Abstract
Stored red blood cells undergo numerous biochemical, structural, and functional changes, commonly referred to as storage lesion. How much these changes impede the ability of erythrocytes to perform their function and, as result, impact clinical outcomes in transfusion patients is unknown. In this study we investigate the effect of the storage on the erythrocyte membrane deformability and morphology. Using optical interferometry we imaged red blood cell (RBC) topography with nanometer sensitivity. Our time-lapse imaging quantifies membrane fluctuations at the nanometer scale, which in turn report on cell stiffness. This property directly impacts the cell's ability to transport oxygen in microvasculature. Interestingly, we found that cells which apparently maintain their normal shape (discocyte) throughout the storage period, stiffen progressively with storage time. By contrast, static parameters, such as mean cell hemoglobin content and morphology do not change during the same period. We propose that our method can be used as an effective assay for monitoring erythrocyte functionality during storage time.
Every year approximately 14 million units of blood are collected, and ~13.9 million units of red blood cells (RBCs) are administered to ~4.8 million patients in the U.S. alone1,2. Present regulations in the U.S. and Canada specify 42 days as the maximum shelf life for stored blood3. Several modifications have been reported in stored RBCs including: 1) slowed metabolism, with a decrease in the concentration of adenosine triphosphate (ATP), 2) oxidative damage, with changes to the structure of band 3 and lipid peroxidation, 3) loss of function (usually transient) of cation pumps and consequent loss of intracellular potassium and accumulation of sodium within the cytoplasm, 4) apoptotic changes with racemisation of membrane phospholipids and loss of parts of the membrane through vesiculation4. The overall degradation of stored RBCs is referred to as storage lesion.
Lacking a 3D cytoskeleton, RBCs maintain their shape and mechanical integrity through a spectrin-dominated, triangular 2D network attached to the cytosolic side of their plasma membrane. This semiflexible filament network contributes to the shear modulus of the composite membrane5. The fluid lipid bilayer is thought to be the principal contributor to its bending or curvature modulus. As normal RBCs are 7–8 μm diameter, slightly wider than some microcapillaries, they must deform to traverse the microvascular network. Less deformable RBCs can either obstruct capillaries or, more commonly, traverse the microcirculation at a significantly increased transit time, resulting in overall diminished O2 delivery to organs6. Their diminished ability to negotiate vascular structures in the spleen ultimately leads to their removal from the circulation. RBC shape changes with increased osmotic fragility and loss of deformability over time. Decreased membrane deformability contributes to increasing red cell-endothelial interaction, with activation of inflammatory pathways7. Previous studies showed that stored spherocytes - spherically shaped cells - had decreased deformability. However, the normal-shaped RBCs (discocytes) were found to maintain a deformability similar to that of fresh RBCs8. Similarly, measurements using ektacytometry indicated that RBCs stored over a 42 day period became less compliant to elongation when stored longer9,10. Recent studies revealed deformability changes of stored RBCs when traveling through human-capillary-like microchannels11.
Optical imaging can provide cell morphology information without contact. Digital holography, a non-contact method, has been used to measure changes in the 3D morphology and mean cell hemoglobin (MCH) of RBCs during storage12. However, information about cell dynamics has not been reported. Previously, it was shown that quantitative phase imaging (QPI) can provide information not only about morphology, but also RBC membrane fluctuations, which in turn informs about cell deformability13,14,15. QPI is an emerging tool for studying weakly scattering and absorbing biological specimens16,17. It has been used to yield useful biological information, including cell mass18, membrane fluctuations19, cell tomography20, intracellular transport21, tissue scattering22, blood cell imaging23,25,26,27, cancer diagnosis24. In QPI, the optical pathlength introduced by the specimen is quantified at each point in the field of view. Recently, a number of QPI methods have been developed for biomedical applications using highly coherent light sources such as lasers32,33,34,28,29,30,31. However, the spatial non-uniformity due to speckles, a phenomenon generated by the random interference of laser light, degrade the contrast in QPI images. Use of broadband sources, such as white-light, can increase the spatial sensitivity of QPI18,35. Spatial light interference microscopy (SLIM) is a highly sensitive white-light QPI method for studying biological samples with nanometer scale sensitivity21,36. SLIM combines Zernike's phase contrast method37 and Gabor's holography38 to provide the spatial uniformity associated with white light and the stability associated with common path interferometry. The RBC membrane fluctuates spontaneously, mainly due to thermal agitation, i.e., Brownian motion39. Stiffer RBCs deform less and, thus, their functionality is impaired. Higher stiffness correlates directly with smaller membrane fluctuation; thus QPI measurements can provide direct access to cell function.
In this paper, we present SLIM measurements of both static and dynamic RBC characteristics, including various morphological parameters, as well as the mean square phase displacement for each individual RBC. We found that the phase displacements decrease with storage time which indicates that the cells become stiffer with time. Further, the results show that the mean cell hemoglobin (MCH), or the total dry mass of the RBC, does not change with time. The same behavior was found for the mean average phase, projected area, maximum phase, and circularity.
Results
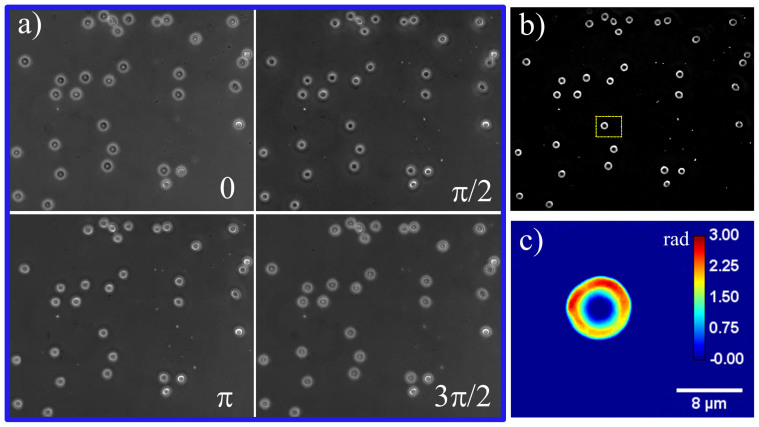
The SLIM instrument is described in more detail elsewhere (see the Methods section and Ref. 36 for detail). We used an advanced version of SLIM which allows for fast phase imaging, up to 12.5 frames/s rate, each frame consisting of 5.5 megapixels. In order to obtain one quantitative phase image, we record 4 intensity images21 (see Fig. 1). Samples from the blood bank (see the Methods section for more on sample preparation) were measured in quadruplicate every week for a period of 42 days. Our study was specifically designed to investigate discocytes, which exhibit no apparent abnormality in morphology. Figure 1a shows the four phase shifted images and Figure 1b shows the resulting quantitative phase image. Normal discocytes are then segmented (N = 110 ± 15 per group) from the field of view and further analyzed. Figure 1c shows such an isolated discocyte, which is selected from Fig. 1b (dotted box).
Figure 1. (a) Phase shifted images, (b) SLIM phase image, (c) selected RBC in (b).
The color bar is in radians.
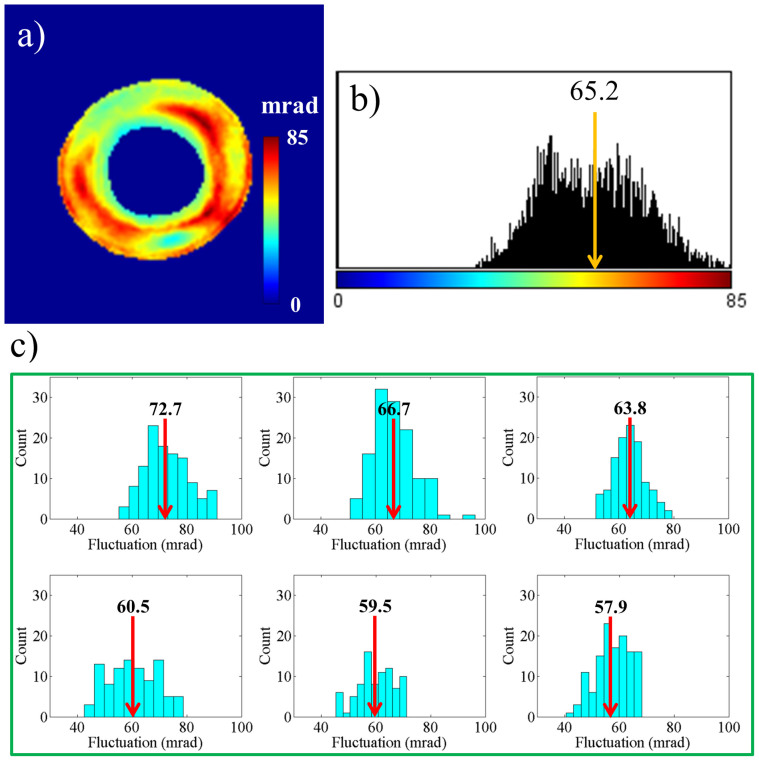
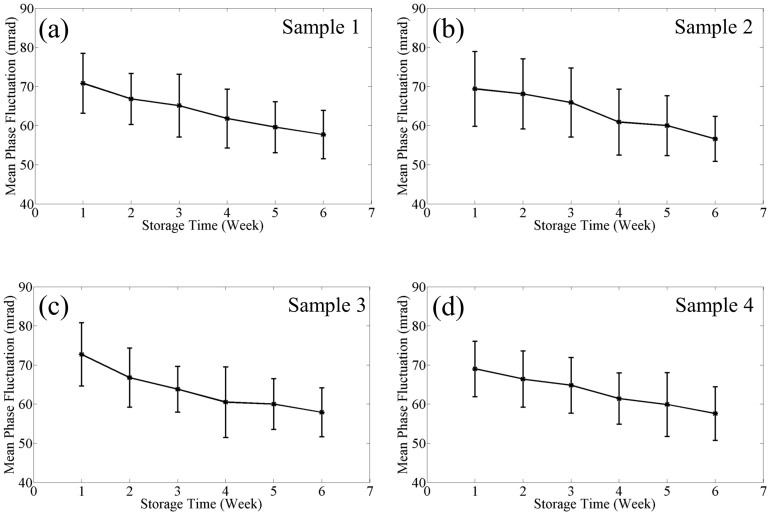
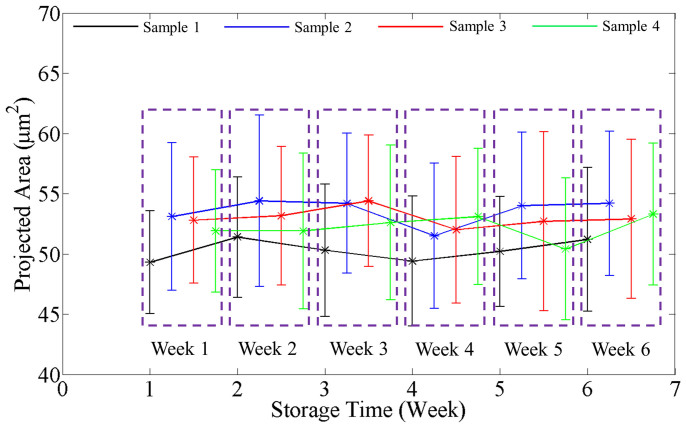
In order to study cell membrane fluctuations, we acquired 128 time-lapse SLIM images at 10 frames per second. We calculated at each pixel the temporal standard deviation of the phase shifts. Figure 2a shows this standard deviation (σT) map from the 128 phase images. Figure 2b shows the histogram of the σT map, calculated from the values across all pixels. The arrow shows the spatial average of σT map, which we use as the representative displacement parameter for the particular RBC. We calculated such average fluctuation for all the cells in each group, during the 42 day period. Figure 2c shows the histograms during the 6 weeks, for one particular blood sample (sample 3). The arrows show the mean phase fluctuation or the mean of the average fluctuations of N = 110 ± 15 cells. Clearly, the position of these arrows consistently shifts toward lower values with time, as shown in Fig. 2c. Figure 3 illustrates this trend for four different blood samples, where the error bars represent twice the standard deviation (SD) of the phase fluctuation calculated over the ensemble of cells (N = 110 ± 15). The decrease of the mean phase fluctuation with storage time indicates that the cell deformability decreases gradually with storage time for all the four samples. It can be seen in Fig. 3(a)–(d) that there exists a variability in the SD of phase fluctuations. This is due to the biological variability present in the samples. Next, using a single quantitative phase image frame from each time sequence, we measured a number of morphological parameters. The projected surface area was obtained by converting the phase image into a binary map40. Figure 4 shows the plots for the mean projected area with storage time for all the samples. The error bars in the plots represent twice the standard deviation of the projected area calculated over the groups of cells (N = 110 ± 15). It can be seen that the mean projected area does not change significantly with storage time. We also calculated the spatially averaged phase shift for each cell. It is known that the dimple region of the RBC can introduce artifacts due to the high curvature15. Thus, in order to avoid such errors, we removed the dimple region from our analysis by applying a threshold to set the dimple to zero (see Fig. 1c). This operation was used previously when imaging red blood cells with quantitative phase imaging15. Figure 5 shows the plots for the mean phase shift calculated over the groups of cells (N = 110 ± 15) as a function of storage time, for all the samples. It can be seen that the mean phase shift does not vary significantly during the storage. The error bars represent twice the standard deviation of the phase shifts.
Figure 2.
RBC fluctuation: (a) Temporal standard deviation (σT) map of a single RBC; colorbar is in mrad, (b) Histogram of the σT map in (a); representative average of the σT map is shown by the arrow, (c) Histogram of the average σT values for 110 ± 15 RBCs at different weeks. The arrows indicate the average of the distribution.
Figure 3. Variation in mean RBC phase fluctuation with storage time for different samples as indicated.
The error bars in the plots are twice the standard deviation of the phase fluctuation calculated over the groups of cells (N = 110 ± 15).
Figure 4. Variation in projected RBC area with storage time.
Data points for different samples are shifted to distinguish them from each other and grouped by the week of measurement. The error bars in the plots are twice the standard deviation of the projected area calculated over the groups of cells (N = 110 ± 15).
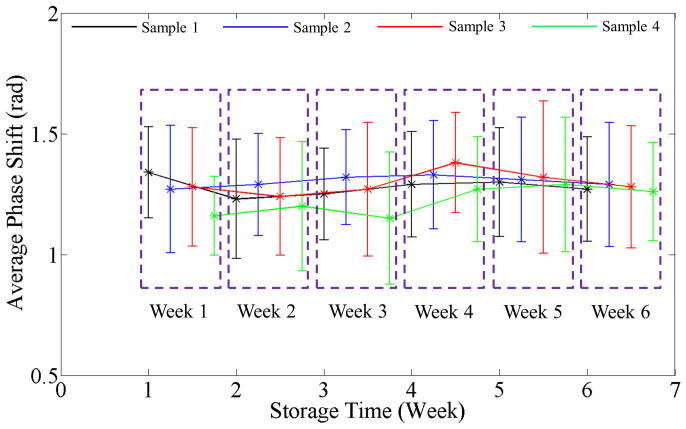
Figure 5. Variation in average RBC phase shift with storage time.
Data points for different samples are shifted to distinguish them from each other and grouped by the week of the measurement. The error bars in the plots are twice the standard deviation of the phase shift calculated over the groups of cells (N = 110 ± 15).
The average phase shift for the whole RBC is proportional to the surface dry mass density (non-aqueous content) of the cell40. The refractive index of a cell exhibits a strong linear dependence with the total cell protein concentration41,42:
 |
where n0 is the refractive index of water, η is the refraction increment (in ml/g) and C is the concentration of dry protein in the solution (in g/ml). Using this relationship, the dry mass surface density (ρ, in pg/μm2) of the cellular matter can be obtained from the measured phase map, ϕ, as follows:
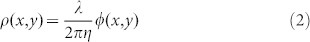 |
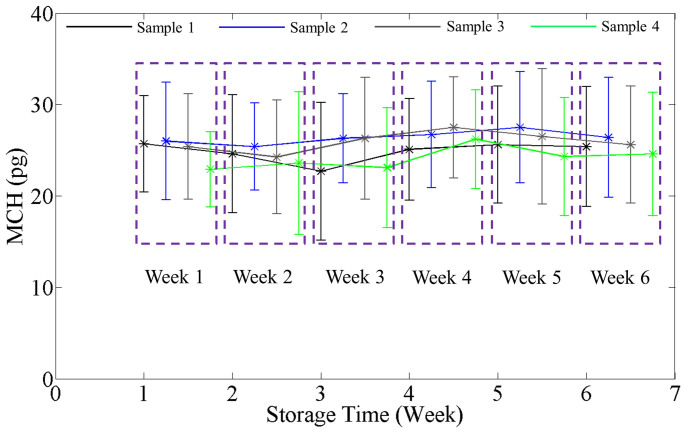
where λ is the center wavelength of the illumination, 550 nm, η = 0.2 ml/g, the wavelength-dependent specific refractive increment of hemoglobin at the center wavelength43. The total dry mass of a RBC can be found by integrating ρ over the entire area of the isolated RBC which is a measure of the total cell hemoglobin. Averaging over the 110 ± 15 cells, we obtained the mean cell hemoglobin (MCH). Figure 6 shows the plots for MCH as a function of storage time for all the samples, where the error bars again represent twice the standard deviation. Note that the MCH does not change with storage time. This result indicates that, while cells can undergo volume changes during storage44, they do not lose hemoglobin into the storage solution. Finally, we calculated the mean cell circularity and maximum phase (Figures S1 and S2), which also remain constant during storage.
Figure 6. Variation in mean cell hemoglobin with storage time.
Data points for different samples are shifted to distinguish them from each other and grouped by the week of the measurement. The error bars in the plots are twice the standard deviation of the MCH calculated over the groups of cells (N = 110 ± 15).
Discussion
Previous studies have shown that RBC morphology affects their stiffness45. Therefore, in order to decouple the morphological and dynamic changes of RBCs, we focused our measurements on discocytes only. These are cells that apparently suffered no modifications during storage. However, quantitative phase imaging has the ability to extract subtle changes in the nanoscale membrane flickering, which are elusive to a common microscope. Our experimental results indicate that the mean phase standard deviation decreases gradually with storage time, indicating that RBC membranes become stiffer with storage time. This is consistent with the previous studies showing the decrease in deformability with storage time2,9,11. However, this is the first time, to our knowledge, when such an assessment was performed via quantifying the membrane fluctuations, i.e., in a noncontact mode. We have shown earlier that stiffer cells show reduction in fluctuation19,45. Further, our results reveal that the MCH does not vary with storage time. In other words, as the total cell dry mass is constant, the MCH does not vary with time. At the same time, the mean phase shift does not vary with time, as this quantity is proportional to the cell dry mass times the projected area. This is consistent with the previous studies, which showed that as the mean cell volume (MCV) increases, the mean cell hemoglobin concentration (MCHC) decreases with storage time, and, as a result MCH, which the product of MCV and MCHC, remains the same44. Note that the absolute quantitative measure of MCH in our work is somewhat underestimated when compared to typical reported values. This is expected, as the analysis scheme discards the data in the dimple region. This underestimation can be calibrated out23. However, for this study, our goal was to measure relative changes during storage time. Our results on projected area measurements indicate that, any cell volume change during storage occurs at constant projected area, meaning that the volume is adjusted via cell thickness changes. Further, decreasing trend in the mean fluctuation is statistically significant as the two-tailed probabilities or the p-values are always less than 0.05 (t-values are between 2 to 5 with around 200 degrees of freedom).
Quantitative phase imaging can be used as non-contact assay for erythrocyte functionality in banked blood. This approach can potentially be applied widely, as the RBC deformability test can be simply performed on an unlabeled fresh smear. This approach will allow us to describe and quantify functional changes in stored cells with apparent normal morphology. Increased stiffness in stored discocytes may be associated with reduction in post-transfusion survival and accelerate removal of transfused cells from the circulation. Availability of this dynamic membrane function assay may allow optimization of storage solution for RBCs with the objective of not only maintaining a normal RBC morphology but also a normal membrane functionality.
Methods
Spatial light interference microscope (SLIM)
The experimental setup is described in more detail in the Supplementary information. The optical layout is essentially a 4f telecentric system (see Fig. S3). SLIM is designed as an add-on module to a commercial phase contrast microscope (Axio Observer Z1, Zeiss). In addition to the conventional π/2 shift introduced between the scattered and unscattered components in phase contrast microscopy, we add further phase shifts in increments of π/2 using a reflective LCPM (Boulder Nonlinear Systems). The lens L1 has a focal length f1 = 150 mm, L2 has f2 = 200 mm, resulting in a further f2/f1 = 4/3 magnification outside the microscope which uses 40× (NA = 0.75, Ph2, Zeiss) objective lens. We used a 5.5 MP scientific-grade complementary metal oxide semiconductor camera (Zyla, Andor), which is capable of imaging at 100 frames/s at full resolution46. Phase shifted images corresponding to π/2 relative phase shifts, are recorded sequentially every 25 ms and combined to produce quantitative phase image at 10 frames per second. Further details on the SLIM system are given in Ref. 21,36.
Blood sample preparation
Packed red cells from donors (collected in anticoagulant and preservation solution) through the apheresis technique were used as source for RBC samples, in accordance with the procedure approved by the Institutional Review Board at University of Illinois at Urbana Champaign (IRB Protocol Number: 10571). The samples (two O+ and two A+) were obtained from saved segments of blood tube which is routinely attached to the blood unit in blood banks. They were maintained at 4°C for the entire measurement period as per the storage guideline. We diluted the RBCs in a Phosphate-Buffered Saline (Life Technologies) solution containing 0.1% Bovine Serum Albumin (Sigma-Aldrich) to achieve concentration of 0.2% RBC by volume. A sample chamber is created by punching a hole in a double sided scotch tape and sticking one side of the tape onto a poly-l-lysine coated cover slip (Neuvitro). The sample is then pipetted into the chamber created by the hole and is sealed on the top using another cover slip. RBCs are allowed to settle for 45 minutes on the poly-l-lysine coated cover slip in order to avoid any cell movement prior to fast RBC imaging.
Imaging and image processing
Fast phase images are continuously recorded at 10 frames per second (corresponding to approximately 400 megabytes per second of throughput, consisting of four 5.5 megapixel 16 bit images for each phase image). Data acquisition and transfer occur in parallel with one thread responsible for triggering and shifting the SLM and a second thread receiving the data. A typical acquisition cycle is divided between SLM stabilization time (~12 ms) and software triggered camera integration time (shutter open, for ~12 ms). A third thread responsible for data serialization (writing tif image files) operates as a producer-consumer queue enabling us to temporarily exceed hard-drive write speeds, with excess images piling into the computer's memory instead of slowing down the acquisition. Software synchronization was accomplished through the use of Windows multimedia timers for repeatable sleep, and synchronization was verified through the camera's on-board timestamp. The acquisition code was written in C++ to meet performance demands, facilitate parallelism, and address synchronization questions. We used the first 128 full-field phase images for phase fluctuation calculations, during which aperture jitter in all frames was under 1 ms. A custom-designed MATLAB™ image processing program was developed for analyzing RBC phase images. First, isolated normal discocytes were selected and cropped from the full field of view in all the 128 images. All the measurements were performed on these cropped RBC images, which occupy 150 × 150 pixels2 in area.
Supplementary Material
Optical Assay of Erythrocyte Function in Banked Blood
Acknowledgments
This research was supported by the National Science Foundation (grants CBET 08-46660 CAREER, CBET-1040462 MRI) and Agilent Laboratories. For more information, visit http://light.ece.uiuc.edu/.
Footnotes
G.P. has financial interest in Phi Optics, Inc., a company developing quantitative phase imaging technology for materials and life science applications, which, however, did not sponsor the research.
Author Contributions B.B. and G.P. conceived the idea, B.B. conducted the experiments, processed the resulting data, B.B., G.P., C.B. and K.T. wrote the manuscript, K.T. provided the samples, and M.K. and B.B. developed the data acquisition software.
References
- Whitaker B. I. & Henry R. A. The 2005 Nationwide Blood Collection and Utilization Survey Report. (National Blood Data Resource Center, Washington, DC, 2005). [Google Scholar]
- Bennett-Guerrero E. et al. Evolution of adverse changes in stored RBCs. P Natl Acad Sci USA 104, 17063–17068 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S. & Gladwin M. T. Bad blood: the risks of red cell storage. Nat Med 16, 381–382 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessandro A., Liumbruno G., Grazzini G. & Zolla L. Red blood cell storage: the story so far. Blood Transfus-Italy 8, 82–88 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier J. B., Lacoste D. & Raphael E. Fluctuation spectrum of fluid membranes coupled to an elastic meshwork: jump of the effective surface tension at the mesh size. Phys Rev Lett 92, 018102 (2004). [DOI] [PubMed] [Google Scholar]
- Secomb T. W., Hsu R. & Pries A. R. Motion of red blood cells in a capillary with an endothelial surface layer: effect of flow velocity. Am J Physiol Heart Circ Physiol 281, H629–636 (2001). [DOI] [PubMed] [Google Scholar]
- Aubron C., Nichol A., Cooper D. J. & Bellomo R. Age of red blood cells and transfusion in critically ill patients. Ann Intensive Care 3, 2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Celle P. L. Alteration of deformability of the erythrocyte membrane in stored blood. Transfusion 9, 238–245 (1969). [DOI] [PubMed] [Google Scholar]
- Frank S. M. et al. Decreased erythrocyte deformability after transfusion and the effects of erythrocyte storage duration. Anesth Analg 116, 975–981 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkelman S. et al. Is red blood cell rheology preserved during routine blood bank storage? Transfusion 50, 941–948 (2010). [DOI] [PubMed] [Google Scholar]
- Zheng Y. et al. Characterization of red blood cell deformability change during blood storage. Lab Chip 14, 577–583 (2014). [DOI] [PubMed] [Google Scholar]
- Moon I. et al. Automated quantitative analysis of 3D morphology and mean corpuscular hemoglobin in human red blood cells stored in different periods. Opt Express 21, 30947–30957 (2013). [DOI] [PubMed] [Google Scholar]
- Park Y. K. et al. Refractive index maps and membrane dynamics of human red blood cells parasitized by Plasmodium falciparum. Proc Natl Acad Sci U S A 105, 13730 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. K. et al. Metabolic remodeling of the human red blood cell membran. Proc. Nat. Acad. Sci. 107, 1289 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. K. et al. Measurement of red blood cell mechanics during morphological changes. Proc. Nat. Acad. Sci. 107, 6731 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu G. Quantitative Phase Imaging of Cells and Tissues. 1st edn, 385 (McGraw-Hill, 2011). [Google Scholar]
- Mir M., Bhaduri B., Wang R., Zhu R. Y. & Popescu G. Quantitative Phase Imaging. Progress in Optics, Vol 57 57, 133–217 (2012). [Google Scholar]
- Bhaduri B., Pham H., Mir M. & Popescu G. Diffraction phase microscopy with white light. Opt. Lett. 37, 1094–1096 (2012). [DOI] [PubMed] [Google Scholar]
- Bhaduri B., Tangella K. & Popescu G. Fourier phase microscopy with white light. Biomed Opt Express 4, 1434–1441 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. et al. White-light diffraction tomography of unlabelled live cells. Nat Photon 8, 256–263, 10.1038/nphoton.2013.350 (2014). [DOI] [Google Scholar]
- Bhaduri B. et al. Cardiomyocyte Imaging Using Real-Time Spatial Light Interference Microscopy (SLIM). PLoS One 8, e56930 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H. et al. Measuring the scattering parameters of tissues from quantitative phase imaging of thin slices. Opt Lett 36, 2281–2283 (2011). [DOI] [PubMed] [Google Scholar]
- Pham H., Bhaduri B., Tangela K., Best-Popescu C. & Popescu G. Real time blood testing using quantitative phase imaging. PLoS One 8, e55676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Tangella K., Balla A. & Popescu G. Tissue refractive index as marker of disease. J Biomed Opt 16, 116017 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anna T., Srivastava V., Mehta D. S. & Shakher C. High-resolution full-field optical coherence microscopy using a Mirau interferometer for the quantitative imaging of biological cells. Appl Optics 50, 6343–6351 (2011). [DOI] [PubMed] [Google Scholar]
- Mehta D. S. & Srivastava V. Quantitative phase imaging of human red blood cells using phase-shifting white light interference microscopy with colour fringe analysis. Appl Phys Lett 101 (2012). [Google Scholar]
- Jang Y., Jang J. & Park Y. Dynamic spectroscopic phase microscopy for quantifying hemoglobin concentration and dynamic membrane fluctuation in red blood cells. Opt Express 20, 9673–9681 (2012). [DOI] [PubMed] [Google Scholar]
- Shaked N. T., Zhu Y. Z., Badie N., Bursac N. & Wax A. Reflective interferometric chamber for quantitative phase imaging of biological sample dynamics. J Biomed Opt 15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquet P. et al. Digital holographic microscopy: a noninvasive contrast imaging technique allowing quantitative visualization of living cells with subwavelength axial accuracy. Opt Lett 30, 468–470 (2005). [DOI] [PubMed] [Google Scholar]
- Popescu G., Ikeda T., Dasari R. R. & Feld M. S. Diffraction phase microscopy for quantifying cell structure and dynamics. Opt Lett 31, 775–777 (2006). [DOI] [PubMed] [Google Scholar]
- Ikeda T., Popescu G., Dasari R. R. & Feld M. S. Hilbert phase microscopy for investigating fast dynamics in transparent systems. Opt Lett 30, 1165–1167 (2005). [DOI] [PubMed] [Google Scholar]
- Popescu G. et al. Fourier phase microscopy for investigation of biological structures and dynamics. Opt Lett 29, 2503–2505 (2004). [DOI] [PubMed] [Google Scholar]
- Barty A., Nugent K. A., Paganin D. & Roberts A. Quantitative optical phase microscopy. Opt Lett 23, 817–819 (1998). [DOI] [PubMed] [Google Scholar]
- Mann C. J., Yu L. F., Lo C. M. & Kim M. K. High-resolution quantitative phase-contrast microscopy by digital holography. Opt Express 13, 8693–8698 (2005). [DOI] [PubMed] [Google Scholar]
- Wang Z. & Popescu G. Quantitative phase imaging with broadband fields. Appl Phys Lett 96 (2010). [Google Scholar]
- Wang Z. et al. Spatial light interference microscopy (SLIM). Opt Express 19, 1016–1026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernike F. Phase contrast, a new method for the microscopic observation of transparent objects. Physica 9, 686–698 (1942). [Google Scholar]
- Gabor D. A new microscopic principle. Nature 161, 777–778 (1948). [DOI] [PubMed] [Google Scholar]
- Park Y. et al. Measurement of red blood cell mechanics during morphological changes. P Natl Acad Sci USA 107, 6731–6736, 10.1073/pnas.0909533107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M., Wang Z., Tangella K. & Popescu G. Diffraction Phase Cytometry: blood on a CD-ROM. Opt Express 17, 2579–2585 (2009). [DOI] [PubMed] [Google Scholar]
- Barer R. Interference microscopy and mass determination. Nature 169, 366 (1952). [DOI] [PubMed] [Google Scholar]
- Davies H. & Wilkins M. Interference microscopy and mass determination. Nature 169, 541 (1952). [DOI] [PubMed] [Google Scholar]
- Friebel M. & Meinke M. Model function to calculate the refractive index of native hemoglobin in the wavelength range of 250–1100 nm dependent on concentration. Appl Optics 45, 2838–2842 (2006). [DOI] [PubMed] [Google Scholar]
- Brugnara C. & Churchill W. H. Effect of irradiation on red cell cation content and transport. Transfusion 32, 246–252 (1992). [DOI] [PubMed] [Google Scholar]
- Popescu G. et al. Optical measurement of cell membrane tension. Phys Rev Lett 97 (2006). [DOI] [PubMed] [Google Scholar]
- Fowler B., C. L., Mims S., Balicki J., Li W., Do H., Appelbaum J. & Vu P. A 5.5 Mpixel 100 Frames/sec Wide Dynamic Range Low Noise CMOS Image Sensor for Scientific Applications. Proc. SPIE 7536, 12 (2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Optical Assay of Erythrocyte Function in Banked Blood