3.35 | 1.15 | |
| 395 | isopropylmalate isomerase small subunit | Bradyrhizobium japonicum USDA 110 | gi|27375606 | Leu biosynthetic pathway and the Met chain elongation | 5.20 | 1.10 | |
| 695 | GroEL protein | Asaia bogorensis | gi|269913095 | proper folding of many proteins | 3.38 | 2.31 | |
| 1252 | replication protein A | Agrobacterium radiobacter K84 | gi|222109068 | DNA replication, repair and recombination | 7.22 | 1.67 | |
| 1860 | ATP synthase FliI/YscN | Phaeobacter gallaeciensis BS107 | gi|163738066 | ATP binding protein | 13.17 | 2.76 | |
| 158 | glutamine synthetase protein | Rhizobium etli CIAT 652 | gi|190892623 | degradation of amino acids | 0.27 | 0.76 | |
| 165 | DNA polymerase III subunit delta | Rhodoferax ferrireducens T118 | gi|89899570 | prokaryotic DNA replication | 0.34 | 0.89 | |
| 209 | DNA-directed RNA polymerase, beta subunit | Neisseria polysaccharea ATCC 43768 | gi|297250922 | transcription of DNA into RNA | 0.46 | 0.97 | |
| 371 | aminoacyl-histidine dipeptidase | Vibrio mimicus VM603 | gi|258627332 | catalyze dipeptides and tripeptides | 0.20 | 0.97 | |
| 459 | hypothetical protein CAMRE0001_2526 | Campylobacter rectus RM3267 | gi|223040483 | Unknown | 0.38 | 0.68 | |
| 518 | glycosyl hydrolase family 3 protein | Escherichia coli MS 21-1 | gi|300936554 | Hydrolyse the glycosidic bond between carbohydrates | 0.19 | 0.74 | |
| 579 | DNA-cytosine methyltransferase | Rhodopseudomonas palustris DX-1 | gi|316931670 | transfer of methyl group to DNA | 0.40 | 0.72 | |
| 820 | Lon-A peptidase | Novosphingobium aromaticivorans DSM 12444 | gi|87199382 | regulates gene expression | 0.20 | 0.55 | |
| 970 | hypothetical protein VMC_13940 | Vibrio alginolyticus 40B | gi|269965870 | Unknown | 0.25 | 0.91 | |
| 1781 | conserved hypothetical protein | Pseudovibrio sp. JE062 | gi|254470700 | Unknown | 0.02 | 0.88 | |
| 2 | 1110 | aldehyde dehydrogenase | Bradyrhizobium japonicum USDA 110 | gi|27377927 | oxidation of aldehydes | 0.40 | 1.43 |
| 1854 | sensor histidine kinase | Geobacter sulfurreducens KN400 | gi|298504392 | signal transduction | 0.65 | 3.78 | |
| 1856 | hypothetical protein Swoo_4462 | Shewanella woodyi ATCC 51908 | gi|170728783 | Unknown | ND | 1.22 | |
| 1858 | putative phosphoribulokinase protein | Bradyrhizobium japonicum USDA 110 | gi|27377693 | Carbon fixation etc. | 0.67 | 1.95 | |
| 3 | 112 | threonine ammonia-lyase, biosynthetic | Vibrio harveyi HY01 | gi|153834318 | biosynthesis of amino acids | Unique | ND |
| 147 | molybdenum ABC transporter, periplasmic molybdate-binding protein | Sideroxydans lithotrophicus ES-1 | gi|291613363 | high affinity molybdate uptake system | 2.43 | 0.95 | |
| 708 | ACT domain-containing protein | Pantoea sp. At-9b | gi|317053226 | amino acid and purine synthesis | 3.30 | 0.49 | |
| 828 | hypothetical protein amb3581 | Magnetospirillum magneticum AMB-1 | gi|83312680 | Unknown | 2.81 | 0.56 | |
| 1087 | putative phage-related protein | Rhizobium etli GR56 | gi|218672483 | Unknown | 1.61 | 0.58 | |
| 1859 | ABC transporter substrate-binding protein | Bradyrhizobium japonicum USDA 110 | gi|27376323 | translocation RNA and DNA etc. repair | 3.29 | 0.31 | |
| 1861 | hypothetical protein xccb100_1606 | Xanthomonas campestris B100 | gi|188991002 | putative peptidase/protease | 5.59 | 0.64 |
*The proteins marked in boldface letters are those up regulated or down regulated by both the WS and Genistein.
Proteins belonging to the first category included nitrogen regulatory protein PII, iron-regulated outer membrane protein, isopropylmalate isomerase small subunit, GroEL protein, replication protein A, and ATP synthase FliI/YscN, which were all up-regulated, and glutamine synthetase protein, DNA polymerase III subunit delta, DNA-directed RNA polymerase, beta subunit, aminoacyl-histidine dipeptidase, hypothetical protein CAMRE0001_2526, glycosyl hydrolase family 3 protein, DNA-cytosine methyltransferase, and Lon-A peptidase which were all down-regulated. Aldehyde dehydrogenase, sensor histidine kinase, hypothetical protein Swoo_4462, and the putative phosphoribulokinase protein which fell into the second category were all up-regulated by genistein but down-regulated by WSHM. Molybdenum ABC transporter, threonine ammonia-lyase, ACT domain-containing protein, hypothetical protein amb3581, putative phage-related protein, ABC transporter substrate-binding protein and hypothetical protein xccb100_1606 in the third category were all up-regulated by WSHM but down-regulated by genistein.
QRT-PCR assay
Quantitative real-time PCR (QRT-PCR) showed that most gene expression data were consistent with the results of the analysis of proteomics, although differences in the rate of gene expression existed between the WSHM treatment and the control (Table 4). For example, nitrogen regulatory protein PII was up-regulated 5.58-fold by WSHM according to the proteomics results (Table 3), but a 17.26-fold difference was observed with RT-PCR (Table 4). Likewise, the apparent expression of the DNA-directed RNA polymerase beta subunit and the putative phosphoribulokinase protein differed significantly with the two methods. These differences might be a reflection of the post-transcriptional and/or post-translational phenomena and required further investigation.
Table 4. Expression of some genes induced by WSHM detected by Real Time PCR.
| Expression* (×10−2) induced by |
||||
|---|---|---|---|---|
| Spot No. | Protein | Genistein | Control | WSHM |
| 39 | nitrogen regulatory protein PII | 1.14 ± 0.0100b | 0.220 ± 0.0600c | 3.76 ± 0.160a |
| 209 | DNA-directed RNA polymerase, beta subunit | 6.35 ± 0.0500b | 3.10 ± 0.0300b | 59.4 ± 3.11a |
| 395 | isopropylmalate isomerase small subunit | 6.26 ± 0.0800b | 1.52 ± 0.0600c | 30.7 ± 2.22a |
| 1858 | putative phosphoribulokinase protein | 86.4 ± 3.99a | 20.4 ± 0.270c | 58.7 ± 1.42b |
| 1859 | ABC transporter substrate-binding protein | 1.95 ± 0.210b | 1.74 ± 0.510b | 39.8 ± 0.800a |
| 1860 | ATP synthase FliI/YscN | 0.610 ± 0.0400b | 0.250 ± 0.0200c | 2.47 ± 0.460a |
Assays were performed in triplicate, and the mean values and SDs are shown. Values in the same line having different letters are significantly different from each other according to LSD test (P < 0.05).
*Fold change in gene expression (vs 16s rDNA).
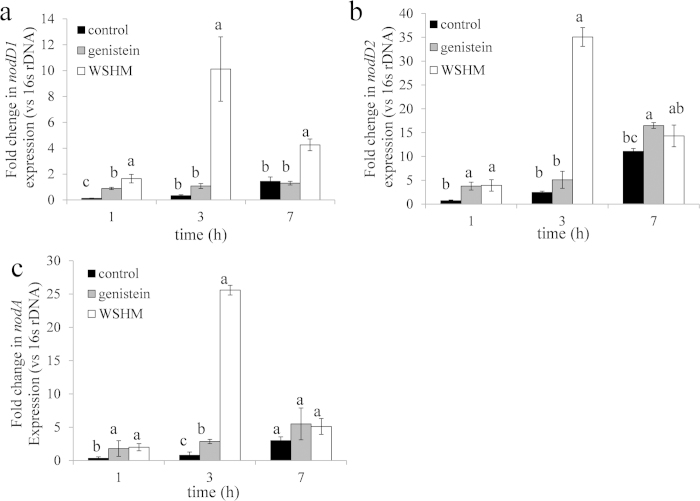
Besides, nod gene expression changes resulted from WSHM or genistein treatments were also investigated. QRT-PCR analyses revealed that the expression of nodD1, nodD2 and nodA increased significantly (P < 0.05) by WSHM treatment (Fig. 3). The expression of these genes reached the highest level 3 hours after the addition of WSHM to the cultures, which were 29.7-, 14.3- and 32.8-fold to those of the control respectively. However, the expression increase came to a halt, except for nodD1, 7 hours after WSHM addition. Similar results were observed in the genistein treatment, but the induction by genistein was significantly lower than that by WSHM.
Figure 3. The expression pattern of nodD and nodA genes in Bradyrhizobium liaoningense CCBAU05525 induced by water-soluble humic materials or genistein.
a: nodD1; b: nodD2; c: nodA. The data are expressed as mean ± SD values (n = 3). The statistical significance among the data set was assessed by LSD test (P < 0.05).
Discussion
Symbiotic nitrogen fixation plays an important role in sustainable agriculture, and this study demonstrated that WSHM, the humic acids, produced by biodegradation of lignite, possesses the ability to enhance symbiotic association between B. liaoningense CCBAU05525 and soybean plants. A WSHM concentration of 500 mg L−1 resulted in the maximal enhancement of cell density and nodulation of B. liaoningense CCBAU05525. According to our other studies, 375 g WSHM per hectare per year would contribute to 12.6% ~ 26.3% increases in the soybean yield (Our unpublished data). Furthermore, the identification of indole-like compounds in WSHM (Table 2) indicated the potential of WSHM in promoting plant growth. Since the WSHM used in this study was water, acid and alkali soluble. Thus, it was not only economically feasible but also profitable to use WSHM as a fertilizer in agriculture.
It was reported that humic acids could not only acting as C or energy source, but also stimulate bacterial growth by regulating cell metabolism20,21. In this study, similar results were observed. As shown in supplementary figure S1, the cell density of B. liaoningense CCBAU05525 in the 500 mg L−1 WSHM solution increased, which meant that WSHM could be used by the bacterium as nutrients; while the eight times of growth increase of CCBAU05525 in the YM broth supplied with 500 mg L−1 WSHM evidenced its stimulation function. Furthermore, the cell density in treatments of 1000 mg L−1 and 1500 mg L−1 WSHM were lower than that in 500 mg L−1 WSHM treatment (Fig. 1), indicating that 500 mg L−1 was the optimum concentration of WSHM in promoting growth of B. liaoningense CCBAU05525. Nonetheless, the 500 mg L−1 WSHM used in this study contained 52.18% and 3.72% of C and N, respectively18, which might account for the diauxic growth pattern of the bacteria in WSHM-treated cultures after the 7th day (Supplementary Fig. S1). These results all suggested that WSHM could serve as nutrients in the growth of B. liaoningense CCBAU05525, but their nutrient effects were negligible. Thus, the stimulation effects of WSHM on B. liaoningense CCBAU05525 growth was mainly attributed to its regulation on cell metabolism and nutrient transport which were demonstrated in the proteomics assay.
In the proteomics assay, proteins of the first category might be involved in the early stages of nodulation, such as Nod factor biosynthesis or the switch from the free-living to the symbiotic living stage. The changes in expression levels (up or down) of these proteins remained the same in both WSHM and genistein treatments, but the WSHM treatment caused a consistent greater increase or decrease than the genistein treatment, indicating that the effects of WSHM were stronger than those of genistein. The differences observed in the second and third categories demonstrated that WSHM had different effects on B. liaoningense CCBAU05525 compared with genistein. Other chemical compounds in WSHM except for flavonoid analogue might also have positive effects on the nodulation of B. liaoningense CCBAU05525 and be even better than genistein.
Belonging to the first category, nitrogen regulatory protein PII was an essential component of a highly efficient system that regulated nitrogen assimilation. This system coordinated the intracellular concentration of glutamine and 2-ketoglutarate to ensure the appropriate regulation of glutamine synthetase (GS) activity and expression of the glnALG operon22,23. Besides, it was a fundamental step to switch off assimilating ammonia during bacteroid development. Furthermore, most aspects of cell growth and division, including the synthesis of nucleic acid were reduced during bacteroid development2, while, ATP synthesis was always essential to nodulation, protein synthesis and active transport. Considering the fact that glutamine synthetase protein, DNA polymerase III subunit delta, DNA-directed RNA polymerase beta subunit and DNA-cytosine methyltransferase were down-regulated and nitrogen regulatory protein PII and ATP synthase FliI/YscN protein were up-regulated by WSHM and genistein, the WSHM (like genistein) probably prepared the bacteria for better bacteroid development.
The chaperonin GroEL which played an essential role in ensuring proteins properly folded, preventing and repairing harmful effects, was highly conserved in evolution24,25. This protein enhances the expression of nod genes26, and was essential to the formation of a functional nitrogenase complex in the bacteroids of B. japonicum27. In addition, it interacted with the NifA polypeptide and enhanced nif gene expression28. Thus the upregulated expression of GroEL demonstrated that WSHM had positive effects on the expression of nod, nif genes and the formation of nitrogenase complex.
It was reported that molybdenum ABC transporter permease protein, molybdenum processing protein, molybdenum transport system permease protein and many ABC transporter substrate-binding proteins were only found to be expressed in 21-day-old bacteroids of Bradyrhizobium japonicum but not in free-living cells29. The molybdenum ABC transporter and iron-regulated outer membrane protein transported molybdenum and iron respectively. Iron and molybdenum were essential to nitrogen fixation because of their necessity as nitrogenase co-factors2. Therefore, up-regulating of these three transporter-associated proteins might be the reason for the increase in nodule fresh weight and nitrogenase activity of soybean by WSHM treatment.
With the proteomic assay, it was concluded that WSHM regulated the nitrogen metabolism, carbon metabolism, energy production, nutrient transport and signaling of B. liaoningense CCBAU05525. On one hand, WSHM had similar but superior effect on the expression of proteins in category 1 compared with genistein. On the other hand, WSHM had different effects on the expression of some proteins related to nitrogen fixation compared with genistein. Specifically, the protein expression pattern changed by WSHM was beneficial to the bacteroid development and the increase in nitrogenase activity.
Flavonoids secreted by host plant induce expression of nodD gene which then regulated the transcription of the structure genes (such as nodABC) to synthesis Nod factor30. Therefore, they were essential to the symbiosis between the legumes and rhizobia31. It was shown in this study that WSHM increased nodD1, nodD2, nodA gene expression of B. liaoningense CCBAU05525, which might be the reason for the increase in the nodule number and nitrogenase activity of soybean in the greenhouse experiments. The enhanced expression of nod genes by humic acids in B. liaoningense had not been reported previously. As expected, genistein also increased the expression of nodD1, nodD2, nodA gene. WSHM contained flavonoid analogue, which might account for its contribution to nodD1, nodD2, nodA gene expression. However, WSHM treatment caused a greater increase in the expression of these genes than genistein did, similar to the tendency of the proteins in category 1 shown in proteomics assay, indicating that the effects of WSHM were stronger than those of genistein. However, the higher concentration of WSHM used in the experiment might partially account for the greater effects. In addition, another explanation might be the complex mixture of components present in the WSHM treatment (Table 2), in which there might be some strong inducers for particular metabolic procedures; since B. liaoningense CCBAU05525 responded to WSHM and genistein differently in category 2 and 3 in proteomics assay. These strong inducers in the WSHM compounds had great potential in the development of new materials to stimulate the symbiotic nitrogen fixation between rhizobium and legume.
The failure to find NodD1, NodD2, and NodA in the analysis of proteomics might be due to their low abundance, since all of them are regulators for the expression of other genes. The function of several hypothetical proteins affected by WSHM treatment would be examined in future work. Also, the mechanism of WSHM in promoting nodulation of soybean with B. liaoningense CCBAU05525 needed to be further investigated, such as the effects of WSHM on the formation of infection thread of B. liaoningense CCBAU05525.
Conclusively, WSHM treatment increased the number of soybean nodules, the nodule fresh weight and the nitrogenase activity, as well as enhanced the cell density of B. liaoningense CCBAU05525. WSHM contained flavonoid analogues and showed similar bioactivities with genistein but had wider effects than genistein such as inducing the expression of nodD1, nodD2, nodA, and up-regulating a range of proteins involved in a variety of metabolic pathways. Furthermore, several nitrogen fixation-associated proteins such as molybdenum ABC transporter were also affected. Therefore, WSHM might prepare the free-living rhizobia for better bacteroid development. This was the first study about the effects of humic acids on rhizobia growth, nod gene expression and proteomics. These results shed new light on the effects of humic material on legumes and demonstrated great application value in improving soybean production.
Materials and methods
Water-soluble humic materials
Lignite was collected from the Huolingele Minerals Administration coal mine, Inner Mongolian Autonomous Region, Northwest China. Water-soluble humic materials (WSHM) was extracted according to a previously described protocol32.
Plant nodulation test
B. liaoningense CCBAU05525 was obtained from the Culture Collection of China Agriculture University, Beijing, China33. This strain was cultured aerobically at 28 °C in YM broth (Mannitol, 10 g; NaCl, 0.1 g; K2HPO4, 0.25 g; KH2PO4, 0.25 g; Yeast Extract, 0.8 g; MgSO4·7 H2O, 0.2 g; pH 6.8–7.0). Bacterial culture with approximately 106 cells and 5 mL of the WSHM solution with different concentration (0, 300, 500 and 1000 mg L−1) were used for inoculating soybean. Soybean seeds were surface-sterilized by successive treatments with 95% ethanol for 30 sec and 0.2% HgCl2 for 5 min, and were then washed for 6 times by sterile water. Then the surface-sterilized seeds were germinated on 0.8% agar-water plates in the dark at 28 °C for 24–48 h. Germinated seeds were planted in vermiculite moisturized with low-N nutrient solution in pots34 and were inoculated with 1 mL of bacterial culture and 5 mL of WSHM solution or 5 mL of sterile water as control per plant. For each treatment, 30 pots were selected and divided into three parallel groups. In each group, there were 10 plants in 10 pots. The average value of each group was taken as one sample value. Plants were grown in greenhouse at 25/17 ± 2 °C for day/night with 60% relative humidity. Pots were rearranged daily to give a random distribution of growth conditions in the greenhouse. After 35 days, all the soybean plants were harvested to detect the number, weight and nitrogenase activity (acetylene reduction assay) of the nodules35.
Effects of WSHM on the growth of B. liaoningense CCBAU05525
It was known that the cell density of rhizobia around the plant roots must reach a threshold level for adequate nodulation. Thus, the effects of WSHM on the growth of B. liaoningense CCBAU05525 were analyzed. The strain was preincubated in 50 mL of YM broth for 4 days at 28 °C under shaking (140 rpm). Then, the CCBAU05525 culture was inoculated at a ratio of 1% (v/v) into 200 mL of YM broth supplied with WSHM at the final concentrations of 0, 100, 200, 500, 1000 and 1500 mg L−1, respectively. The flasks containing the inoculated YM broth were incubated at 28 °C under shaking (140 rpm) for four days up to the middle exponential phase and culture samples were taken to evaluate the rhizobial cell density by dilution-plating procedure. The results were then used to choose the suitable WSHM concentration for the study of the effects of WSHM on the growth curve (up to 10 days of incubation) and the nod gene expression and proteomics of rhizobia.
TMAH-py-GC/MS of WSHM samples
The Py-GC/MS system used in this study was a combination of a PY-2020S pyrolyzer (Frontier Laboratories Ltd., Japan) and Shimadzu GCMS-QP2010 gas chromatograph mass spectrometer (Shimadzu, Milan, Italy) equipped with commercial mass spectral libraries (Nist107). All the WSHM samples (ca. 200 μg) were placed on a ferromagnetic pyrofoil, and 2 μL of tetramethyl ammonium hydroxide (TMAH) aqueous solution (10%, w/v) were added to the samples. The pyrolysis temperature was 550 °C, and the total heating time was 10 sec. The pyrolysis products were transported to the GC/MS, and separated on the GC equipped with a UA-5MS capillary column (Frontier Laboratories Ltd., Japan, 30 m × 0.25 mm ID, 0.25 μm thickness). The temperature protocol used was 6 °C min−1 from 40 °C (3 min) to 300 °C (10 min) with a heating rate of 10 °C min−1. The mass spectra of compounds were measured at 70 eV. The products released from pyrolysis were differentiated based on a search in the mass spectral library (Nist107) comparing the relative retention time and other Py-GC/MS data of humic acids (HA) and fulvic acids (FA)36,37.
Protein preparation and extraction
B. liaoningense CCBAU05525 was pre-cultured aerobically at 28 °C to the early exponential phase (OD600 = 0.4 ~ 0.5) in 200 mL YM broth. Then, 100 μL of 1 mM (final concentration of 0.5 μM) genistein, which was a known flavonoid specific for the Nod factor induction in soybean rhizobia38,39; or 5 mL WSHM (at a concentration of 20 g L−1) were added and the incubation continued to the mid-exponential phase (OD600 = 0.7 ~ 0.8). Cultures without addition of the inducers were included as a control. Cells were harvested by centrifugation (8,000 × g, 10 min) at 4 °C, the cell pellet was washed with 50 mL NaCl solution (0.85%), resuspended in the same solution at a concentration of 0.5 g mL−1, and frozen at −80 °C in aliquots of 50–100 μL for Proteins extraction.
Proteins were extracted using the phenol/ammonium acetate method as described previously40 with slight modifications. Cell samples (1 g) were homogenized in 3 mL extraction buffer, and the protein concentration was determined using 2D Quant Kit (GE Healthcare) according to the instructions of the manufacturer.
2-D electrophoresis, imaging and data analysis
For 2-D electrophoresis, proteins were separated by isoelectric focusing (IEF) in the first dimension and SDS-PAGE in the second dimension. Samples were adjusted to a final concentration of 1.2 mg of protein in 450 μL DeStreak rehydration solution (GE Healthcare). Isoelectric point gel (IPG) strips (24 cm, diameter of 0.5 cm, pH 3–10; (GE Healthcare) were rehydrated with the sample and cover fluid and electrophoresed as described41. After IEF, the strip was equilibrated and placed on a large format 13% (w/v) polyacrylamide gel (26 × 20 cm) for SDS-PAGE as described42. After electrophoresis, the gel was stained with colloidal CBB (20% ethanol, 1.6% phosphoric acid, 8% ammonium sulfate, 0.08% CBB G-250) and imaged using a UMAX imagescanner. The image was analyzed using ImageMaster 2D Platinum v 5.0 software. At least three replicate gels of three different protein preparations were analyzed for consistency. If the change in the relative area of a protein spot was greater than 1.5-fold and shown to be statistically significant by the t-test (P < 0.05) using SPSS software, the protein expression level was considered to be modified by the treatment.
In-gel digestion and protein identification
Protein spots that showed a significant difference (P < 0.05) in the 2-D electrophoresis experiment were excised manually, and in-gel digested overnight at 37 °C with sequencing grade modified trypsin (Promega, USA). Tryptic peptides (0.5 mL) were mixed with a saturated solution of α-Cyano-4-hydroxycinnamic acid (CHCA) in 50% (w/v) acetonitrile containing 0.1% trifluoroacetic acid. The mixture was spotted onto a MALDI sample plate and crystallized at room temperature. The same procedure was used for the standard peptide calibration mixture (Bruker Daltonics, Germany). Mass spectra were acquired using a MALDI-TOF-MS Autoflex spectrometer (Bruker Daltonics, Germany) operating in the PMF fully automated mode, or manually in the LIFT mode in the case of MALDI-TOF/TOF. PMFs and MS/MS ions were searched against the NCBI nr/Proteobacteria database using MASCOT software (Matrix Science, UK). The Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/) covering a total of 2270 B. japonicum genes and showing a complete or nearly complete pathway in several cases43 was used to identify the possible pathways affected by WSHM treatment. Enzymes and proteins annotated under the global KEGG category metabolism were identified.
Quantitative Real-time PCR assay
Quantitative real-time PCR (QRT-PCR) was used to confirm the results of the proteomics experiments and detect the effects of WSHM on nodD and nodA gene expression. Cells were cultured as described above, except that the cells were collected at 1, 3 and 7 h after WSHM or genistein was added as inducer for nod gene expression. Total RNAs were isolated using TRIzol reagent (TIANGEN, Beijing) according to the manufacturer’s protocol. First strand cDNAs were synthesized using PrimeScript Reverse Transcriptase (RT) (TaKaRa Code: D2680S) according to the manufacturer’s instructions. RT samples were used for quantitative RT-PCR with the primers shown in Table 5, which were designed using the program Premier 5.0 based on the B. liaoningense CCBAU05525 genome sequence33. Expression of 16S rRNA was used as an internal control for normalization. Each reaction contained 10 μL of Power SYBR Green master Mix (ABI, USA) in a final volume of 20 μL. The PCR program was as follows: (i) 95 °C for 10 min, 40 cycles of 95 °C for 30 sec, 60 °C for 35 sec. PCR was performed on an ABI 7500 Thermo cycler and data were analyzed using ABI software (Foster City, USA).
Table 5. Primers used in this study.
| Name of primers | Primer sequence (5’-3’) | Corresponding gene | Tm (°C) |
|---|---|---|---|
| 16S rRNA gene (341f) | CCTACGGGAGGCAGCAG | 16S rRNA | 58.1 |
| 16S rRNA gene (534r) | ATTACCGCGGCTGCTGG | 58.6 | |
| 39F | TCGGCGTTCACGGTCTCA | glnK | 61.1 |
| 39R | TGGCGTCGATGGTCTTGT | 57.2 | |
| 209F | TCGGCGAACTCATGGAGAA | Gene for DNA-directed RNA polymerase, beta subunit | 60.5 |
| 209R | GCTGCGAGGAACCGAAGAA | 61.1 | |
| 1859F | CTCTACGAAGGCACCGACTG | Gene for ABC transporter substrate-binding | 60.0 |
| 1859R | CGGGAATGACCTGCCAGTAG | 60.3 | |
| 395F | CGATCCGCGTCAGCCAGGAA | leuD | 63.9 |
| 395R | AGGCAGTGCTTGCGGAACG | 61.8 | |
| 1858F | ATTCGGACCTGCTGTTCTACG | cbbP | 59.4 |
| 1858R | TCCGCCGCAGAATGGTGT | 62.7 | |
| 1860F | GCTCGCGGTCGCCGAATAT | Gene for ATP synthase FliI/YscN | 65.6 |
| 1860R | CGGCAACTCGGTGAAGACG | 62.1 | |
| nodAF | GAGCCTTCGTTGGGAAAGTG | Gene for acyltransferase | 60 |
| nodAR | CGCAGTAGCCCGATGTGAG | 60 | |
| nodD1F | CGCACTGAGACCGAACCT | Gene for a member of the NodD family of LysR-type transcriptional regulators | 60 |
| nodD1R | CAGTCGTAAGGGCATCGT | 60 | |
| nodD2F | TTGGGACGAACCGCATAG | Gene for a regulator of Nod factor production | 60 |
| nodD2R | GGGTCGCTGTTGTGAAGTG | 60 |
Additional Information
How to cite this article: Gao, T.G. et al. Nodulation Characterization and Proteomic Profiling of Bradyrhizobium liaoningense CCBAU05525 in Response to Water-Soluble Humic Materials. Sci. Rep. 5, 10836; doi: 10.1038/srep10836 (2015).
Supplementary Material
Acknowledgments
This research was supported by the National High Technoligy Research and Development Programme of China (No. 2011AA10A206) and Special Fund for Agro-scientific Research in the Public Interest (201403048-2).
Footnotes
The authors declare no competing financial interests.
Author Contributions TGG, FJ, JSY and HLY conceived and designed the study. TGG and YYX performed the experiments. TGG and YYX wrote the manuscript text. TGG prepared samples for proteomic analysis, and performed the statistical analysis and YYX performed the QRT-PCR assay. YYX, FJ and BZL participated in the preparation of water-soluble humic materials. ETW helped to design the experiments and draft the manuscript. All authors read and approved the final manuscript.
References
- Oldroyd G. E., Murray J. D., Poole P. S. & Downie J. A. The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45, 119–144 (2011). [DOI] [PubMed] [Google Scholar]
- Udvardi M. & Poole P. S. Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol 64, 781–805 (2013). [DOI] [PubMed] [Google Scholar]
- Erman M. et al. Effects of Rhizobium, arbuscular mycorrhiza and whey applications on some properties in chickpea (Cicer arietinum L.) under irrigated and rainfed conditions 1—Yield, yield components, nodulation and AMF colonization. Field Crop Res 122, 14–24 (2011). [Google Scholar]
- Siczek A., Lipiec J., Wielbo J., Szarlip P. & Kidaj D. Pea growth and symbiotic activity response to Nod factors (lipo-chitooligosaccharides) and soil compaction. Appl Soil Ecol 72, 181–186 (2013). [Google Scholar]
- Siczek A., Lipiec J., Wielbo J., Kidaj D. & Szarlip P. Symbiotic activity of pea (Pisum sativum) after application of Nod factors under field conditions. Int J Mol Sci 15, 7344–7351 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik Y., Joseph C. M. & Phillips D. A. Flavone limitations to root nodulation and symbiotic nitrogen fixation in alfalfa. Plant Physiol 84, 1193–1196 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D. A., Joseph C. M. & Maxwell C. A. Trigonelline and stachydrine released from alfalfa seeds activate NodD2 protein in Rhizobium meliloti. Plant Physiol 99, 1526–1531 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S., Stacey G. & Yu O. Distinct, crucial roles of flavonoids during legume nodulation. Trends Plant Sci 12, 282–285 (2007). [DOI] [PubMed] [Google Scholar]
- Hayes M. & Wilson W. Humic substances, peats and sludges: Health and environmental aspects. (Elsevier, 1997). [Google Scholar]
- Traversa A. et al. Comparative evaluation of compost humic acids and their effects on the germination of switchgrass (Panicum vigatum L.). J Soil Sediment 14, 432–440 (2013). [Google Scholar]
- Jindo K. et al. Root growth promotion by humic acids from composted and non-composted urban organic wastes. Plant Soil 353, 209–220 (2011). [Google Scholar]
- Canellas L. P. et al. Chemical composition and bioactivity properties of size-fractions separated from a vermicompost humic acid. Chemosphere 78, 457–466 (2010). [DOI] [PubMed] [Google Scholar]
- Jannin L. et al. Microarray analysis of humic acid effects on Brassica napus growth: Involvement of N, C and S metabolisms. Plant Soil 359, 297–319 (2012). [Google Scholar]
- Tahir M. M., Khurshid M., Khan M. Z., Abbasi M. K. & Kazmi M. H. Lignite-Derived Humic Acid Effect on Growth of Wheat Plants in Different Soils. Pedosphere 21, 124–131 (2011). [Google Scholar]
- Tan K. H. & Tantiwiramanond D. Effect of humic acids on nodulation and dry matter production of soybean, peanut, and clover. Soil sci soc Am. J. 47, 1121–1124 (1983). [Google Scholar]
- Til’ba V. A. & Sinegovskaya V. T. Role of symbiotic nitrogen fixation in increasing photosynthetic productivity of soybean. Russ Agric Sci 38, 361–363 (2012). [Google Scholar]
- Yuan H. L., Yang J. S., Wang F. Q. & Chen W. X. Degradation and solubilization of Chinese lignite by Penicillium sp. P6. Appl Biochem Micro 42, 52–55 (2006). [PubMed] [Google Scholar]
- Gao T. G., Jiang F., Yang J. S., Li B. Z. & Yuan H. L. Biodegradation of Leonardite by an alkali-producing bacterial community and characterization of the degraded products. Appl Microbiol Biotechnol 93, 2581–2590 (2012). [DOI] [PubMed] [Google Scholar]
- Dong L., Córdova-Kreylos A. L., Yang J., Yuan H. & Scow K. M. Humic acids buffer the effects of urea on soil ammonia oxidizers and potential nitrification. Soil Biol Biochem 41, 1612–1621 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard A. Kirschner Jr., Bruce C. Parker & III, J. O. F. Humic and fulvic acids stimulate the growth of Mycobacterium avium. FEMS Microbiol Ecol 30, 327–332 (1999). [DOI] [PubMed] [Google Scholar]
- Tikhonov V., Yakushev A., Zavgorodnyaya Y. A., Byzov B. & Demin V. Effects of humic acids on the growth of bacteria. Eurasian Soil Sci 43, 305–313 (2010). [Google Scholar]
- Dixon R. & Kahn D. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol 2, 621–631 (2004). [DOI] [PubMed] [Google Scholar]
- Huergo L. F., Chandra G. & Merrick, (II) signal transduction proteins: nitrogen regulation and beyond. FEMS Microbiol Rev 37, 251–283 (2013). [DOI] [PubMed] [Google Scholar]
- Ranson N., White H. & Saibil H. Chaperonins. Biochem. J 333, 233–242 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes D. F., Batista J. S. d. S., Schiavon A. L., Andrade D. S. & Hungria M. Proteomic profiling of Rhizobium tropici PRF 81 identification of conserved and specific responses to heat stress. BMC Microbiol 12, 1–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wais R. J., Wells D. H. & Long S. R. Analysis of differences between Sinorhizobium meliloti 1021 and 2011 strains using the host calcium spiking response. Mol plant microbe In 15, 1245–1252 (2002). [DOI] [PubMed] [Google Scholar]
- Fischer H.-M., Schneider K., Babst M. & Hennecke H. GroEL chaperonins are required for the formation of a functional nitrogenase in Bradyrhizobium japonicum. Arch microbiol 171, 279–289 (1999). [Google Scholar]
- Govezensky D., Greener T., Segal G. & Zamir A. Involvement of GroEL in nif gene regulation and nitrogenase assembly. J bacteriol 173, 6339–6346 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriella Pessi et al. Genome-Wide Transcript Analysis of Bradyrhizobium japonicum Bacteroids in Soybean Root Nodules. Mol Plant Microbe In 20, 1353–1363 (2007). [DOI] [PubMed] [Google Scholar]
- Taylor L. P. & Grotewold E. Flavonoids as developmental regulators. Curr Opin Plant Biol 8, 317–323 (2005). [DOI] [PubMed] [Google Scholar]
- Subramanian S., Stacey G. & Yu O. Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum. Plant J 48, 261–273 (2006). [DOI] [PubMed] [Google Scholar]
- Dong L., Yuan Q. & Yuan H. Changes of chemical properties of humic acids from crude and fungal transformed lignite. Fuel 85, 2402–2407 (2006). [Google Scholar]
- Tian C. F. et al. Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations. Proc Natl Acad Sci USA 109, 8629–8634 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. High-resolution transcriptomic analyses of Sinorhizobium sp. NGR234 bacteroids in determinate nodules of Vigna unguiculata and indeterminate nodules of Leucaena leucocephala. PloS one 8, 1–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio Suganuma et al. Bacteroids Isolated from Ineffective Nodules of Pisum sativum Mutant E135 (syml3) Lack Nitrogenase Activity but Contain the Two Protein Components of Nitrogenase. Plant Cell Physiol 39, 1093–1098 (1998). [Google Scholar]
- Lu X. Q., Hanna J. V. & Johnson W. D. Source indicators of humic substances: an elemental composition, solid state 13C CP/MAS NMR and Py-GC/MS study. Appl Geochem 15, 1019–1033 (2000). [Google Scholar]
- Xiaoli C., Shimaoka T., Qiang G. & Youcai Z. Characterization of humic and fulvic acids extracted from landfill by elemental composition, 13C CP/MAS NMR and TMAH-Py-GC/MS. Waste manage 28, 896–903 (2008). [DOI] [PubMed] [Google Scholar]
- Loh J. & Stacey G. Nodulation Gene Regulation in Bradyrhizobium japonicum: a Unique Integration of Global Regulatory Circuits. Appl Environ Microb 69, 10–17 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. & Smith D. L. Preincubation of Bradyrhizobium japonicum with genistein accelerates nodule development of soybean at suboptimal root zone temperatures. Plant Physiol 108, 961–968 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurkman W. J. & Tanaka C. K. Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol 81, 802–806 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süß C. et al. Identification of genistein-inducible and type III-secreted proteins of Bradyrhizobium japonicum. J biotechnol 126, 69–77 (2006). [DOI] [PubMed] [Google Scholar]
- Herbert B. Advances in protein solubilisation for two‐dimensional electrophoresis. Electrophoresis 20, 660–663 (1999). [DOI] [PubMed] [Google Scholar]
- Delmotte N. et al. An integrated proteomics and transcriptomics reference data set provides new insights into the Bradyrhizobium japonicum bacteroid metabolism in soybean root nodules. Proteomics 10, 1391–1400 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.