HBV sub-genotype A1 has a severe virologic expression among HIV-positive Malawians, contrasting with the mild outcomes observed elsewhere. By deep sequencing, HBV drug-resistance was universal in viremic patients receiving lamivudine/stavudine/nevirapine, with faster emergence than previously estimated by Sanger sequencing.
Keywords: sub-genotype A1, viral load, resistance, Africa, lamivudine
Abstract
Background. It has been proposed that hepatitis B virus (HBV) sub-genotype A1 infections have mild outcomes and a low risk of drug-resistance among patients infected with human immunodeficiency virus (HIV) receiving lamivudine-containing antiretroviral therapy (ART) without tenofovir in Africa.
Methods. The virologic expression of HBV sub-genotype A1 coinfection was studied over 12 months in HIV-positive adults starting stavudine/lamivudine/nevirapine in Malawi, using Sanger, deep, clonal, and single full-genome sequencing for the sensitive characterization of HBV resistance-associated mutations (RAMs).
Results. Among 1117 subjects, 133 (12%) tested HBsAg-positive. After starting ART, retention rates were 96/133 (72%) at 6 months and 54/133 (41%) at 12 months. Based upon the last available follow-up, 92/96 (96%) subjects achieved HIV-1 RNA <40 copies/mL, 48/96 (50%) showed HBV DNA <14 IU/mL, and 24/96 (25%) acquired HBV RAMs. At 6 months, M204I was detected in 8/46 (17%) and 16/17 (94%) subjects using Sanger and deep sequencing, respectively. At 12 months, all viremic patients had multiple resistance and compensatory mutations coexisting on the same HBV genomes. Comparing HBeA-positive (67/133, 50%) with HBeAg-negative subjects, 64/67 (96%) vs 35/66 (55%) showed baseline HBV DNA >2000 IU/mL (P = .0006), 39/47 (17%) vs 9/49 (82%) had persistent HBV DNA detection during follow-up (P < .0001), and 23/47 (49%) vs 2/49 (4%) acquired HBV RAMs (P < .0001). Baseline HBV DNA levels were median 8.1 vs 5.3 log10 IU/mL in subjects with vs those without treatment-emergent RAMs (P < .0001).
Conclusions. HBV sub-genotype A1 infections showed a severe virologic expression in HIV-positive Malawians. The findings strengthen the urgency of interventions to improve ascertainment and management of chronic hepatitis B in the region.
Chronic hepatitis B virus (HBV) infection is common in patients who are positive for human immunodeficiency virus (HIV) [1]. HIV promotes HBV replication and markedly increases the risk of liver-related disease, with the greatest impact seen in patients with low CD4 cell counts and high HBV DNA levels [2]. Guidelines recommend HBV screening for all HIV-infected patients and indicate that effective management of co-infection requires maximal suppression of HIV and HBV replication, typically with antiretroviral therapy (ART) regimens containing tenofovir plus lamivudine or emtricitabine [3, 4].
Although regional differences are marked, high HBV coinfection rates—in some areas exceeding 10%—have been observed in HIV-positive adults in sub-Saharan Africa [5–16], mirroring high-endemicity levels in the general population [17]. Yet, HBV screening is not systematically performed and ART remains “HBV-blind” across much of the region [8, 12, 15, 16, 18, 19]. Recommended ART regimens have traditionally comprised zidovudine or stavudine with lamivudine plus nevirapine or efavirenz. Although access to tenofovir is increasing [20], it remains far from universal. As a result, large numbers of HIV/HBV coinfected patients in sub-Saharan Africa have for years received lamivudine as the sole HBV-active agent [8, 12, 15, 16, 18].
Treating HBV with lamivudine alone carries a risk of virologic breakthrough with selection of HBV drug resistance, high HBV DNA levels, and progression of liver damage [3]. In Western cohorts of HIV/HBV coinfected patients the incidence rate of HBV resistance is approximately 20% per year [21–23]. Data from sub-Saharan Africa are conflicting. Some investigators have suggested that treatment outcomes in the region are distinct from those observed in Western cohorts [11, 12, 15, 16], possibly reflecting the circulation of HBV variants such as sub-genotype A1 that are unique to Africa [24]. Studies to date have used Sanger sequencing for detecting HBV drug-resistant mutants, which can underestimate resistance rates. To gather more evidence, we investigated the virologic expression of HBV sub-genotype A1 infection in HIV-positive adults starting lamivudine-containing ART without tenofovir in Malawi and used multiple sequencing methodologies for the sensitive characterization of treatment-emergent drug resistance.
METHODS
Study Population
Subjects starting fixed-dose stavudine/lamivudine/nevirapine in 2007–2009 at the Queen Elizabeth Central Hospital (QECH) of Blantyre entered a prospective study evaluating ART outcomes. Indications for starting ART were a CD4 count <250 cells/mm3 or World Health Organisation (WHO) stage 3/4 disease. Sampling was scheduled at baseline and after 6 and 12 months of ART. Nonattendances at 6 months were traced by telephoning the patient or the patient's named next of kin and checking medical records. Reasons for nonattendance at 12 months were not recorded. CD4 cell counts were measured in the QECH diagnostic laboratory as part of routine care. Alanine transaminase (ALT) levels were not part of routine testing and were measured in the local Malawi-Liverpool Wellcome Trust research laboratory. Serum and plasma were stored at −80°C prior to transport to the United Kingdom for HBV serology and HIV and HBV virology testing. The College of Medicine Research and Ethics Committee and the Liverpool School of Tropical Medicine Ethics Board approved the study. Written informed consent was obtained from participants; the study was conducted in accordance with the principles of the Declaration of Helsinki. Patients with jaundice were excluded.
Serology
HBV testing was not part of routine care at QECH. Hepatitis B surface antigen (HBsAg) was tested retrospectively with the Bioelisa HBsAg assay (Biokit, Barcelona, Spain). HBVe antigen (HBeAg) and anti-HBe antibody were tested by Architect (Abbott Diagnostics, Maidenhead, UK).
Viral Load
HBV DNA was quantified by real-time polymerase chain reaction (PCR) as previously described [9]; the assay lower limit of quantification is 14 IU/mL. Human immunodeficiency virus type 1 (HIV-1) RNA was quantified by the Real Time HIV-1 assay (Abbott Diagnostics); the assay lower limit of quantification is 40 copies/mL.
Population and Clonal Sequencing
Plasma samples with HBV DNA levels >50 IU/mL underwent Sanger sequencing of the HBV polymerase reverse transcriptase domain (amino acids 1–344) and genotyped as previously described [9]. For clonal sequencing, PCR products were purified using the Geneclean kit (MP Biomedicals, Santa Ana, California) and cloned in TOPO XL vectors (Life Technologies).
Deep Sequencing
Following extraction (QIAamp UltraSens Virus Kit, Qiagen, Crawley, UK), HBV DNA was amplified by Phusion High-Fidelity DNA Polymerase (Finnzymes, Vantaa, Finland) using primers shown in the Supplementary Table. The 623 bp amplicon was purified (QIAquick PCR Purification Kit, Qiagen), analyzed for purity (Bioanalyzer DNA 7500 kit, Agilent Santa Clara, California), and quantified (Qubit dsDNA HS Assay, Life Technologies), prior to randomly shearing by sonication (median length 300 bp) and sequencing by Illumina GAII (San Diego, California). Sequence reads were adaptor-trimmed. Reads with quality scores <40 were discarded using the Quality Assessment of Short Read (QUASR) Pipeline [25]. Analysis (>3000 reads per nucleotide) was performed using Segminator II [26].
Full-length Single Genome Sequencing
Following extraction, HBV DNA plated under limiting dilution was subjected to nested PCR amplification by Expand High FidelityPLUS PCR System (Roche, Basel, Switzerland) to produce a 3220 bp product. Amplicon-positive wells were identified by gel electrophoresis, and their DNA concentration was analyzed by Nanodrop 8000 (Thermo Fisher Scientific, Waltham, Massachusetts), prior to sequencing. By Poisson distribution, where 30% of PCR reactions yielded an amplicon there was an 80% likelihood that amplification had occurred from a single molecule. Amplification and sequencing primers are shown in the Supplementary Table.
Analysis
HBV resistance-associated mutations (RAMs) were classed as major (M204I/V/S, A181T/V/S, A194T, N236T) and compensatory (L80I/V, I169T, V173L, L180M/C, T184A/G/I/S, S202C/G/I, M250L/V) [27]. Deep sequences were screened for APOBEC-driven G-to-A hypermutation using an in-house HYPERMUT algorithm (Supplementary file). Phylogenetic analyses (heuristic maximum likelihood) were performed using PhyML 3.0 (1000 bootstrap replicates) with tree reconstruction by FigTree v1.4. The Fisher exact test was used to compare the proportion of HBeAg-positive and HBeAg-negative subjects showing (a) HBV DNA >14 IU/mL, (b) HBV DNA >2000 IU/mL, and (c) HBV RAMs. The Wilcoxon–Mann–Whitney test was used to compare (a) HBV DNA levels in HBeAg-positive vs HBeAg-negative subjects, (b) HBV DNA levels in subjects with vs those without HBV RAMs, and (c) M204I frequencies in deep sequencing reads of samples with vs those without M204I/M by Sanger sequencing. The crude incidence of resistance was estimated as cases over person-time at risk, censored at the last available sample (42 at 6 months; 54 at 12 months). Statistical analyses were performed with SPSS version 21 (IBM, New York).
RESULTS
Baseline Characteristics
Among 1117 consecutive adults starting stavudine/lamivudine/nevirapine, 133 (11.9%; 95% confidence interval [CI]: 10.0%–13.8%) tested HBsAg-positive (Table 1). HBeAg-positive subjects (67/133, 50.4%) had higher HBV DNA detection rates (P = .0014), proportions with HBV DNA levels >2000 IU/mL (P = .0006), and median HBV DNA levels (P < .0001) than HBeAg-negative subjects (Table 2). HBV genotypes were predominantly sub-genotype A1 (110/111, 99.1%) and rarely E (1/111, 0.9%).
Table 1.
Baseline Characteristics of the Hepatitis B Virus Surface Antigen Positive Population Starting Lamivudine-based Antiretroviral Therapy in Blantyre
| Characteristics | ||
|---|---|---|
| Subjects, n (%) | 133 | (100) |
| Female gender, n (%) | 67 | (50.4) |
| Age, median years (IQR) | 36 | (31, 42) |
| CD4 counts, median cells/mm3 (IQR) | 109 | (53, 192) |
| HIV-1 RNA, median log10 copies/mL (IQR) | 4.0 | (3.5, 4.7) |
| Alanine transaminase, median U/L (IQR) | 23 | (15, 36) |
| HBV e-antigen positive, n (%) | 67 | (50.4) |
| HBV DNA <14 IU/mL, n (%) | 9 | (6.8) |
| HBV DNA >14 IU/mL, n (%) | 124 | (93.2) |
| HBV DNA >2000 IU/mL, n (%) | 99 | (74.4) |
| HBV DNA, median log10 IU/mL (IQR) | 6.4 | (3.1, 8.1) |
Abbreviations: HBV, hepatitis B virus; HIV-1, human immunodeficiency virus type 1; IQR, interquartile range.
Table 2.
Virologic Outcomes During 12 Months of Lamivudine Exposure According to Baseline Hepatitis B Virus e-Antigen Status
| HBeAg Positive |
HBeAg Negativea
|
|||||
|---|---|---|---|---|---|---|
| Months of lamivudine exposure | 0 | 6 | 12 | 0 | 6 | 12 |
| Total number attending | 67 | 47 | 26 | 66 | 49 | 28 |
| HIV-1 RNA <40 copies/mL, n (%)b | 67 (100) | 45 (95.7) | 24 (92.3) | 66 (100) | 48 (98.0) | 27 (96.4) |
| HBV DNA <14 IU/mL, n (%) | 0 (0) | 5 (10.6) | 7 (26.9) | 9 (13.6) | 34 (69.4) | 27 (96.4)c |
| HBV DNA >14 IU/mL, n (%) | 67 (100) | 42 (89.4) | 19 (73.1) | 57 (86.4) | 15 (30.6) | 1 (3.6) |
| HBV DNA >2000 IU/mL, n (%) | 64 (95.5) | 24 (51.1) | 18 (69.2) | 35 (53.0) | 0 (0) | 0 (0) |
| Median HBV DNA log10 IU/mL (IQR) | 8.0 (7.2, 8.5) | 3.5 (2.0, 4.3) | 6.1 (UD, 8.1) | 3.5 (2.2, 5.5) | UD (UD, 1.3) | UD |
| Sanger sequences, n (%)d | 67 (100) | 36 (76.6) | 18 (69.2) | 44 (66.7) | 4 (8.2) | 1 (3.6) |
| Sanger RAMs, n (%)e | 0 (0) | 7 (14.9) | 17 (65.4) | 0 (0) | 1 (2.0) | 1 (3.6) |
| M204I/M | … | 7 | … | … | 1 | … |
| M204V L180M | … | … | 10 | … | … | … |
| M204V V173L L180M | … | … | 3 | … | … | 1 |
| M204V L180M A181S | … | … | 2 | … | … | … |
| M204V L80I | … | … | 1 | … | … | … |
| M204V L80I L180M | … | … | 1 | … | … | … |
Abbreviations: HBV, hepatitis B virus; HBeAg, hepatitis B virus e-antigen; HIV-1, human immunodeficiency virus type 1; IQR, interquartile range; PCR, polymerase chain reaction; RAM, resistance-associated mutation; UD, below detection limit (<14 IU/mL).
a HBeAg-negative subjects comprised 43 subjects with and 23 subjects without anti-HBe antibodies.
b HIV-1 RNA was detected in 3/96 (3.1%) subjects at 6 months (43, 77, and 219 copies/mL), and 3/54 (5.6%) subjects at 12 months (3673, 7300, and 11 538 copies/mL).
c The 1 subject with detectable HBV DNA (2.8 log10 IU/mL) at 12 months showed a suppressed HIV-1 RNA.
d Samples with HBV DNA levels >50 IU/mL underwent Sanger sequencing of HBV polymerase; success rates were 111/120 (92.5%) at baseline, 40/46 (87.0%) at 6 months, and 19/19 (100%) at 12 months. HBV DNA levels in samples that failed to yield a PCR product in ≥2 attempts were median 2.2 (IQR 2.0, 2.4) log10 IU/mL.
e HBV RAMs were more prevalent in HBeAg-positive than in HBeAg-negative subjects at both 6 months (P = .029) and 12 months (P < .0001).
Responses to Starting ART
Overall 96/133 (72.2%) subjects attended follow-up at 6 months and 54 (40.6%) also attended at 12 months. Reasons for nonattendance at 6 months comprised death (n = 10, 7.5%), transfer of care (n = 10. 7.5%), loss to follow-up (n = 9, 6.8%), treatment interruption (n = 7, 5.3%), and hospitalization (n = 1, 0.8%). Over the entire cohort of 1117 subjects, by 6 months 103 (9.2%) had died, 44 (3.9%) had transferred care, and 50 (4.5%) were lost to follow-up. Reasons for nonattendance at 12 months were not recorded. Based upon the last available follow-up after starting ART, 92/96 (95.8%) patients achieved HIV-1 RNA suppression <40 copies/mL, and 48/96 (50.0%) showed HBV DNA suppression <14 IU/mL; among subjects with persistent HBV DNA detection 47/48 (97.9%) had suppressed HIV-1 RNA levels. HBeAg-positive subjects continued to have higher HBV DNA detection rates, proportions with HBV DNA levels >2000 IU/mL, and median HBV DNA levels than HBeAg-negative subjects (P < .0001 for all comparisons) (Table 2). In this group, HBV DNA levels declined by median 4.5 log10 IU/mL at 6 months but rebounded at 12 months. In contrast, most HBeAg-negative subjects achieved HBV DNA suppression.
HBV Drug Resistance
Sanger Sequencing
At baseline, 0/120 subjects had HBV RAMs. At 6 months, 8/46 (17.4%) subjects showed the major RAM M204I/M; median HBV DNA levels were similar in subjects with vs those without detectable resistance (3.2 vs 3.8 log10 IU/mL; P = .691). At 12 months, 18/19 (94.7%) subjects showed M204V accompanied by ≥1 compensatory RAM (L80I, V173L, L180M) and in 2 subjects by the major RAM A181S. Resistance rates among all patients in follow-up were 0/133 (0%), 8/96 (8.3%), and 18/54 (33.3%) at baseline, 6 months, and 12 months, respectively. Based upon 42 subjects with 6 months of follow-up (6 with resistance) and 54 subjects with 12 months of follow-up (2 with resistance at 6 months, 16 with resistance at 12 months), the crude incidence rate of resistance was 32.4 per 100 person-years.
Deep Sequencing
At baseline, 0/5 subjects had HBV RAMs. At 6 months, 16/17 (94.1%) subjects showed M204I, including 6 with M204I/M by Sanger sequencing and 10 without RAMs by Sanger sequencing (Table 3). M204V was detected at low frequency in 7/17 (41.2%) samples containing higher frequencies of M204I. Other RAMs were less common and occurred always with M204I, comprising the major RAM A181T in 4/17 (23.5%) subjects and the compensatory RAM V173L in 2/17 (11.8%) subjects. One month-12 sample that lacked RAMs by Sanger sequencing showed M204I by deep sequencing. None of the deep sequences had evidence of G-to-A hypermutation. Results of deep sequencing increased HBV drug-resistance rates in the population to 18/96 (18.8%) at 6 months and 19/54 (35.2%) at 12 months.
Table 3.
Hepatitis B Virus Drug-Resistance Patterns by Sanger and Deep Sequencing at 6 and 12 Months of Lamivudine Exposure
| IDa | Month | HBV DNA log10 IU/mL | Resistance-associated Mutations |
|
|---|---|---|---|---|
| Sanger Sequence | Deep Sequences (Frequency)b | |||
| 84* | 6 | 3.9 | M204I/M | M204I (20) M204V (1) |
| 12 | 7.8 | M204V L180M | … | |
| 97 | 6 | 3.6 | None | M204I (7) |
| 12 | 6.5 | M204V L80I L180M | … | |
| 117 | 6 | 3.6 | None | M204I (32) M204V (2) |
| 12 | 6.1 | M204V L180M | … | |
| 136* | 6 | 2.3 | M204I/M | M204I (11) |
| 200 | 6 | 2.5 | None | M204I (3) |
| 12 | 8.3 | M204V L180M | … | |
| 212 | 6 | 3.9 | None | M204I (20) M204V (6) |
| 12 | 8.7 | M204V L180M A181S | … | |
| 221 | 6 | 2.3 | None | M204I (27) |
| 12 | 5.4 | M204V L180M | … | |
| 243 | 6 | 3.2 | None | M204I (9) M204V (1) A181T (1) |
| 12 | 8.8 | M204V L180M A181S | … | |
| 448 | 6 | 4.5 | None | M204I (3) |
| 12 | 8.0 | M204V L180M | … | |
| 457 | 6 | 4.9 | None | M204I (7) M204V (2) A181T (3) |
| 12 | 7.4 | M204V L180M | … | |
| 509 | 6 | 5.1 | None | M204I (8) M204V (2) A181T (1) |
| 12 | 4.4 | M204V L180M | … | |
| 592* | 6 | 5.0 | M204I/M | M204I (6) M204V (3) A181T (2) |
| 595* | 6 | 2.6 | M204I/M | M204I (14) |
| 668 | 6 | 4.3 | M204I/M | M204I (16) |
| 729* | 6 | 2.5 | M204I/M | M204I (14) V173L (2) |
| 12 | 7.6 | M204V V173L L180M | … | |
| 854 | 6 | 4.3 | None | M204I (6) M204V (3) V173L (46) |
| 12 | 6.3 | M204V V173L L180M | … | |
| 996 | 6 | 8.2 | None | None |
| 12 | 4.8 | None | M204I (2) | |
Abbreviations: HBV, hepatitis B virus; ID, identification number; RAM, resistance-associated mutation.
a Each subject is indicated by an ID number.
b Subjects with HBV DNA levels ≥300 IU/mL and sufficient stored sample available underwent deep sequencing (≥1% sensitivity), including five subjects at baseline (indicated by *), 17 at 6 months and one at 12 months. None of the baseline samples showed RAMs. The frequency (%) of mutants in the deep sequencing reads is indicated in brackets. The frequency of M204I in Sanger positive vs Sanger negative samples was median 14% vs 7% (P = .05).
Combining Sanger and deep sequencing data, among subjects with at least 1 follow-up visit, 24/96 (25.0%) acquired HBV RAMs, with significantly higher rates in HBeAg-positive vs HBeAg-negative subjects (23/47, 48.9% vs 2/49, 4.1%; P < .0001) and in patients with higher baseline HBV DNA load. Baseline HBV DNA levels were median 8.1 (interquartile range 7.1, 8.6) vs 5.3 (2.4, 7.7) log10 IU/mL in subjects with vs those without treatment-emergent HBV RAMs, respectively (P < .0001).
Co-evolution of HBV RAMs
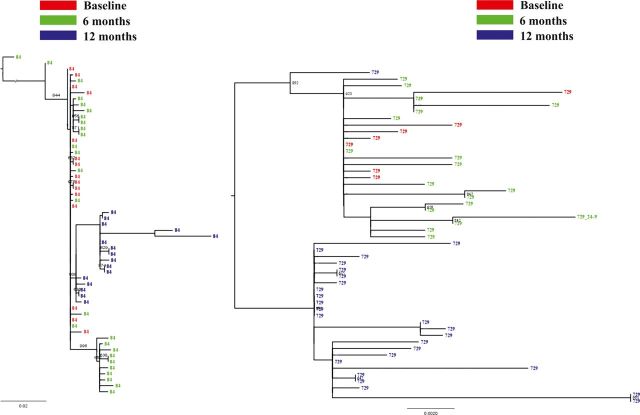
Clonal polymerase sequences and single full-genome sequences were obtained in 3 subjects at baseline and after 6 and 12 months of ART (Table 4). HBV DNA levels declined in the 3 subjects after starting ART coinciding with the emergence of M204I and rebounded at 12 months coinciding with the detection on the same viral genomes of M204V plus compensatory RAMs (L180M, V173L). There was excellent correlation between Sanger and other sequencing results (Table 4). Phylogenetic analysis of clonal sequences indicated replacement of M204I variants with M204V variants in 96%–100% of clones at 12 months (Figure 1).
Table 4.
Coevolution of Hepatitis B Virus Resistance-associated Mutations During Lamivudine Therapy Determined by Clonal Polymerase Sequencing and Single Full Genome Sequencing in Three Subjectsa
| Subject | Months of Lamivudine Exposure |
||
|---|---|---|---|
| 0 | 6 | 12 | |
| ID 84 | |||
| HBV DNA log10 IU/mL | 8.6 | 3.9 | 7.8 |
| Sanger sequence RAMs | None | M204I/M | M204V L180M |
| Deep sequences RAMs | None | M204I (20) M204V (1) | ND |
| Clones, n | 15 | 26 | 16 |
| Clonal sequences RAMs | 15 None | 19 None | 0 None |
| 7 (27) M204I | 16 (100) M204V L180M | ||
| ID 729 | |||
| HBV DNA log10 IU/mL | 7.0 | 2.5 | 7.6 |
| Sanger sequence RAMs | None | M204I/M | M204V V173L L180M |
| Deep sequences RAMs | None | M204I (14) V173L (2) | ND |
| Clones, n | 7 | 18 | 26 |
| Clonal sequences RAMs | 7 None | 16 None | 25 (96) M204V V173L L180M |
| 2 (11) M204I | 1 (4) M204V V173L T184S | ||
| Single genomes, n | 1 | 14 | 12 |
| Single genomes RAMs | 1 None | 12 None | 0 None |
| 2 (14) M204I | 11 (92) M204V V173L L180M | ||
| 1 (8) M204V V173L T184S | |||
| ID 221 | |||
| HBV DNA log10 IU/mL | 6.4 | 2.3 | 5.4 |
| Sanger sequence RAMs | None | None | M204V L180M |
| Deep sequences RAMs | ND | M204I (27) | ND |
| Single genomes, n | 7 | 9 | 1 |
| Single genomes RAMs | 7 None | 8 None | 1 M204V L180M |
| 1 (11) M204I | |||
Abbreviations: HBV, hepatitis B virus; ID, identification number; ND, not done; RAM, resistance-associated mutation.
a The frequency (%) of the mutants is indicated in brackets.
Figure 1.
Phylogenetic analysis of HBV clonal polymerase sequences obtained in 2 subjects (ID 84 and ID 729) over 12 months of lamivudine therapy. Numbers above the branches indicate bootstrap values. Abbreviations: HBV, hepatitis B virus; ID, identification number.
DISCUSSION
This study undertook a prospective characterization of the virologic expression of HBV infection in HIV-positive Malawians starting lamivudine-containing ART without tenofovir and is the first to our knowledge to employ deep sequencing to detail the emergence of HBV resistance to lamivudine in the context of HIV coinfection. At 12% HBsAg prevalence was among the highest in sub-Saharan Africa [5, 9–16], particularly for the Eastern region, and consistent with rates observed in HIV-positive and HIV-negative subjects in Malawi [6–8, 28]. Coinfected patients showed a HBV virologic profile indicative of a risk of liver disease: at study entry, half tested HBeAg-positive and half of those who tested HBeAg-negative had HBV DNA levels >2000 IU/mL. Over 12 months of stavudine/lamivudine/nevirapine, patients who attended follow-up showed excellent HIV-1 RNA suppression, consistent with good adherence to ART. In contrast, HBV outcomes were poor among HBeAg-positive patients: HBV DNA levels initially declined by 4.5 log10 IU/mL, then rebounded, coinciding with the rapid emergence of major resistance and compensatory mutations coexisting on the same HBV genomes. Recent reports from Uganda [29] and Nigeria [30] indicate that HIV/HBV coinfection and high HBV DNA levels are predictive of severe liver fibrosis by transient elastography. Together with observations from southern Africa [10, 31] and Tanzania [14], this implies that a substantial proportion of HIV-infected patients in Blantyre were at risk of progressive liver fibrosis and excess mortality due to poorly controlled HBV coinfection. Suboptimal HBV suppression and emergence of resistance were most evident in subjects that tested HBeAg-positive and had high HBV DNA load at treatment initiation. Although HBeAg-negative subjects generally achieved HBV DNA suppression over 6–12 months of ART, longer follow-up is required to determine their long-term risk of rebound.
The concern that widespread use of lamivudine-containing ART without tenofovir in HBV endemic settings may lead to high rates of HBV drug resistance and poor outcomes has been raised previously [3, 5, 10, 32]. Recent studies, however, have reported that HBV drug resistance is infrequent in sub-Saharan Africa and proposed that the outcomes of lamivudine treatment in the region are distinct from those in Western cohorts. In 4 studies from Cameroon, Zambia, South Africa, and Kenya, the crude incidence of HBV drug resistance (by Sanger sequencing) during lamivudine-containing ART without tenofovir ranged from 3 to 11.5 per 100 person-years [11, 12, 15, 16]. These data are in contrast with those from Malawi, where baseline HBeAg seroprevalence and HBV DNA levels were higher than in the other cohorts, and the incidence of HBV drug resistance (by Sanger sequencing) was 32.4 per 100 person-years. In agreement with our findings, a small study found that 6/21 (29%) HIV/HBV coinfected pregnant women in Malawi had evidence of HBV drug-resistance (by Sanger sequencing) after median 37 weeks of lamivudine-containing ART without tenofovir [19]. There is a preponderance of HBV sub-genotype A1 infections in Malawi [19, 28], as also observed in Kenya, Zambia, and South Africa [11, 15, 16]. Sub-genotype A1 also circulates in Uganda, Tanzania, Somalia, Yemen, India, Nepal, the Philippines, and Brazil [24], and the factors modulating its variable virologic expression warrant investigation. Host-related immunological and genetic factors impacting on HBV replication may influence such differences. Of note, in the studies from Kenya, Zambia, and South Africa median CD4 counts at ART initiation ranged between 97 and 132 cells/mm3 [11, 15, 16] and were therefore similar to those in our cohort.
Partly due to the overlapping genomic structure, HBV resistance to lamivudine emerges more slowly than with HIV [33]. Using deep sequencing we obtained a more sensitive assessment of the kinetics of HBV drug resistance than allowed by Sanger sequencing. After 6 months of therapy, M204I was detected in nearly all subjects with HBV DNA levels >300 IU/mL. Improved detection of HBV resistance by deep sequencing has been previously reported and shown to carry significance despite initial concerns that some low-frequency variants may be technical artifacts or replication defective APOBEC-induced hypermutants [34, 35]. To reduce misinterpretation, we set the sensitivity of mutant detection at 1%, applied a high read quality cutoff score, and screened deep sequences for evidence of G-to-A hypermutation. In agreement with previous observations [34], no M204I was found in lamivudine-naive subjects by deep sequencing. At 6 months, M204I frequencies were predictably higher in subjects that also showed M204I by Sanger sequencing than in those lacking the mutation. Consistent with ongoing drug selective pressure however, subjects with both low- and high-frequency M204I at month 6 went on to acquire M204V by Sanger sequencing at month 12. Importantly, we obtained clonal and single full-genome sequences over time in 3 subjects and showed excellent correlation between the different sequencing methods, which has not been previously documented. Thus, the data indicate that after just 6 months of therapy, lamivudine resistance was nearly universal in subjects with persistent HBV viremia, providing a revised estimate of the speed of emergence of HBV resistance to lamivudine in the context of HIV infection.
The evolution of resistance followed classic pathways: M204I emerged first (at low HBV DNA load) and was then replaced by fitter variants carrying M204V plus compensatory RAMs (at high HBV DNA load). These mutants have reduced susceptibility to lamivudine, emtricitabine, telbivudine, and entecavir [36]. Two subjects showed the less common pathway M204V + L180M + A181S, which also confers resistance to adefovir and to a lesser extent tenofovir [37]. By deep sequencing, 4 subjects harbored A181T, which reduces susceptibility to tenofovir [36]; mutant frequencies were low, however, and A181T did not emerge in the Sanger sequences obtained at 12 months, although 1 subject acquired A181S. Thus, most patients in the Malawi cohort are expected to respond virologically to tenofovir [38], although the outcomes of introducing tenofovir in HIV/HBV coinfected populations in sub-Saharan Africa have been poorly studied.
There are limitations to this study. As previously reported in sub-Saharan Africa [8, 14], missed follow-up was common, and after the second visit we had no formal tracing of nonattendance. We lacked a formal measure of adherence and inferred that nearly all subjects who attended for follow-up had good adherence to fixed-dose stavudine/lamivudine/nevirapine based on a suppressed HIV-1 RNA. As sample volumes were small, we were unable to perform deep sequencing in all subjects with HBV DNA >300 IU/mL and to repeat HBeAg testing during follow-up. Due to the very limited local diagnostic infrastructure and the retrospective nature of HBsAg testing, we were unable to measure indices of liver disease other than baseline ALT and correlate the virologic findings with clinical parameters. A more extensive evaluation of liver function including noninvasive measures of liver fibrosis (eg, by transient elastography) is planned.
Recent WHO guidelines have recommended the inclusion of tenofovir in first-line ART in sub-Saharan Africa [20], and this is now part of national policy in Malawi: at the end of September 2013, 73% of adults accessing first-line ART were receiving tenofovir. These high rates are not universal across sub-Saharan Africa however. Our data provide clear evidence in support of the notion that HBsAg testing should be introduced as part of routine HIV care in sub-Saharan Africa in order to guide ART selection and early adoption of tenofovir and to identify patients that require follow-up of liver disease [39, 40]. Although HBV DNA testing is not routinely available, the introduction of HIV-1 RNA monitoring may provide a platform for expanded access to molecular assays [20, 39]. HBeAg offers a more easily accessible marker for identifying subjects with the highest risk of virologic breakthrough, for whom it is most urgent to abandon the use lamivudine as the sole HBV-active agent.
There has traditionally been poor attention paid to liver disease across sub-Saharan Africa, despite high prevalence of life-long HBV carriage, and repeated calls have been made to improve ascertainment and management of chronic hepatitis B in the region, targeting people with and without HIV infection [39, 40]. Our data strengthen the case for a rapid implementation of such recommendations.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online (http://cid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. The authors wish to thank Dr John Kenny (Dept of Functional and Comparative Genomics, Institute of Integrative Biology, University of Liverpool), Dr Daniel Depledge (Div of Infection and Immunity, Faculty of Medical Sciences, University College London), and Dr Ian Harrison and Dr Ana Garica (Dept of Virology, Royal Hampstead Foundation Trust, London) for valuable technical and analytical assistance.
Financial support. This work was supported by a University College London PhD studentship (S. A.), a University of Liverpool/Malawi Liverpool Wellcome Trust PhD Fellowship (M. C.), and a Leverhulme Royal Society Africa Award.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Lacombe K, Rockstroh J. HIV and viral hepatitis coinfections: Advances and challenges. Gut. 2012;61(suppl 1):i47–58. doi: 10.1136/gutjnl-2012-302062. [DOI] [PubMed] [Google Scholar]
- 2.Chun HM, Roediger MP, Hullsiek KH, et al. Hepatitis B virus coinfection negatively impacts HIV outcomes in HIV seroconverters. J Infect Dis. 2012;205:185–93. doi: 10.1093/infdis/jir720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gish R, Jia JD, Locarnini S, Zoulim F. Selection of chronic hepatitis B therapy with high barrier to resistance. Lancet Infect Dis. 2012;12:341–53. doi: 10.1016/S1473-3099(11)70314-0. [DOI] [PubMed] [Google Scholar]
- 4.Wilkins E, Nelson M, Agarwal K, et al. British HIV Association guidelines for the management of hepatitis viruses in adults infected with HIV 2013. HIV Med. 2013;14(suppl. 4):1–71. doi: 10.1111/hiv.12106. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann CJ, Thio CL. Clinical implications of HIV and hepatitis B co-infection in Asia and Africa. Lancet Infect Dis. 2007;7:402–9. doi: 10.1016/S1473-3099(07)70135-4. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed SD, Cuevas LE, Brabin BJ, et al. Seroprevalence of hepatitis B and C and HIV in Malawian pregnant women. J Infect. 1998;37:248–51. doi: 10.1016/s0163-4453(98)91983-1. [DOI] [PubMed] [Google Scholar]
- 7.Sutcliffe S, Taha TE, Kumwenda NI, Taylor E, Liomba GN. HIV-1 prevalence and herpes simplex virus 2, hepatitis C virus, and hepatitis B virus infections among male workers at a sugar estate in Malawi. J Acquir Immune Defic Syndr. 2002;31:90–7. doi: 10.1097/00126334-200209010-00012. [DOI] [PubMed] [Google Scholar]
- 8.Moore E, Beadsworth MB, Chaponda M, et al. Favourable one-year ART outcomes in adult Malawians with hepatitis B and C co-infection. J Infect. 2010;61:155–63. doi: 10.1016/j.jinf.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Geretti AM, Patel M, Sarfo FS, et al. Detection of highly prevalent hepatitis B virus coinfection among HIV-seropositive persons in Ghana. J Clin Microbiol. 2010;48:3223–30. doi: 10.1128/JCM.02231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthews GV, Manzini P, Hu Z, et al. Impact of lamivudine on HIV and hepatitis B virus-related outcomes in HIV/hepatitis B virus individuals in a randomized clinical trial of antiretroviral therapy in southern Africa. AIDS. 2011;25:1727–35. doi: 10.1097/QAD.0b013e328349bbf3. [DOI] [PubMed] [Google Scholar]
- 11.Kim NH, Scott J, Cent A, et al. HBV lamivudine resistance among hepatitis B and HIV coinfected patients starting lamivudine, stavudine, and nevirapine in Kenya. J Viral Hepat. 2011;18:e447–452. doi: 10.1111/j.1365-2893.2011.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouanfack C, Aghokeng AF, Mondain AM, et al. Lamivudine-resistant HBV infection in HIV-positive patients receiving antiretroviral therapy in a public routine clinic in Cameroon. Antivir Ther. 2012;17:321–6. doi: 10.3851/IMP1911. [DOI] [PubMed] [Google Scholar]
- 13.Ive P, Macleod W, Mkumla N, et al. Low prevalence of liver disease but regional differences in HBV treatment characteristics mark HIV/HBV co-infection in a South African HIV clinical trial. PLoS One. 2013;8:e74900. doi: 10.1371/journal.pone.0074900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawkins C, Christian B, Ye J, et al. Prevalence of hepatitis B co-infection and response to antiretroviral therapy among HIV-infected patients in Tanzania. AIDS. 2013;27:919–27. doi: 10.1097/QAD.0b013e32835cb9c8. [DOI] [PubMed] [Google Scholar]
- 15.Day SL, Odem-Davis K, Mandaliya KN, et al. Prevalence, clinical, and virologic outcomes of hepatitis B virus co-infection in HIV-1 positive Kenyan women on antiretroviral therapy. PLoS One. 2013;8:e59346. doi: 10.1371/journal.pone.0059346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamers RL, Zaaijer HL, Wallis CL, et al. HIV-HBV coinfection in Southern Africa and the effect of lamivudine- versus tenofovir-containing cART on HBV outcomes. J Acquir Immune Defic Syndr. 2013;64:174–82. doi: 10.1097/QAI.0b013e3182a60f7d. [DOI] [PubMed] [Google Scholar]
- 17.Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30:2212–9. doi: 10.1016/j.vaccine.2011.12.116. [DOI] [PubMed] [Google Scholar]
- 18.Chadwick D, Ankcorn M, Sarfo F, et al. Outcomes of starting first-line antiretroviral therapy in hepatitis B virus/HIV-coinfected patients in Ghana. J Antimicrob Chemother. 2012;67:2939–42. doi: 10.1093/jac/dks333. [DOI] [PubMed] [Google Scholar]
- 19.Galluzzo C, Liotta G, Andreotti M, et al. Emergence of lamivudine resistance hepatitis B virus mutations in pregnant women infected with HBV and HIV receiving antiretroviral prophylaxis for the prevention of mother-to-infant transmission in Malawi. J Med Virol. 2012;84:1553–7. doi: 10.1002/jmv.23365. [DOI] [PubMed] [Google Scholar]
- 20.World Health Organization (WHO) 2013 consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. Available at: http://www.who.int/hiv/pub/guidelines/arv2013/en/index.html. Accessed 15 July 2014. [PubMed]
- 21.Benhamou Y, Bochet M, Thibault V, et al. Long-term incidence of hepatitis B virus resistance to lamivudine in human immunodeficiency virus-infected patients. Hepatology. 1999;30:1302–6. doi: 10.1002/hep.510300525. [DOI] [PubMed] [Google Scholar]
- 22.Matthews GV, Bartholomeusz A, Locarnini S, et al. Characteristics of drug resistant HBV in an international collaborative study of HIV-HBV-infected individuals on extended lamivudine therapy. AIDS. 2006;20:863–70. doi: 10.1097/01.aids.0000218550.85081.59. [DOI] [PubMed] [Google Scholar]
- 23.Ramos B, Nunez M, Martin-Carbonero L, et al. Hepatitis B virus genotypes and lamivudine resistance mutations in HIV/hepatitis B virus-coinfected patients. J Acquir Immune Defic Syndr. 2007;44:557–61. doi: 10.1097/QAI.0b013e3180314b46. [DOI] [PubMed] [Google Scholar]
- 24.Kramvis A, Kew MC. Molecular characterization of subgenotype A1 (subgroup Aa) of hepatitis B virus. Hepatol Res. 2007;37(suppl 1):S27–32. doi: 10.1111/j.1872-034X.2007.00100.x. [DOI] [PubMed] [Google Scholar]
- 25.Watson SJ, Welkers MR, Depledge DP, et al. Viral population analysis and minority-variant detection using short read next-generation sequencing. Philos Trans R Soc Lond B Biol Sci. 2013;368 doi: 10.1098/rstb.2012.0205. 20120205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Archer J, Baillie G, Watson SJ, Kellam P, Rambaut A, Robertson DL. Analysis of high-depth sequence data for studying viral diversity: a comparison of next generation sequencing platforms using Segminator II. BMC Bioinformatics. 2012;13:47. doi: 10.1186/1471-2105-13-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhee SY, Margeridon-Thermet S, Nguyen MH, et al. Hepatitis B virus reverse transcriptase sequence variant database for sequence analysis and mutation discovery. Antiviral Res. 2010;88:269–75. doi: 10.1016/j.antiviral.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Candotti D, Mundy C, Kadewele G, Nkhoma W, Bates I, Allain JP. Serological and molecular screening for viruses in blood donors from Ntcheu, Malawi: High prevalence of HIV-1 subtype C and of markers of hepatitis B and C viruses. J Med Virol. 2001;65:1–5. [PubMed] [Google Scholar]
- 29.Stabinski L, Reynolds SJ, Ocama P, et al. High prevalence of liver fibrosis associated with HIV infection: A study in rural Rakai, Uganda. Antiviral Ther. 2011;16:405–11. doi: 10.3851/IMP1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hawkins C, Agbaji O, Ugoagwu P, et al. Assessment of liver fibrosis by transient elastography in patients with HIV and hepatitis B virus coinfection in Nigeria. Clin Infect Dis. 2013;57:189–92. doi: 10.1093/cid/cit564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin NK, Devine A, Eaton JW, et al. Modeling the impact of early antiretroviral therapy for adults coinfected with HIV and hepatitis B or C in South Africa. AIDS. 2014;28(suppl 1):S35–46. doi: 10.1097/QAD.0000000000000084. [DOI] [PubMed] [Google Scholar]
- 32.Di Bisceglie AM, Maskew M, Schulze D, Reyneke A, McNamara L, Firnhaber C. HIV-HBV coinfection among South African patients receiving antiretroviral therapy. Antivir Ther. 2010;15:499–503. doi: 10.3851/IMP1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pallier C, Castera L, Soulier A, et al. Dynamics of hepatitis B virus resistance to lamivudine. J Virol. 2006;80:643–53. doi: 10.1128/JVI.80.2.643-653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solmone M, Vincenti D, Prosperi MC, Bruselles A, Ippolito G. Use of massively parallel ultradeep pyrosequencing to characterize the genetic diversity of hepatitis B virus in drug-resistant and drug-naive patients and to detect minor variants in reverse transcriptase and hepatitis B S antigen. J Virol. 2009;83:1718–26. doi: 10.1128/JVI.02011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margeridon-Thermet S, Svarovskaia ES, Babrzadeh F, et al. Low-level persistence of drug resistance mutations in hepatitis B virus-infected subjects with a past history of lamivudine treatment. Antimicrob Agents Chemother. 2013;57:343–9. doi: 10.1128/AAC.01601-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Locarnini S, Zoulim F. Molecular genetics of HBV infection. Antivir Ther. 2010;15(suppl 3):3–14. doi: 10.3851/IMP1619. [DOI] [PubMed] [Google Scholar]
- 37.Karatayli E, Karayalcin S, Karaaslan H, et al. A novel mutation pattern emerging during lamivudine treatment shows cross-resistance to adefovir dipivoxil treatment. Antivir Ther. 2007;12:761–8. [PubMed] [Google Scholar]
- 38.Baran B, Soyer OM, Ormeci AC, et al. Efficacy of tenofovir in patients with lamivudine failure is not different from that in nucleoside/nucleotide analogue-naive patients with chronic hepatitis B. Antimicrob Agents Chemother. 2013;57:1790–6. doi: 10.1128/AAC.02600-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiersma ST, McMahon B, Pawlotsky JM, et al. World Health Organization Department of Immunization, Vaccines and Biologicals. Treatment of chronic hepatitis B virus infection in resource-constrained settings: Expert panel consensus. Liver Int. 2011;31:755–61. doi: 10.1111/j.1478-3231.2010.02373.x. [DOI] [PubMed] [Google Scholar]
- 40.Lemoine M, Thursz M, Njie R, Dusheiko GM. Forgotten, not neglected: Viral hepatitis in resource-limited settings, recall for action. Liver Int. 2014;34:12–5. doi: 10.1111/liv.12283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.