Abstract
Development of the mammalian lung is predicated on cross-communications between two highly interactive tissues, the endodermally-derived epithelium and the mesodermally-derived pulmonary mesenchyme. While much attention has been paid the lung epithelium, the pulmonary mesenchyme, partly due to lack of specific tractable markers remains under-investigated. The lung mesenchyme is derived from the lateral plate mesoderm and is the principal recipient of Hedgehog (Hh) signaling, a morphogenetic network that regulates multiple aspects of embryonic development. Using the Hh-responsive Gli1-creERT2 mouse line, we identified the mesodermal targets of Hh signaling at various time points during embryonic and postnatal lung development. Cell lineage analysis showed these cells serve as progenitors to contribute to multiple lineages of mesodermally-derived differentiated cell types that include parenchymal or interstitial myofibroblasts, parabronchial and perivascular smooth muscle as well as rare populations of cells within the mesothelium. Most importantly, Gli1-creERT2 identified the progenitors of secondary crest myofibroblasts, a hitherto intractable cell type that plays a key role in alveolar formation, a vital process about which little is currently known. Transcriptome analysis of Hh-targeted progenitor cells transitioning from the pseudoglandular to the saccular phase of lung development revealed important modulations of key signaling pathways. Amongst these, there was significant down-regulation of canonical WNT signaling. Ectopic stabilization of β-Catenin via inactivation of Apc by Gli1-creERT2 expanded the Hh-targeted progenitor pools, which caused the formation of fibroblastic masses within the lung parenchyma. The Gli1-creERT2 mouse line represents a novel tool in the analysis of mesenchymal cell biology and alveolar formation during lung development.
Introduction
Development of vertebrate organs is initiated by specification of a primordium within the early embryo and usually requires contributions from more than one germ layer. Ontogeny and development of the mammalian lung is no exception and requires contributions from at least two highly interactive embryonic tissues, the endodermally-derived epithelium and the mesodermally-derived pulmonary mesenchyme. Epithelial-mesenchymal interactions are centerpiece in both structural development of the lung as well as differentiation of its many highly specialized cell types.
While the last two decades have witnessed extensive analysis of the lung epithelium, the pulmonary mesoderm, partly due to lack of specific markers has been less tractable. The pulmonary mesenchyme is derived from the lateral plate mesoderm, which forms in the early embryo subsequent to gastrulation. One of the earliest mesodermal cell types to differentiate in the embryonic lung is distinguished by ACTA2 expression. In the adult lung, the ACTA2-expressing lineages can be viewed as belonging to two large classes of mesodermally-derived cell populations; smooth muscle cells and myofibroblasts. As early as embryonic day E11.5, ACTA2-expressing smooth muscle cells are found as distinct cell layers around the nascent airways and the mainstem bronchi that are formed by the first endodermal bifurcation. As development of the airways proceeds in a proximo-distal direction, the ACTA2-expressing smooth muscle lineage contribute to parabronchial & perivascular smooth muscle fibers (PBSM & PVSM respectively) and possibly cells known as pericytes. Abnormalities in these structures have profound consequence on normal airway and vascular function and lead to diseases such as asthma and pulmonary hypertension.
The lung mesoderm also serves as the source of interstitial myofibroblasts (IMF), the contractile fibroblasts that express ACTA2. During early lung development (before saccular stage) progenitors of IMFs are scattered in the parenchyma of the lung. In these cells, ACTA2 is undetectable or absent, and no marker has been reported to distinguish them from other fibroblast progenitors. However, PDGFRα was reported as a marker for IMF progenitors in saccular lungs 1, 2. In the adult lung, IMFs appear as ACTA2pos cells embedded in the alveolar parenchyma but in much reduced numbers3. The function of IMF in the adult lung remains entirely unknown but the IMFs in the perinatal lung are the source of alveolar or secondary crest myofibroblasts (SCMFs). SCMFs are located at the tip of secondary crest structures during the saccular and alveolar phases of lung development. SCMFs have remained a highly intractable, elusive cell type and there is urgent need to gain a better understanding of their biology. SCMFs play a key role in alveolar formation. In human preterm neonates, interruption in alveogenesis underlies the pathogenesis of the chronic lung disease known as bronchopulmonary dysplasia or BPD. In adults, destruction of alveoli is a hallmark of emphysema and COPD. Both the neonatal and adult manifestations of alveolar defects are highly morbid and can be lethal.
During embryonic development, the lung mesenchyme is the principal recipient of Hedgehog (Hh) signaling, an evolutionarily highly conserved morphogenetic network that regulates multiple aspects of development in vertebrate and invertebrate alike. Sonic hedgehog (Shh) is expressed by the lung epithelium and it is thought to activate signaling in what has been named the “sub-epithelial mesenchyme” 4 to initiate a cascade of gene expression. Shh(-/-) lungs are profoundly abnormal, failing to undergo normal branching morphogenesis 5. Importantly, Shh(-/-) lungs are reported to be entirely devoid of ACTA2-expressing cells. This places Hh signaling at the top of a putative regulatory cascade that controls the commitment and differentiation of multipotential mesenchymal cell progenitors along the ACTA2pos cell lineages.
Hh signaling is initiated upon binding of secreted Hh ligands to a receptor, Patched1 (Ptch1). In the absence of Hh ligands, Ptch1 inhibits the signaling activity of Smoothened (Smo), a seven trans-membrane protein that shares sequence homology with G-protein coupled receptors. Binding the ligand releases Ptch1-mediated repression of Smo, activating its function at the cell surface that leads to transcriptional induction of the Hh signaling cascade. In Drosophila, Hh signaling is mediated solely by the zinc finger containing transcription factor family cubitus interruptus, ci/Gli 6, 7. In vertebrates, three Gli molecules, Gli1, Gli2 and Gli3 function downstream of Hh signaling. That Gli1 is a direct transcriptional target of Hh signaling is supported by the results of many studies. For example, ectopic Shh induces Gli1 expression 8-11 and in the absence of Shh, Gli1 is not expressed 12. Thus, Hh activity is both necessary and sufficient for Gli1 transcription. Recently Ahn and Joyner 13 generated mice carrying a knockin allele of Gli1-creERT2. Gli1-CreERT2 is expressed exclusively in cells that have received positive Hh signaling and only in the presence of tamoxifen (Tam). In these mice, addition of Tam defines the time point at which Gli1-expressing cells are marked. Thereafter, the progeny of such cells can be followed with GFP using ROSAmTmG reporter mice. Thus, Gli1-CreERT2 represents an inducible, highly reliable tool in genetic fate-mapping strategy to identify Hh-responding cells and their descendents during the process of development and postnatally only in the presence of Tam.
In the present work, we have utilized the ability of Gli1-CreERT2 to permanently label progenitor cell populations that are recipients of Hh signaling during early embryonic and postnatal development of the murine lung. By cell lineage analysis, we show that the targets of Hh signaling in the early lung serve as progenitors to contribute to a number of mesodermally-derived differentiated cell types. The Gli1-CreERT2-tagged cells contribute to the IMF cell population and are found in the PBSM and PVSM layers as well as within the mesothelium. Most importantly, we demonstrate that Hh-tageted mesodermal cells in early embryonic lung (E10.5-E11.5) serve as progenitors to SCMFs. Gene array analysis of Gli1-creERt2-tagged cells in early embryonic lungs revealed the involvement of multiple key signaling pathways as such progenitors underwent differentiation during two critical periods of lung development. Ectopic activation of one such pathway, WNT signaling, by targeted genetic inactivation of Apc resulted in expansion of the SCMF progenitor cell population and subsequently lead to formation of multiple myofibroblast masses in the perinatal lungs. The present findings pave the way towards isolation and characterization of SCMF progenitors, an admittedly important cell type whose understanding may be key to elucidating the mechanisms of alveolar formation in normal lung development and, failure in both neonatal and adult lung diseases such as BPD and COPD respectively.
Materials & Methods
Mouse Breeding and Genotyping
All animals were maintained and housed in pathogen-free conditions according to the protocol approved by The University of Southern California Institutional Animal Care and Use Committee (IACUC) (Los Angeles, CA, USA). Gli1-creERT2; ROSAmTmG mice were generated by breeding the Gli1-creERT2 13 and the ROSAmTmG mice [Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J, The Jackson Laboratory] on 129S6/SuEuTac genetic background. The Gli1-creERT2; ROSAmTmG mice were then bred with the Apcflox/flox to generate the ApcGli1mice. Genotyping of the transgenic mice were determined by PCR with genomic DNA isolated from mouse tails or embryo tissue. The forward (F) and reverse primers (R) for transgenic mouse genotyping are listed below.
Gli1-creERT2: (forward) 5′-TAA AGA TAT CTC ACG TAC TGA CGG TG-3′ and (reverse) 5′-TCT CTG ACC AGA GTC ATC CTT AGC-3′. Apcflox/flox: (Forward) 5′-GAGAAACCCTGTCTCGAAAAAA-3′ and (Reverse) 5′-AGTGCTGTTTCTATGAGTCAAC-3′.
Immunofluorescent staining
Immunofluorescent staining was performed as previously described 14. Primary antibodies used are: mouse anti-GFP (Santa Cruz, CA); mouse anti-ACTA2 (Sigma); rabbit anti-ACTA2 (Abcam, MA); rabbit anti-PDGFRα (Cell Signaling). Nuclei were counterstained with Dapi. For quantitative analyses, multiple images (n>8, each contains 300 to 600 cells) were used to count the ratio of labeled cells. Quantitative data are presented as average values+/-standard error of the mean. Sections from at least three lungs were analyzed for each data point.
For Oil red O staining, frozen sections of neonatal lungs (8 um) were air dried, washed with tap water, rinsed with 60% isopropanol and then stained with freshly prepared Oil Red O solution for 15 minutes. The stained sections were rinsed with 60% isopropanol followed by distilled water and then preserved in VECTASHELD mounting medium with Dapi (to visualize nuclei).
Tamoxifen administration
Tamoxifen was administered by oral gavages. For cell fate of neonatal lungs, two doses of Tamoxifen were administered to neonates at postnatal day 5 and 6 (PN5 and PN6). For cell fate of embryonic lungs, timed-pregnant Gli1-creERT2; ROSAmTmG females received two doses of Tamoxifen at embryonic day 10.5 and 11.5 (E10.5 and E11.5). Animals were sacrificed at E12.5 to determine the pattern of Gli1-creERT2 labeled cells. For lineage-tracing from embryonic stage to postnatal stage, Tamoxifen treated pregnant females were sacrificed and the pups were extracted by cesarean section at around E18.5. The pups were cleaned, dried and fed by a foster mother that had delivered her own pups within a 2 day period.
Cell isolation
Gli1-creERT2; ROSAmTmG embryonic lungs at stage of E14.5 or E18.5 were dissected in HBSS (GIBCO24020-117). Lung lobes were cut into small pieces, treated in 0.25% Trypsin-EDTA (GIBCO 25200-056) at 37°C for 10min, and transferred to 3 ml of DMEM/F12 with 1 mg/ml DNase I (Roche10104159001), followed by repeated pipetting. After adding 20 ml of DMEM/F12 containing 10%FBS, the mixture was pipetted up and down until tissue was completely dissociated. Dissociated cells were filtered through 100um and 40um cell strainers, centrifuged at 4°C at 1250 rpm for 5 min and re-suspended in DMEM/F12. GFPpos cells were isolated by cell sorting (BD FACSAria III). GFPneg cells from ROSAmTmG (crenegative) lungs isolated under the same condition were used as blank control for cell sorting. Relative abundance of GFPpos cells between E14.5 and E18.5 was calculated by comparison of the number of GFPpos cells from each whole lung, collected by cell sorting. Data represent average of four ratios from four E14.5 and four E18.5 isolations. p-value was calculated by one-sample t-test (hypothetical mean is 1).
Gene Expression Profiling
Gli1-creERT2; ROSAmTmG mouse embryonic lungs, which received Tam at E10.5 and E11.5, were dissected at E14.5 and E18.5. GFPpos cells were sorted (BD FACSAria III) and isolated for RNA extraction. RNA was subjected to Whole Mouse Genome Oligo Microarrays (Agilent). Raw intensities were log2-transformed and quantile normalized (Partek Genomics Suite 6.6). We selected probe sets with absolute fold change greater than 2 and a false discovery rate (FDR) value less than 0.05. Genes with raw intensity less than 50 in both stages are not included. Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems, Redwood city, CA) was used to analyze functional relationships of differentially expressed genes.
Realtime Polymerase Chain Reaction
Expression of selected genes was quantified by Realtime PCR using a LightCycler with LightCycler Fast Start DNA Master SYBR Green I Kit (Roche Applied Sciences, IN) as previously described 15. Relative ratio of a target gene transcript in GFPpos cells between E18.5 and E14.5 was calculated with the ΔΔCt method 14. All primers for Realtime PCR were designed by using the program of Universal ProbeLibrary Assay Design Center from Roche Applied Sciences (IN). Sequences of the primers are listed in supplemental Table ST1.
Results
Labeling of Hedgehog targets in Embryonic Lungs
To determine an optimal dose of Tamoxifen (Tam) that would activate Gli1-creERT2 in the embryonic lung cells receiving Hh signaling, without causing abortion of the fetus, we tested two strategies. First we delivered a single dose of 4 mg of Tam per 30 g of body weight by gavage to pregnant Gli1-creERT2; ROSAmTmG mice at gestational day E10.5 and examined their embryos within 12, 18-20, and 24 hours of exposure. Figure 1 shows that within the first 12 hours of Tam exposure only a minority of cells of distinctly mesenchymal lineage (i.e. NKX2.1neg, Figure 1, A) are GFPpos. The number of GFPpos cells increased in the 18-20, and the 24 hour post-Tam embryonic lungs (Figure 1, B and C, respectively). In both the 18-20 and 24 hour windows, GFPpos cells were localized around the branching epithelium within a region that has been referred to as the “sub-epithelial mesenchyme” 4. Scant GFPpos cells were localized close to or within the mesothelium of the lung (arrows). As an alternative to maximize the labeling potential of the Gli1-creERT2 we also administered two doses of Tam, 24 hours apart, delivered between E10.5 & E11.5. This regimen significantly increased the number of GFPpos cells in E12.5 lungs (Figure 1, D-F). Examination of multiple embryonic lung samples showed that GFPpos cells tagged on E10.5-E11.5 made up nearly 27 % (+/- 0.075) of the total (Dapi positive) mesenchymal cell population in E12.5 lungs. The majority of these cells were undifferentiated interstitial cells in which ACTA2 was undetectable (Figure 1, D-F). There were also few GFPpos cells near or within the peribronchial smooth muscle or perivascular smooth muscle layers (only PBSM shown). Importantly, these were ACTA2pos (Figure 1, D-F).
Figure 1.
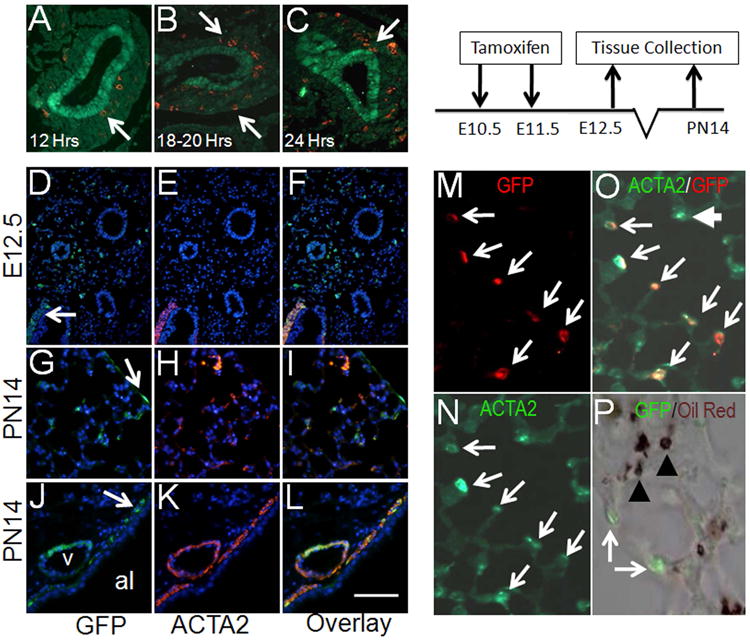
Lineage tracing of Gli1-creERT2;ROSAmTmG labeled cells in embryonic lungs. (A-C) Co-immunofluorescent staining of NKX2.1 (green) and GFP (red) in lungs of E11.5 Gli1-creERT2;ROSAmTmG embryos, treated with one dose of Tam at E10.5. (D-F) Co-immunofluorescent staining of GFP (green) and ACTA2 (red) in E12.5 lungs. (G-L) Co-immunofluorescent staining of GFP (green) & ACTA2 (red) in PN14 Gli1-creERT2;ROSAmTmG lungs. Arrows in panels G and J indicate GFPpos cells in mesothelial and PBSM layer, respectively. (M-N) Co-immunofluorescent staining of ACTA2 (green) and GFP (red) in secondary crest myofibroblast cells of PN14 pups that were treated with Tam at E10.5 and E11.5. (P) Distribution of GFP (green) and Oil-red-O (brown) are mutually exclusive in the Tam-treated PN14 lungs. Chart illustrates the experimental plan for panels D-P. “v” indicates blood vessel. “al” indicates airway. Arrows indicate GFPpos cells. Arrowheads indicate Oil red O positive staining. Scale bar: 85 um for D-F; 50 um for A-C, G-L; 15 um for M-P.
Postnatal Fate of Early Embryonic Hedgehog Targets
Tam remains active for approximately 30 hours 13, 16. Therefore, we adopted the “two-dose” Tam strategy (described above) and used the observations on E12.5 lungs as baseline for comparison in lineage analyses of E10.5-E11.5-tagged cells with later stages of lung development in the remainder of this project. Lineage tracing of cells labeled in E10.5-E11.5 embryos to postnatal day 14 (PN14), showed GFPpos cells can be found as interstitial myofibroblasts (IMF), and a very rare population of cells in the mesothelium. Immunohistohemistry for ACTA2 revealed that the majority (94.5+/-5.9%) of the GFPpos IMFs had now differentiated to ACTA2pos (Figure 1, G-I, M-O). As expected, the GFPpos cells within the mesothelium showed no detectable expression of ACTA2 (arrow in Figure 1, G-I). GFPpos cells were also found in the two structures composed of smooth muscle cells in PN14 lungs. Both PVSM and PBSM layers in PN14 lungs contained GFPpos cells (Figure 1, J-L).
Progenitors of Secondary Crest Myofibroblasts are Committed in Early Lung Mesenchyme
We also examined the contribution of (Gli1pos) GFPpos cells to another class of ACTA2-expressing cells; the SCMFs. These are ACTA2pos cells appearing at the tip of the secondary crest found during the process of alveolization and thought to have a major role in this process. Unexpectedly, co-immunostaining with anti-GFP and anti-ACTA2 antibodies revealed that GFPpos cells labeled in E10.5-11.5 lungs or their daughters also contributed to SCMFs, which appear only during alveolar formation. In PN14 Gli1-creERT2;ROSAmTmG lungs there were large number of GFPpos areas, within which upwards of nearly 95% of the SCMFs were double positive for both GFP and ACTA2 (Figure 1, M-O). Occasionally, ACTA2pos cells were observed that were not at first sight GFPpos. However, careful examination showed that they were weakly stained with GFP antibody. Finally, due to spatial proximity to SCMFs we also examined whether any of the GFPpos cells identified in PN14 lungs were lipofibroblasts. Using Oil-red-O, it is clear that GFPpos and Oil-red-Opos cells are indeed distinct (Figure 1, P). This evidence shows that the SCMF cell fate is already established in E10.5-E11.5 embryonic lungs, a surprisingly early commitment for cells whose functional role is required during late and postnatal lung development.
Gli1-creERT2 labeled myofibroblast progenitors progressively commit to PDGFRα expression
PDGFRα has been proposed as an early marker of smooth muscle and myofibroblast cell lineages 1, 2. In PdgfA(-/-) lungs, parenchymal and scattered cells expressing PDGFA's sole receptor, PDGFRα were absent, but expression in PBSM and PVSM appeared normal 1. Correspondingly, myofibroblast cell differentiation and alveolization in PdgfA(-/-) lungs were blocked. To ascertain the temporal relationship between Hh signaling as assessed by GFPpos status and PDGFRα expression, we examined the lungs of embryos and newborns from Gli1-creERT2;ROSAmTmG pregnant mice that received Tam on E10.5-E11.5. In E18.5, during the saccular stage of lung development, approximately 60% overlap was observed between GFPpos cells and those identified by immunostaining as PDGFRαpos (Figure 2, A-I). In contrast, as the lungs developed to PN14 (alveolar stage) when extensive overlap between ACTA2pos and GFPpos cells was clearly evident (Figure 1) nearly all (97.2+/-4.4%) GFPpos cells were also PDGFRαpos by immunostaining (Figure 2, J-R). The increase in GFP/PDGFRα double positive cells is unlikely caused by different proliferation rate of PDGFRαpos and PDGFRαneg cells, because it was reported that PDGFRαpos cells show decreased proliferation as compared to PDGFRαneg lung fibroblast17. Thus, there is progressive acquisition of PDGFRα expression by E10.5-E11.5 mesenchymal targets of Hh signaling as lung development proceeds. These data demonstrate that activation of Pdgfrα expression occurs subsequent to commitment of the progenitor cells to ACTA2pos lineage.
Figure 2.
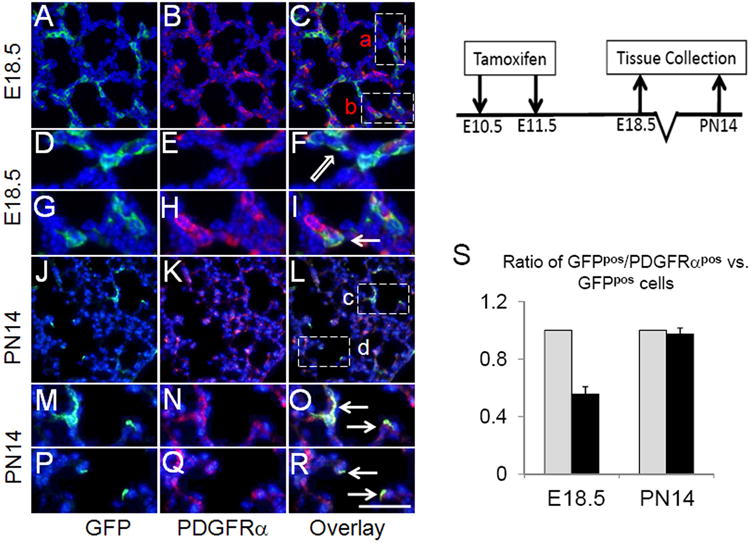
Timed pregnant females were treated with two doses of Tam at E10.5 and E11.5. Gli1-creERT2;ROSAmTmG lungs of E18.5 embryos and PN14 pups were collected and analyzed by co-immunofluorescent staining of GFP (green) & PDGFRα (red). (D-F) Higher magnification of boxed area “a” rotated counterclockwise. (G-I) Higher magnification of boxed area “b”. (M-O &P-R) Higher magnification of boxed areas “c” & “d”, respectively. Arrows indicate GFPpos/PDGFRαpos cells. Block arrows indicate GFPpos/PDGFRαneg cells. (S) Calculated ratios of GFPpos/PDGFRαpos cells vs. GFPpos cells (black bar). Total GFPpos cells were arbitrarily set as 100% (grey bar). Scale bar: 50 um for A-C, J-L; 20 um for D-I, M-R.
Hedgehog Signaling Targets in Postnatal Lung
Hh signaling in the postnatal period of lung development has not been adequately studied. Therefore, we also examined the fate of cells receiving Hh signaling during the first week of life, a period during which alveolar formation is initiated. Newborn mice were treated with two doses of Tam on P5 and P6 and lungs were examined at two subsequent time points. Within a short window of only 5 days (P11) subsequent to Tam administration, GFPpos cells appeared in 4 compartments including PBSM and PVSM layers (Figure 3, D-I). Rare but definitively GFPpos cells were also found in the mesothelium of P11 lungs (Figure 3, A-C). The GFPpos cells in the PBSM and PVSM compartments were ACTA2pos (Figure 3, D-I). In addition, SCMFs were uniformly GFPpos indicating that they were recipients of Hh signaling in early postnatal lung development. The source of the Hh ligand at this time period in lung development remains unexplored 18. GFPpos SCMFs were also uniformly ACTA2pos and PDGFRαpos (Figure 3, J-O, R). Importantly, as we found in Figure 3, none of the GFPpos SCMFs labeled on P5-P6 was positive for Oil red O staining (Figure 3, P-R).
Figure 3.
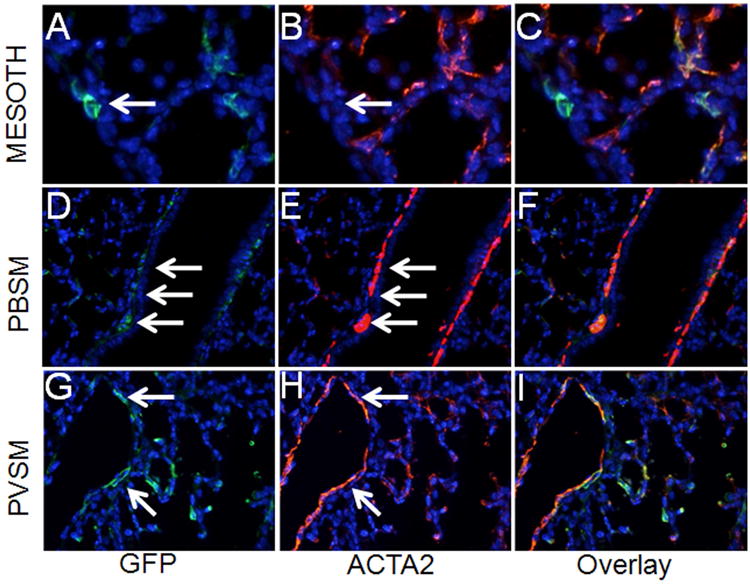
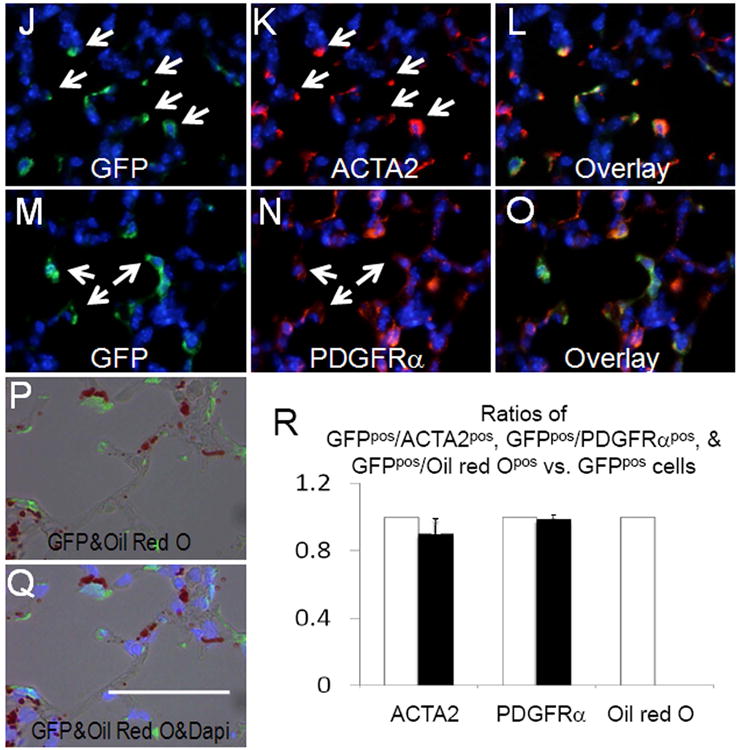
Distribution of GFPpos cells in PN11 lungs treated with Tam on PN5 and PN6. (A-L) Co-immunofluorescent staining of GFP (green) and ACTA2 (red) in areas around mesothelium (A-C), epithelial airways (D-F), blood vessels (G-I), and alveoli (J-L), respectively. (M-O) Co-immunofluorescent staining of GFP (green) and PDGFRα (red) in alveolar interstitium. (P-Q) Distribution of GFP and Oil-red-O in the Tam-treated PN11 lungs. (R) Ratios of GFPpos/ACTA2pos, GFPpos/PDGFRαpos, and GFPpos/Oil-red-Opos cells in total GFPpos cells. Black bars represent the ratio of double positive cells. Total GFPpos cells were arbitrarily set as 100% (white bars). Scale bar: 50 um for A-C, J-Q; 100 um for D-I.
Previous studies demonstrated that myofibroblasts are present specifically during alveolization and mostly disappear in normal adult lungs, reflected as loss of ACTA2pos expression 3. This raised an interesting question regarding the fate of the GFPpos cells labeled during P5-P6 period; whether the GFPpos cells simply disappeared or survived but turned off Acta2 expression? We therefore determined the fate of the cells labeled on P5-P6 after a prolonged period extending to 3 months of age. Here, GFPpos cells or their progeny were found to survive within the PBSM (Figure 4, A-C) and the PVSM layers (Figure 4, D-F). There were also rare GFPpos cells in the mesothelial layer (Figure 4, G-I). Importantly, only a few GFPpos cells (less than 1% of total Dapi positive cells) were detected in the adult lung interstitium (Figure 4, J-L). Whether these surviving GFPpos cells are derived from a sub-population of the interstitial myofibroblast or other unidentified interstitial cells remains to be determined. Further characterization of the surviving GFPpos cells revealed they express a number of differentiated mesodermal markers including Desmin, NG2 and PDGFRβ (Figure 4, M-U). These have been shown to be expressed in “pericyte-like-cells”19. The GFPpos cells also express tropoelastin (data not shown). However, expression of S100A4 [also known as FSP1, 19] was not detected. Thus mesenchymal cells that are the target of Hh signaling on P5-P6 are found in multiple lung compartments with various differentiated mesenchymal phenotypes and survive to adulthood.
Figure 4.
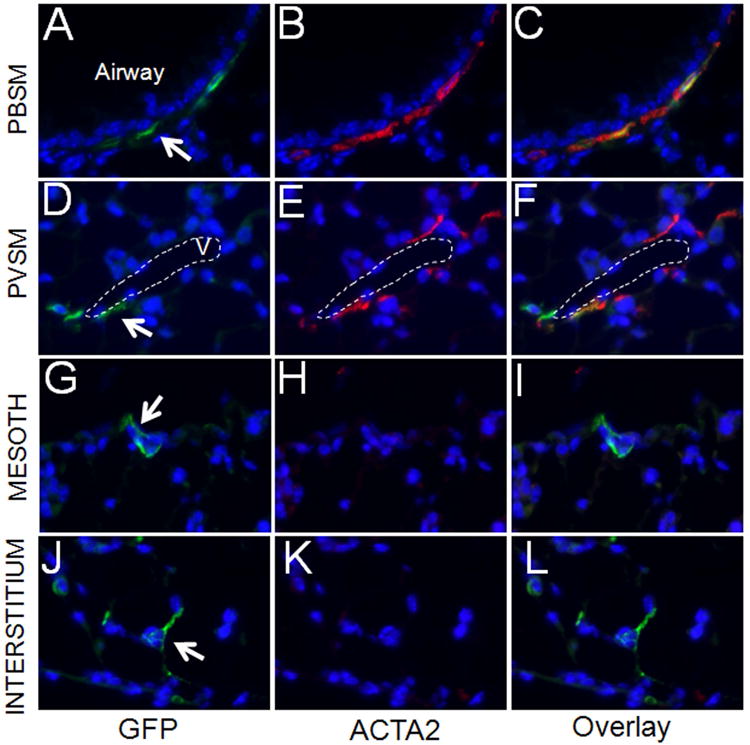
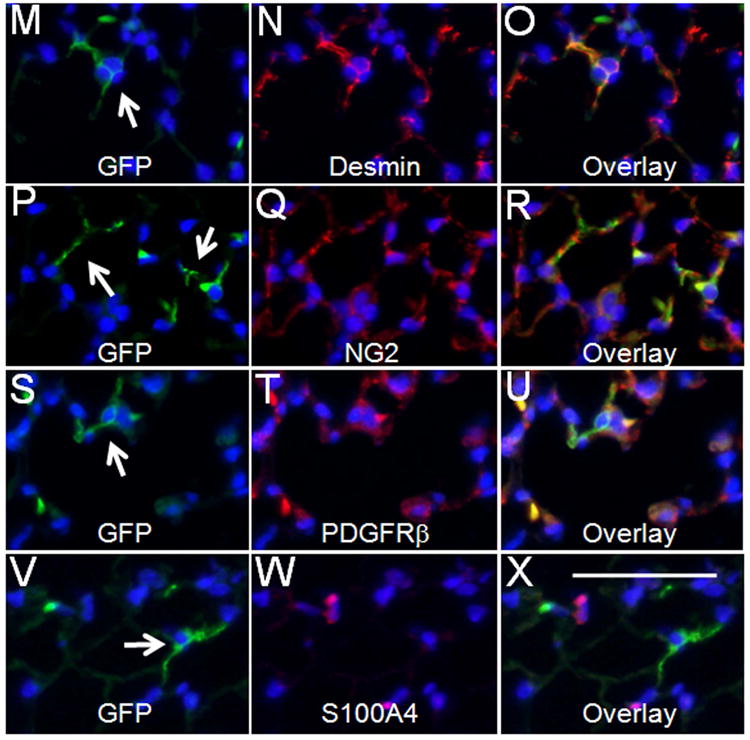
Distribution of GFPpos cells in lungs of 3 month-old mice treated with Tam on PN5 and PN6. (A-L) Co-immunofluorescent staining of GFP (green) and ACTA2 (red) in the areas around epithelial airways (A-C), blood vessels (D-F), mesothelium (G-I), and interstitium (J-L), respectively. Only small numbers of GFPpos cells were present in alveolar interstitium. These cells were analyzed by co-immunofluorescent staining of GFP (green) and Desmin (red) (M-O), GFP (green) and NG2 (red) (P-R), GFP (green) and PDGFRβ (red) (S-U), and GFP (green) and S100A4 (red) (V-X). Arrows indicate GFPpos cells. “v” indicates blood vessel. Scale bar: 50 um.
Transcriptomic Analysis of Myofibroblast Cell Differentiation
Based mostly on histologic criteria, embryonic lung development has been divided into somewhat distinct periods 20. Transition from the pseudoglandular stage that ends the process of branching morphogenesis (E14.5) to the canalicular/saccular stage (E18.5), which precedes alveolization (PN5-PN30) encompasses dramatic anatomical and cellular changes. Surprisingly, the molecular and genetic bases of this transition are poorly understood and represent a major challenge. In particular, multiple signaling pathways are thought to be involved in this transition 20. The utilization of Gli1-creERT2 presented an opportunity to label and therefore isolate a highly specific pool of mesodermal progenitors and examine the dynamics of gene expression in such cells as they underwent pseudoglandular to saccular transition. Accordingly, Hh-targeted mesodermal progenitors were tagged in Gli1-creERT2;ROSAmTmG lungs by 2 doses of Tam at E10.5-E11.5 and their GFPpos progeny were isolated from E14.5 and E18.5 lungs and purified by flow cytometry. The relative abundance of GFPpos cells between E14.5 and E18.5 lungs is around 0.45 (+/-0.25, p=0.02). Gene expression profiles of purified GFPpos cells were obtained on the Agilent gene array platform. The results were analyzed by comparative bio-informatics using the E14.5 versus E18.5 data sets. The microarray data were filtered for expression of various key functional categories of genes including levels of major signaling pathways. These analyses showed clear dynamic and profound changes (4722 differentially expressed genes with >2fold change and FDR<0.05) in genes belonging to all functional categories including transcription factors, signaling molecules and extracellular matrix components (complete bio-informatic analysis of this study will be reported separately). In the signaling category, robust changes occurred in the WNT pathway whereby positive and negative regulators of the canonical WNT pathway were dynamically modulated (please see below and Table 1). Also, both TGFβ and PDGFRA/PDGFRα pathways which are critical for myofibroblast differentiation increased significantly as cells transitioned from the pseudoglandular to the cannalicular/saccular phases of lung development. Thus, transition of hedgehog targeted cells tagged in E10.5-E11.5 from the pseudoglandular to cannalicular/saccular phase of lung development, a period of major changes that has remained little understood is accompanied by robust modulation of many components of various signaling pathways, among which there are extensive functional interactions or crosstalk (supplemental Figure SF1). We propose that it is the total sum of the interactions amongst these signaling engines that ultimately controls the profound anatomical, cellular and molecular changes that occur during this critical period of embryonic lung development.
Table 1.
Differentially expressed genes (E18.5 vs. E14.5 Gli1-creERT2;ROSAmTmG labeled cells) related to signaling pathways of PDGF, Wnt, TGFβ, EGF, IGF, Shh, and FGF. Transcriptomic analyses of Gli1-creERT2;ROSAmTmG labeled cells were conducted with Whole Mouse Genome Oligo Microarrays (Agilent) and IPA software. Differentially expressed genes (identified by IPA with greater than 2 fold changes and an FDR value less than 0.05) are listed. The left column shows genes of each signaling pathway that decrease from E14.5 to E18.5. The right column shows genes of each signaling pathway that increase from E14.5 to E18.5.
| Genes with decreased expression in E18.5 | Genes with increased expression in E18.5 |
|---|---|
| PDGFsignaling (14 genes) | |
|
| |
| SRF(serum response factor) | FOS(FBJ murine osteosarcoma viral oncogene homolog |
| ACP1(acid phosphatase1) | JUN(jun proto-oncogene) |
| MYC(V-myc avian myelocytomatosis viral oncogene homolog) | SPHK1(sphingosine kinase1) |
| PIK3R3(phosphoinositidde-3-kinase) | PIK3R1(phosphoinositide-3-kinase, regulatory subunit3(gamma) |
| PDGFRA(platelet-derived growth factor,alpha polypeptide) | |
| INPP5K(inositol polyphosphate-5-phosphatase K) | |
| STAT3(signal transducer and activator of transcription3) | |
| PRKCA(protein kinase C, alpha) | |
| RRAS(related RAS viral(r-ras) oncogene homolog) | |
| RRAS2(related RAS viral(r-ras) oncogene homolog2) | |
|
| |
| Wnt/beta-catenin signaling (16 genes) | |
|
| |
| CCND1(cyclin D1) | WIF1(WNT inhibitory factor 1) |
| MYC(V-myc avian myelocytomatosis viral oncogene homolog) | DKK3(dikkopf WNT signaling pathway inhibitor 3) |
| RSP02 (Mus musculus R-spondin 2 homolog) | WNT5A(wingless-type MMTV integration site family, member5A) |
| SFRP2(secreted frizzled-related protein2) | WNT5B(wingless-type MMTV integration site family, member 5B) |
| CDH2(cadherin 2) | TLE1(transducin-like enhancer of split1) |
| RUVBL2(RuvB-like AAA ATPase 2) | ILK(integrin-linked kinase) |
| JUN(jun proto-oncogene) | |
| LRP1(low density lipoprotein receptor-related protein 1) | |
| APPL2(adaptor protein, phsphotyrosine interation, PH domain and leucine zipper container 2) | |
| FZD4(frizzled family receptor 4) | |
|
| |
| TGFbeta signaling (12 genes) | |
|
| |
| MAP2K6(mitogen-activated protein kinase kinase 6) | TGFB1(transforming growth factor, beta1) |
| MAPK11(mitogen-attivated protein kinase 11) | BMP4(bone morphogenetic protein 4) |
| TGFBR2(transforming growth factor, beta receptor II) | |
| TGFBR3(transforming growth factor, beta receptor III) | |
| JUN(jun proto-oncogene) | |
| FOS(FBJ murine osteosarcoma viral oncogene homolog | |
| INHA(inhibin, alpha) | |
| RRAS(related RAS viral(r-ras) oncogene homolog) | |
| PMEPA1(prostate transmembrane protein, androgen induced 1) | |
| RRAS2(related RAS viral(r-ras) oncogene homolog2) | |
|
| |
| EGF signaling (9 genes) | |
|
| |
| SRF(serum response factor) | FOS(FBJ murine osteosarcoma viral oncogene homolog |
| MAPK11(mitogen-activated protein kinase 11) | JUN(jun proto-oncogene) |
| PIK3R3(phosphoinositidde-3-kinase) | PIK3R1(phosphoinositide-3-kinase, regulatory subunit3(gamma) |
| PRKCA(protein kinase C, alpha) | |
| STAT3(signal transducer and activator of transcription3) | |
| ITPR2(inositol,1,4,5-trisphosphate receptor, type2) | |
|
| |
| IGF signaling (17 genes) | |
|
| |
| SRF(serum response factor) | CVR61(cysteine-rich, angiogenic inducer,61) |
| PRKAR2B(protein kinase,cAMP-dependent, regulatory, type II, beta) | FOS(FBJ murine osteosarcoma viral oncogene homolog |
| PIK3R3(phosphoinositdde-3-kinase) | IGFBP3(insulin-like growth factor binding protein 3) |
| IGFBP6(insulin-like growth factor binding protein 6) | |
| JUN(jun proto-orcogene) | |
| PIK3R1(phosphoinositide-3-kinase, regulatory subunit3(gamma) | |
| SOCS2(suppressor of cytokine signaling 2) | |
| STAT3(signal transducer and activator of transcription 3) | |
| RRAS(related RAS viral(r-ras) oncogene homolog) | |
| lRS2(insulin receptor substrate 2) | |
| IGF1(insulin-like growth factor 1, somatomedin C) | |
| CTGF(connective tissue growth factor) | |
| RRAS2(related RAS viral(r-ras) oncogene homolog2) | |
| FOX01(forkhead box 01) | |
|
| |
| Sonic Hedgehog signaling (3 genes) | |
|
| |
| PRKAR2B(protein kinase, cAMP-dependent, type II, beta) | HHIP(hedgehog interacting protein) |
| PRKAG2(protein kinase, AMP-activated, gamma 2 non-catalytic subunit | |
|
| |
| FGF signaling (12 genes) | |
|
| |
| MAP2K6(mitogen-activated protein kinase kinase6) | FGF7(fibroblast growth factor 7) |
| MAPK11(mitogen-activated protein kinase 11) | FGFR4(fibroblast growth factor receptor 4) |
| PIK3R3(phosphoinositidde-3-kinase) | PIK3R1(phosphoinositide-3-kinase, regulatory subunit3(gamma) |
| MAP3K5(mitogen-activated protein) | FGFR3(fibroblast growth factor receptor 3) |
| STAT3(signal transducer and activator of transcription3) | |
| FGFl8(fibroblast growth factor 18) | |
| PRKCA(protein kinase C, alpna) | |
| Creb5(cAMP responisive element: binding protein 5) | |
Ectopic Activation of Canonical WNT Signaling in myofibroblast progenitors by Apc Inactivation
A major finding of the transcriptomic analysis revealed alterations in a number of mediators of the WNT pathway with an overall decrease in canonical WNT activity as Hh-targeted mesodermal progenitors underwent transition from the pseudoglandular to the saccular phase of lung development (Table1). For example, the canonical WNT signaling target CCND1 was significantly reduced (8.7 fold decrease, p=1.04E-06). In contrast, inhibitors of the canonical WNT signaling were highly increased. These included WNT5a (increased 7.6 fold, p=2.77E-06), WIF (increased 23.8 fold, p=6.72E-05), DKK3 (increased 6.8 fold, p=1.03E-05), and TLE1 (increased 2.0 fold p=5.8E-04). These changes were validated by Realtime PCR (supplemental Table ST2). To determine the functional significance of decreased WNT signaling specifically in the Hh-targeted progenitors, we used Gli1-creERT2 and induced ectopic activation of WNT signaling by inactivating Apc, which encodes a component of the destructive complex for CTNNB1. Inactivation of Apc by Tam at E10.5 and E11.5 lead to increased canonical WNT activity represented by accumulation of CTNNB1 (Figure 5) and increased LEF1 and AXIN2 expression (Supplemental Figure SF2). As a consequence, there was significant expansion of the mesodermal progenitor pools leading to formation of GFPpos focal masses, scattered within the lung parenchyma (Supplemental Figure SF3). At E14.5, the expanded progenitor pools with accumulated CTNNB1 were PDGFRαneg and expressed no detectable ACTA2, indicating they remain as early progenitors of IMF (Figure 5). Proliferation of these cells is reduced as compared to the control lungs (Supplemental Figure SF2). Intriguingly, as the Gli1-creERT2;Apcflox/flox (ApcGli1) lungs underwent further morphogenesis to E18.5, the expanded progenitor cells differentiated to a myofibroblast phenotype displaying robust expression of PDGFRα, ACTA2 and FN (fibronectin) and eventually appearing as fibroblastic foci in the perinatal lungs (Figure 5).
Figure 5.
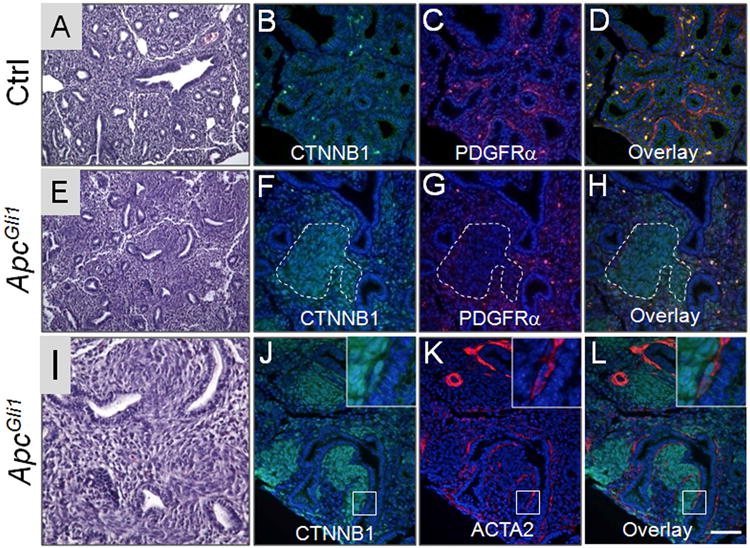
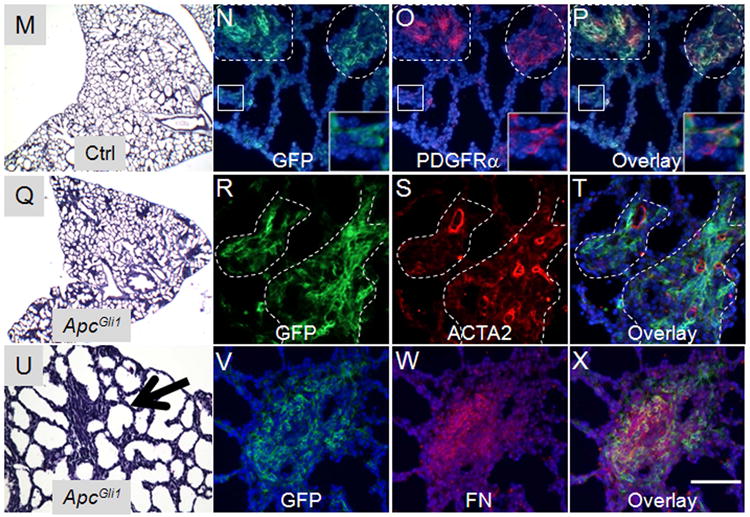
The Gli1-creERT2;Apcflox/flox (ApcGli1) mice were treated with Tam at E10.5 and E11.5 and the lungs were characterized at E14.5 (A-L) and E18.5 (M-X). (A, E, I) H&E staining of E14.5 control (A) and ApcGli1 (E, I) lungs. (B-D & F-H) Co-immunofluorescent staining of CTNNB1 (green) and PDGFRα (red) in E14.5 control (B-D) and ApcGli1 (F-H) lungs. (J-L) Co-immunofluorescent staining of CTNNB1 (green) and ACTA2 (red) in E14.5 ApcGli1 lungs. Inset shows higher magnification of boxed area, which contains PBSM with accumulated CTNNB1. (M, Q) H&E staining of E18.5 control (M) and ApcGli1 (Q) lungs. (U) Higher magnification of ApcGli1 lungs. Arrow indicates the area with formation of myofibroblast colonies. (N-P, R-T, V-X) Co-immunofluorescent staining of GFP and PDGFRα, GFP and ACTA2, GFP and FN in E18.5 ROSAmTmG;ApcGli1 lungs, respectively. Inset in N-P shows higher magnification of boxed area. Dotted lines in N-P and R-T indicate myofibroblast colonies. Scale bar in panel L: 50 um for B-D, F-H, J-L; 100um for A, E; 40 um for I. Scale bar in Panel X: 50 um for N-P, R-T, V-X; 150 um for M, Q; 90 um for U.
Discussion
Cross-communication between the epithelium and its mesenchymal counterpart, a requirement in development of the lung and differentiation of its highly specialized cell types is predicated on key signaling pathways including FGF10, BMP4, TGFβ, Wnt and Hh. Shh is produced by the lung epithelium early in its morphogenesis and imparts specific instructional cues to the developing mesenchyme which in turn directs epithelial morphogenesis. Shh(-/-) lungs are profoundly abnormal in structure and are reported to lack the entire lineage of ACTA2pos cells indicating absolute requirement of Hh signaling in either commitment or differentiation of these cell lineages.
The ACTA2pos cell lineage comprises the fused smooth muscle cells of the PBSM and PVSM, and the scattered single cell interstitial myofibroblasts (IMF), the progenitors of SCMF. While starting in early embryonic development, PBSM and PVSM are for the large part ACTA2pos, IMFs become ACTA2pos only in late embryonic or postnatal lung development and get lost in adult lungs. Owing to lack of specific markers, little is known regarding the lineage of IMFs. This study demonstrates that Gli1-creERT2 represents a useful in vivo genetic tool to label, trace, and target IMF. Even though some PBSM and PVSM cells are also labeled by Gli1-creERT2, these cells are easily distinguishable in the embryonic stage from IMF progenitors based on their anatomic location and gene expression (i.e. ACTA2). Accordingly, we used Gli1-creERT2 mice to first identify and then lineage trace Hh signaling targets during embryonic and postnatal lung development. Identification of Hh targets was based on GFP expression resulting from activation of Gli1 in Gli1-creERT2;ROSAmTmG lungs by two doses of Tamoxifen (TAM), 24 hours apart. This was done either during early embryonic development at E10.5-E11.5 or during postnatal lung development on P5-P6. Given the approximate 30 hour duration of TAM activity, we used the observations on E12.5 as baseline.
The majority of cells labeled by Gli1-creERT2 in E10.5-E11.5 embryonic lungs were localized to the sub-epithelial mesoderm (Figure 1). Shh is expressed by the pseudoglandular distal epithelium and acts on the adjacent sub-epithelial mesenchyme 21. Consistent with the known range of Hh signaling, this implies that only the mesenchyme immediately proximal to the branching epithelium receives signaling. However, we also found GFPpos cells distantly localized in the sub-mesothelium and also within the mesothelium (Figure 1). Hh ligands are secreted proteins whose spatial range of activity can be expanded by moieties such as cholesterol on its carboxyl terminus. Hh signaling is also dependent on proteoglycans and heparan sulfate synthesis by the target cells (on the range of hedgehog signaling). We cannot rule out the possibility of cell migration as the basis for the distantly localized GFPpos cells.
Because SCMFs appear specifically at the alveolar stage, it is obvious that E10.5-E11.5, Gli1-creERT2-labeled IMFs serve as their progenitors. In contrast, whether cells tagged in E10.5-E11.5 lungs serve as progenitors to GFPpos/ACTA2pos cells localized to the PBSM or PVSM of E12.5 lungs remains questionable. ACTA2pos cells are found around the bifurcation of mainstem bronchi as early as E11.5 22. Therefore, we cannot rule out the likely possibility that Gli1-creERT2 targets a select group of differentiated PBSM cells, which remain Hh-responsive. Progenitors of PBSM are proposed to be derived at least partly from FGF10pos cells in the sub-mesothelial mesoderm 4. These cells are thought to migrate along the branching airways in a distal to proximal direction and undergo differentiation to become ACTA2pos in response to SHH 23. The origin of PVSM of the intralobular blood vessels remains to be determined. Wnt2pos;Gli1pos;Isl1pos cardiopulmonary mesoderm progenitors tagged at E8.5, have been shown to give rise to pulmonary PVSM, PBSM as well as cardiomyocytes 24. In contrast to the progressive (or sequential) pattern of PBSM differentiation, SCMF progenitors differentiate to ACTA2pos cells in a synchronous manner (Figure 1). These progenitor cells, which are first identified as interstitially scattered Gli1pos with no detectable expression of ACTA2, display PDGFRα in mid-canalicular development 2, but subsequently become ACTA2pos in near unison in the postnatal lung (Figures 1 and 2). In sum, the evidence strongly supports a model that cells labeled by Gli1-creERT2 in E10.5-E11.5 represent progenitors of IMFs, which subsequently undergo differentiation to SCMFs during alveolization. This conclusion is further supported by the transcriptomic analysis (please see below).
The finding that Hh signaling as early as E10.5 establishes the commitment of SCMFs, a key and the least understood cell type in the lung is an important advancement that will undoubtedly facilitate the study of alveolar formation. Alveogenesis is the last phase of lung development during which functional units of gas exchange are generated. In humans alveogenesis commences in utero, but the process continues postnatally. In mice the postnatal period spanning P5 to P30 marks the process of alveolar formation. During alveogenesis secondary septa subdivide the saccular space, thereby significantly expanding the gas exchange area. Septation requires elastic fibers deposited in the extracellular matrix by SCMFs whose identity and characteristics had hitherto remained poorly defined 20. Topographically, SCMFs are localized to the alveolar septa. A major obstacle in characterization of SCMFs has been lack of technical ability to isolate them for detailed analysis. Lineage tracing of GFPpos cells tagged on E10.5-E11.5, a period of at least 7-10 days prior to the onset of alveogenesis clearly shows that this population harbors the progenitors of SCMFs (Figure 1). Indeed we also found that SCMFs continue to receive Hh signaling during alveogenesis (Figure 3). This result is consistent with the observation using Gli1nlacZ model by Liu et al 18. The source of Hh ligand in the postnatal lung remains unexplored.
Based on collective and currently available data, we propose a model (Figure 6) in which Hh signaling is the key determinant of the multipotential mesenchymal cell commitment to the ACTA2pos lineage, as there is a purported lack of ACTA2pos cell types in Shh(-/-) lungs 5. This decision point, regulated by Shh, is marked by activation of Gli1 and using this information, we have now labeled and traced the progenitors and descendants of this lineage using the Gli1-creERT2 mice 13. Subsequent to Shh, PDGFA is key in regulating the decision that separates the fate of smooth muscle cells (PBSM & VSM) from that of SCMFs. In PdgfA(-/-) lungs, SCMFs are absent and alveogenesis is blocked, while the PBSM and PVSM appear intact 1. In these lungs, elastin is significantly diminished and expression of PDGFAs' sole receptor Pdgfrα on scattered IMFs, normally observed in wild type embryonic lungs is missing. Interestingly, PDGFRαpos cells and elastin fibers within the PBSM and PVSM are intact. This suggests that Pdgfrα activation occurs subsequent to Shh & Gli1 and, in the smooth muscle lineage is independent of PDGFA. In contrast, Pdgfrα expression in progenitors of SCMF is dependent on PDGFA signaling. Thus, PDGFA is required for establishment of SCMF, but not PBSM or PVSM from a common Gli1pos progenitor cell population. Consistent with this model, reduction of PDGFRα does not significantly disrupt PBSM cells in E14.5 Gli1-creERT2;Apcflox/flox lungs (Figure 5).
Figure 6.
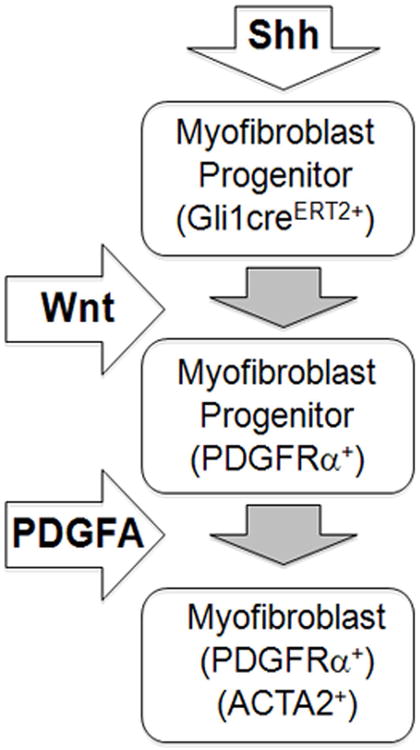
A simplified model illustrating the roles of Shh and Wnt signaling in alveolar myofibroblast differentiation. Shh signaling is activated in early myofibroblast progenitors, which express Gli1-creERT2. During myofibroblast differentiation, Wnt signaling controls the size of the progenitor pool. During saccular stage of lung development, PDGFA signaling is thought to be required for migration and distribution of myofibroblast progenitors to the lung interstitium to facilitate secondary crest formation, a key step in alveogenesis. Differentiated myofibroblasts localize to the tip of secondary crests and areas of the primary septa, and express ACTA2 and PDGFRα.
PDGFRα has been hitherto recognized as the earliest marker of SCMF progenitors 2. Our findings demonstrate that Hh signaling precedes PDGFRα expression. In E18.5 lungs we found only partial overlap between GFPpos and PDGFRαpos cells (Figure 2). However, by P14, nearly all GFPpos cells had become both PDGFRαpos and ACTA2pos (Figures 1 and 2). This observation was validated by the microarray analysis, which showed that Pdgfrα transcripts significantly increased (5.67 fold, P<0.005) in the Gli1-creERT2-labeled cells transitioning from E14.5 to E18.5 phases of lung development.
Of note, in E18.5 Gli1-creERT2;ROSAmTmG lungs, a large portion (60.2+/-9.8%) of PDGFRαpos cells are GFPneg. The origin and lineage of these cells remain unknown. One possibility is that not all SCMF progenitors are exposed to Shh signaling around E10.5 and E11.5. Another possibility is that not all PDGFRαpos cells serve as SCMF progenitors, even though PDGF-A signaling is required for SCMF differentiation. In agreement with this possibility, Chen et al., (2012) reported that during compensatory lung growth, only a subset of PDGFRαpos cells (the dim PDGFRα expressing cells) serve as myogenic precursors25.
Smooth muscle differentiation is also dependent on canonical WNT (WNT/CTNNB1) signaling 26-28. Wnt7b broadly regulates the development of smooth muscle 28. In Wnt7b (-/-) mice PVSM development is disrupted causing perinatal pulmonary hemorrhage 29. Wnt2 acts upstream of Fgf10 and the critical transcription factors myocardin and Mrtf-B to regulate early smooth muscle cell differentiation 30. The role of Wnt2 or Wnt7b in SCMF has not been investigated. In the current study, we found the role of canonical WNT signaling during IMF differentiation to be more complex. In early stages (i.e. E14.5), WNT appears to be critical for regulating the pool of IMF. Ectopic activation of WNT signaling lead to expansion of this progenitor pool, which resulted in perinatal appearance of mesodermal cellular masses. Therefore, reduced WNT signaling observed in the transcriptomic analysis (Table 1) is likely related to controlling the size of the IMF progenitor pool.
Disruption of a number of signaling pathways causes abnormal alveolar development. For example, IGF1 deficiency in mice disrupts alveologenesis and leads to neonatal death 31. Transcriptomic analysis in Igf1(-/-) lungs revealed reduced expression of two major genes; Klf2 and Egr1 suggesting their potential involvement 31. Our microarray analysis showed increased transcripts for multiple mediators of IGF pathway during the transition of Hh-targeted wild type mesodermal cells from E14.5 to E18.5 (Table 1). Klf2 increased by 17.29 (p<0.005) and Egr1 by 18.91 (p<0.005) fold prior to the onset of alveogenesis. Of interest, treatment of Igf1(-/-) lungs with IGF1 increased distal lung maturation and expression of Klf2 and Cyr61. Our array analysis also showed an 11.4 fold increase in Cyr61 (p<0.005). Collectively these findings support the assertion that the GFPpos cells tagged on E10.5-E11.5 represent a cell population with properties consistent with those of SCMFs and their role in alveologenesis
The mechanisms that regulate late-stage myofibroblast differentiation may involve interactions of multiple pathways. For example, transcripts for several mediators of TGFβ, IGF, and PDGF signaling are increased during the E14.5 → E18.5 transition (Table1). The precise “signaling stoicheometery” between Wnt and TGFβ, IGF and PDGF may be critical in achieving normal SCMF differentiation, which in turn is essential for alveogenesis. But ultimately the dynamics, and precisely “regulated” interactions amongst multiple signaling pathways appears necessary for proper differentiation of myofibroblast progenitors. In the present study, alterations in a single signaling pathway, that of WNT alone in Gli1-creERT2;Apcflox/flox lungs caused a phenotype similar but not identical to the fibroblastic masses found in interstitial pulmonary fibrosis or IPF. Not identical, as the clinical IPF is a highly complex pathology that includes not only fibroblastic masses (as in Gli1-creERT2;Apcflox/flox lungs) but also epithelial malfunction 32, 33. Increased canonical WNT signaling has been observed in human and experimental IPF, but it is usually combined with alterations in multiple other signaling pathways, such as TGFβ, PDGF and IGF 34, 35. As the change in WNT signaling via inactivation of Apc by Gli1-creERT2 was induced at a specific time and only in a specific subpopulation of mesodermal cells, these findings raise the intriguing possibility that this population may be particularly sensitive to alterations in Wnt signaling and prone to contributing to mesenchymal masses like those found in IPF. Whether as such this cell population may contribute to and thus represent a new and potential therapeutic target in IPF remains a future challenge.
Supplemental Table ST1. Sequences of realtime PCR primers.
Supplemental Table ST2. Realtime PCR validation of select genes from the microarray analysis (E18.5 vs. E14.5 Gli1-creERT2;ROSAmTmG labeled cells). In array fold changes, positive numbers indicate increases and negative numbers indicate decreases in expression level of each gene from E14.5 to E18.5. Realtime PCR fold changes show relative ratio of each gene transcript between E18.5 and E14.5 GPFpos cells. Increased expression is represented by fold changes larger than 2. Decreased expression is represented by fold change less than 1.
Supplementary Material
Supplemental Figure SF1. A network generated by IPA shows the functional interactions amongst the differentially expressed genes listed in Table 1. Red color indicates increased expression from E14.5 to E18.5. Green color indicates decreased expression from E14.5 to E18.5. Solid lines indicate direct, and dashed lines indicate indirect interactions.
Supplemental Figure SF2. (A) Relative expression levels of genes related to WNT signaling (LEF1 and AXIN2), proliferation (Ki67 and PCNA), and migration (IQGAP1 and CDC42) in E14.5 ApcGli1 lungs (black bars) as compared to the control lungs (grey bars). Data represents average+/- standard error of four independent experiments. (B&C) Co-immunofluorescent staining of CTNNB1 (green) and Ki67 (red) in E14.5 control (B) and ApcGli1 (C) lungs. Scale bar: 50 um.
Supplemental Figure SF3. Immunofluorescent staining of GFP in E14.5 control (Gli1-creERT2;ROSAmTmG) and ApcGli1 lungs. GFPpos colonies expanded in E14.5 ApcGli1 lungs. Dotted lines outline the lumen of epithelial airways. Scale bar: 50 um.
Acknowledgments
We thank Dr. Raju Kucherlapati (Harvard Medical School, MA) for the Apcflox mice. We thank Dr. Alexandra Joyner (Sloan Kettering Institute, NY) for providing and Dr. Arturo Alvarez-Buylla (UCSF, CA) for making available the Gli1-creERT2 mice.
Supported by NIH/NHLBI and generous funds from the Hastings Foundation. [HL107307 to P.M.; HL092967 to S.D.L.; HL112638-01A1 to Z.B.]. ZB is Ralph Edgington Chair in Medicine.
Footnotes
Author contributions: C. Li and P. Minoo: conceived and designed the study, C. Li, M. Li, S. Li, Y. Xing, C. Y. Yang and A. Li: performed the experiments and collected data, C. Li and P. Minoo: interpreted data, C. Li, P. Minoo, Z. Borok and S. De Langhe: helped with interpretation of data and wrote the manuscript
References
- 1.Bostrom H, Willetts K, Pekny M, et al. PDGF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis. Cell. 1996;85:863–873. doi: 10.1016/s0092-8674(00)81270-2. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl P, Karlsson L, Hellstrom M, et al. Alveogenesis failure in PDGF-A-deficient mice is coupled to lack of distal spreading of alveolar smooth muscle cell progenitors during lung development. Development. 1997;124:3943–3953. doi: 10.1242/dev.124.20.3943. [DOI] [PubMed] [Google Scholar]
- 3.Yamada M, Kurihara H, Kinoshita K, et al. Temporal expression of alpha-smooth muscle actin and drebrin in septal interstitial cells during alveolar maturation. J Histochem Cytochem. 2005;53:735–744. doi: 10.1369/jhc.4A6483.2005. [DOI] [PubMed] [Google Scholar]
- 4.White AC, Xu J, Yin Y, et al. FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development. 2006;133:1507–1517. doi: 10.1242/dev.02313. [DOI] [PubMed] [Google Scholar]
- 5.Pepicelli CV, Lewis PM, McMahon AP. Sonic hedgehog regulates branching morphogenesis in the mammalian lung. Curr Biol. 1998;8:1083–1086. doi: 10.1016/s0960-9822(98)70446-4. [DOI] [PubMed] [Google Scholar]
- 6.Bai CB, Stephen D, Joyner AL. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev Cell. 2004;6:103–115. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 7.Methot N, Basler K. An absolute requirement for Cubitus interruptus in Hedgehog signaling. Development. 2001;128:733–742. doi: 10.1242/dev.128.5.733. [DOI] [PubMed] [Google Scholar]
- 8.Marigo V, Johnson RL, Vortkamp A, et al. Sonic hedgehog differentially regulates expression of GLI and GLI3 during limb development. Dev Biol. 1996;180:273–283. doi: 10.1006/dbio.1996.0300. [DOI] [PubMed] [Google Scholar]
- 9.Grindley JC, Bellusci S, Perkins D, et al. Evidence for the involvement of the Gli gene family in embryonic mouse lung development. Dev Biol. 1997;188:337–348. doi: 10.1006/dbio.1997.8644. [DOI] [PubMed] [Google Scholar]
- 10.Hynes M, Stone DM, Dowd M, et al. Control of cell pattern in the neural tube by the zinc finger transcription factor and oncogene Gli-1. Neuron. 1997;19:15–26. doi: 10.1016/s0896-6273(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 11.Lee J, Platt KA, Censullo P, et al. Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development. 1997;124:2537–2552. doi: 10.1242/dev.124.13.2537. [DOI] [PubMed] [Google Scholar]
- 12.Bai CB, Auerbach W, Lee JS, et al. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- 13.Ahn S, Joyner AL. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 2004;118:505–516. doi: 10.1016/j.cell.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 14.Li C, Li A, Li M, et al. Stabilized beta-catenin in lung epithelial cells changes cell fate and leads to tracheal and bronchial polyposis. Dev Biol. 2009;334:97–108. doi: 10.1016/j.ydbio.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C, Hu L, Xiao J, et al. Wnt5a regulates Shh and Fgf10 signaling during lung development. Dev Biol. 2005;287:86–97. doi: 10.1016/j.ydbio.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 16.Ahn S, Joyner AL. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature. 2005;437:894–897. doi: 10.1038/nature03994. [DOI] [PubMed] [Google Scholar]
- 17.McGowan SE, McCoy DM. Platelet-derived growth factor-A regulates lung fibroblast S-phase entry through p27(kip1) and FoxO3a. Respir Res. 2013;14:68. doi: 10.1186/1465-9921-14-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, Kugler MC, Loomis CA, et al. Hedgehog signaling in neonatal and adult lung. Am J Respir Cell Mol Biol. 2013;48:703–710. doi: 10.1165/rcmb.2012-0347OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rock JR, Barkauskas CE, Cronce MJ, et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci U S A. 2011;108:E1475–1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bellusci S, Furuta Y, Rush MG, et al. Involvement of Sonic hedgehog (Shh) in mouse embryonic lung growth and morphogenesis. Development. 1997;124:53–63. doi: 10.1242/dev.124.1.53. [DOI] [PubMed] [Google Scholar]
- 22.Hines EA, Jones MK, Verheyden JM, et al. Establishment of smooth muscle and cartilage juxtaposition in the developing mouse upper airways. Proc Natl Acad Sci U S A. 2013;110:19444–19449. doi: 10.1073/pnas.1313223110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mailleux AA, Kelly R, Veltmaat JM, et al. Fgf10 expression identifies parabronchial smooth muscle cell progenitors and is required for their entry into the smooth muscle cell lineage. Development. 2005;132:2157–2166. doi: 10.1242/dev.01795. [DOI] [PubMed] [Google Scholar]
- 24.Peng T, Tian Y, Boogerd CJ, et al. Coordination of heart and lung co-development by a multipotent cardiopulmonary progenitor. Nature. 2013;500:589–592. doi: 10.1038/nature12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, Acciani T, Le Cras T, et al. Dynamic regulation of platelet-derived growth factor receptor alpha expression in alveolar fibroblasts during realveolarization. Am J Respir Cell Mol Biol. 2012;47:517–527. doi: 10.1165/rcmb.2012-0030OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Langhe SP, Sala FG, Del Moral PM, et al. Dickkopf-1 (DKK1) reveals that fibronectin is a major target of Wnt signaling in branching morphogenesis of the mouse embryonic lung. Dev Biol. 2005;277:316–331. doi: 10.1016/j.ydbio.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 27.De Langhe SP, Carraro G, Tefft D, et al. Formation and differentiation of multiple mesenchymal lineages during lung development is regulated by beta-catenin signaling. PLoS One. 2008;3:e1516. doi: 10.1371/journal.pone.0001516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen ED, Ihida-Stansbury K, Lu MM, et al. Wnt signaling regulates smooth muscle precursor development in the mouse lung via a tenascin C/PDGFR pathway. J Clin Invest. 2009;119:2538–2549. doi: 10.1172/JCI38079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shu W, Jiang YQ, Lu MM, et al. Wnt7b regulates mesenchymal proliferation and vascular development in the lung. Development. 2002;129:4831–4842. doi: 10.1242/dev.129.20.4831. [DOI] [PubMed] [Google Scholar]
- 30.Goss AM, Tian Y, Cheng L, et al. Wnt2 signaling is necessary and sufficient to activate the airway smooth muscle program in the lung by regulating myocardin/Mrtf-B and Fgf10 expression. Dev Biol. 2011;356:541–552. doi: 10.1016/j.ydbio.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pais RS, Moreno-Barriuso N, Hernandez-Porras I, et al. Transcriptome analysis in prenatal IGF1-deficient mice identifies molecular pathways and target genes involved in distal lung differentiation. PLoS One. 2013;8:e83028. doi: 10.1371/journal.pone.0083028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selman M, Pardo A. Idiopathic pulmonary fibrosis: an epithelial/fibroblastic cross-talk disorder. Respir Res. 2002;3:3. doi: 10.1186/rr175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li M, Krishnaveni MS, Li C, et al. Epithelium-specific deletion of TGF-beta receptor type II protects mice from bleomycin-induced pulmonary fibrosis. J Clin Invest. 2011;121:277–287. doi: 10.1172/JCI42090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeda A, Hiyama K, Yamakido H, et al. Increased expression of platelet-derived growth factor A and insulin-like growth factor-I in BAL cells during the development of bleomycin-induced pulmonary fibrosis in mice. Chest. 1996;109:780–786. doi: 10.1378/chest.109.3.780. [DOI] [PubMed] [Google Scholar]
- 35.Trojanowska M. Role of PDGF in fibrotic diseases and systemic sclerosis. Rheumatology (Oxford) 2008;47(Suppl 5):v2–4. doi: 10.1093/rheumatology/ken265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure SF1. A network generated by IPA shows the functional interactions amongst the differentially expressed genes listed in Table 1. Red color indicates increased expression from E14.5 to E18.5. Green color indicates decreased expression from E14.5 to E18.5. Solid lines indicate direct, and dashed lines indicate indirect interactions.
Supplemental Figure SF2. (A) Relative expression levels of genes related to WNT signaling (LEF1 and AXIN2), proliferation (Ki67 and PCNA), and migration (IQGAP1 and CDC42) in E14.5 ApcGli1 lungs (black bars) as compared to the control lungs (grey bars). Data represents average+/- standard error of four independent experiments. (B&C) Co-immunofluorescent staining of CTNNB1 (green) and Ki67 (red) in E14.5 control (B) and ApcGli1 (C) lungs. Scale bar: 50 um.
Supplemental Figure SF3. Immunofluorescent staining of GFP in E14.5 control (Gli1-creERT2;ROSAmTmG) and ApcGli1 lungs. GFPpos colonies expanded in E14.5 ApcGli1 lungs. Dotted lines outline the lumen of epithelial airways. Scale bar: 50 um.


