Abstract
The function of the tumor suppressor p53 is universally compromised in cancers. It is the most frequently mutated gene in human cancers (reviewed). In cases where p53 is not mutated, alternative regulatory pathways inactivate its tumor suppressive functions. This is primarily achieved through elevation in the expression of the key inhibitors of p53: Mdm2 or Mdmx (also called Mdm4) (reviewed). In breast cancer (BrCa), the frequency of p53 mutations varies markedly between the different subtypes, with basal-like BrCas bearing a high frequency of p53 mutations, whereas luminal BrCas generally express wild-type (wt) p53. Here we show that Mdmx is unexpectedly highly expressed in normal breast epithelial cells and its expression is further elevated in most luminal BrCas, whereas p53 expression is generally low, consistent with wt p53 status. Inducible knockdown (KD) of Mdmx in luminal BrCa MCF-7 cells impedes the growth of these cells in culture, in a p53-dependent manner. Importantly, KD of Mdmx in orthotopic xenograft transplants resulted in growth inhibition associated with prolonged survival, both in a preventative model and also in a treatment model. Growth impediment in response to Mdmx KD was associated with cellular senescence. The growth inhibitory capacity of Mdmx KD was recapitulated in an additional luminal BrCa cell line MPE600, which expresses wt p53. Further, the growth inhibitory capacity of Mdmx KD was also demonstrated in the wt p53 basal-like cell line SKBR7 line. These results identify Mdmx growth dependency in wt p53 expressing BrCas, across a range of subtypes. Based on our findings, we propose that Mdmx targeting is an attractive strategy for treating BrCas harboring wt p53.
The p53 tumor suppressor protein is a key factor in the cellular stress response.1, 2 Functional p53 prevents the progression of cancer by mounting growth inhibition in the form of apoptosis, senescence and/or autophagy.3 The exact tumor suppressive functions of p53 that prevent cancer are currently the subject of extensive studies (reviewed in Bieging et al.4). These growth inhibitory functions are frequently lost during tumorigenesis, most commonly through direct mutations in the p53 gene, which occur in ~50% of all human cancer cases.5 However, in the remaining cases, p53 status remains wild type (wt) and its function and/or expression is compromised by other mechanisms. The two major nonredundant inhibitors of p53 are the Mdm proteins: Mdm2 and Mdmx (also called Mdm4).6, 7
Mdm2 is the major E3 ligase of p53, promoting its ubiquitination and proteasomal degradation.8, 9 Mdmx in contrast, inhibits the transcriptional activity of p53 and enhances the ability of Mdm2 to target p53 for degradation, although it does not have an E3 ligase activity of its own.10 Both Mdm2 and Mdmx expression are elevated in various cancer types. For example, Mdm2 is amplified in the majority (70%) of well-differentiated liposarcomas,11 whereas the Mdmx protein is elevated in most melanomas and retinoblastoma (~70%).12, 13 In these cases, elevation of these Mdm proteins directly correlates with wt p53 status. In contrast, Yu et al.14 reported elevated protein levels of Mdm2 (38%) and Mdmx (65%), and low-level Mdmx gene amplification (56.5% identified by fluorescence in situ hybridization (FISH)) in an apparently wt p53 context (as suggested by an absence of allelic loss and no increased protein detection) in primary breast cancers (BrCas). This contrasted with the far more modest levels (5%) previously described.15 The discrepancy between these findings is apparently due to what is considered amplification, where the former included low-level amplifications.14
An overall p53 mutation rate approaching 30% defines it as the most common genetic alteration in BrCa. However, the mutational frequency is highly dependent on the cancer subtype. Specifically, p53 mutations have been reported in 88% of basal-like BrCas, ~70% of apocrine carcinomas and in ~50% of HER2-amplified tumors. In the more common luminal subtypes, p53 mutations are reported in ~15% of luminal A and ~40% of luminal B subtype. Moreover, the nature of p53 mutations also differ between subtypes, with basal-like BrCa and apocrine cancers having complex p53 mutations characterized by ‘insertion/deletion polymorphisms', whereas luminal tumors are generally simpler base substitutions.16, 17, 18
In this study we tested whether elevated Mdmx expression can account for the tolerance of wt p53 in BrCas, and whether downregulation of Mdmx is an efficient approach to targeting BrCas bearing wt p53. We found that Mdmx is highly expressed in BrCas. We showed that ablation of Mdmx impeded the growth of luminal BrCa cell line MCF-7 in culture, in a p53-dependent manner. Growth inhibition in response to Mdmx knockdown (KD) was replicated in an additional wt p53 expressing luminal cell line MPE600. In an orthotopic model of human luminal BrCa using MCF-7 cells, Mdmx KD was demonstrated to both prevent tumor initiation and also to inhibit progression in established tumors. Importantly, our findings extend beyond luminal BrCas and identified that in the wt p53 basal-like cell line SKBR7, Mdmx KD is also growth inhibitory. Our study strongly supports the notion that targeting Mdmx has a therapeutic potential across a range of BrCa subtypes expressing wt p53.
Results
Mdmx levels are elevated in luminal BrCa corresponding with low p53 levels
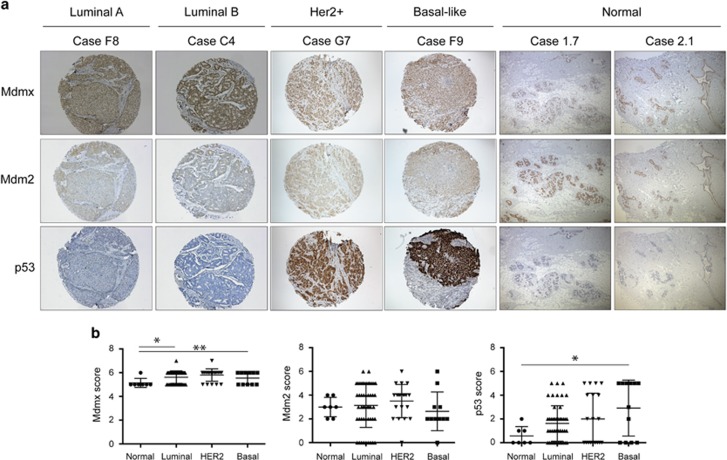
Wt p53 status in cancers is often associated with elevated Mdm proteins (reviewed in Wade et al.6). To explore whether gene copy number of Mdm2 and/or Mdmx are abnormal in BrCas, we measured gene copy amplification by FISH in a BrCa tissue micro-array (TMA). The TMA comprised 81 BrCa biopsies: luminal (n=50; 62%), Her2 (n=20; 25%), basal (n=11; 13%) and a panel of normal breast tissues. As shown in Supplementary Figure 1A, none of these BrCa samples had Mdm2 probe amplification and only a single luminal sample had Mdmx probe amplification (Supplementary Figure 1B). Immunohistochemistry (IHC) staining of Mdm2, Mdmx and p53 protein levels were compared in BrCa TMA samples relative to normal controls. Unexpectedly, we observed high Mdmx and Mdm2 expression levels in the normal breast ductal epithelial cells relative to neighboring cells (Figure 1, see Discussion). Mdm2 staining was detected in both the nucleus and the cytoplasm of many of the BrCa samples (representative luminal, Her2+ and basal-like samples in Figure 1a) and high levels were quantified in the nucleus (Figure 1b). Mdm2 protein in the luminal tumors was present at similar levels to the normal controls, consistent with no amplification of the mdm2 gene. The majority of the luminal samples exhibited low levels of p53 protein, suggestive of wt p53 status and were consequently chosen as the main focus of our study. Significantly higher levels of Mdmx protein were identified in the luminal tumor samples compared with the normal samples (Student's t-test P=0.0258; Figure 1b). Strong nuclear Mdmx expression was detected in the majority of the cancer cells in the tumor biopsies, but not in the adjacent normal tissue, or the stroma (Figure 1a). Most luminal samples with high Mdmx expression had low p53 expression, suggestive of wt p53 status. Luminal samples were further stratified according to Ki67 staining19 into luminal A (<14% Ki67 positive) and B subtype (≥14% Ki67 positive), and Mdmx was high across both (i.e., Mdmx staining did not discriminate luminal A and B subtypes; as evident in Figure 1a). Taken together, these analyses of luminal BrCa biopsies show a strong correlation between low p53 expression levels and elevated levels of Mdm proteins. These findings are consistent with previous studies demonstrating elevated expression of the Mdm proteins in BrCa cell lines with wt p53 status.20
Figure 1.
Elevated Mdmx expression inversely correlated with p53 levels. TMA from luminal, Her 2-positive (Her2+), basal-like BrCas and normal breast were stained for Mdmx (top row), Mdm2 (middle row) and p53 (lower row). Representative samples from the 81 tumor biopsies (for luminal A (Case F8) and luminal B (Case C4), Her2+ and Basal subtypes) and 6 normal breast tissues are shown in a. Tissues were scored according to the proportion of cells stained and the overall intensity in the nucleus, and the findings are presented in b according to BrCa subtypes. Staining intensity ranged from minimum to maximum of 0 to 3, whereas the proportion of cells stained was according to an elevating scale of 0 to 4 (0 to >80%, respectively, as described in the text) and scores were summed to a final possible maximum score of 8. Statistical significance discriminated the Mdmx normal and luminal samples (S.D., P=0.0258) and also normal and Her2 samples (P=0.0055). p53 staining only differed significantly from normal levels in the basal samples (P=0.0216). All the experiments were repeated at least three times. Errors bars represent S.D. of the mean. P-values were calculated by Student's t-test
Downregulation of Mdmx in luminal BrCa MCF-7 cells impedes growth in vitro in a p53-dependent manner
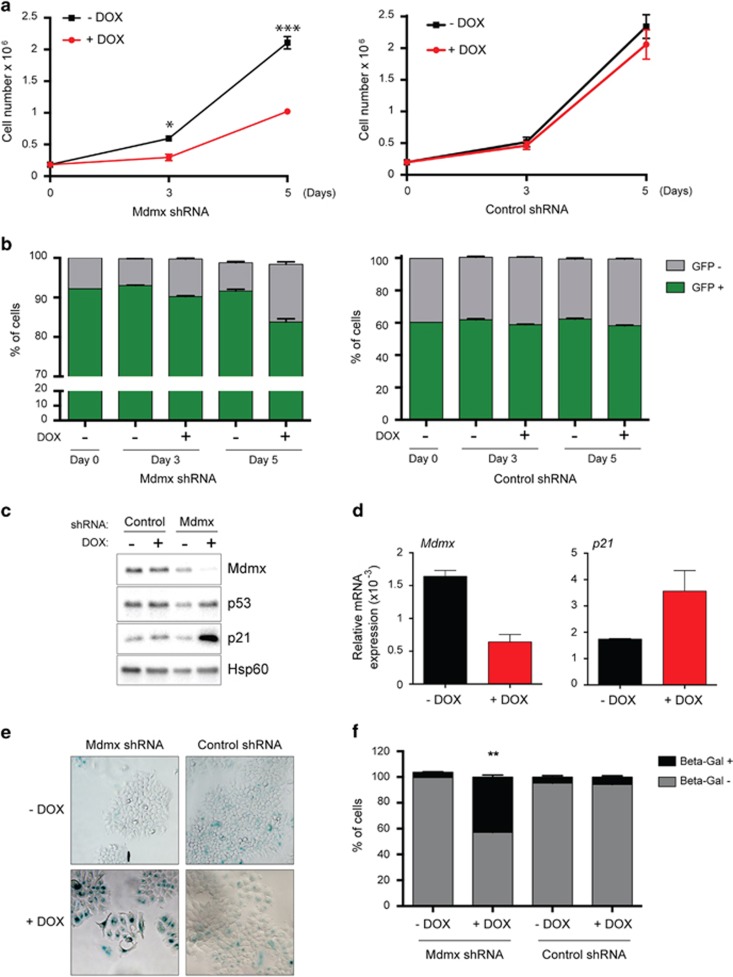
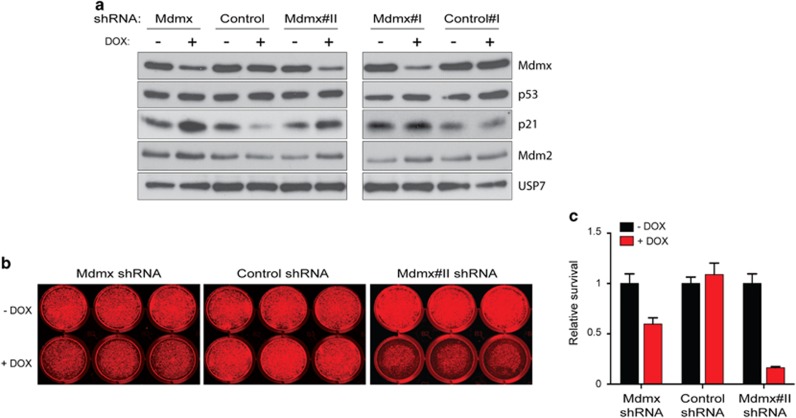
To examine whether Mdmx expression promotes the growth of luminal wt p53 BrCa cells, we chose MCF-7 as the primary model system, as it has wt p53 and relatively high Mdmx levels. The expression of Mdmx was knocked down by lentiviral-mediated doxycycline (DOX)-inducible shRNA expression. Controls included an empty vector and a wobble shRNA (identical sequence but with two substitutions) to Mdmx. Mdmx KD was induced with DOX, and in MCF-7 cells reduced cell numbers were evident after 3 days and further exacerbated after 5 days (>50% Figure 2a). As this lentiviral vector also expresses GFP, we used fluorescence to discriminate how transduced cells were effected by Mdmx KD (where nontransduced cells in the population were GFP negative). This analysis revealed that Mdmx KD (after 5 days) reduced viability of GFP-positive cells by <10% (indicating that cell death was not the major cause of the reduced cell numbers (Figure 2a)), whereas the control shRNA had no significant effect (Figure 2b). DOX treatment of MCF-7 parental cells or cells infected with the lentiviral vector control also had no effect on cell growth (Supplementary Figure 2A and Figure 2b, respectively). The growth inhibitory effects of Mdmx KD were reiterated using additional Mdmx shRNA sequences: shRNA 1 with 3 and 5 days of DOX induction (Supplementary Figures 3A–D, western blot, densitometric quantification, reduction in live cell numbers, respectively) and shRNA II with 3 and 6 days of DOX induction (Supplementary Figures 3E–G, western blot, densitometric quantification, reduction in live cell numbers, respectively, without significant increase in death).
Figure 2.
KD of Mdmx in MCF-7 cells attenuated cell proliferation by activating p53. MCF-7 cells expressing Mdmx shRNA or control shRNA (shMdmx wobble control) were treated with DOX (80 ng/ml) and cell numbers were quantified at 3 and 5 days using volumetric counting on the FACs VERSE flow cytometer. Statistical significance was only measured after DOX treatment of Mdmx shRNA samples (3 days: P=0.0041; 5 days P=0.0004) (a). The proportion of cells expressing the shRNA constructs were determined by GFP positivity at different days following treatment using flow cytometry (b). Protein expression of Mdmx, p53 and p21, PML and the loading control HSP60 were determined by western blotting at day 3 of treatment (c). RNA expression of Mdmx and p21 were determined at day 3 of treatment by RT-PCR. Each experiment in a, b and d represents triplicate data. All the experiments were repeated at least three times. Errors bars represent S.D. of the mean. P-values were calculated by Student's t-test. Senescence after 5 days of DOX treatment led to significant senescence only in shMdmx cells and is indicated by β-gal staining (e), and quantified as significant by Student's t-test P-value<0.0001 (f)
To measure the effect of DOX treatment on Mdmx levels, cells were treated for 3 days with DOX, or controls were left untreated, before Mdmx protein staining by western blotting. Mdmx levels were significantly reduced in the presence of DOX (Figure 2c), which correlated with elevated expression of the p53 target gene p21, which is growth inhibitory. p53 levels remained relatively unchanged compared with the shRNA control (Figure 2c). Reduction in Mdmx messenger RNA (mRNA) levels and concurrent induction of p21 mRNA levels were confirmed by quantitative PCR (QPCR) (Figure 2d). Senescence growth inhibition was suggested by the elevation of levels of p21 protein in response to shMdmx induction, together with β-galactosidase (β-gal) staining of cells (Figures 2e and f).
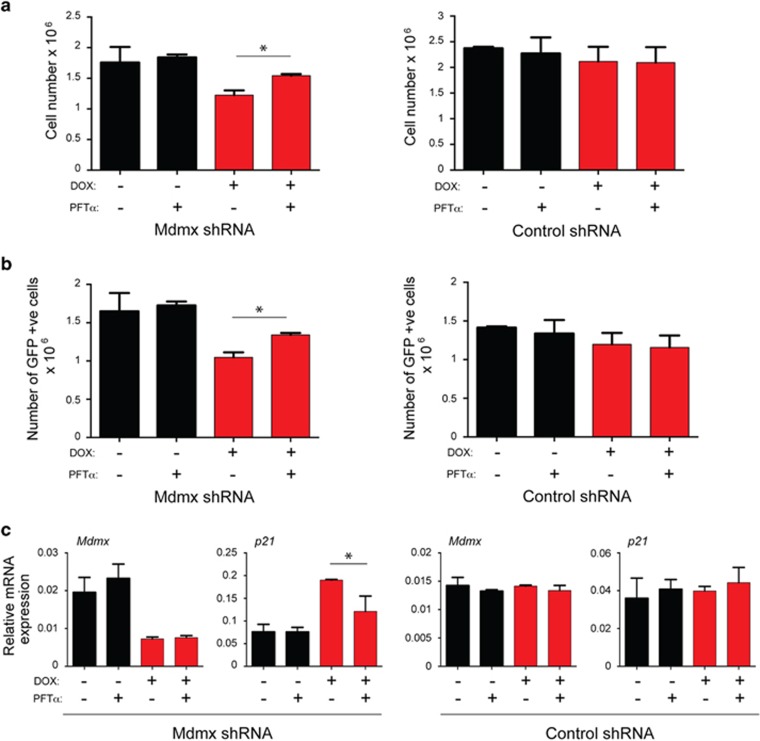
To demonstrate more directly the role of p53 in the effect of Mdmx KD, the p53 inhibitor pifithrin-α (PFTα) was included. PFTα partially relieved the effect of Mdmx on total cell viability (Figure 3a), as evident from the proportion of viable GFP-positive cells (Figure 3b) and the unaltered expression of the p53 target gene p21 (Figure 3c). Of note, treatment of cells with PFTα did not interfere with the Mdmx KD nor did it affect p53 levels (data not shown). Together, these experiments demonstrate that treatment of cells with PFTα partially rescued the cells from the effect of Mdmx KD, which supports the role of p53 in the observed effects of Mdmx KD.
Figure 3.
KD of Mdmx impeded MCF-7 cells in a p53-dependent manner. MCF-7 cells expressing Mdmx shRNA or control shRNA (shMdmx wobble control) were treated and analyzed as described in Figure 2, with the exception that half the samples were also treated with the p53 inhibitor pifithrin-α (2.5 μM PFTα). For the analysis of cell numbers (a) and GFP positivity (b), cells were treated for 5 days. PFTα offered statistically significant protection against Mdmx shRNA, both for cell numbers (a, P=0.0027) and also for numbers of GFP positive cells (b, P=0.0018). For the measurement of Mdmx and p21 expression, cells were treated for 3 days and analyzed by RT-PCR (c). All the experiments represent triplicates. Errors bars represent S.D. of mean. P-values were calculated by Student's t-test
Downregulation of Mdmx impedes growth of BrCa cells in mice
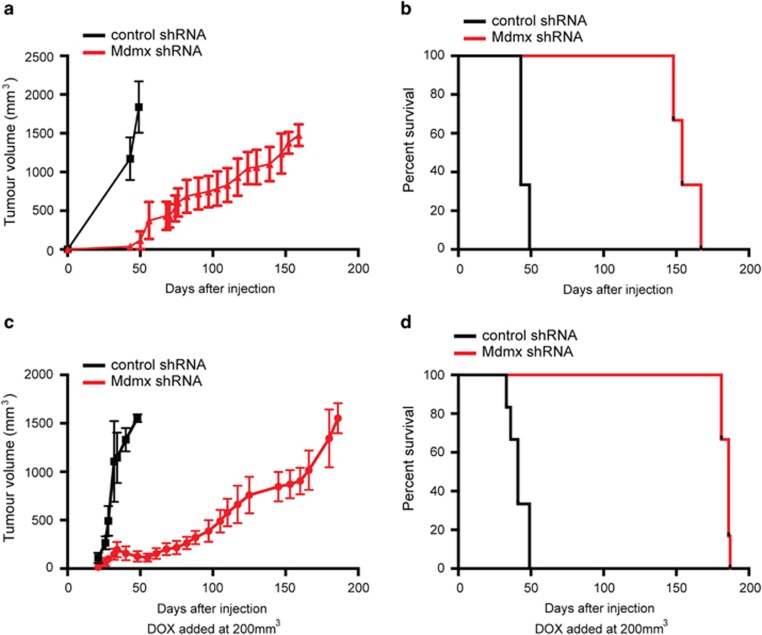
To determine whether the effects of Mdmx KD on cell growth and reactivation of p53 that were observed in cultured cells in vitro also occur in vivo, we employed the orthotopic xenograft transplantation approach in NOD/SCID/common−γchain knockout (NSG) mice. Initially, we examined the effect of Mdmx KD on the establishment of the tumors. For this purpose, MCF-7 cells expressing either the conditional Mdmx KD or control lentiviral vector were orthotopically injected into contralateral mammary fat pads (numbers 4 and 9) of six NSG females and mice were treated with DOX through an IP injection followed by DOX-supplemented drinking water. Tumors in the control mice grew rapidly, with the sum of the two tumors per mouse reaching ethical size of 1500 mm3 by ~50 days post injection (Figure 4a) and were duly terminated (Figure 4b). Remarkably, by this time point none of the Mdmx KD mice developed tumors to any significant size (Figure 4a) and survived past 150 days (Figure 4b). These results demonstrate that downregulation of Mdmx dramatically extends survival of mice orthotopically transplanted with luminal BrCa cells. Therefore, Mdmx promotes establishment of wt p53-harboring BrCa tumors in this orthotopic xenograft model.
Figure 4.
KD of Mdmx attenuated the development and progression of MCF-7-derived xenograft tumors in mice. MCF-7 cells expressing Mdmx shRNA or control shRNA (vector alone) were injected into two mammary fat pads of each female mouse (n=6 per group). For the assay assessing prevention of tumor development, mice were pretreated 3 days before cell injection then again on the day of injection with DOX (0.4 mg/ml) intraperitoneally, followed by DOX in drinking water (2 mg/ml). Tumor volume was measured every 3 days following the first appearance (a) and mice were culled and analyzed when tumor volume reached ethical size (b). The median survival for the control group was 43 days, whereas survival for the group expressing Mdmx shRNA was 154 days (Mantel–Cox test, P=0.0006). For the treatment assay, the DOX treatment started when tumor size reached 200 mm3. Tumor volume (c) and survival of mice are plotted (d). The median survival for the control group was 41 days, whereas that of Mdmx shRNA-expressing group was 186 days (Mantel–Cox test, P=0.0008)
To mimic a more clinical setting, we examined the effect of Mdmx KD on established tumors. For this purpose, the same orthotopic xenograft model described above was employed, with the exception that DOX treatment was started when tumors reached 200 mm3. The DOX treatment had no effect on the volume of the control tumors, which grew to the ethical end point size by ~50 days post injection. In stark contrast, DOX treatment of Mdmx KD tumors resulted initially in tumor regression, followed by extreme growth retardation compared with the controls. Importantly, the Mdmx KD tumors reached their ethical end point at ~180 days post injection (Figure 4c), 130 days after control mice termination (Figure 4d). This dramatic result clearly demonstrates that downregulation of Mdmx efficiently impedes the growth of an established tumor of transplanted MCF-7 cells. This provides the first demonstration that, at least in this model, downregulation of Mdmx can be used to treat established BrCa luminal tumors.
Downregulation of Mdmx induces cellular senescence in vivo
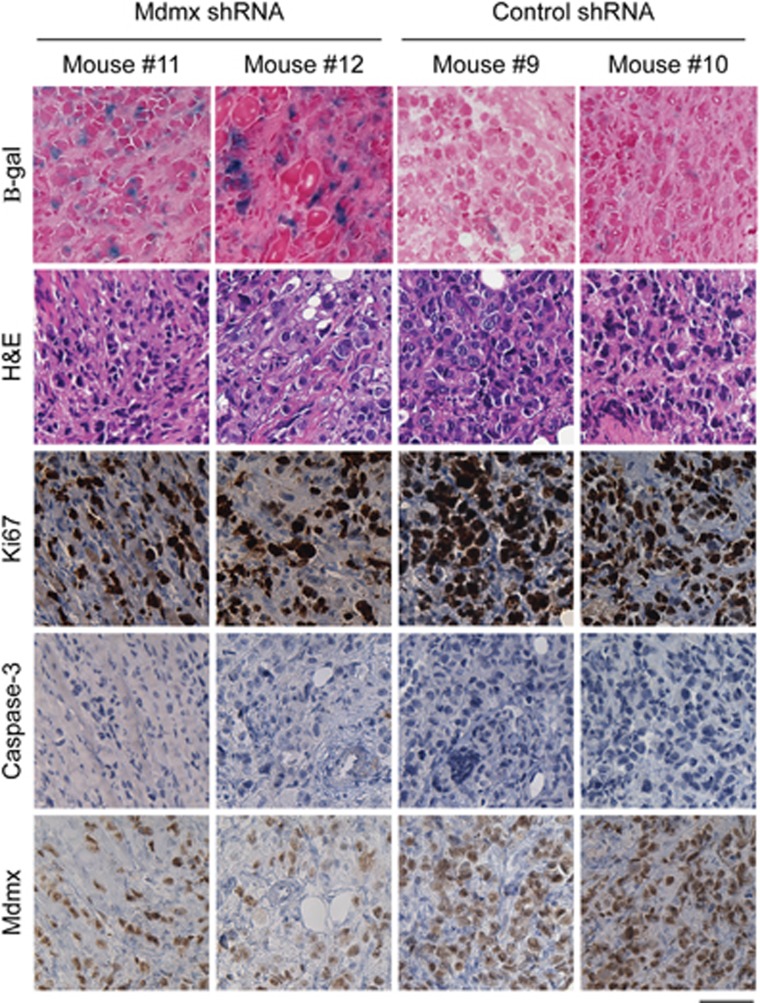
To define the cellular mechanism by which Mdmx KD impedes the growth of MCF-7 cells in vivo, the treatment experiment described in Figures 4c and d was repeated with the exception that mice were subjected to DOX treatment for only 7 days before tumor tissues were collected for analyses. This time point was chosen, as it defined the narrow time window when the tumor size in both cohorts was sufficiently small for DOX infiltration, but large enough for accurate measurement. To identify the nature of the cellular response to Mdmx KD, samples were subjected to analysis of Ki67 as a proliferation marker, activated caspase-3 as a marker for apoptosis and β-gal for identification of cellular senescence. A distinct difference in the cellular response between control mice and the Mdmx KD cohort was evident after 7 days of DOX treatment. Notably, the Mdmx KD tumors revealed a significantly stronger staining for β-gal than the control group, implicating cellular senescence as the major cellular response to Mdmx KD (Figure 5). This notion is also supported by elevation in p21 expression (as seen in vitro in Figures 2c and d), an important marker for cellular senescence.21 Consistently, reduction in Ki67 staining in Mdmx KD tumors was more evident than in the control group. Low levels of apoptosis, as defined by active caspase-3 staining, was measured in all samples, with no significant difference between the groups. This is consistent with senescence (as indicated by β-gal staining) as the major outcome in response to Mdmx KD. The morphology of the transplanted tumors was visualized by hematoxylin and eosin staining and, as expected, it was very similar between groups at this early time point. Reduction in Mdmx levels was identified as reduced staining intensity (not cellular proportion) in response to KD after 7 days, indicating that DOX was penetrating the tumors. We therefore conclude that Mdmx KD in MCF-7 cells induces cellular senescence in vivo.
Figure 5.
KD of Mdmx promoted cellular senescence in tumor xenografts in vivo. MCF-7 cells expressing Mdmx shRNA or control shRNA (shMdmx wobble control) were injected orthotopically into the mammary fat pads of NSG females (n=6) as described in Figure 4c treatment assay. Following 7 days of DOX treatment, mice were killed and tumors were either cryopreserved in preparation for β-gal staining (senescence marker) or formalin fixed and paraffin embedded for staining with hematoxylin and eosin (H&E; for morphological characterization) or IHC staining for Ki67 (proliferation marker), activated caspase-3 (apoptosis marker) and Mdmx, respectively. Mdmx KD was found to correspond with increased senescence and reduced ki67, even after only 7 days of treatment. The scale bar is 50 μm. Representative tumors from two mice of each group mice are shown
Growth inhibition through Mdmx KD is corroborated in wt p53 luminal MPE600 cell line
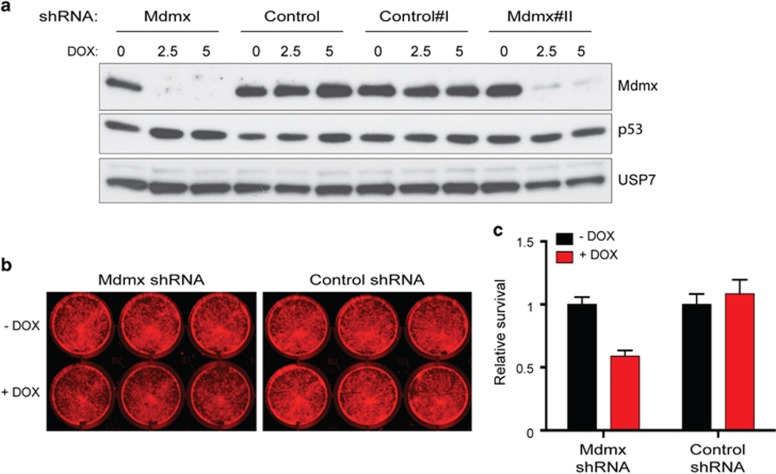
Reiteration of the growth inhibitory potency of Mdmx KD in an additional luminal BrCa cell line was considered important to establish its potential across this BrCa subtype. The wt p53 cell line MPE600 was subjected to Mdmx KD for 8 days (western blotting, Figure 6a). Mdmx KD was coincident with elevation of p21 and Mdm2 proteins levels, consistent with p53 transcriptional activity, where USP7 was used as a housekeeping control. Cell growth inhibition in response to 2.5 ng/ml DOX for 8 days was demonstrated in a colony assay (Figure 6b) and was identified to be statistically significant (Figure 6c).
Figure 6.
KD of Mdmx induced growth inhibition also in wt p53 luminal BrCa cell line MPE600. MPE600 cells transduced with either shMdmx constructs or shRNA controls were either not treated or treated with DOX (2.5 ng/ml) for 8 days and analyzed by western blotting (a) with protein staining for Mdmx, p53, p21, Mdm2 and USP7. Cell growth inhibition under the same conditions was demonstrated by a colony assay, representative plates are shown in b and the results are summarized in c
Basal-like cell line SKBR7 expressing wt p53 was also growth inhibited by Mdmx KD
To identify whether high levels of Mdmx in additional BrCa subtypes could account for wt p53 tolerance, the basal-like BrCa cell line SKBR7 was subjected to Mdmx KD (2.5 ng/ml DOX, 8 days; western blotting, Figure 7a), causing profound growth inhibition (Figure 7b), in a statistically significant manner (Figure 7c). Together, these findings imply that Mdmx KD has a capacity to inhibit the growth of BrCas expressing wt p53, independent of BrCa subtype.
Figure 7.
KD of Mdmx growth inhibited wt p53 basal-like SKBR7. Wt p53 basal-like SKBR7 cells transduced with either shMdmx constructs or shRNA controls were either not treated or treated with DOX (2.5 and 5 ng/ml) for 8 days and analyzed by western blotting (a) with protein detection of Mdmx, p53 and USP7. A colony assay of 2.5 ng/ml DOX-treated samples and controls demonstrated significant growth inhibition with Mdmx KD as visualized in b and as quantified in c
Discussion
The molecular explanation for the relatively low p53 mutational frequency in luminal BrCas and increased incidence in basal-like subtypes in contrast16, 18 has only been partially described. The major molecular mechanisms underlying wt p53 status in cancer cells are elevation in the Mdm proteins levels (reviewed in Wade et al.6 and Marine et al.10) or a loss of the key activator ARF (reviewed in Sherr22). Screening for alterations in the Mdm genes in luminal BrCa revealed rare amplification of the Mdm2 gene14, 23 and no selection for Mdm2 SNP309 in primary tumors.24
Similar to our findings, Mdm2 nuclear staining was relatively frequent in BrCa biopsies (>50%)25 and also in ~40% of benign BrCa cases.26 However, deregulated Mdm2 linked to estrogen in this BrCa context cannot account for low p53 levels (as reported for MCF7, where estrogen-induced elevated Mdm2 did not correlate with reduced p53).27
As for Mdmx, there is no evidence for its involvement in heritable BrCa,28 and in somatic BrCa reports of amplifications of the Mdmx gene vary14, 15 (apparently dependent on the designated threshold). Our results are consistent with low-frequency involvement of Mdm2 in luminal BrCa, but revealed high Mdmx protein levels in all the luminal BrCa cases (Figure 1). Our FISH analyses identified only one sample with amplification of the Mdmx gene (Supplementary Figure 1); hence, we conclude that elevation in Mdmx expression is not due to gene amplification. Danovi et al.15 showed that the majority of BrCa cases did not express high Mdmx RNA levels, which support the notion that much of the elevation in Mdmx expression occurs at the protein level.
Unexpectedly, we found that normal breast ductal epithelial cells express elevated levels of Mdmx and Mdm2, correlating with low p53 levels. In contrast, the myoepithelial cells, stroma and fatty tissue lack Mdmx staining. Importantly, the breast ductal epithelial cells are the origin of malignant growth. Our study therefore demonstrates that during the transition to luminal malignancy, the levels of Mdmx and Mdm2 increase from an already high expression. This is consistent with the lack of gene amplification in most BrCa samples that we analyzed. The finding of elevated Mdm proteins in normal breast was unexpected, as it stands in sharp contrast to melanoma13 and gastric29 cancers, for example, where normal skin and the stomach have near to no detectable Mdmx levels.
As p53 is activated by oncogenic stresses, we argue that the maintenance of high levels of Mdm protein expression in the breast is selected for during tumorigenesis, to suppress the growth inhibition by p53. A central question raised by these observation is why are such elevated levels of the Mdm proteins required and why exclusively in the epithelial cells? High Mdm levels in the normal ductal epithelial cells have not been duly considered to our knowledge in studies of BrCa to date (despite indication of staining in the normal breast in The Protein Altas30). These findings are likely to have important consequences for therapy as discussed below.
Mdmx has been targeted in MCF-7 cells by RNAi15, 20, 31 and by small-molecule inhibitors,32 causing significantly reduced cell proliferation. Our in vitro results (Figure 2) are consistent with these findings and support the p53 dependence (Figure 3). Importantly, by reiterating growth inhibition induced by Mdmx KD in an additional wt p53 luminal BrCa line (Figure 6), our findings demonstrate the broader implications of this phenomena across this BrCa subtype.
We have extended the in vitro findings to a mouse model demonstrating that Mdmx KD efficiently attenuates the establishment of orthotopic transplantation of MCF-7 cells (Figure 4), regresses established tumors and attenuates tumor regrowth, which together substantially prolongs life by almost 19 weeks (Figure 4). Validating the effect of Mdmx KD in patient-derived xenografts (PDXs) is desirable; however, the ‘take-rate' of luminal BrCA PDXs is very low,33 having an impact on the feasibilities of such experiments. The predominant response in these tumors is cellular senescence (Figure 5), which is consistent with elevated p21 expression following Mdmx KD (Figure 2). Previous studies targeting Mdmx in MCF-7 cells using small-molecule inhibitors resulted in the activation of p53-induced cell death32, 34 or cell cycle arrest.31 However, in both studies the assays were performed in vitro, whereas our analysis was performed on in vivo-derived tumors. Therefore, we provide the first demonstration of the role of cellular senescence in response to targeting Mdmx in BrCa in vivo.
Significantly, these studies for the first time extended beyond the luminal subtype to basal-like BrCa and demonstrated that in a wt p53 background, Mdmx KD caused potent growth inhibition (SKBR7; Figure 7). These findings are profound, as they demonstrate that the growth inhibitory capacity of Mdmx KD is not restricted by BrCa subtype and thus has potential therapeutic relevance to all wt p53 BrCas.
Approximately 50% of human cancers express wt p53. The proof-of-concept of restoration of wt p53 tumor-suppressive function has been demonstrated in a series of elegant papers, using multiple mouse models with conditional p53 expression.35, 36, 37 As Mdm2 and Mdmx are the key inhibitors of p53, cancers in which either or both Mdm proteins are deregulated opens an attractive opportunity to restore p53 activity by targeting either one or both inhibitors. Targeting Mdm2 as a means to restore p53 function has attracted the major efforts to date.38 However, clinical trials revealed significant side effects due to on-target effects of Mdm2 inhibition in driving hematological toxicity as well as toxicity in the digestive system (e.g., see Ray-Coquard et al.39). These issues lend strong support for targeting Mdmx as an alternative approach to restore p53 activities (reviewed in Marine40). The proof-of-concept for targeting Mdmx has been successfully demonstrated in multiple cultured cell culture models and, importantly, in an in vivo mouse model, which demonstrated tolerance of mouse tissues to Mdmx deletion.6, 40, 41 We have now extended it to an in vitro and in vivo experimental model for wt p53 BrCa cells. The important distinction between BrCa cell sensitivity to Mdmx KD (as seen in this study and in others15, 31, 32, 34) compared with the apparent tolerance of normal cells41 implies that BrCa and other cancer cells with elevated Mdmx are hypersensitized to p53 activation. This phenomenon has been demonstrated in a wide range of cancers, where p53 has been restored (see Martins et al.,35 Ventura et al.36 and Xue et al.37) and supports the implications of our study that targeting Mdmx is a valid approach for treating BrCas harboring wt p53.
Multiple approaches have been developed to protect p53 from the inhibitory effects of Mdmx (reviewed in Wade et al.6 and Khoo et al.38). Targeting the p53–Mdmx interaction predominates using the following: small molecules (SJ-172550, WK298 and RO-5963), stapled peptides (SAH-p53-8, sMTide-02/O2a and ATSP-7041) and other modified peptides (pDI, PMI and PMI-N8A). Alternative approaches to reduce Mdmx expression levels include small molecules such as NSC207895, which is related to DNA-damaging agents, or 17-AAG, an HSP-90 inhibitor that destabilizes Mdmx by unknown mechanisms (reviewed in Wade et al.6 and Khoo et al.38). Some of these small molecules and peptides have a dual specificity for Mdm2 and Mdmx, albeit often with different affinities. The potency of dual targeting in BrCa is suggested by an in vitro study,42 but awaits testing in vivo with precision therapeutic agents.
Conclusions
Our study demonstrates that Mdmx is highly expressed in ductal epithelial breast cells, but not in other breast cells, and is further elevated in BrCa. Downregulation of Mdmx alone is sufficient to significantly attenuate the establishment of tumor growth and the progression of established tumors. Overall, our findings suggest that targeting Mdmx is an attractive strategy for the treatment of BrCas that express wt p53 (~70% of all cases).
Materials and Methods
All chemicals and reagents used in this study were purchased from Sigma-Aldrich, NSW, Australia, unless otherwise indicated. Analytical grade solvents and double-distilled water were used for all experiments, which were performed in triplicate and undertaken at least three times.
Cell culture
The BrCa cell line MCF-7 was acquired from ATCC (www.atcc.org) and maintained in Dulbecco's modified Eagle's medium (Gibco, Invitrogen, VIC, Mount Waverley, Australia) supplemented with 10% fetal calf serum (FCS) (Gibco) and 0.1% penicillin/streptomycin. BrCa cell lines SKBR7 and MPE600 were a gift from Mieke Schutte/John Martens (Erasmus MC, Rotterdam, The Netherlands) and were maintained in RPMI 1640 (Invitrogen) supplemented with 10% FCS and antibiotics. All cells were incubated at 5% CO2, 37 °C. All cells express wt p53 and elevated levels of Mdmx were previously demonstrated.19
Inducible lentiviral shRNA constructs and viral production
A third-generation lentiviral vector, FH1t with GFP tag43 was harnessed to generate shRNA to KD Mdmx (shMdmx: forward (Fwd) strand: 5′-TCCC ACAGTCCTTCAGCTATTTCAT TTCAAGAGA ATGAAATAGCTGAAGGACTGT TTTTTC 3′ and reverse (Rev) strand: 5′-TCGAGAAAAA ACAGTCCTTCAGCTATTTCAT TCTCTTGAA ATGAAATAGCTGAAGGACTGT-3′) and for the control shRNA (shMdmxWobble: Fwd strand: 5′-TCCC ACCGTCCGCAAGCTATGTCAT TTCAAGAGA ATGACATAGCTTGCGGACGGT TTTTTC-3′ and Rev strand: 5′-TCGAGAAAAA ACCGTCCGCAAGCTATGTCAT TCTCTTGAA ATGACATAGCTTGCGGACGGT-3′). Confirmation was provided by additional Mdmx shRNA constructs (shMdmx#I: Fwd strand: 5′-TCCC GTGCAGAGGAAAGTTCCAC TTCAAGAGA GTGGAACTTTCCTCTGCAC TTTTTC-3′ and Rev strand: 5′-TCGAGAAAAA GTGCAGAGGAAAGTTCCAC TCTCTTGAA GTGGAACTTTCCTCTGCAC-3′ shMdmx#II: Fwd strand: 5′-TCCC AGTCAAGACCAACTGAAGC TTCAAGAGA GCTTCAGTTGGTCTTGACT TTTTTC-3′ and Rev strand: 5′-TCGAGAAAAA AGTCAAGACCAACTGAAGC TCTCTTGAA GCTTCAGTTGGTCTTGACT-3′ shMdmx#III: Fwd strand: 5′-TCCC GAATCTCTTGAAGCCATGT TTCAAGAGA ACATGGCTTCAAGAGATTC TTTTTC-3′ and Rev strand: 5′-TCGAGAAAAA GAATCTCTTGAAGCCATGT TCTCTTGAA ACATGGCTTCAAGAGATTC-3′). The additional shControl I was directed toward mouse Mdmx sequence and does not knock down human Mdmx Fwd strand: 5′-TCCC GAATCTTGTTACATCAGCT TTCAAGAGA AGCTGATGTAACAAGATTC TTTTTC-3′ and Rev strand: 5′-TCGAGAAAAA AGCTGATGTAACAAGATTC TCTCTTGAA ACATGGCTTCAAGAGATTC-3′. Viruses were generated and cells transduced essentially as described,43 with CaPO4 transfection replaced by PEI (as per the manufacturer's instructions).
Western blot analysis
Western blot analysis was undertaken as previously described.44 Immunoblotting was performed with antibodies to human as follows: p53 (DO-1; 1801; a kind gift of Sir D. Lane), Mdmx (mouse clone 8C6; Millipore, Billerica, MA, USA; BL1285 Bethyl Laboratories, Inc., Montgomery, TX, USA), Mdm2 (SMP14; Santa Cruz Biotechnology, Inc., Dallas, TX, USA; 3G9 and Millipore), HSP 60 (rabbit; sc-13966; Santa Cruz Biotechnology, Inc.), USP7 (BL1285 Bethyl Laboratories) and p21 (sc-397; Santa Cruz Biotechnology, Inc.; CP64 Millipore).
Flow cytometry analyses
Cells were cultured in doxycyclin (DOX) for induction of the shRNAs and were collected for live cell counts essentially as described previously,45 with the exception that To-pro-3-iodide (Life Technologies, Thermo Scientific, VIC, Mulgrave, Australia) was employed for DNA staining (according to the manufacturer's instructions) and volumetric cell enumeration was undertaken on the FACSVERSE (BD Biosciences, North Ryde, NSW, Australia). Cell cycle analyses were undertaken using the same facilities following the addition of 0.1% Triton X-100, to permeabilize the plasma membranes.
Quantitative real-time RT-PCR
Total RNA was extracted using TRIzol (Life Technologies, Thermo Scientific) according to the manufacturer's protocol. mRNA was reverse transcribed using M-mlv Reverse transcriptase (Promega, Alexandria, NSW, Australia) and QPCR was performed on the cDNA using SYBR green-reagent (Applied Biosystems, Scoresby, VIC, Australia) and designated primer sets.
Hu UBC Fwd: 5′-ATTTGGGTCGCGGTTCTTG-3′
Hu UBC Rev: 5′-TGCCTTGACATTCTCGATGGT-3′
Hu p21 Fwd: 5′-GAGGCCGGGATGAGTTGGGAGGAG-3′
Hu p21 Rev: 5′-CAGCCGGCGTTTGGAGTGGTAGAA-3′
Hu Mdmx Fwd: 5′-CAGCAGGTGCGCAAGGTGAA-3′
Hu Mdmx Rev: 5′-CTGTGCGAGAGCGAGAGTCTG-3′
The expression of each gene was calculated based on the cycle threshold (CT), set within the linear range of DNA amplification. The expression (arbitrary units) was calculated as the relative transcript abundance (RTA) by: RTA=10 000/(2ΔCT), where ΔCT=CT (gene of interest)−CT(UBC).
Colony assays
For colony assay, 10 000 (SKBR7) or 20 000 (MPE600) cells were seeded into 12-well plates and allowed to recover overnight before the addition of DOX (2.5 ng/ml). Nine days later, cells were fixed with 4% paraformaldehyde and stained with crystal violet (0.05% aqueous; 30 min). Plates were rinsed, dried and scanned on the Licor Odyssey (Mulgrave, VIC, Australia). Intensity of the signal was determined with the Image Studio software (Mulgrave, VIC, Australia).
Orthotopic tumor studies
Female NOD SCID γ-IL2R γ-chain (NSG) mice at ~6–8 weeks were orthotopically injected into contralateral mammary fat pads (numbers 4 and 9). The gene KD was affected by DOX administration initially through IP injection (40 μg/100 μl, with simultaneous DOX supplementation in the drinking water (2 mg/ml). Mice were aged during which time their tumors were measured, and at their ethical endpoints they were killed per institutional guidelines. Necropsies were performed, and tumor and control tissues were collected and divided for protein and RNA analysis and microscopy. All procedures were conducted in accordance with the Institute for Laboratory Animal Research Guide: The Care and Use of Laboratory Animals. All animal work was performed with approval from the Peter MacCallum Cancer Centre Animal Experimentation Ethics Committee.
Microscopy analyses
In preparation for examination of fixed tissues, tumors were preserved in 10% neutral buffered formalin. Fixed tissues were processed for paraffin embedding, sectioned and stained. Immunohistochemical staining was performed with antibodies to human p53 (Leica Biosystems, Newcastle Upon Tyne, UK; Novocastra mouse monoclonal DO-7), Mdmx (Bethyl Laboratories, Inc.; A300 287A), Mdm2 (Thermo Scientific, Neo Markers, Fremont, CA, USA), caspase-3 (R&D Systems, Minneapolis, MN, USA; Monoclonal Mouse 84803) and ki67 (Cell Marque, Sigma-Aldrich company, Carlsbad, CA, USA; rabbit monoclonal, SP6). Histopathological analyses were performed by an onco-pathologist (JMP). Photographs were taken using a using a BX-50 microscope with Leica DFC290HD camera (Olympus, Notting Hill, VIC, Australia) and Leica Application Suite V3.8.0 software. IHC scoring was undertaken using BX-41 microscope (Olympus).
Fresh-frozen tissues were prepared from OCT-embedded tissues snap frozen in isopentane liquid nitrogen vapors and cryostat sections were stained for β-gal (essentially as in Dimri et al.46). Images were visualized using a BX- 51 microscope (Olympus). β-Gal staining of cultured cells was performed as published.46 Images were visualized using a Zeiss Axio Vert.A1 microscope (Zeiss, Peabody, MA, USA) and acquired using SPOT Version 4.7 software (Diagnostic Instruments).
Tissue microarray
BrCa biopsies from archived patient samples from the Peter MacCallum Cancer Institute, Melbourne, were compiled into a BrCa TMA. Each tissue core was of 1-mm diameter47 and was classified into luminal, basal, HER2 and null subtypes according to immunohistochemical profiling.48 TMA were immunostained using the antibodies listed above. Samples were scored for both the proportion of cells stained and the intensity of staining. Staining intensity was evaluated on an escalating scale ranging from 0 to 3, whereas the proportion of cells stained was according to the following scale: 0 is negative; 1: <10% stained; 2: 10–50% stained; 3: 50–80% stained; and 4: at least 80%. A combined histoscore was calculated by adding the value of the intensity to the proportion of cells stained with a final maximum score of 8. The TMA consisted for 81 biopsies samples: 50 luminal (62%), 20 Her2 (25%), 11 basal (13%) and a panel of normal breast tissues; these were analyzed for the IHC staining.
Fluorescence in situ hybridization
Analyses were undertaken essentially as published.49 Target detection used were as follows: Vysis LSI MDM2 Spectrum Orange Probe (Abbott Molecular, Des Plaines, IL, USA) and Vysis CEP 12 (D12Z3) Spectrum Green Probe (Abbott Molecular) or Poseidon MDM4 (1q32) and SE 1 Control Probe (Kreatech Diagnostics, Amsterdam, The Netherlands). Fifty tumor nuclei per case were assessed for each case using the following criteria: a case was considered amplified if the ratio was >2.0 and not amplified if the ratio was≤2.0. This study was carried out according to the provisions of the Helsinki Declaration of 1975 and was reviewed and approved by the Peter MacCallum Cancer Centre Ethics Committee. For FISH analysis, Mdm2 gene amplification analyses 76 biopsy cores were analyzed: luminal n=36; Her2 n=12; and basal n=9. For Mdmx gene amplification, 80 biopsy cores were analyzed: luminal n=49; Her2 n=19; and basal n=12 (where numbers differ slightly from IHC due to core dropout during FISH staining).
Acknowledgments
This work was supported by NHMRC project grants (1026990, 1026988, 1049179 and 1063389) and NHMRC Fellowship to YH (9628426), by the CCV grant (1085154) and by the CASS Foundation. MJH was supported by an NHMRC project grant (APP1049720).
Author contributions
SH and YH conceived that study, designed experiments and wrote the paper. AJ, J-CM and SBF gave advice and helped interpret the results and designed the experiments. DB performed and designed experiments. CG helped drafting the paper and performed experiments. J-MP, PJP, SH, ET, T-MR, YYL, JP and AFAST performed experiments. MJH provided reagents and advice on experimental design. All authors have read and approved the final version of the manuscript.
Glossary
- β-gal
β-galactosidase
- CT
cycle threshold
- DOX
doxycycline
- FISH
fluorescence in situ hybridization
- Fwd
forward
- KD
knockdown
- NSG
NOD SCID γ-IL2R γ-chain
- PFTα
pifithrin-α
- Rev
reverse
- RTA
relative transcript abundance
- TMA
tissue micro-array
- Wt
wild type
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Melino
Supplementary Material
References
- 1Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer 2009; 9: 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Zuckerman V, Wolyniec K, Sionov RV, Haupt S, Haupt Y. Tumor suppression by p53: the importance of apoptosis and cellular senescence. J Pathol 2009; 219: 3–15. [DOI] [PubMed] [Google Scholar]
- 3Selivanova G. Wild type p53 reactivation: from lab bench to clinic. FEBS Lett 2014; 588: 2628–2638. [DOI] [PubMed] [Google Scholar]
- 4Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumor suppression. Nat Rev Cancer 2014; 14: 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Soussi T, Wiman KG. Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell 2007; 12: 303–312. [DOI] [PubMed] [Google Scholar]
- 6Wade M, Li YC, Wahl GM. MDM2 MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013; 13: 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Marine JC. MDM2 and MDMX in cancer and development. Curr Top Dev Biol 2011; 94: 45–75. [DOI] [PubMed] [Google Scholar]
- 8Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature 1997; 387: 296–299. [DOI] [PubMed] [Google Scholar]
- 9Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature 1997; 387: 299–303. [DOI] [PubMed] [Google Scholar]
- 10Marine JC, Dyer MA, Jochemsen AG. MDMX: from bench to bedside. J Cell Sci 2007; 120: 371–378. [DOI] [PubMed] [Google Scholar]
- 11Ito M, Barys L, O'Reilly T, Young S, Gorbatcheva B, Monahan J et al. Comprehensive mapping of p53 pathway alterations reveals an apparent role for both SNP309 and MDM2 amplification in sarcomagenesis. Clin Cancer Res 2011; 17: 416–426. [DOI] [PubMed] [Google Scholar]
- 12Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C et al. Inactivation of the p53 pathway in retinoblastoma. Nature 2006; 444: 61–66. [DOI] [PubMed] [Google Scholar]
- 13Gembarska A, Luciani F, Fedele C, Russell EA, Dewaele M, Villar S et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nat Med 2012; 18: 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Yu Q, Li Y, Mu K, Li Z, Meng Q, Wu X et al. Amplification of Mdmx and overexpression of MDM2 contribute to mammary carcinogenesis by substituting for p53 mutations. Diagn Pathol 2014; 9: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol 2004; 24: 5835–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Dumay A, Feugeas JP, Wittmer E, Lehmann-Che J, Bertheau P, Espie M et al. Distinct tumor protein p53 mutants in breast cancer subgroups. Int J Cancer 2013; 132: 1227–1231. [DOI] [PubMed] [Google Scholar]
- 17Bertheau P, Lehmann-Che J, Varna M, Dumay A, Poirot B, Porcher R et al. p53 in breast cancer subtypes and new insights into response to chemotherapy. Breast 2013; 22: S27–S29. [DOI] [PubMed] [Google Scholar]
- 18Walerych D, Napoli M, Collavin L, Del Sal G. The rebel angel: mutant p53 as the driving oncogene in breast cancer. Carcinogenesis 2012; 33: 2007–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 2009; 101: 736–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Lam S, Lodder K, Teunisse AF, Rabelink MJ, Schutte M, Jochemsen AG. Role of Mdm4 in drug sensitivity of breast cancer cells. Oncogene 2010; 29: 2415–2426. [DOI] [PubMed] [Google Scholar]
- 21Munoz-Espin D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol 2014; 15: 482–496. [DOI] [PubMed] [Google Scholar]
- 22Sherr CJ. Ink4-Arf locus in cancer and aging. Wiley Interdiscip Rev Dev Biol 2012; 1: 731–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Quesnel B, Preudhomme C, Fournier J, Fenaux P, Peyrat JP. MDM2 gene amplification in human breast cancer. Eur J Cancer 1994; 30A: 982–984. [DOI] [PubMed] [Google Scholar]
- 24Kulkarni DA, Vazquez A, Haffty BG, Bandera EV, Hu W, Sun YY et al. A polymorphic variant in human MDM4 associates with accelerated age of onset of estrogen receptor negative breast cancer. Carcinogenesis 2009; 30: 1910–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Marchetti A, Buttitta F, Girlando S, Dalla Palma P, Pellegrini S, Fina P et al. mdm2 gene alterations and mdm2 protein expression in breast carcinomas. J Pathol 1995; 175: 31–38. [DOI] [PubMed] [Google Scholar]
- 26Baunoch D, Watkins L, Tewari A, Reece M, Adams L, Stack R et al. MDM2 overexpression in benign and malignant lesions of the human breast. Int J Oncol 1996; 8: 895–899. [DOI] [PubMed] [Google Scholar]
- 27Graves B, Thompson T, Xia M, Janson C, Lukacs C, Deo D et al. Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc Natl Acad Sci USA 2012; 109: 11788–11793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Reincke S, Govbakh L, Wilhelm B, Jin H, Bogdanova N, Bremer M et al. Mutation analysis of the MDM4 gene in German breast cancer patients. BMC Cancer 2008; 8: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Busuttil RA, Zapparoli GV, Haupt S, Fennell C, Wong SQ, Pang JM et al. Role of p53 in the progression of gastric cancer. Oncotarget 2014; 5: 12016–12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Ponten F, Schwenk JM, Asplund A, Edqvist PH. The Human Protein Atlas as a proteomic resource for biomarker discovery. J Intern Med 2011; 270: 428–446. [DOI] [PubMed] [Google Scholar]
- 31Heminger K, Markey M, Mpagi M, Berberich SJ. Alterations in gene expression and sensitivity to genotoxic stress following HdmX or Hdm2 knockdown in human tumor cells harboring wild-type p53. Aging (Albany NY) 2009; 1: 89–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Wang H, Ma X, Ren S, Buolamwini JK, Yan C. A small-molecule inhibitor of MDMX activates p53 and induces apoptosis. Mol Cancer Ther 2011; 10: 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Vaillant F, Merino D, Lee L, Breslin K, Pal B, Ritchie ME et al. Targeting BCL-2 with the BH3 mimetic ABT-199 in estrogen receptor-positive breast cancer. Cancer Cell 2013; 24: 120–129. [DOI] [PubMed] [Google Scholar]
- 34Brekman A, Singh KE, Polotskaia A, Kundu N, Bargonetti J. A p53-independent role of Mdm2 in estrogen-mediated activation of breast cancer cell proliferation. Breast Cancer Res 2011; 13: R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell 2006; 127: 1323–1334. [DOI] [PubMed] [Google Scholar]
- 36Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L et al. Restoration of p53 function leads to tumor regression in vivo. Nature 2007; 445: 661–665. [DOI] [PubMed] [Google Scholar]
- 37Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V et al. Senescence and tumor clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007; 445: 656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Khoo KH, Verma CS, Lane DP. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Discov 2014; 13: 217–236. [DOI] [PubMed] [Google Scholar]
- 39Ray-Coquard I, Blay JY, Italiano A, Le Cesne A, Penel N, Zhi J et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol 2012; 13: 1133–1140. [DOI] [PubMed] [Google Scholar]
- 40Marine JC. Pharmacological rescue of p53 in cancer therapy: widening the sensitive tumor spectrum by targeting MDMX. Cancer Cell 2010; 18: 399–400. [DOI] [PubMed] [Google Scholar]
- 41Garcia D, Warr MR, Martins CP, Brown Swigart L, Passegue E, Evan GI. Validation of MdmX as a therapeutic target for reactivating p53 in tumors. Genes Dev 2011; 25: 1746–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42Hu B, Gilkes DM, Farooqi B, Sebti SM, Chen J. MDMX overexpression prevents p53 activation by the MDM2 inhibitor Nutlin. J Biol Chem 2006; 281: 33030–33035. [DOI] [PubMed] [Google Scholar]
- 43Herold MJ, van den Brandt J, Seibler J, Reichardt HM. Inducible and reversible gene silencing by stable integration of an shRNA-encoding lentivirus in transgenic rats. Proc Natl Acad Sci USA 2008; 105: 18507–18512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44Louria-Hayon I, Grossman T, Sionov RV, Alsheich O, Pandolfi PP, Haupt Y. The promyelocytic leukemia protein protects p53 from Mdm2-mediated inhibition and degradation. J Biol Chem 2003; 278: 33134–33141. [DOI] [PubMed] [Google Scholar]
- 45Haupt S, di Agostino S, Mizrahi I, Alsheich-Bartok O, Voorhoeve M, Damalas A et al. Promyelocytic leukemia protein is required for gain of function by mutant p53. Cancer Res 2009; 69: 4818–4826. [DOI] [PubMed] [Google Scholar]
- 46Dimri G, Lee X, Basile G, Acosta M, Scott G, Roskelley C et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. PNAS 1995; 92: 9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Giltnane JM, Rimm DL. Technology insight: Identification of biomarkers with tissue microarray technology. Nat Clin Pract Oncol 2004; 1: 104–111. [DOI] [PubMed] [Google Scholar]
- 48Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 2004; 10: 5367–5374. [DOI] [PubMed] [Google Scholar]
- 49Takano EA, Rogers TM, Young RJ, Rayoo M, Kostos P, Ferguson R et al. The molecular characterisation of unusual subcutaneous spindle cell lesion of breast. J Clin Pathol 2012; 65: 746–750. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.