We describe host genetic factors determining susceptibility to rotavirus infection and report that Lewis-negative and secretor-negative children are resistant to P[8] rotavirus strains, but not to P[6] rotavirus strains, providing a plausible explanation to reduced vaccine efficacy in some populations.
Keywords: rotavirus, histo–blood group antigen, Lewis, susceptibility, vaccine
Abstract
Background. The live oral rotavirus (RV) vaccines have shown a reduced efficacy in Africa. Recent in vitro studies have shown binding of the RV surface protein (VP4) to histo–blood group antigens (HBGAs) in an RV genotype–dependent manner, suggesting them to be putative receptors for RV. The diversity of HBGA phenotypes in different ethnic populations, combined with prevalence/absence of specific RV genotypes, led us to hypothesize whether the genetic variations in HBGAs in a population limit susceptibility to certain RV genotypes, plausibly leading to reduced vaccine efficacy.
Methods. Association between HBGAs status and susceptibility to RV P genotypes was investigated in children in Burkina Faso and Nicaragua. In total, 242 children with diarrhea in Burkina Faso and Nicaragua were investigated, 93 of whom were RV positive.
Results. In Burkina Faso, the P[8] RV strains (n = 27) infected only Lewis- and secretor-positive children (27/27; P < .0001), but no Lewis-negative children. In contrast, the P[6] strains (n = 27) infected predominantly Lewis-negative children (n = 18; P < .0001) but also Lewis-positive children, irrespective of their secretor status. The results from Nicaragua confirmed that all P[8]-infected children (n = 22) were secretor Lewis positive.
Conclusions. As VP4 of genotype P[8] is a component of current RV vaccines, our finding that Lewis-negative children are resistant to P[8] strains provides a plausible explanation for the reduced vaccine efficacy in populations with a high percentage of Lewis-negative individuals, such as in Africa. Furthermore, our findings provide a plausible explanation as to why P[6] RV strains are more common in Africa.
Rotavirus (RV) is the leading cause of severe acute gastroenteritis in infants and young children and is responsible for approximately 450 000 deaths annually, the majority occurring in children from low- and middle-income countries, with an especially high burden in sub-Saharan Africa [1, 2]. The World Health Organization recommends inclusion of RV vaccines in the national immunization program of countries highly affected by RV. Currently, 2 vaccines, Rotarix and RotaTeq, are licensed and available in many countries. Although the vaccines have demonstrated high efficacy in industrialized countries, vaccine trials from sub-Saharan Africa have shown reduced efficacy [3, 4]. The reasons for this are yet unknown, but malnutrition, concomitant infections, RV strain diversity, and host factors have been proposed. The genetic makeup of RV strains circulating in Africa is different from those in high-income countries. In addition to a higher degree of animal or animal-derived strains, the genotype P[6] is also relatively more prevalent in Africa than in Europe and North America [5–7].
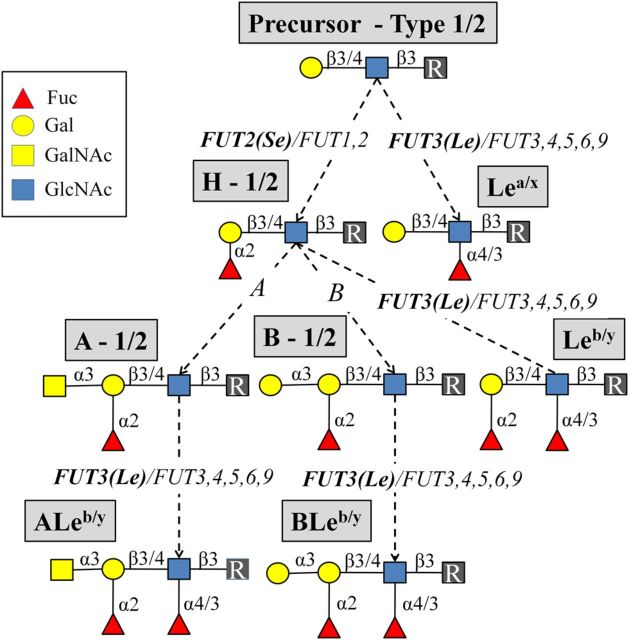
The VP4 (P genotype) protein mediates RV attachment and entry into the mature enterocytes through its glycan binding domain, VP8* [8]. Previous in vitro studies have shown RV infection to be sialic acid dependent. Animal rotaviruses recognize terminal sialic acids of the host cell (sensitive to sialidase), whereas many human rotaviruses recognize internal sialic acid residues (insensitive to sialidase) [9, 10]. However, an association of these specificities for susceptibility to RV infection in vivo has not been observed to date. Recent in vitro studies, using various glycan binding assays, have demonstrated that the most common human RVs recognize human histo–blood group antigens (HBGAs) in a P genotype–specific manner [9, 11–13]. The type 1 chain HBGAs are primarily expressed in the gut and present on most epithelial cells in the intestinal and respiratory tracts as well as in secretions such as saliva. These glycans are recognized as susceptibility and cell attachment factors for several enteric pathogens such as Helicobacter pylori and norovirus [14–16]. For the biosynthesis of HBGAs, the secretor gene (FUT2) encoding enzyme transfers fucose into an α1-2 linkage to the terminal Gal of the type 1 chain precursor (Galβ3GlcNAcβ), making the H type 1 antigen (Fucα2Galβ3GlcNAcβ) (Figure 1). Interestingly, a recent in vivo study suggested the secretor-positive genotype to confer susceptibility to P[8] RV genotypes [17]. The Lewis gene (FUT3)–coded enzyme, on the other hand, transfers fucose into an α1-4 linkage to the type 1 chain precursor or to the H type 1 antigen, generating Lewis a (Galβ3[Fucα4]GlcNAcβ) or Lewis b antigens (Fucα2Galβ3[Fucα4]GlcNAcβ), respectively (Figure 1). The expression of H type 1 and Lewis HBGAs is thus genetically determined based on polymorphisms in the FUT2 and FUT3 genes. The frequency of these polymorphisms varies markedly between different ethnic populations; for instance, only 4%–6% of white populations do not express Lewis antigens, whereas the prevalence of this Lewis-negative phenotype can reach >30% in certain African and Latin American populations [18–20].
Figure 1.
Biosynthetic pathways of histo–blood group antigens. The precursors from type 1/2 chain are considered. The antigen names (in gray boxes), genetic loci of glycosyltransferases (in italics), and the glycosidic linkages (in solid lines) are separated by a forward slash (/) to specify the pathway for different chains. The FUT2 and the FUT3 genetic loci driving the biosynthesis of the secretor (Se) and the Lewis (Le) antigens, respectively, are highlighted (in bold).
Here, we report that the Lewis (FUT3) and secretor (FUT2) genes mediate susceptibility for symptomatic RV infection, with both Lewis-negative and secretor-negative phenotypes being strong restriction factors for RV genotype P[8] infections. We further show that P[6] RVs mainly infect Lewis-negative children, a phenotype more common in African populations, providing a plausible explanation for the relatively high frequency of the P[6] genotype in Africa. Moreover, we hypothesize that the resistance to P[8] strains in Lewis-negative children could be an important contributing factor to the modest RV vaccine efficacy in sub-Saharan Africa.
MATERIALS AND METHODS
Study Population and Sample Collection
Fecal and saliva samples analyzed for association of RV P genotypes and host HBGA phenotypes in children with diarrhea were collected from 2 different populations, Burkina Faso in West Africa and Nicaragua in Central America. The RV-positive samples from Burkina Faso included in this study (n = 59) were collected during the RV season (December 2009–March 2010) at Saint Camille Medical Center CERBA/LABIOGENE in Ouagadougou, Burkina Faso [21]. In brief, stools, saliva, and buccal swabs were collected from children <5 years of age who presented with diarrheal illness. The G and P genotypes of the infecting RVs were determined by multiplex polymerase chain reaction (PCR) and/or sequencing as described previously [7]. The 59 RV strains represent all RV-positive samples among the 208 children included in the study. The study protocol was approved by the institutional ethics committee of Saint Camille Medical Center (2012CR/X-25/i135).
In addition to the Burkina Faso material, a total of 34 children from León, Nicaragua (with RV infection confirmed by enzyme-linked immunosorbent assay [ELISA] and PCR in previous studies) were also included. These children were previously enrolled in studies of childhood diarrhea etiologies involving >1000 diarrheic cases from the community, primary care clinics, and the hospital in León that were carried out in 2005 [22], 2008–2009, and 2010–2011. Saliva samples from the children included in the present study (n = 34) were collected by a nurse during home visits. Prior to sampling, informed consent was signed by a parent or legal guardian. All children were <5 years of age when initially examined for RV. This study was approved by the local ethical committee for biomedical research (Acta No. 18, 2012). The G and P types of the RV-positive samples, if not previously determined, were assessed by multiplex PCR and confirmed with sequencing as described previously [22].
Saliva Phenotyping Assay
A detailed description of the saliva phenotyping assay is available in the Supplementary Materials and Methods 1. In brief, presence of the HBGAs (A, B, Lewis a, and Lewis b) in saliva was determined by ELISA assay, as previously described [20]. A subset of saliva samples were also investigated with Ulex europaeus agglutinin (UEA-I), which recognizes Fucα1-2Gal-R present in saliva of secretors, with an ELISA assay as previously described [20].
Genotypic Characterization of the FUT3 Gene From Children in Burkina Faso
To confirm the Lewis phenotype, DNA was extracted from buccal cells collected from a subset of children from Burkina Faso (n = 35) with Whatman OmniSwab (GE Healthcare, Uppsala, Sweden), and the coding region of the FUT3 gene was sequenced. All Lewis-negative children from whom buccal swabs were available were included (n = 31), together with 4 randomly selected Lewis-positive children from Burkina Faso. MagAttract DNA Mini M48 kit (Qiagen, Hilden, Germany) was used for extraction, and DNA was stored at −20°C for subsequent sequencing.
The messenger RNA sequence of the FUT3 gene (accession number NM_000149) was obtained from the University of California, Santa Cruz Genome browser (available at: http://genome.ucsc.edu). ExonPrimer (available at: http://ihg.gsf.de/ihg/ExonPrimer.html) was used to design primers to cover the coding sequence (CDS). These primers, covering the FUT3 CDS region for exon 3, were designed in 4 fragments; for the FUT3_Ex3_2 fragment, the pair (EL-3s and III-l2as) was adapted from Elmgren et al [23]. The sequences of the PCR primers for FUT3 are listed in Supplementary Table 1, together with detailed description of the analytical procedure (Supplementary Materials and Methods 2).
Statistical Analysis
Fisher exact test and unadjusted odds ratios were used to compare frequencies of qualitative variables between dependent groups using GraphPad Prism version 5.0 (GraphPad Software, San Diego, California). Differences were considered to be statistically significant when the level of 2-tailed significance was <.05.
RESULTS
Lewis-Negative Children Were Resistant to P[8] RV Strains in Burkina Faso
In a previous study, we observed a high prevalence of the Lewis-negative phenotype (Lea− b−) in Burkina Faso [20]. Among the 208 children with diarrhea enrolled, the phenotypic HBGA distribution was Lea− b+ (54%), Lea+ b− (14%), and Lea− b− (32%), with 80% and 20% being secretors and nonsecretors, respectively. To determine the association between HBGA and susceptibility to RV infections and genotypes, 59 RV-positive children infected with P[4] (n = 2), P[6] (n = 27), P[8] (n = 27), and P[6]/[P8] (n = 3) were investigated. Whereas the frequency of the Lewis-negative children was almost identical between the population and the RV-infected groups (32% vs 31%; Table 1), none of the RV-positive, Lewis-negative children were infected with P[8] RV genotypes (Table 1; P < .0001). P[8] RV strains (n = 27) were only found in Lewis-positive children, who were all secretors (27/27; P < .0001; Tables 1 and 2). The 2 P[4] RV genotypes as well as the 3 mixed P[6]/P[8] RV infections were observed only in children with the Lewis- and secretor-positive phenotype (Lea− b+).
Table 1.
Association Between Lewis Phenotype and Infecting P Genotype Among Rotavirus-Positive Children in Burkina Faso
| Lewis Phenotype | Lewis Phenotype in the Study Population, n = 208 (%) | Lewis Phenotype in RV Positives, n = 59a (%) | P[6] |
P[8] |
||||
|---|---|---|---|---|---|---|---|---|
| RV P[6], n = 27 (%) | Odds Ratio (95% CI)b | P Valuec | P[8], n = 27 (%) | Odds Ratio (95% CI)b | P Valuec | |||
| Lea− b+ | n = 113 (54) | 38 (64) | 6 (22) | 0.2 (.1–.5) | .0004 | 27 (100)d | NA | <.0001 |
| Lea− b− | n = 66 (32) | 18 (31) | 18 (67) | 5.5 (2.3–13.2) | <.0001 | 0 (0) | 0.0 (NA) | <.0001 |
| Lea+ b− | n = 29 (14) | 3 (5) | 3 (11) | 0.7 (.2–2.7) | 1.0 | 0 (0) | 0.0 (NA) | .0176 |
Abbreviations: CI, confidence interval; Le, Lewis; NA, not applicable; RV, rotavirus.
a Two P[4] RV infections were found in Lea− b+ children, and 3 mixed P[6]/P[8] infections were found in Lea− b+ children. In the table, only P[6] or P[8] single-genotype infections are presented.
b Unadjusted odds ratio.
c Fisher exact test with 2-tailed significance compared with Lewis phenotype distribution in study population.
d One P[8]-infected Lewis b–positive, Ulex europaeus agglutinin I–negative child was classified as a secretor based on FUT2 genotyping.
Table 2.
Association Between Rotavirus P Genotypes to Lewis and Secretor Status in Burkina Faso
| Phenotype | Study Population (n = 208) (%) | P[6] |
P[8] |
||||
|---|---|---|---|---|---|---|---|
| RV P[6] (n = 27) (%) | Odds Ratio (95% CI)a | P Value | RV P[8] (n = 27) (%) | Odds Ratio (95% CI)a | P Value | ||
| Lewis positive | n = 142 (68) | n = 9 (33) | 0.2 (.08–.4) | <.0001b | n = 27 (100) | NA | <.0001b |
| Secretor | n = 113 (80) | 6 (67) | 0.5 (.1–2.1) | .389c | 27d (100) | NA | .0013c |
| Nonsecretor | n = 29 (20) | 3 (33) | 2.1 (.5–8.8) | .389c | 0 (0) | 0.0 (NA) | .0013c |
| Lewis negative | n = 66 (32) | 18 (67) | 5.5 (2.3–13.2) | <.0001b | 0 (0) | 0.0 (NA) | <.0001b |
| Secretor | n = 52 (79) | 12 (67) | 0.4 (.1–1.4) | .180c | 0 (0) | NA | NA |
| Nonsecretor | n = 14 (21) | 6 (33) | 2.5 (.7–8.6) | .180c | 0 (0) | NA | NA |
Abbreviations: CI, confidence interval; NA, not applicable; RV, rotavirus.
a Unadjusted odds ratio.
b Fisher exact test with 2-tailed significance, compared with Lewis phenotype distribution in the study population.
c Fisher exact test with 2-tailed significance, comparing secretor vs nonsecretor, stratified by Lewis positive or Lewis negative, respectively.
d One P[8]-infected Lewis b–positive, Ulex europaeus agglutinin I–negative child was classified as a secretor based on FUT2 genotyping.
RV Strains of Genotype P[6] Infected Mainly Lewis-Negative Children in Burkina Faso
Two-thirds of the P[6] RV genotypes infected Lewis-negative children (18/27; odds ratio, 5.5; P < .0001; Table 1). The P[6] RV strains were significantly more frequent in Lewis-negative children, but independent of the secretor status as determined by Ulex europeus binding to saliva or presence of Lewis antigens (Table 2). Among the secretor-positive children, P[6] RVs were observed more often in children of blood type O compared with P[8] RVs (61% vs 23%; P = .015), while being less frequent in children of blood type A (17% vs 31%; P > .05) and blood type B (17% vs 46%; P > .05), compared with P[8] RVs.
Sequencing of the FUT3 Open-Reading Frame of Lewis-Negative Children Confirmed the Saliva Phenotypes
To confirm the Lewis-negative phenotype, the entire coding sequence of the FUT3 gene was sequenced from 31 children who were Lewis-negative by ELISA. Thirty of 31 were genotypically confirmed as Lewis-negative for previously published alleles (available at: http://www.ncbi.nlm.nih.gov/projects/gv/mhc/xslcgi.cgi?cmd=bgmut/systems).
None of those individuals was homozygous for only a single allele. In total, 15 Lewis-negative alleles were identified (Supplementary Table 2), and the 3 dominating were the 59T>G; 508G>A; 858A>G, the 13G>A; 484G>A; 667G>A and the 202T>C; 314C>T alleles, corresponding to 18 (30%), 8 (13%), and 6 (10%) of the alleles, respectively. One RV-negative child was phenotyped as Lewis negative but was by sequencing heterozygous, with only 1 known negative FUT3 allele (59T>G, 508G>A, 858A>G); the other had a point mutation of unknown significance and could thus not be completely explained. Sequencing was performed for 4 Lewis-positive children (3 Lea+ b− and 1 Lea− b+), and they were genetically heterozygous with only 1 Lewis-negative allele.
P[8] RVs Infected Only Lewis- and Secretor-Positive Children in Nicaragua
To confirm that the host genetic pattern observed in Burkina Faso was not limited to a particular ethnic population in Africa, saliva samples from RV-positive children in Nicaragua were also collected and analyzed. The Lewis distribution in RV-positive children was Lea− b+ (94%), Lea+ b− (0%), and Lea− b− (6%), The P genotype distribution of the 34 RV-positive samples was P[4] (n = 7), P[6] (n = 4), P[8] (n = 22), and P[4]/P[6]/P[8] mix (n = 1). RV of genotype P[8] (n = 22) was only observed in Lewis- and secretor-positive (Lea− b+ children) and not in Lewis-negative children, nor in Lewis-positive nonsecretors (Supplementary Table 3). Also, RV of genotype P[4] was only observed in Lewis- and secretor-positive children, but their small number warrants a cautious interpretation (Supplementary Table 3). Furthermore, as in Burkina Faso, the Lewis-negative children were infected only with genotype P[6] (Supplementary Table 3); however, the number of Lewis-negative children in this cohort (n = 2) was too small to draw any conclusions. The 2 Lewis-negative children infected with RV P[6] were also nonsecretors as determined by the UEA-I lectin. Even though the prevalence of the Lewis-positive no-secretor (Lea+ b−) phenotype is low (<5%) in Nicaragua [24], no such child was infected with any RV genotype (Supplementary Table 3).
DISCUSSION
It was recently described that human RVs recognize HBGAs [9, 11–13]. In this study from Burkina Faso and Nicaragua, we found that P[8] RVs infected exclusively Lewis- and secretor-positive children. In contrast, P[6] RV strains infected mainly Lewis-negative children, independent of their secretor status.
Our observation that RV P[6] genotypes preferably infected Lewis-negative children is complementary to the in vitro findings by Huang et al [12] showing binding of recombinant VP8* protein of P[6] genotypes to synthetic H type 1 antigen (present in epithelial surfaces of Lewis-negative secretors), but not to the Lewis b antigen (present in the epithelial surfaces of Lewis-positive secretors). In our study, we also observed P[6] RV strains infecting 6 Lewis-negative nonsecretors, suggesting that the type 1 chain precursor could also act as a mediating factor for susceptibility to the P[6] genotypes. This antigen was not tested for its binding capacity by Huang et al [12]. The core structure of the type 1 chain glycans, Galβ3GlcNAc, may thus be essential for cell-mediated binding and entry, or more important, for productive infection by P[6] strains. However, we also observed that 33% of all P[6] infections were found in Lewis-positive children. It is important to note that all type 1 chain precursors are not always entirely converted into Lewis a, H, or Lewis b antigens by the fucosyltransferase encoded by the FUT3 and FUT2 genes; thus, core structures may coexist together with the ABH and Lewis HBGAs [25, 26], acting as possible attachment factors. Interestingly, a recent in vivo study from Vietnam reported low Lewis b expression in P[6]-infected children [27], indicative of a lower conversion rate and higher presence of such core structures.
We further observed that Lewis-negative children were infected neither with P[8] nor with P[4] RV; however, the number of P[4] genotypes in this study were few, thus warranting careful interpretation. All P[8] and P[4] RVs infected Lewis- and secretor-positive children. Of note, among P[8]-infected children, one child had, in addition to Lewis b, high levels of Lewis a and was UEA-I lectin negative. Nucleotide sequencing of the FUT2 gene for this child showed a heterozygous secretor genotype and was thus considered to be a secretor. Huang et al [12] showed an in vitro binding of recombinant VP8* proteins of the P[8] and P[4] genotypes to both H type 1 and Lewis b antigens, but not to Lewis a, which partially agrees with our findings. However, in our study, no Lewis-negative secretor (expressing the H type 1 antigen) was infected with P[8] strains, suggesting that presence of H type 1 antigen was not sufficient for establishing a P[8] genotype–specific symptomatic infection. Our results thus indicate that the additional α1-4 linked fucose on the H type 1 antigen (making the Lewis b antigen) is important for susceptibility of genotype P[8]. Interestingly, Huang et al [12] observed that purified triple layer particles of the Wa strain (P[8] genotype) exhibited in vitro binding to Lewis-positive saliva but not to Lewis-negative saliva of secretors, which is in line with our findings.
Moreover, our in vivo observations together with previous binding studies [12] favor that RV P[4] and P[8] genotypes use similar cell attachment factors; although the number of P[4] genotype infections in this study were few, thus further studies are warranted. As previously suggested [12], this would explain why these 2 P genotypes occur in a cyclic pattern in white populations in Europe and North America having a high prevalence of individuals with the Lewis- and secretor-positive phenotype.
Recent in vivo studies from France and Vietnam observed that RVs of genotype P[8] infected only secretor-positive children [17, 27]. Both studies focused on the role of secretor status in RV infection. The vast majority of secretor-positive individuals among white populations express the Lewis b antigen. However, in Burkina Faso, which has a much higher percentage of Lewis-negative individuals; we could study the role of both FUT2 and FUT3 genes in relation to RV strain–specific host susceptibility and thus suggest the importance of functionality of both these genes in susceptibility to P[8] RV infections.
Our observation that only P[6] but not P[8]/P[4] genotypes infected Lewis-negative children could further provide an explanation to why the P[6] genotype is observed in the vast majority of neonatal RV infections [28, 29], but rarely in older children [6, 30]. Support for this hypothesis comes from previous studies reporting that the expression of Lewis antigens on the erythrocytes develop with age and that young children usually are Lewis negative until 1–2 months of age, after which the Lewis antigens are expressed [31]. Thus, neonatal children having a Lewis-negative phenotype could be susceptible to RV P[6] genotypes, but not to genotypes P[4] and P[8]. However, it is important to note that these studies were performed on erythrocytes, which do not directly correspond to Lewis antigens present in the intestinal tract. Glycosphingolipids carrying the HBGA type 1 chain ABH and Lewis antigens are enriched in fetal gut epithelium and in meconium of neonates [32]. The type 1 chain precursor (Galβ3GlcNAc) is typically present in high amounts in neonatal gut together with all the HBGAs on glycolipids carrying the ABH, secretor, and Lewis gene end products [26]. This pattern contrasts to that of the adult small intestine, where the type 1 chain precursor is usually not detectable and has only been described in 1 Lewis- and secretor-negative individual [33] and in a proximal resection of jejunum taken from a patient with gastric cancer [25]. Although only minor structural changes of fucosylated glycosphingolipids occur along the villus axis of an adult intestine [34], little is known about the cell-specific, temporal, and longitudinal changes of HBGAs in intestinal tissues in early childhood [35], especially in relation to changes in the gut microflora [36]. It is important to note that discrepancies between saliva and gut HBGA expression can occur, for example, due to infection with pathogens such as H. pylori as recently reported [37].
The fact that the Lewis-negative phenotype is more common in many African populations [19, 20] may explain why P[6] RV infections are much more common in Africa than in white populations in Europe and North America where the Lewis-negative phenotype is rare [18, 23]. In view of our findings, host genetic susceptibility could also have implications for vaccine efficacy. The P[8] genotype is a major component of 2 licensed RV vaccines whose efficacy has been proven modest in Africa compared with other regions [3, 4], with African populations having a high frequency of Lewis-negative individuals [19, 20]. Because a productive infection by the live attenuated vaccines would stimulate an immune response, it is reasonable to suggest vaccine efficacy to be lower for Lewis-negative individuals. Moreover, as expression of HBGAs is developmentally regulated, it is important to consider both the composition of the vaccine strain and the optimal age of immunization.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online (http://cid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Financial support. This work was supported by the Swedish Research Council (grant numbers 8266 to G. L., 3485 to L. S., and dnr-348-2011-7420 to L. S. and F. B.). All genotyping was performed at the Genomics Platform, Core Facilities, Sahlgrenska Academy at the University of Gothenburg.
Potential conflicts of interest. All authors: No potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12:136–41. doi: 10.1016/S1473-3099(11)70253-5. [DOI] [PubMed] [Google Scholar]
- 2.Parashar UD, Burton A, Lanata C, et al. Global mortality associated with rotavirus disease among children in 2004. J Infect Dis. 2009;200(suppl 1):S9–15. doi: 10.1086/605025. [DOI] [PubMed] [Google Scholar]
- 3.Madhi SA, Cunliffe NA, Steele D, et al. Effect of human rotavirus vaccine on severe diarrhea in African infants. N Engl J Med. 2010;362:289–98. doi: 10.1056/NEJMoa0904797. [DOI] [PubMed] [Google Scholar]
- 4.Armah GE, Sow SO, Breiman RF, et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;376:606–14. doi: 10.1016/S0140-6736(10)60889-6. [DOI] [PubMed] [Google Scholar]
- 5.Todd S, Page NA, Duncan Steele A, Peenze I, Cunliffe NA. Rotavirus strain types circulating in Africa: review of studies published during 1997–2006. J Infect Dis. 2010;202(suppl):S34–42. doi: 10.1086/653555. [DOI] [PubMed] [Google Scholar]
- 6.Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol. 2005;15:29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- 7.Nordgren J, Nitiema LW, Sharma S, et al. Emergence of unusual G6P[6] rotaviruses in children, Burkina Faso, 2009–2010. Emerg Infect Dis. 2012;18:589–97. doi: 10.3201/eid1804.110973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pesavento JB, Crawford SE, Roberts E, Estes MK, Prasad BV. pH-induced conformational change of the rotavirus VP4 spike: implications for cell entry and antibody neutralization. J Virol. 2005;79:8572–80. doi: 10.1128/JVI.79.13.8572-8580.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Huang P, Tan M, et al. Rotavirus VP8*: phylogeny, host range, and interaction with histo-blood group antigens. J Virol. 2012;86:9899–910. doi: 10.1128/JVI.00979-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isa P, Arias CF, Lopez S. Role of sialic acids in rotavirus infection. Glycoconjugate J. 2006;23:27–37. doi: 10.1007/s10719-006-5435-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu L, Crawford SE, Czako R, et al. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature. 2012;485:256–9. doi: 10.1038/nature10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang P, Xia M, Tan M, et al. Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J Virol. 2012;86:4833–43. doi: 10.1128/JVI.05507-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramani S, Cortes-Penfield NW, Hu L, et al. The VP8* domain of neonatal rotavirus strain G10P[11] binds to type II precursor glycans. J Virol. 2013;87:7255–64. doi: 10.1128/JVI.03518-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Pendu J, Ruvoen-Clouet N, Kindberg E, Svensson L. Mendelian resistance to human norovirus infections. Semin Immunol. 2006;18:375–86. doi: 10.1016/j.smim.2006.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rydell GE, Kindberg E, Larson G, Svensson L. Susceptibility to winter vomiting disease: a sweet matter. Rev Med Virol. 2011;21:370–82. doi: 10.1002/rmv.704. [DOI] [PubMed] [Google Scholar]
- 16.Boren T, Falk P, Roth KA, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262:1892–5. doi: 10.1126/science.8018146. [DOI] [PubMed] [Google Scholar]
- 17.Imbert-Marcille BM, Barbe L, Dupe M, et al. A FUT2 gene common polymorphism determines resistance to rotavirus A of the P[8] genotype. J Infect Dis. 2013;209:1227–30. doi: 10.1093/infdis/jit655. [DOI] [PubMed] [Google Scholar]
- 18.Larsson MM, Rydell GE, Grahn A, et al. Antibody prevalence and titer to norovirus (genogroup II) correlate with secretor (FUT2) but not with ABO phenotype or Lewis (FUT3) genotype. J Infect Dis. 2006;194:1422–7. doi: 10.1086/508430. [DOI] [PubMed] [Google Scholar]
- 19.Corvelo TCO, Aguiar DCF, Sagica FES. The expression of ABH and Lewis antigens in Brazilian semi-isolated black communities. Genet Mol Biol. 2002;25:259–63. [Google Scholar]
- 20.Nordgren J, Nitiema LW, Ouermi D, Simpore J, Svensson L. Host genetic factors affect susceptibility to norovirus infections in Burkina Faso. PLoS One. 2013;8:e69557. doi: 10.1371/journal.pone.0069557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nitiema LW, Nordgren J, Ouermi D, et al. Burden of rotavirus and other enteropathogens among children with diarrhea in Burkina Faso. Int J Infect Dis. 2011;15:e646–52. doi: 10.1016/j.ijid.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Bucardo F, Karlsson B, Nordgren J, et al. Mutated G4P[8] rotavirus associated with a nationwide outbreak of gastroenteritis in Nicaragua in 2005. J Clin Microbiol. 2007;45:990–7. doi: 10.1128/JCM.01992-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elmgren A, Borjeson C, Svensson L, Rydberg L, Larson G. DNA sequencing and screening for point mutations in the human Lewis (FUT3) gene enables molecular genotyping of the human Lewis blood group system. Vox Sang. 1996;70:97–103. doi: 10.1111/j.1423-0410.1996.tb01300.x. [DOI] [PubMed] [Google Scholar]
- 24.Bucardo F, Kindberg E, Paniagua M, et al. Genetic susceptibility to symptomatic norovirus infection in Nicaragua. J Med Virol. 2009;81:728–35. doi: 10.1002/jmv.21426. [DOI] [PubMed] [Google Scholar]
- 25.Bjork S, Breimer ME, Hansson GC, Karlsson KA, Leffler H. Structures of blood group glycosphingolipids of human small intestine. A relation between the expression of fucolipids of epithelial cells and the ABO, Le and Se phenotype of the donor. J Biol Chem. 1987;262:6758–65. [PubMed] [Google Scholar]
- 26.Karlsson KA, Larson G. Molecular characterization of cell surface antigens of fetal tissue. Detailed analysis of glycosphingolipids of meconium of a human O Le(a–b+) secretor. J Biol Chem. 1981;256:3512–24. [PubMed] [Google Scholar]
- 27.Van Trang N, Vu HT, Le NT, Huang P, Jiang X, Anh DD. Association between norovirus and rotavirus infection and histo-blood group antigen types in Vietnamese children. J Clin Microbiol. 2014;52:1366–74. doi: 10.1128/JCM.02927-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shim JO, Son DW, Shim SY, Ryoo E, Kim W, Jung YC. Clinical characteristics and genotypes of rotaviruses in a neonatal intensive care unit. Pediatr Neonatol. 2012;53:18–23. doi: 10.1016/j.pedneo.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Ray P, Sharma S, Agarwal RK, et al. First detection of G12 rotaviruses in newborns with neonatal rotavirus infection at all India Institute of Medical Sciences, New Delhi, India. J Clin Microbiol. 2007;45:3824–7. doi: 10.1128/JCM.01288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iturriza-Gomara M, Dallman T, Banyai K, et al. Rotavirus genotypes co-circulating in Europe between 2006 and 2009 as determined by EuroRotaNet, a pan-European collaborative strain surveillance network. Epidemiol Infect. 2011;139:895–909. doi: 10.1017/S0950268810001810. [DOI] [PubMed] [Google Scholar]
- 31.Ameno S, Kimura H, Ameno K, et al. Lewis and Secretor gene effects on Lewis antigen and postnatal development of Lewis blood type. Biol Neonate. 2001;79:91–6. doi: 10.1159/000047073. [DOI] [PubMed] [Google Scholar]
- 32.Karlsson KA, Larson G. Molecular characterization of cell-surface antigens of human fetal tissue: meconium, a rich source of epithelial blood-group glycolipids. FEBS Lett. 1978;87:283–7. doi: 10.1016/0014-5793(78)80352-4. [DOI] [PubMed] [Google Scholar]
- 33.Henry S, Jovall PA, Ghardashkhani S, et al. Structural and immunochemical identification of Le(a), Le(b), H type 1, and related glycolipids in small intestinal mucosa of a group O Le(a-b-) nonsecretor. Glycoconjugate J. 1997;14:209–23. doi: 10.1023/a:1018541821819. [DOI] [PubMed] [Google Scholar]
- 34.Breimer ME, Hansson GC, Karlsson KA, Larson G, Leffler H. Glycosphingolipid composition of epithelial cells isolated along the villus axis of small intestine of a single human individual. Glycobiology. 2012;22:1721–30. doi: 10.1093/glycob/cws115. [DOI] [PubMed] [Google Scholar]
- 35.Larson G, Watsfeldt P, Falk P, Leffler H, Koprowski H. Fecal excretion of intestinal glycosphingolipids by newborns and young children. FEBS Lett. 1987;214:41–4. doi: 10.1016/0014-5793(87)80009-1. [DOI] [PubMed] [Google Scholar]
- 36.Garrido D, Dallas DC, Mills DA. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology. 2013;159(pt 4):649–64. doi: 10.1099/mic.0.064113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruvoen-Clouet N, Magalhaes A, Marcos-Silva L, et al. Increase in genogroup II.4 norovirus host spectrum by CagA-positive Helicobacter pylori infection. J Infect Dis. 2014;210:183–91. doi: 10.1093/infdis/jiu054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.