Abstract
Green fluorescent protein (GFP) has been used as a proof of concept for a novel “leave-one-out” biosensor design in which a protein that has a segment omitted from the middle of the sequence by circular permutation and truncation binds the missing peptide and reconstitutes its function. Three variants of GFP have been synthesized that are each missing one of the 11 β-strands from its β-barrel structure, and in two of the variants, adding the omitted peptide sequence in trans reconstitutes fluorescence. Detailed biochemical analysis indicates that GFP with β-strand 7 “left out” (t7SPm) exists in a partially unfolded state. The apo form t7SPm binds the free β-strand 7 peptide with a dissociation constant of ~0.5 µM and folds into the native state of GFP, resulting in fluorescence recovery. Folding of t7SPm, both with and without the peptide ligand, is at least a three-state process and has a rate comparable to that of the full-length and unpermuted GFP. The conserved kinetic properties strongly suggest that the rate-limiting steps in the folding pathway have not been altered by circular permutation and truncation in t7SPm. This study shows that structural and functional reconstitution of GFP can occur with a segment omitted from the middle of the chain, and that the unbound form is in a partially unfolded state.
Removal of a segment from a protein chain is often disastrous to its folding and stability, but in many cases, it is possible to reconstitute the structure and function by adding back the complementary sequence as an autonomous peptide (1–9). Analogous to “leave-one-out” experiments in statistics, where data omitted from a training set is nonetheless accurately predicted by modeling the remaining data, the truncated sequence sometimes retains enough information about its native structure to form a specific binding pocket that is complementary to, and binds tightly to, the missing piece. If the reconstituted protein has a self-reporting signal such as fluorescence, then the leave-one-out protein is a sensor for its missing piece. In this study, we use green fluorescent protein (GFP)1 to prove this concept.
GFP, a small globular protein consisting of 238 amino acids, was first found in the bioluminescent system of the jellyfish Aequorea victoria in 1962 (10). Its well-known stability (11, 12) originated from the unique barrel-like structure (Figure 1) that is composed of a series of 11 β-strands connected by loops (13, 14), and its tightly encapsulated chromophore that is derived from the post-translational and intramolecular cyclization and oxidation of the tripeptide motif Ser65-Tyr66-Gly67 (15) makes GFP one of the most important proteins to biological and biomedical research. The fluorescence spectrum of GFP has been reengineered, and its many color variants (16–20) have been used widely as markers for gene expression (21) and protein dynamics and localization (22, 23). Recent studies have confirmed the utility of GFP variants as reporters for intracellular pH (24, 25), halide (18) and calcium ion concentration (26–28), and protein-protein interactions (4–8). GFP also has been used as a biosensor for sensing ligands such as Gram-negative bacteria (29, 30) and other metal ions (31).
Figure 1.
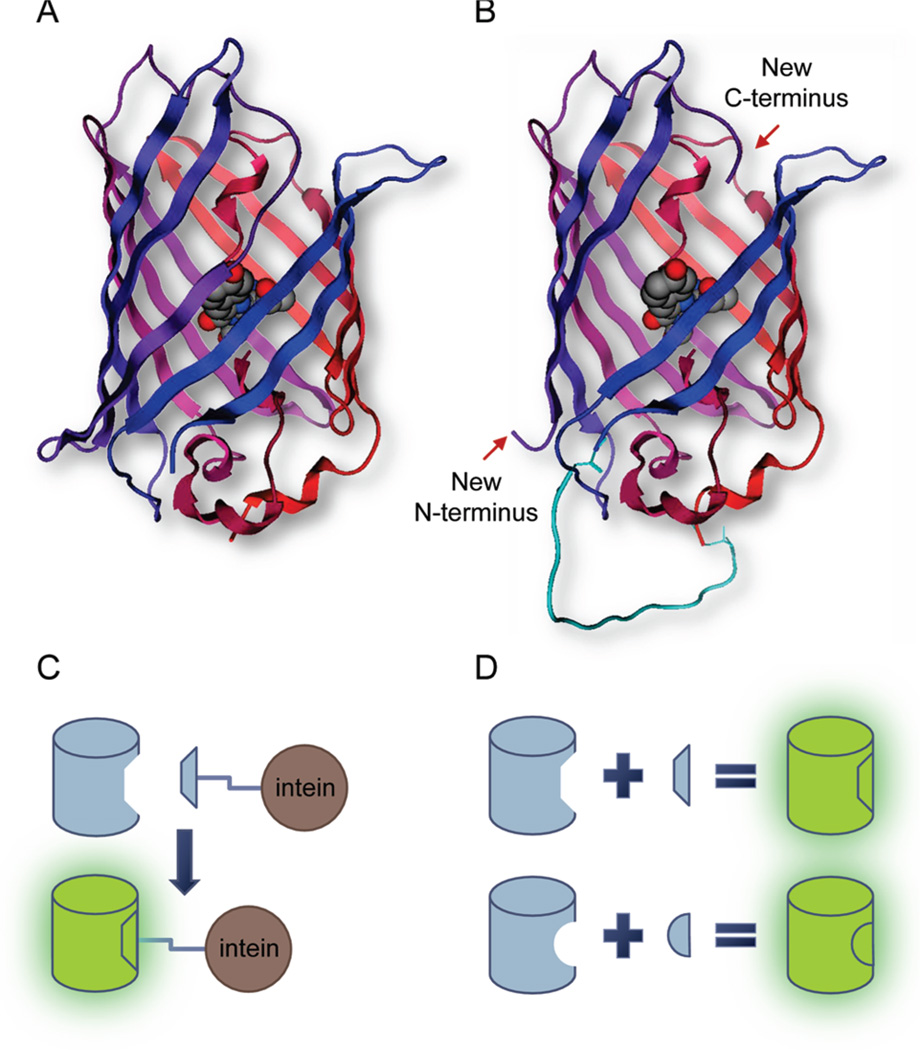
Concept of leave-one-out peptide biosensors. (A) Three-dimensional β-barrel structure of GFP. The chromophore shown in space-fill mode is located in the geometric center of the cylinder composed of 11 β-strands. Notice that the Tyr66 side chain moiety of the chromophore points to and interacts with β-strand 7 through hydrogen bounding. Coordinates were obtained from PDB entry 1EMA(13). The image was made using MOE (Chemical Computing Group). (B) Schematic diagram of the circularly permuted and truncated leave-one-out GFP (t7SP). The short linker peptide (cyan) is modeled to connect the original N- and C-termini, while β-strand 7 is removed. Coordinates were obtained from PDB entry 1EMA(13), and the computational model was created with MOE. (C) Diagram for the complementation between the leave-one-out GFP and peptide fusions. Complementation between the leave-one-out GFP and peptide fusions in vivo was examined through the reconstitution of the fluorescence in this study. (D) Diagram for the leave-one-out GFP peptide biosensors. The fluorescence can be reconstituted from a permuted, truncated GFP construct by adding back a left-out peptide strand.
The slow folding kinetics (32) of GFP and its tendency to misfold and aggregate limit its use in many applications. Because of this, extensive efforts have been made to engineer GFP variants with improved folding properties. Mutants such as Ser147Pro (33), Val163Ala/Ser175Gly (34), and Phe99Ser/ Met153Thr/Val163Ala (35) (known as the cycle 3 mutant) improve solubility and refolding efficiency. Using the in vitro evolution technique, a “superfolder” GFP which has six additional mutations to the cycle3 mutant has been identified and exhibits not only a strong folding efficiency but also a faster folding and a higher denaturant resistance (36). The improvement in folding is mainly due to the reduction of total hydrophobicity of GFP, an increase in β-strand propensity, and the increase in the number of favorable hydrogen bonding and electrostatic interactions. Superfolder GFP has been further evolved in vitro by DNA shuffling (37) to optimize its solubility and to develop a novel protein tagging technique based on protein fragment complementation (38). The resulting sequence, called superfolder GFP OPT (OPT), was the starting point for this work (Figure 1A of the Supporting Information).
Protein complementation systems have been developed as detectors of protein–protein interactions, using reporter proteins such as β-galactosidase (1), dihydrofolate reductase (2), CyaA adenylate cyclase (3), and GFP (4–8). In these cases, a pair of interacting proteins, fused to the split segments, were required to reassociate the active reporter protein. For example, each half of a bisected GFP was fused with one-half of an antiparallel leucine zipper domain (4). The additional binding energy of the leucine zipper reconstituted the fluorescent state from the fragments. The energetic contribution of the interacting pair surmounts the entropic barrier to reconstitution of the unfolded components.
If component parts could be made that exist in a near-native state before association, their binding affinity would be substantially higher. Although this would defeat the purpose of reconstitution as a reporter of protein-protein association, it would open up the possibility of using reconstitution as a means of designing new receptors or biosensors. For example, truncated OPT (GFP1 −10, residues 1–214), which is missing β-strand 11 (S11, residues 215–230), was used to detect proteins fused with S11. Detection of the proteins tagged with S11 relies solely on the binding of S11 to GFP1 –10 and reconstitution of fluorescence. However, the recovered fluorescence needed several hours to reach a plateau upon complementation (38), probably corresponding to a slow chromophore maturation. The slowness of the chromophore maturation may hamper the usefulness of the GFP complementation system as a rapid sensor. A solution to this problem was found as part of our studies, using preincubation with the peptide ligand. The differences between the preincubated and nonpreincubated biosensor constructs are discussed.
Many proteins will tolerate circular permutation of the sequence by linking the N- and C-termini and cutting the chain at any other position to make new termini (39, 40). The evidence that GFP can fold and fluoresce despite circular rearrangement (27, 36), combined with the evidence of functional complementation using a short terminal segment (38), suggests that GFP may work as a biosensor for internal segments.
By first circularly permuting the sequence and then truncating it, we explore the tolerance of GFP to the removal of internal segments of the chain. There are three possible outcomes of each experiment: (a) the permuted and truncated protein does not fold; (b) it folds but does not bind the “missing piece” peptide, or (c) it folds and binds the missing piece. By exploring which parts of a protein may be successfully omitted and added in trans, we may learn something about the folding pathway of GFP (44). Parts of the protein that fold late in the folding pathway are likely to be the least detrimental to folding when removed, leaving an only partially unfolded state, minimizing the entropic barrier to reconstitution. In this paper, we explore the removal of three segments of GFP and the properties of one of the resulting leave-one-out variants as a sensor of its missing piece.
MATERIALS AND METHODS
Leave-One-Out Biosensor Design
Exposure of the GFP chromophore to the solvent can quench its fluorescence. After inspection of the GFP crystal structures [PDB entries 1EMA (13), 1GFL (14), and 2B3P (36)], β-strands 7, 10, and 11 were selected as the potential peptide binding sites for the leave-one-out biosensor design (Figure 1B of the Supporting Information). Removal of one of these strands exposes the chromophore to solvent and quenches fluorescence, as we show in this study.
H–D exchange NMR studies (45) and molecular dynamics simulations (46) have led to speculation that the closure of the cleft between β-strands 7 and 8 is one of the last events in the β-barrel folding pathway. If so, then leaving out β-strand 7, by circular permutation and truncation, should minimally interfere with folding, and refolding upon reconstitution with β-strand 7 should be efficient. Two other β-strands, β-strand 10 and β-strand 11, were also selected as leave-one-out targets that would be potentially the least detrimental to folding (45). Removal of β-strand 11 and reconstitution of fluorescence upon addition of the peptide in trans were demonstrated in a previous study (38) and were repeated as a control in this study.
Molecular modeling was carried out using MOE (Chemical Computing Group) with GFP structures [PDB entries 1EMA (13), 1GFL (14), and 2B3P (36)]. The original N- and C-termini were connected by the linker peptide with the sequence GGTGGS, as previously described (27, 47), to make a truncated and circularly permuted version of GFP.
Sequence Construction
Oligonucleotide sequences that covered the complete OPT gene were designed automatically using DNA Works (48). In this design, the universal annealing temperature for the oligonucleotides was set to 62 ± 1 °C, and the oligonucleotide length was set to 50 nucleotides. The resulting oligonucleotide sequences were then synthesized and linked using assembly PCR as described previously (49) to produce the full-length OPT gene with half of the GGTGGS linker encoded on either terminus. Oligonucleotide sequences that cover the full-length OPT gene are given in Table 1 of the Supporting Information. All oligonucleotides in this study were synthesized by Integrated DNA Technologies.
Table 1.
Characteristics of GFP Variants
| folding kineticsa |
||||||||
|---|---|---|---|---|---|---|---|---|
| variant | k1 (s−1) | k2 (s−1) | k3 (s−1) | A1 (%) | A2 (%) | A3 (%) | quantum yieldb (%) | pKac |
| t7SP | 0.19997 ± 0.05186 | 0.01403 ± 0.00122 | 0.00137 ± 0.00040 | 35.17 ± 0.95 | 43.87 ± 0.71 | 20.96 ± 1.32 | 17.99 ± 1.40 | 8.08 ± 0.04 |
| t7SPm | 0.15993 ± 0.00133 | 0.01327 ± 0.00050 | 0.00122 ± 0.00005 | 35.37 ± 1.28 | 43.70 ± 1.00 | 20.92 ± 0.76 | 39.50 ± 0.27 | 8.22 ± 0.01 |
| OPT | 0.34307 ± 0.03827 | 0.02357 ± 0.00080 | 0.00289 ± 0.00001 | 56.96 ± 0.23 | 34.92 ± 0.46 | 8.12 ± 0.37 | 100 ± 0.17 | 5.00 ± 0.01 |
| c7SP | 0.36067 ± 0.06859 | 0.04897 ± 0.00225 | 0.01213 ± 0.00012 | 44.18 ± 1.95 | 17.87 ± 1.90 | 37.95 ± 0.57 | 78.63 ± 2.37 | 7.87 ± 0.03 |
Refolding of each acid-denatured protein was performed three times independently, and the time-dependent fluorescence recovery (F) was fitted to the triple-exponential equation F =A1(1 – e−tk1) + A2(1 – e−tk2) + A3(1 – e−tk3), with six fitting parameters. Amplitudes of exponential terms (A1, A2, and A3) are in units of relative percentage (±SD), and rate constants (k1, k2, and k3) are given (±SD).
The fluorescence intensity at 508 nm with excitation at 485 nm was measured three times independently and normalized by the OD485 to present the quantum yield of each protein. The relative quantum yield compared to OPT is provided as a percentage (±SD).
The fluorescence intensity at 508 nm with excitation at 485 nm and in different pH buffers was measured three times independently, and the transition curve was fitted to the Henderson–Hasselbalch equation to obtain the pKa (±SD).
After assembly PCR, the full-length linear gene was purified and self-ligated by T4 DNA ligase (New England Biolabs) to form the circular template for circular permutation and truncation, and candidate permuted and truncated sequences were then synthesized using PCR. Briefly, permuted and/or truncated sequences were amplified from the circular DNA templates by selected primers (Table 2 of the Supporting Information) to omit the desired segment. The 5′ primers also encoded six-His tags for the protein purification purpose. Amplified candidate genes were cloned into pET-28a (Novagen) via NcoI and EcoRI restriction sites and verified by dideoxynucleotide sequencing. The constructed proteins were named t7SP, t10SP, and t11SP to denote the truncation at β-strand 7 (residues 146–157), 10 (residues 198–216), and 11 (residues 214–230) in OPT, respectively. Circularized OPT, called c7SP, with new termini between F145 and N146 of β-strand 7, without leaving out the β-strand, was generated simultaneously using a different set of primers (Table 2 of the Supporting Information), as a covalent mimic of the t7SP-β-strand 7 binding complex. In addition, full-length and unpermuted OPT genes were also made, to serve as a positive control. The arrangements of β-strands in primary sequences of GFP variants are shown in Figure 2 of the Supporting Information.
Table 2.
Concentration-Dependent Kinetics of in Vitro Complementation of t7SPm
| rate constantb (s−1) |
amplitudeb (%) |
|||||
|---|---|---|---|---|---|---|
| [s7 peptide]a (µM) | k1 | k2 | k3 | A1 | A2 | A3 |
| 14.40 | 0.12805 ± 0.02581 | 0.03045 ± 0.00460 | 0.00190 ± 0.00053 | 41.64 ± 10.33 | 66.66 ± 8.85 | −8.29 ± 1.48 |
| 7.20 | 0.08270 ± 0.00057 | 0.01830 ± 0.00113 | 0.00149 ± 0.00048 | 45.72 ± 3.05 | 66.19 ± 3.51 | −11.92 ± 0.46 |
| 3.60 | 0.06235 ± 0.00276 | 0.01025 ± 0.00007 | 0.00219 ± 0.00033 | 43.77 ± 0.96 | 71.83 ± 1.60 | −15.60 ± 0.64 |
| 1.80 | 0.03810 ± 0.00170 | 0.00630 ± 0.00096 | 0.00158 ± 0.00177 | 51.05 ± 3.15 | 82.56 ± 2.96 | −33.61 ± 0.18 |
| 0.90 | 0.02475 ± 0.00346 | 0.00621 ± 0.00079 | NDc | 39.54 ± 6.07 | 60.46 ± 6.07 | NDc |
| 0.45 | 0.01680 ± 0.00042 | 0.00619 ± 0.00007 | NDc | 29.95 ± 0.10 | 70.05 ± 0.10 | NDc |
| 0.36 | NDc | 0.00770 ± 0.00009 | NDc | NDc | 100.00 ± 0.00 | NDc |
| 0.24 | NDc | 0.00584 ± 0.00005 | NDc | NDc | 100.00 ± 0.00 | NDc |
| 0.18 | NDc | 0.00496 ± 0.00017 | NDc | NDc | 100.00 ± 0.00 | NDc |
| 0.09 | NDc | 0.00452 ± 0.00057 | NDc | NDc | 100.00 ± 0.00 | NDc |
In vitro complementation assays with 0.1 µM t7SPm under native conditions and various concentrations of s7 were performed independently. Time course studies of fluorescence intensity changes were recorded every 5 s for 20 min.
The kinetics of in vitro complementation were determined by fitting time-dependent changes in fluorescence F to equations with various exponential terms {F = ∑1→i[Ai(1 – e−tki)], where i = 1, 2, or 3} and then reporting the optimal fitting. Rate constants (ki) and amplitudes (Ai) are provided (±SD; n = 2), the amplitudes in units of relative percentage.
Not detected.
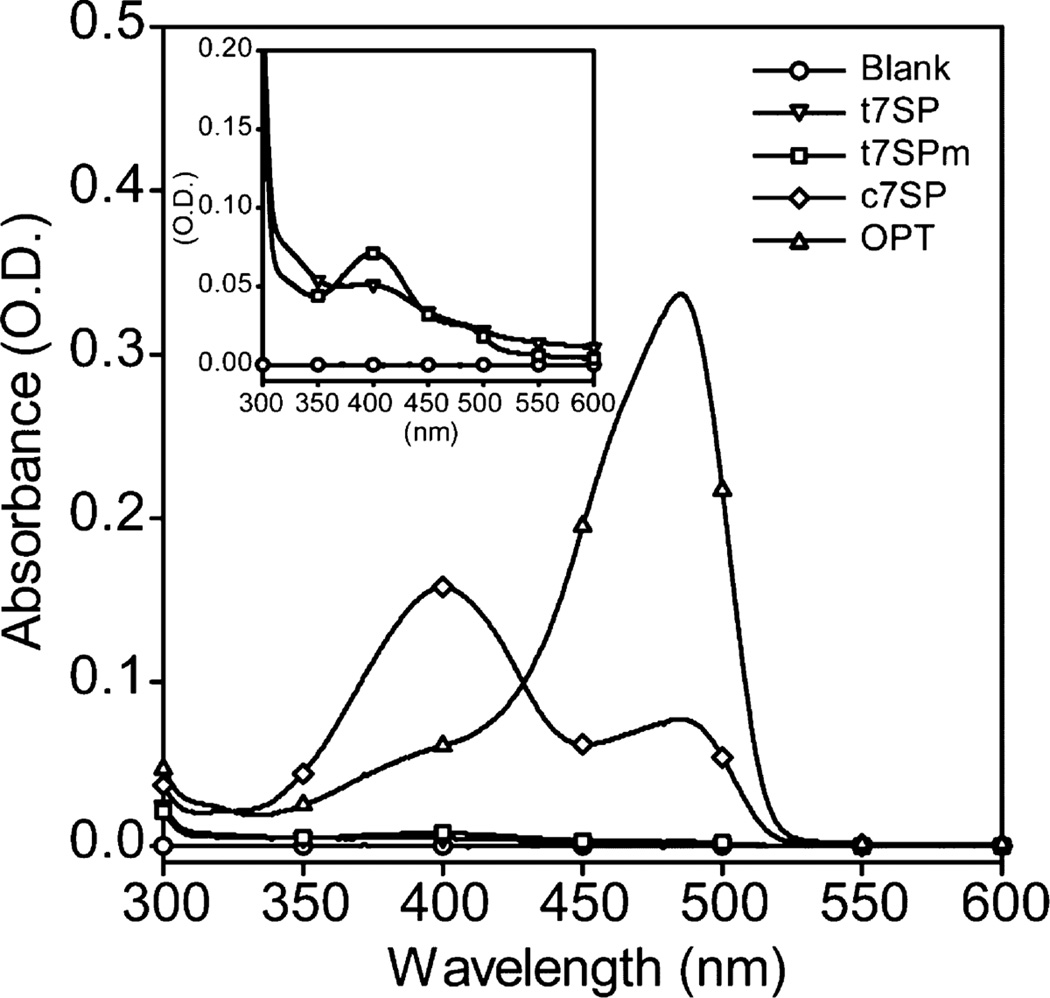
Figure 2.
UV–visible spectra of native GFP variants. Purified t7SP, t7SPm, OPT, and c7SP were diluted to 10 µM in TN buffer (pH 7.5), and absorbance spectra between 300 and 600 nm were recorded. The UV–visible spectra of t7SP and t7SPm were also measured with a protein concentration of 80 µM to increase the absorbance signal (inset).
Construction of Target Peptide–Intein Fusion Proteins
The recovery of the fluorescence upon complementation can be examined in vitro and in vivo. To achieve a quick assessment of complementation in vivo, additional constructs were made to express peptide–intein fusion proteins. The pTWIN1 vector in the IMPACT-TWIN system (New England Biolabs) was used as the template for PCR amplification of the intein gene. An ~1.5 kb intein cassette including the multiple clone site and intein genes was amplified using 5′ and 3′ primers (Table 3 of the Supporting Information) carried BspHI and PstI restriction enzyme sites, respectively, followed by the digestion on the resultant PCR product with BspHI and SapI. BspHI–SapI restriction fragments containing the desired intein gene were purified for the succeeding cloning. Complementary DNA sequences containing SapI and PstI sticky ends and encoding the left-out segment of the OPT sequence (i.e., the residues removed from β-strands 7 and 11 of OPT) were prepared by annealing two respective oligonucleotides (Table 4A of the Supporting Information). The fragments encoding the intein gene and the annealed oligonucleotides were cloned into the NcoI and PstI sites of expression vector pCDF-1b (Novagen) which is compatible with pET-28a in coexpression (50). All identified constructs (so-called s7NT and s11NT) were verified by dideoxynucleotide sequencing. The peptide–intein fusion construct for β-strand 10 (so-called s10NT) was made by PCR using specific primers (Table 4B of the Supporting Information) and identified s7NT plasmids as templates. The peptide–intein fusions (s7NT, s10NT, and s11NT) were expressed under the control of the T7 promoter in vector pCDF-1b and were used to express the intein genes fused with the peptide sequences that were originally omitted from β-strand 7 (NSHNVYITADKQ), β-strand 10 (NHYLSTQTVLSKDP-NEKRD), and β-strand 11 (KRDHMVLLEFVTAAGIT) of OPT, respectively.
Table 3.
Concentration-Dependent Amplitude Shifts of in Vitro Complementation of t7SPm
| amplitudea (%) |
|||
|---|---|---|---|
| [s7 peptide] (µM) | A1 | A2 | A3 |
| 14.40 | 112.10 ± 0.57 | −8.07 ± 0.93 | −4.02 ± 1.50 |
| 7.20 | 96.48 ± 0.60 | 21.53 ± 1.47 | −18.01 ± 0.86 |
| 3.60 | 70.48 ± 0.08 | 55.34 ± 0.44 | −25.82 ± 0.36 |
| 1.80 | 51.05 ± 3.15 | 82.56 ± 2.96 | −33.61 ± 0.18 |
In vitro complementation assays of 0.1 µM t7SPm were performed with various concentrations of s7 under native conditions. Time-dependent changes in fluorescence were best fitted to the triple-exponential equation with fixed rate constants derived from the same measurement with 1.8 µM s7 peptide (i.e., k1 = 0.03810, k2 = 0.00630, and k3 = 0.00158). Amplitudes are provided in relative percentage (±SD; n = 2).
Protein Expression and Purification
Identified constructs (t7SP, t10SP, t11SP c7SP, OPT, s7NT, s10NT, and s11NT) were transformed and expressed under control of the T7 promoter/lac operator in Escherichia coli strain BL21(DE3) (Novagen). Plasmids for leave-one-out constructs and peptide fusion constructs were also cotransformed into BL21(DE3) for in vivo complementation. Freshly transformed cells were grown to an OD600 of 0.6 at 37 °C in LB medium containing 30 µg/mL kanamycin when expressing leave-one-out constructs alone, or 15 µg/mL kanamycin and 15 µg/mL streptomycin when coexpressing leave-one-out constructs and peptide fusions. After the induction with 0.5 mM IPTG, cells were further grown for 24 h at 25 °C with 250 rpm shaking and then harvested.
Cells from growth cultures were precipitated by the centrifugation, washed with PBS buffer [137 mM NaCl, 2.7 mM KCl, 1.47 mM KH2PO4, and 4.3 mM Na2HPO4•7H2O (pH 7.0)], resuspended with lysis buffer [300 mM NaCl, 50 mM Na2HPO4, and 50 mM Tris-HCl (pH 7.5); 8 mL per gram of cell pellets], and then frozen (−80 °C for 10 min) and thawed (37 °C for 5 min) three times. The resultant cell suspension was lysed with 50 mg/mL lysozyme, 10 mg/mL DNaseI, 10 mg/mL RNaseA, and 1% Triton X-100 and incubated at 37 °C for 30 min. After centrifugation at 27000g for 30 min, the supernatant was collected for further purifications. Six-His tagged proteins from single expression and coexpression were purified with the Ni-NTA purification system (Invitrogen) at room temperature under hybrid conditions according to the manufacturer’s instruction manual and the on-column protein refolding protocols (51). Briefly, the supernatant was first loaded onto the Ni-NTA resin column equilibrated with the lysis buffer under native conditions. The column was then washed with wash buffer 1 [300 mM NaCl, 50 mM Na2HPO4, 50 mM Tris-HCl, and 10 mM imidazole (pH 7.5)] and wash buffer 2 [300 mM NaCl, 50 mM Na2HPO4, 50 mM Tris-HCl, and 20 mM imidazole (pH 7.5)] sequentially. Proteins were eluted with elution buffer [300 mM NaCl, 50 mM Na2HPO4, 50 mM Tris-HCl, and 250 mM imidazole (pH 7.5)], and eluted fractions were pooled and dialyzed against TN buffer [100 mM NaCl and 50 mM Tris-HCl (pH 7.5)] at 4 °C overnight.
The protein purification was continued under denaturing conditions after dialysis. To denature proteins, the dialyzed solution was first titrated to near pH 2.0 using lysis buffer (pH 2.0) and then supplemented with SDS and β-mercaptoethanol to final concentrations of 1% and 100 mM, respectively. The resultant protein solution was then titrated back to near pH 8.5 with lysis buffer (pH 8.5) at a final dilution ratio of 1:10. The SDS concentration was maintained at 0.1% to keep proteins unfolded. Denatured proteins were then bound to the Ni-NTA resin equilibrated with the lysis buffer (pH 8.5) by the batch absorption at room temperature overnight. The on-column refolding and purification were then started by packing the resin into a column and washing the column with wash buffer 1 and 2 containing 0.1% SDS and 10 mM β-mercaptoethanol. This was followed by washing the column with wash buffer 2 containing 0.1% CHAPS and 10 mM β-mercaptoethanol to remove SDS from denatured proteins and to trigger protein refolding. Finally, refolded proteins were eluted with elution buffer containing 0.1% CHAPS and 10 mM β-mercaptoethanol, and eluted fractions were dialyzed against TN buffer (pH 7.5) at 4 °C for 8 h and repeated twice. After dialysis, the protein concentration was determined with a BCA protein assay (Pierce) with the UV–visible absorbance spectrometer (UV-1650PC, Shimadzu) as described in the manufacturer’s instruction manual. The purity of proteins was determined by SDS–PAGE and was quantified by laser-based densitometry (Typhoon TRIO+, Amersham Biosciences). All purified proteins were aliquoted and stored at −80 °C before use. The leave-one-out GFP (t7SP) purified from the coexpression was denoted as t7SPm (m for mature), whereas the construct purified from the single expression was denoted t7SP.
Optical Characterization and Fluorescence Measurement
To determine the optical characteristics of native and denatured GFP variants, purified proteins were diluted to 10 or 80 µM in TN buffer (pH 7.5) or TN buffer (pH 2.0) and absorption spectra between 300 and 600 nm were recorded using the UV–visible absorbance spectrometer. The spectra were recorded at 25 °C with 10 mm light path quartz cuvettes, and the spectral bandwidth of the spectrometer was set to 2 nm. The acid-denatured proteins were incubated at room temperature for at least 1 h before the measurement. The absorption of acid-denatured proteins under 382.5 nm (OD382.5) was used to quantify the chromophore (12, 52).
The fluorescence spectra of purified proteins were measured with a Fluorolog-3 TAU fluorometer (HORIBA Jobin Yvon). Samples were prepared in TN buffer (pH 7.5) at a protein concentration of 1 µM for t7SP and t7SPm or 0.01 µM for OPT and c7SP. In the emission spectra, emission intensities were collected from 495 to 550 nm with a 0.5 nm increment while excited under 485 nm. The excitation spectra were recorded by collecting excitation intensities from 350 to 500 nm with a 0.5 nm increment under 508 nm emission. All the emission and excitation spectra were recorded at room temperature with a slit setting of 5 nm and an integration time of 0.5 s.
The pH-dependent transition curves for protein folding were determined by the fluorescence intensity using a Fluorolog-3 TAU fluorometer with excitation at 485 nm and emission at 508 nm. Protein stock solutions were prepared in TN buffer (pH 7.5) with a protein concentration of 10 µM and then diluted into TN buffer with different pH values that ranged from 2.0 to 10.0 and with 1 mM DTT. The final concentration of each protein was 0.1 µM. After the sample had been incubated for 30 min, the fluorescence intensity was measured at room temperature with a slit setting of 5 nm and an integration time of 0.5 s. The pH-dependent transition curve was fitted to the Henderson-Hasselbalch equation using SigmaPlot (Systat Software Inc.) to evaluate the pKa of each protein.
Measurement of Refolding Kinetics
The refolding kinetics of purified GFP variants were measured with a Fluorolog-3 TAU fluorometer by manual mixing. Briefly, each protein (10 µM) was first denatured in TN buffer (pH 2.0) containing 1 mM DTT for 1 h. The protein refolding was then triggered by diluting denatured protein solutions into TN buffer (pH 7.5). The final solutions contained proteins at a concentration of 0.01 µM for OPT and c7SP or 0.1 µM for t7SP and t7SPm, and 1 mM DTT (pH 7.5). The fluorescence intensity was collected every 5 s for 30 min with excitation at 485 nm and emission at 508 nm, and at room temperature with a slit setting of 5 nm and an integration time of 0.5 s. The refolding curve of each GFP variant was best fitted to the triple-exponential equation with six fitting parameters using SigmaPlot, and rate constants and amplitudes for three kinetic phases were identified. Using a double-exponential fit was insufficient to model the data, showing correlated residuals.
Complementation Assay
After the IPTG induction, in vivo complementation of engineered GFP variants and peptide fusions was observed directly by illuminating cell pellets with the blue light transilluminator (λ = 400–500 nm, Dark reader, Clare Chemical Research) or measured with a Fluorolog-3 TAU fluorometer.
The complementation was also performed in vitro with the purified GFP variants and chemically synthesized peptides, and the resultant fluorescence intensity was collected with a Fluorolog-3 TAU fluorometer. Chemically synthesized strand 7 peptides (s7) with the amino acid sequence NSH-NVYITADKQ were prepared with a purity of >75% (Biopeptide) or with a purity of >95% (GenScript). Briefly, s7 (>75% pure) and purified t7SP and t7SPm were first freshly prepared in native solution [TN buffer (pH 7.5)] or denatured solution [TN buffer (pH 2.0) with 1 mM DTT]. The final concentrations of proteins and peptides were 10 and 360 µM, respectively. To inspect the in vitro complementation starting from native conditions, proteins prepared in native solution were first diluted to 0.1 µM in TN buffer (pH 7.5) and equilibrated for 60 s before peptide solutions were added. The s7 peptide in native solution was then added to a final concentration of 3.6 µM. The fluorescence intensity upon complementation was recorded every 5 s for 30 min with excitation at 485 nm and emission at 508 nm, and at room temperature with a slit setting of 5 nm and an integration time of 0.5 s. To investigate the in vitro complementation that started from the refolding condition, equal volumes of the protein solution and the peptide solution from the denaturing condition were first mixed together and then diluted 50-fold into TN buffer (pH 7.5) to initiate protein refolding and complementation. The fluorescence intensity was measured as described above.
To determine the binding affinity between the leave-one-out GFP and synthesized strand 7 peptides, in vitro complementation was also performed under native conditions as described above but with different s7 concentrations. Synthesized s7 peptides with a purity of >95% were used in these assays. The recovered fluorescence intensity (F) was measured every 5 s for 20 min as described above and normalized by the initial fluorescence intensity (F0) to express the relative intensity increase upon complementation. The Kd was determined by fitting the binding curve with nonlinear regression fitting parameters for a one-site binding model using SigmaPlot.
RESULTS
In Vivo Complementation
The full-length, unpermuted superfolder GFP OPT (OPT), three peptide fusions (s7NT, s10NT, and s11NT), three leave-one-out GFP variants (t7SP, t10SP, and t11SP), and the full-length, circularly permuted OPT (c7SP) were first expressed individually in BL21(DE3) to inspect for any possible background fluorescence from each construct. Freshly transformed cells were allowed to express recombinant proteins with 0.5 mM IPTG induction at 25 °C for 24 h. The fluorescence emission was observed directly by illuminating cell pellets with the blue light transilluminator or measured with a Fluorolog-3 TAU fluorometer. OPT and c7SP exhibited the highest fluorescence emission among these constructs, while no significant fluorescence was observed from cells expressing three peptide fusions, t10SP or t11SP (data not shown). To our surprise, a detectable fluorescence emission was obtained directly from t7SP, although the intensity was not as high as that of OPT or c7SP (Figure 3 of the Supporting Information). N-Terminal fragments of enhanced GFP (residues 1 – 158) have been shown to fold and allow a significant level of chromophore formation in a previous study (53). Thus, the background fluorescence of t7SP indicated not only the formation of the chromophore but also at least a partial folding of t7SP.
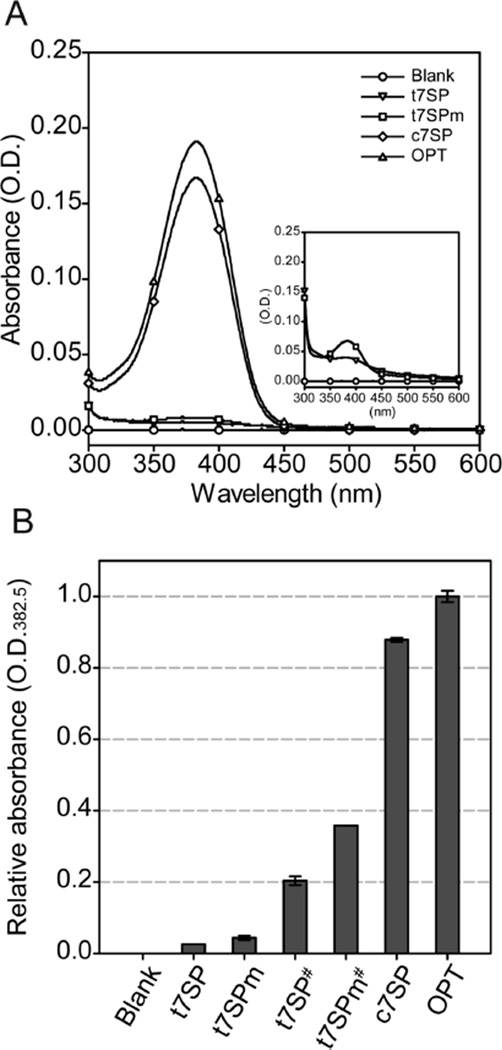
Figure 3.
Optical characteristics of denatured GFP variants. (A) Optical spectra of purified GFP variants under denaturing conditions. Purified t7SP, t7SPm, OPT, and c7SP were diluted to 10 µM in TN buffer (pH 2.0) and incubated for at least 1 h to ensure a complete denaturation. Absorbance spectra between 300 and 600 nm were recorded. Measurements were also performed for 80 µM purified t7SP and t7SPm with identical protocols (inset). (B) Relative quantity of the mature chromophore in purified GFP variants. The absorbances of 10 µM denatured t7SP, t7SPm, OPT, and c7SP or 80 µM (#) denatured t7SP and t7SPm under 382.5 nm were measured three times independently and normalized by the average reading of OD382.5 from OPT. The average of relative OD382.5 values is shown, and error bars indicate the two standard deviations obtained from triplicate measurements.
When s7NT with t7SP were coexpressed, the fluorescence was increased ~2-fold compared with cells expressing t7SP alone, indicating the in vivo complementation of the β-strand 7 peptides and the leave-one-out t7SP had occurred. In vivo complementation was also observed in coexpression of t11SP and s11NT, and this was consistent with a previous study (38). Fluorescence was not recovered from coexpression of t10SP and s10NT, indicating that removal of the β-strand 10 segment was fatal to folding. No fluorescence was observed for the mispaired coexpression of t11SP with s7NT. No detectable additional fluorescence was observed for the mispaired coexpression of t7SP with s11NT, beyond the intensity observed for t7SP alone (Figure 3 of the Supporting Information).
Absorbance and Fluorescence Features of GFP Variants
To further study the optical characteristics of GFP variants, the proteins OPT, c7SP, and t7SP were purified by nickel-charged affinity resin (Invitrogen) as described in Experimental Procedures and were used to investigate the optical characteristics via an UV–visible absorbance spectrometer and the emission and excitation spectra with a fluorometer. Proteins from each construct were purified under hybrid conditions with detergent exchange to increase the purity and to eliminate any protein bound to the leave-one-out GFP variant. OPT purified from the hybrid method exhibited a fluorescence intensity comparable to the one purified without the unfolding–refolding process, indicating that the mature chromophore remained intact throughout the purification in the hybrid method. The purity of prepared proteins was ~90% as determined by densitometry quantification from SDS–PAGE gels. The refolded leave-one-out GFP variants from coexpression of t7SP and s7NT were denoted as t7SPm (m for mature).
To compare the formation of chromophores from purified GFP variants, absorbance spectra of each protein were measured. Purified proteins were diluted to 10 µM, and absorbance spectra between 300 and 600 nm were recorded under native conditions (Figure 2). The absorbance spectrum of OPT exhibited a major peak near 480 nm, indicating a dominance of the deprotonated and anionic chromophore (54), while the cyclized version of OPT (c7SP) exhibited two major peaks near 400 and 480 nm, indicating the coexistence of a protonated and deprotonated chromophore, respectively, which agreed with the finding in the previous study (47). Although the absorbance feature of the chromophore from t7SP and t7SPm was only observed at a higher concentration [80 µM (Figure 2 inset)], both exhibited similar signature peaks of the protonated and deprotonated chromophore.
To investigate the quantity of the mature chromophore in each purified protein, absorbance spectra were also recorded under denaturing conditions. Fully denatured GFP has signature absorbance peaks under acid conditions (~383 nm) and under base conditions (~447 nm), and it has been concluded that the spectra resulted from the interactions between the chromophore and the solvent (12). After denaturation with TN buffer (pH 2.0), all purified proteins exhibited an absorbance peak around 380 nm to a different extent (Figure 3A). As shown in Figure 3B, the relative quantity of the chromophore indicated only a small fraction of purified t7SP and t7SPm possessed the chromophore, and the quantity of chromophore was correlated with the observation of the difference in absorbance amplitudes under native conditions (Figure 2). A small fraction of t7SP contained the mature chromophore, indicating the background fluorescence of t7SP was from the limited chromophore maturation. The in vivo complementation of t7SP and s7NT increased the quantity of the chromophore to nearly 80% when compared with the quantity of t7SP alone. This result shows that an intact β-barrel structure promotes the maturation of chromophore, resulting in the increased fluorescence intensity observed in vivo.
To understand the difference in each protein in terms of quantum yield, the fluorescence spectra were recorded and normalized by the corresponding OD485. All purified proteins exhibited an emission of 508 nm with different excitation wavelengths (Figure 4). OPT was excited primarily by wavelengths near 485 nm, while c7SP, t7SP, and t7SPm all had excitation peaks near 400 and 485 nm.
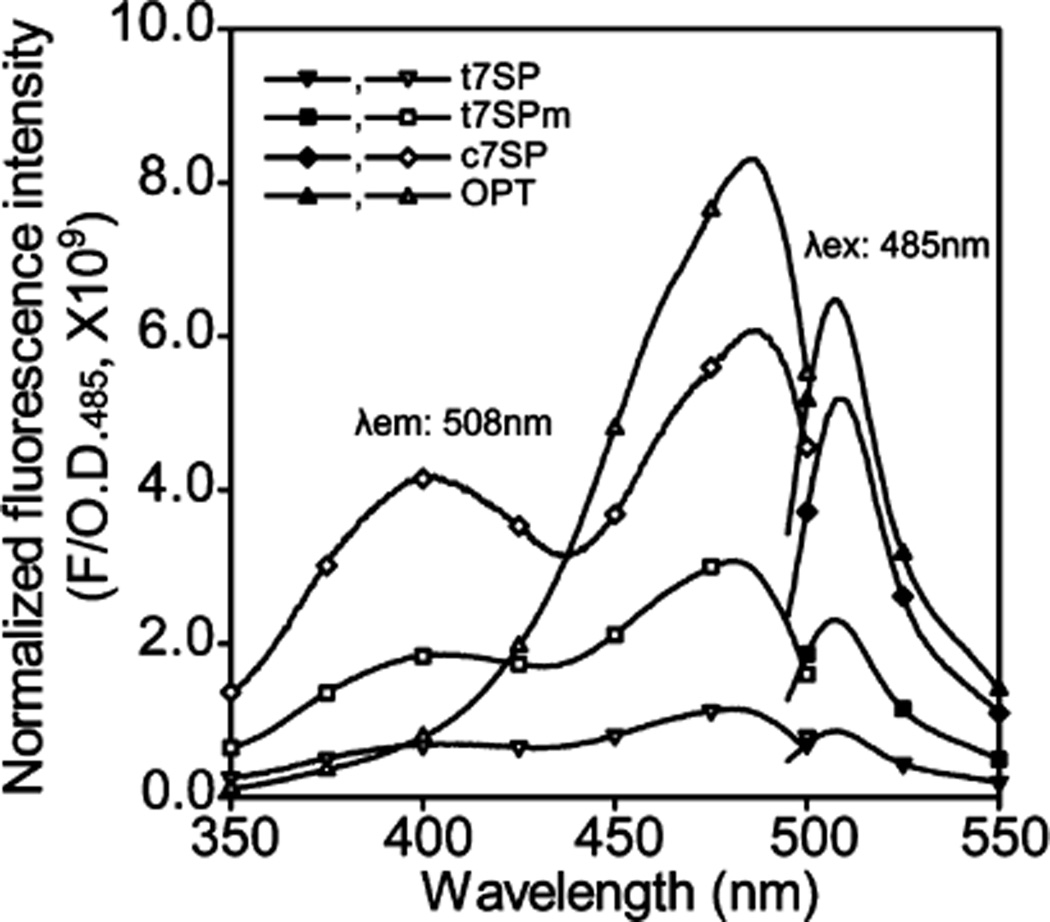
Figure 4.
Fluorescence emission and excitation spectra of purified GFP variants. Purified GFP variants were diluted to 1 µM for t7SP and t7SPm or 0.01 µM for OPT and c7SP in TN buffer (pH 7.5). The emission spectra (black symbols) and the excitation spectra (white symbols) were recorded with excitation at 485 nm and emission at 508 nm and normalized by the corresponding OD485 of each variant.
We observed significant fluorescence intensity from t7SP and t7SPm in the absence of the target peptide, though at a lower quantum yield. At an excitation wavelength of 485 nm, OPT exhibited the highest quantum yield at the emission wavelength of 508 nm. c7SP had only ~78% of the quantum yield of OPT (Table 1), while t7SP and t7SPm revealed relative quantum yields of ~18 and ~40%, respectively. The t7SPm protein was distinguishable from the singly expressed t7SP by its higher quantum yield of fluorescence. The unexpected fluorescence from the leave-one-out constructs suggests that the fully closed β-barrel is not completely necessary for blocking the full quenching effect of the solvent. Modeling studies have shown that 10 strands are not sufficient to close the β-barrel around the central chromophore (data not shown); however, the results suggest that there must be a significant degree of exclusion of solvent from the chromophore in the 10-stranded, partially open β-barrel.
In Vitro Complementation
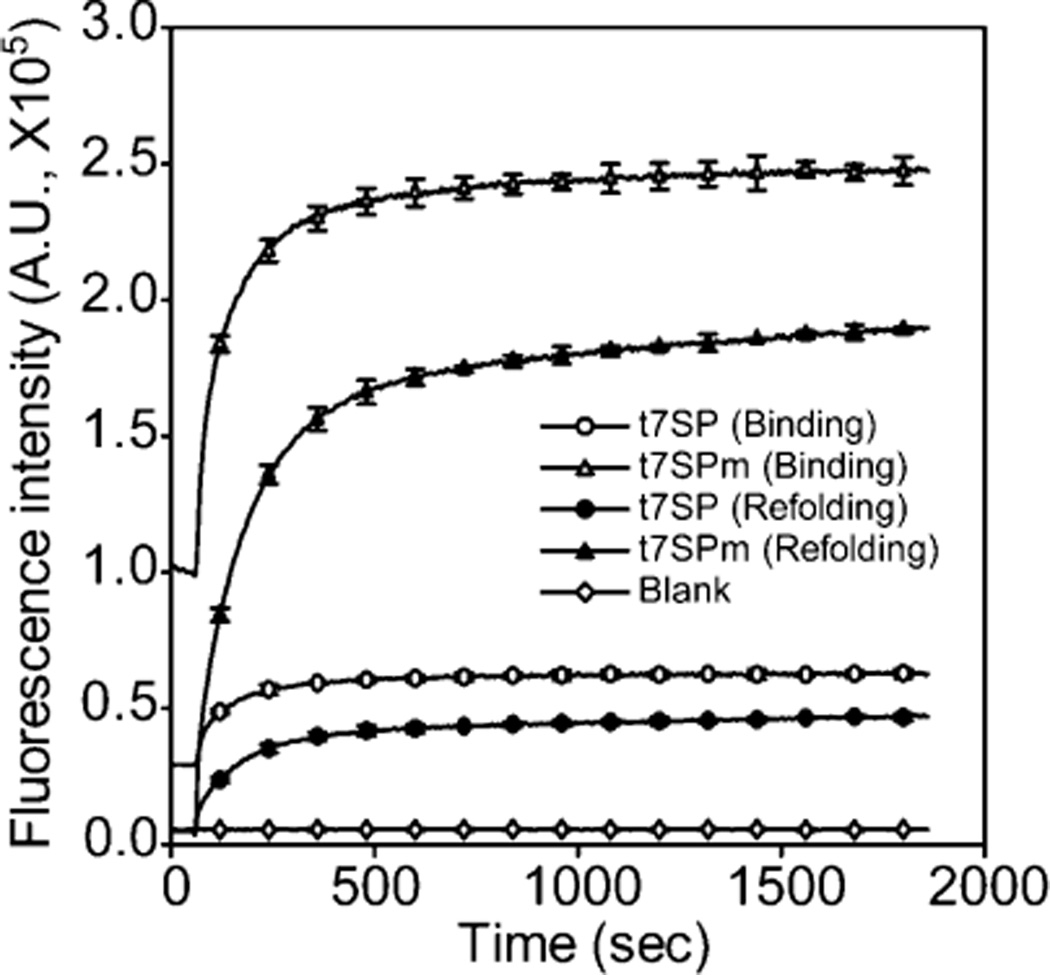
The complementation of chemically synthesized β-strand 7 peptides (>75% pure) with the purified t7SP–t7SPm protein was further inspected in vitro. Two different strategies were tested to initiate the complementation: (a) starting from native conditions and (b) starting from pH 2.0, unfolded conditions. The complementation was triggered by adding the synthesized β-strand 7 peptides after equilibrium for 60 s in native experiments or by adding the denatured protein and peptide mixture to TN buffer (pH 7.5).
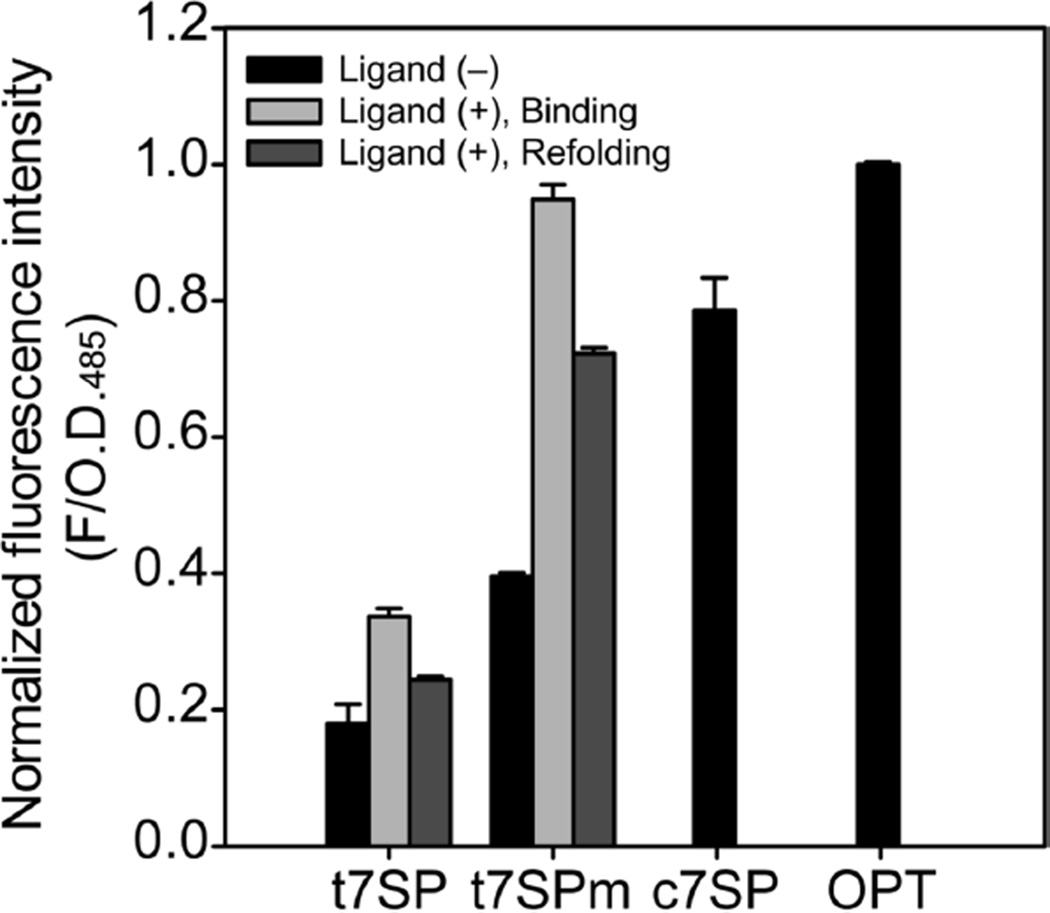
As shown in Figure 5, the fluorescence reached ~90% of its final value within 10 min, in both peptide binding and binding–refolding experiments. The complementation also recovered the relative quantum yield upon the binding of s7, although it recovered less in refolding experiments (Figure 6). Purified t7SPm that possessed more of the mature chromophore, as measured by UV absorbance, regained more fluorescence intensity upon complementation when compared with t7SP, but even when fluorescence was normalized by UV absorbance, the fluorescence was relatively higher for t7SPm than for t7SP. The results when considered together suggest a hypothesis that GFP loses some of its capacity for fluorescence when unfolded in vivo and that the degree to which this capacity is lost is roughly proportional to the time spent in the unfolded state. The chromophore is exposed to oxygen in the unfolded state, and this might lead to an uncharacterized oxidized form of the chromophore or of its surroundings in proteins that have been exposed for longer durations. This hypothesis remains to be tested. Thus far, we have observed only a qualitative correlation between the loss of quantum yield and time spent in the unfolded state. Quantum yield is not recovered upon exposure to DTT. Oxidation byproducts, including carbon–carbon bond cleavages, have been seen in crystal structures of other GFP variants (55).
Figure 5.
Time-dependent fluorescence recovery of the leave-one-out GFP. The fluorescence recovery of the leave-one-out GFP upon in vitro complementation is shown. Chemically synthesized peptides (s7) with a purity of >75% were used. The complementation was initiated by adding peptide solutions (pH 7.5) to equilibrated native t7SP and t7SPm (white symbols) or by bringing the pH of protein/ peptide mixtures from 2.0 to 7.5 (black symbols). Final concentrations of the leave-one-out GFP (t7SP and t7SPm) and s7 in both experiments were 0.1 and 3.6 µM, respectively. The fluorescence intensity at 508 nm emission upon the complementation was recorded and plotted every 5 s for 30 min with excitation at 485 nm at room temperature. Error bars indicate the two standard deviations from triplicate measurements and were provided only every 120 s.
Figure 6.
Recovery of the quantum yield upon complementation. The fluorescence intensity at 508 nm emission with excitation at 485 nm was measured for purified t7SP, t7SPm, OPT, and c7SP and normalized by individual OD485 to give the quantum yield. Three different conditions were used, and chemically synthesized peptides (s7) with a purity of >75% were used: (a) without the s7 peptide ligand (black), (b) 30 min after complementation with the s7 peptide ligand (light gray), and (c) 30 min after pH refolding in the presence of the s7 peptide ligand (dark gray). Error bars indicate the two standard deviations from triplicate measurements.
Target Peptide Binding Affinity
To evaluate the target peptide detection sensitivity of t7SPm, in vitro complementation was performed with serially diluted s7 peptides prepared with a purity of >95% as described in Experimental Procedures, covering a > 100-fold range of molar ratios from 0.36:1 to 144:1. Fluorescence intensity was collected for 1200 s (Figure 4 of the Supporting Information).
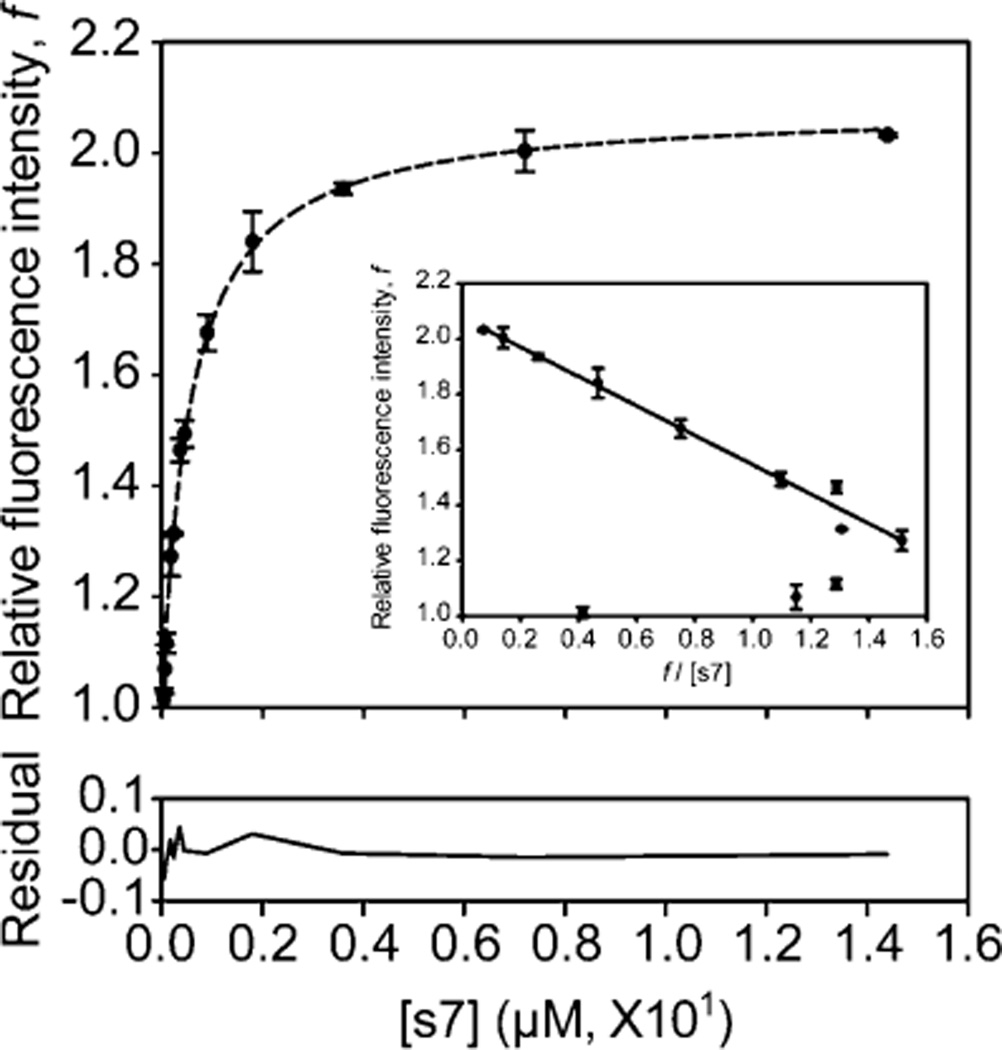
The recovery of relative fluorescence intensity f was plotted versus peptide concentration [s7] and fitted to a simple one-site binding isotherm (Figure 7) to determine the dissociation constant (Kd) for the peptide, giving a value of approximately 5.51 × 10−7 M. An Eadie–Hofstee style plot of the relative fluorescence intensity f versus f/[s7] (Figure 7, inset) shows the expected linear relationship with the exception of three outliers with lower peptide concentrations, due to the depletion of unbound ligand at these concentrations. After outliers had been eliminated, the corrected Kd was estimated to be 5.31 × 10−7 M.
Figure 7.
Concentration-dependent fluorescence recovery of the leave-one-out GFP. The in vitro complementation assays were performed with 0.1 µM t7SPm and with variable concentrations of the s7 peptide ligand under native conditions. The fluorescence intensity at 508 nm emission upon complementation was recorded every 5 s for 20 min with excitation at 485 nm at room temperature, and the normalized fluorescence (F/F0) was plotted as the relative fluorescence intensity, f (Figure 4 of the Supporting Information). Different molar ratios of t7SPm to the s7 peptide ligand (>95% pure) covering a >100-fold range from 1:0.36 to 1:144 were tested, and the average of relative fluorescence intensity f (n = 2) at the end point (i.e., 1200 s after the initiation of complementation) was plotted and fitted to the one-site binding equation (dashed line) using SigmaPlot to evaluate the dissociation constant (Kd). The residual of the fitting was provided. Relative fluorescence intensity f and f/[s7] were also plotted (inset) and fitted to the linear regression using SigmaPlot (solid line) after excluding three outliers. Error bars indicate the two standard deviations obtained from two independent measurements.
For comparison, the binding affinity of the src-SH3 domain for its target peptide is 100-fold worse at 3.0 × 10−5 M (56), while human leukocyte antigen (HLA) systems bind peptides with Kd values in the range of 10−8 M (57). The higher affinity for t7SPm as compared to src-SH3 can be attributed to the larger amount of buried surface area in the peptide binding interface.
Refolding Kinetics
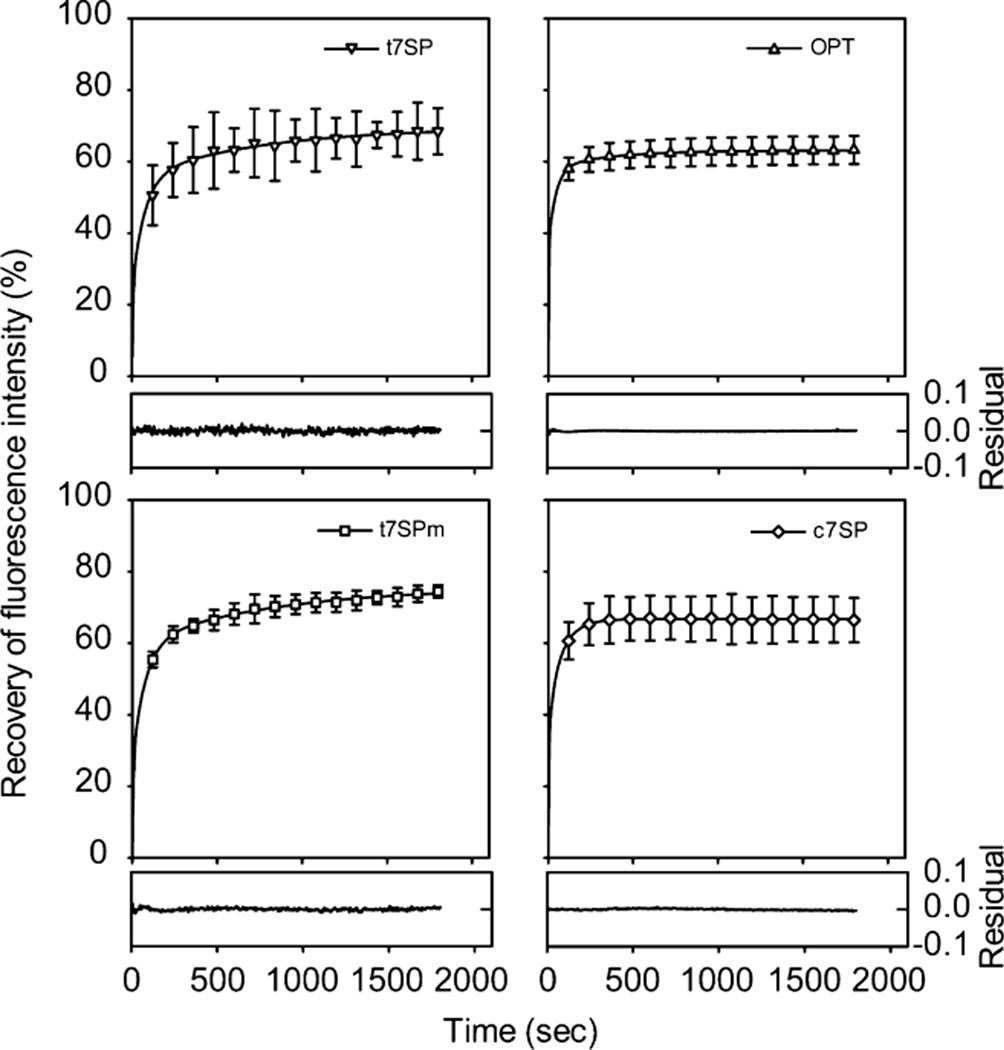
Purified proteins were acid denatured in TN buffer (pH 2.0) containing 1 mM DTT, and protein refolding was initiated by dilution into TN buffer (pH 7.5) containing 1 mM DTT. The fluorescence recovery was recorded and normalized by the fluorescence intensity under native conditions. In all GFP variants, more than 50% of the fluorescence was recovered within 5 min, indicating a similar kinetics of refolding (Figure 8). Single- and double-exponential fits to the data each showed correlated residuals (data not shown), leading to a triple-exponential fit as the simplest kinetic model, yielding three different rate constants and relative amplitudes (Table 1).
Figure 8.
Refolding kinetics of GFP variants. Refolding of acid-denatured GFP variants was initiated by dilution into TN buffer (pH 7.5) in the presence of 1 mM DTT. The fluorescence intensity at 508 nm emission was collected every 5 s for 30 min with excitation at 485 nm at a final concentration of 0.01 µM (OPT and c7SP) or 0.1 µM (t7SP and t7SPm) and presented as the percent recovery of the fluorescence. The measurements were performed at room temperature. Error bars indicate the two standard deviations from triplicate measurements and were only provided every 120 s. The refolding curve of each GFP variant was then fitted to the triple-exponential equation using SigmaPlot, and the residual of each fitting was provided. Identified rate constants and amplitudes for three kinetic phases are listed in Table 1.
Similar multiphase refolding kinetics were reported for cycle 3 GFP by Enoki et al. (58), who determined that the slow phase (k3 = 0.001–0.012 s−1) was due to proline isomerization. The variable amplitude (A3) for the slow phase in our experiments might be due to the variable aging time in the denatured state, since this was not controlled in our experiments.
A medium phase (k2 = 0.01–0.05 s−1) was found with a range similar to that of the medium phase of cycle 3 folding, but with variable amplitude (A2). Part of the variability could be due to the variable aging time (i.e., time spent in pH 2.0 buffer), since the cycle 3 medium phase amplitude depended partially on aging time (58). However, it is likely that the permutation of the polypeptide chain and the absence (in t7SP and t7SPm) or presence (in c7SP) of β-strand 7 played a role in preferentially stabilizing one of the folding intermediates. For example, in c7SP, the low amplitude of the medium phase (A2) relative to OPT implies the destabilization of intermediate I2 relative to intermediate I1 (see Figure 10) by the circular permutation at β-strand 7. Meanwhile, in t7SP and t7SPm, omitting β-strand 7 in the context of c7SP led to an increased stability of I2 relative to I1. Both observations point to β-strand 7 being in transition between I2 and I1. This is consistent with speculation that the cleft between strands 7 and 8 is a late-forming feature in GFP folding (45, 46).
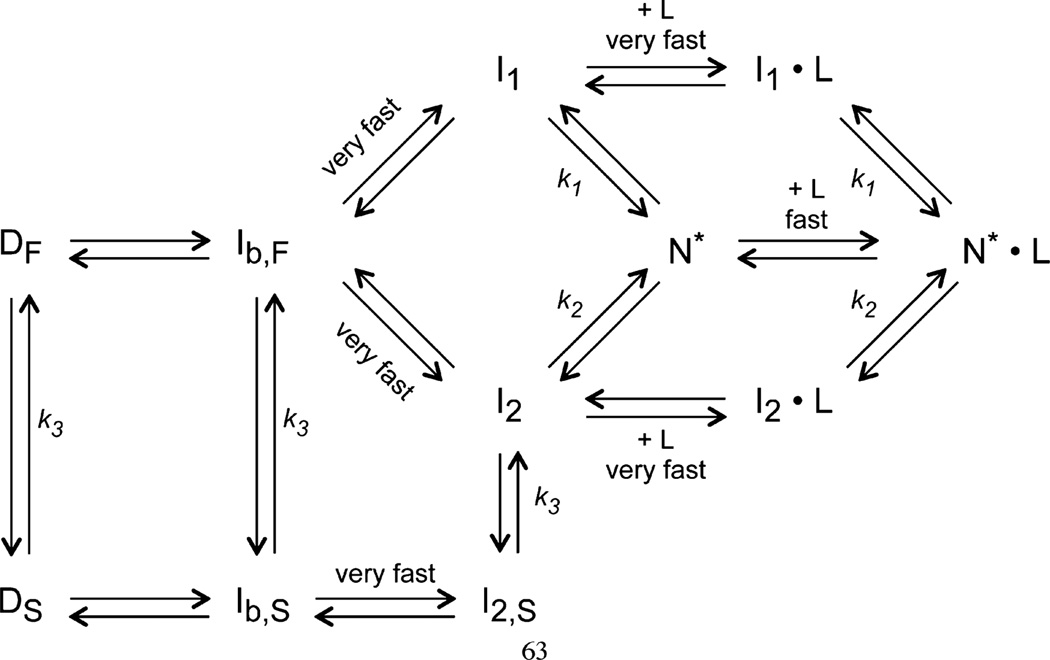
Figure 10.
Folding–ligand binding kinetics of the leave-one-out GFP. Proposed model of the folding–ligand binding mechanism of the leave-one-out GFP, modified from ref 58 by the addition of the three ligand binding steps. Two unfolded species (DF and DS) are equilibrated through a slow prolyl isomerization and fold to the intermediate states (I1 and I2) after the initiation of refolding. In the presence of ligands (L), both intermediate species form complexes (I1•L and I2•L) rapidly and further fold to the native state (N*•L). Notice that both intermediates can also fold to a different native state (N*) without the presence of ligands, and N* can further bind to ligands and convert to the N*•L state. Both N* and N*•L are the fluorescent species, by definition.
A fast phase (k1 = 0.16–0.36 s−1) was observed that is in the range of the previously reported fast phase for cycle 3 GFP. The fast phase was faster for OPT and c7SP than for t7SP and t7SPm, but indistinguishable in rate between OPT and c7SP. This is consistent with the view that β-strand 7 is folded in I1 but the β-barrel is not yet closed (since I1 is not fluorescent). Since the presence or absence of β-strand 7 affects the rate of the final β-barrel-closing step, that step must be driven by interactions involving β-strand 7 on the remaining side, either the β-strand 8 side or the β-strand 10 side.
We were not able to measure the previously reported burst phase with a small negative amplitude in the chromophore fluorescence (58). Our data showed no burst phase within experimental error.
Peptide Binding Kinetics
The time course studies of fluorescence recovery from in vitro complementation assays with various s7 concentrations (Figure 4 of the Supporting Information) were further used to analyze the peptide binding kinetics of t7SPm. The kinetic parameters for each s7 concentration were determined by first fitting the curve to various exponential equations with different numbers of kinetic phases ranging from one to three and then reporting the optimal fit (Table 2).
At low concentrations of the s7 peptide (0.09–0.36 M), only one kinetic phase with a medium rate constant was detected. However, at higher concentrations of the s7 peptide (0.45 and 0.9 M), an additional kinetic phase with a fast rate constant was identified. Further increasing the concentration of the s7 peptide to 1.8–14.4 M resulted in a three-phase kinetic fit with an additional slow rate constant which gave a negative amplitude and was responsible for the slow decrease in the fluorescence intensity with time. This result showed s7 peptide concentration-dependent binding kinetics. Interestingly, all the identified rates were with the same approximation in both refolding studies (Table 1) and binding studies (Table 2), and no other measurable phase was observed, demonstrating that the peptide binding step is fast relative to the peptide-independent folding steps. Peptide binding should in effect fold the protein by shifting the equilibrium toward the folded state, using its higher binding affinity for the native state.
The decrease in fluorescence intensity over time in in vitro complementation was especially noticeable at higher concentrations of the s7 peptide (Figure 4 of the Supporting Information). It is known that the excitation of chromophore and fluorophore generates reactive oxygen species (ROS) which deactivate the function of targeted proteins (59, 60), and this technique, so-called chromophore-assisted light inactivation (CALI), has been used for acute protein inactivation in cell biology studies (61, 62) or to serve as photosensitizers (63). Although GFP is inefficient in generating ROS possibly due to the protection of the barrel-like structure, it is likely that the same CALI mechanism would explain the fluorescence decay of t7SPm upon complementation. It is possible that the leave-one-out design of GFP makes it more vulnerable to the ROS attack, or it could presumably produce more ROS than wild-type GFP, since the chromophore of t7SPm is exposed to solvent and is not fully protected.
To improve our understanding of the trend of kinetic phases with changes in s7 peptide concentrations, fluorescence recovery curves from higher concentrations of s7 (14.4, 7.2, and 3.6 M) were fitted to the triple-exponential equation with fixed rate constants derived from fits to the 1.8 M s7 measurements, and the relative amplitudes are reported (Table 3). The effect of peptide concentration on the relative amplitudes of the fast and medium phases suggests that the peptide binds more tightly to I2 than to I1 because the fastest phase (associated with I1) increases in amplitude only at higher concentrations of the s7 peptide. This is consistent with the finding that at low concentrations of the s7 peptide, only the medium phase (k2), corresponding to the folding of I2, is observed (Table 2). We might speculate that one of these intermediates has s7 hydrogen-bonded to β-strand 8 (I1 perhaps) and the other has s7 hydrogen-bonded to β-strand 10 (I2 perhaps, since β-strand 7 makes more hydrogen bonds to β-strand 10 than to β-strand 8 in GFP crystal structures). The higher amplitude of A1 in c7SP would then be explained by the lower entropic cost of binding when β-strand 7 is covalently connected to β-strand 8.
Chromophore Formation in Leave-One-Out GFP
We observed on the basis of UV absorbance that only a small fraction of the leave-one-out variants, t7SP and t7SPm, possessed the mature chromophore, but this fraction was observed to increase over time, on the time scale of weeks to months (data not shown). Even t7SP that had never been exposed to the target peptide was observed to synthesize the chromophore, although very slowly.
In vivo complementation of t7SP and s7NT increased the level of maturation of chromophore (Figure 3). More importantly, in vitro complementation increased the fluorescence immediately and the fluorescence recovery reached the plateau within 10 min (Figure 5). Preincubation eliminated the slow de novo formation of the chromophore after the peptide binding and showed the chromophore remained intact after the modified preparation procedure. Moreover, the quantum yield was comparable to that of OPT after complementation for 30 min (Figure 6), which indicated the chromophore was completely buried after the binding of the strand 7 peptide. However, the recovery of quantum yield upon in vitro complementation in refolding experiments was not as high as the one in native experiments (Figure 6). This might be simply due to the loss of the chromophore by misfolding.
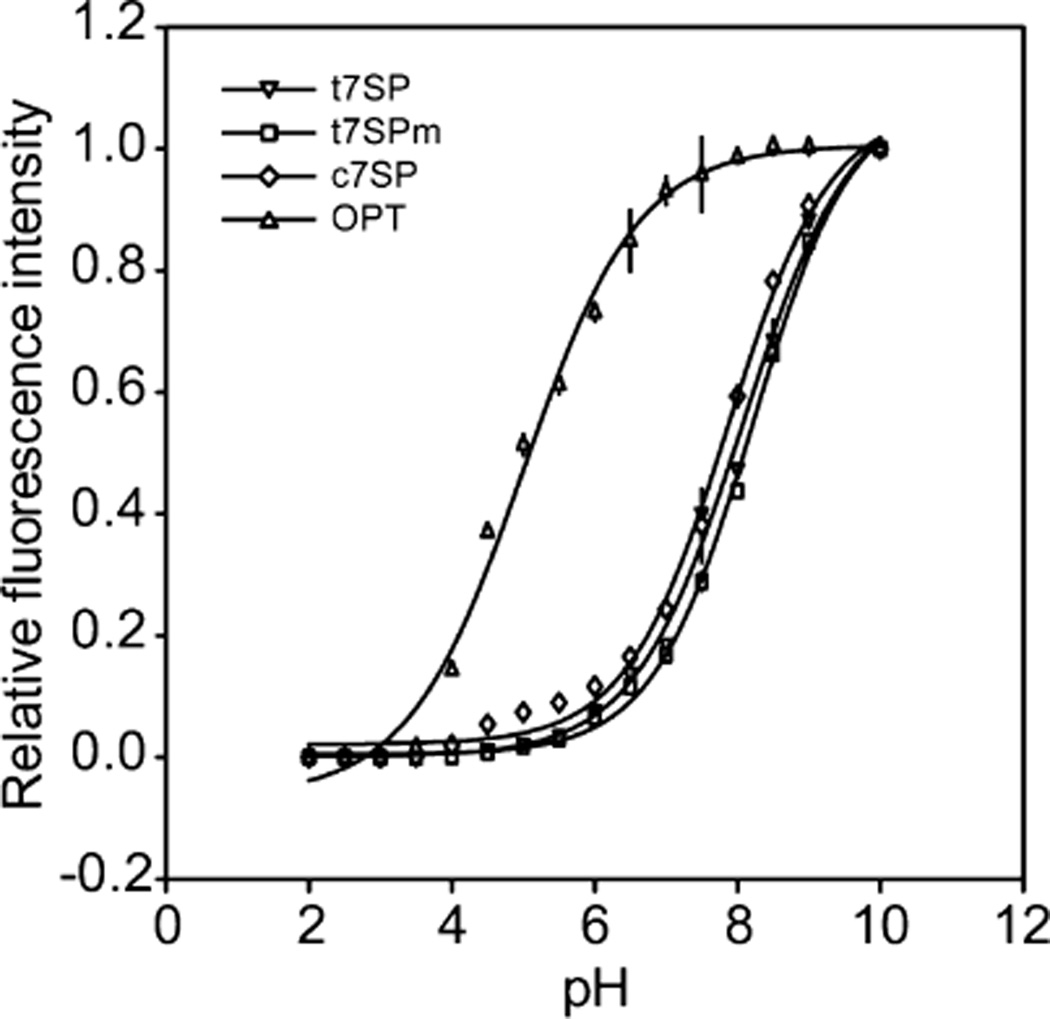
pH Dependence
The pH dependence of acid unfolding and refolding was determined for all purified proteins by plotting the fluorescence intensity relative to native conditions (Figure 9). The resulting curves were fit to the Henderson– Hasselbalch equation to evaluate the pKa of each protein. As shown in Table 1, the pKa of OPT was near 5.0 while all others were elevated to near 7.8–8.2, indicating a loss of stability to acid unfolding in all permutants that have the termini at the β-strand 7 position.
Figure 9.
Unfolding and refolding of GFP variants in pH titration. Each purified GFP variant (0.1 µM) was prepared in TN buffer with different pH values from 2.0 to 10.0 and in the presence of 1 mM DTT. After incubation for 30 min at room temperature, the fluorescence intensity of 508 nm emission was measured with excitation at 485 nm. The measurement was blanked with the fluorescence intensity under pH 2.0, normalized by the fluorescence intensity under pH 10.0 for individual protein, and plotted as the relative fluorescence intensity. The pH-dependent transition curve was then fitted to the Henderson–Hasselbalch equation (solid line) using SigmaPlot to evaluate the pKa of each protein. Error bars indicate two standard deviations from mean triplicate measurements.
DISCUSSION
Removal of three segments of GFP was explored, one of these in detail. Cyclizing and truncating OPT at β-strand 7 (t7SP/t7SPm) produced a sensor molecule that bound the strand 7 peptide with submicromolar affinity (Kd ~ 0.5 µM) and recovered the fluorescence upon binding. Weak fluorescence and slow formation of the chromophore were observed for t7SP in the absence of the peptide ligand, suggesting that β-strand 7 plays a fairly weak role in defining the environment of the chromophore. Removal of β-strand 11 obliterated fluorescence, but the truncated protein (t11SP) folded well enough to detect the free peptide or a peptide fusion, as previously described (38). When incubated with the target peptide in vivo, the complex efficiently synthesized its chromophore. Removal of β-strand 10 produced a cyclized and truncated molecule (t10SP) that was unable to fold or to synthesize its chromophore, even when expressed in the presence of the target peptide as an intein fusion (s10NT).
Kinetic Mechanism of Folding and Binding
If we assume that the phases we observed upon binding represent a pathway of refolding intermediates and we assume that these intermediates are most likely the same intermediates that were described for cycle 3 GFP (58), then the permutation and binding experiments described here should help us improve our understanding of the structure of these intermediates with regard to the role of β-strand 7. Figure 10 presents our conclusions based on the kinetic fits.
Our model differs slightly from that of Enoki et al., who hypothesize a pathway from a cis-Pro intermediate to I1 (I1F in ref 58), the fast phase intermediate, but this pathway is not necessary to explain the observed negative dependence of the fast phase on aging time. Therefore, we have omitted the cis-Pro I1 intermediate in our model, equivalent to omitting I1S in ref 58.
Consistent with the Enoki model, we find two parallel folding pathways. The data are not consistent with the alternative sequential pathway where both intermediates, I1 and I2, are obligatory. Both kinetic phases are observed after hydrophobic collapse (58). The higher amplitude A1 for c7SP says that it folds more through intermediate I1 than does t7SP/ t7SPm. In binding experiments, increasing the concentration of the s7 peptide ligand increased the folding traffic through I1 relative to I2. We propose that β-strand 7 is bound more tightly in I2 than in I1.
Binding of the s7 peptide is fast relative to the fastest protein folding phase since no additional phase was detected on the kinetics fits of the peptide binding. Also, chromophore formation, with a t1/2 of ~76 min for wild-type GFP (64), is expected to have a negligible contribution to the kinetic measurements described here.
Differences between Early and Late Chromophore Maturation
The quantum yield of t7SPm is consistently higher than that of t7SP over the entire range of the emission and excitation spectra (Figure 4). Although t7SPm has a more mature chromophore than t7SP, the normalization of fluorescence should cancel out this difference. The only remaining explanation for the difference in quantum yield is that t7SPm and t7SP have structural differences, despite having the same amino acid sequence and the same chromophore. t7SPm, which was grown in the presence of its target peptide, spent less time in the unfolded state than t7SP, which was exposed to its peptide target only after purification.
It stands to reason, therefore, that t7SP could have been chemically modified in the unfolded state, for example, by oxidation of an internal side chain, and that the chromophore is more buried in t7SPm than in t7SP. The difference in the UV–visible spectra from native t7SPm and t7SP hints at structural differences, though it is not obvious. The structural difference related to the time in the unfolded state may also be the cause of lost fluorescence recovery upon refolding.
Structural State of t7SP
Because t7SP/t7SPm is soluble and fluorescent, it is most likely trapped as one of the structural intermediates present in the folding pathway, in equilibrium with the native, fluorescent state. Others have shown that mechanical unfolding passed through a structural intermediate lacking one β-strand (65), although a different β-strand was used, and the mechanical denaturation is unusual. An unpublished result from Jackson’s group (66) also showed an intermediate state where GFP β-strands 7–9 had unfolded, while the rest remained intact, and exhibited 10% of the native GFP fluorescence.
CONCLUSIONS
In this study, a β-strand in the middle of the GFP sequence was left out and the function of the protein was reconstituted when it was added back in the form of a synthetic peptide or a fusion protein. The results of in vitro complementation experiments prove that the leave-one-out GFP immediately recovers fluorescence upon complementation with the target peptide, as long as its chromophore is mature. After pH 2.0 denaturation, the refolded leave-one-out GFP still retains its ability to reconstitute fluorescence upon peptide binding, showing monomeric and efficient refolding to the active form in the absence of strand 7. Our findings underscore the potential for the leave-one-out GFP to be used as a self-reporting biosensor for the missing piece of its sequence. Circular permutations provide a larger repertoire of potential leave-one-out sites.
Our kinetic analysis of peptide binding to the permuted and truncated GFP may help dissect the GFP folding pathway and the chromophore formation process. For example, the chromophore forms very slowly in the absence of β-strand 7, and significant fluorescence intensity is observed with this strand omitted. Further studies aimed at improving our understanding of the contributions of various parts of the GFP structure to the chromophore formation are ongoing.
Our studies of the peptide concentration dependence on the folding kinetics have led to hypotheses about the structures of the two folding intermediates. Leave-one-out constructs could be a new way to study folding intermediates in proteins.
Supplementary Material
ACKNOWLEDGMENT
Special thanks goes to Dr. Donna Crone for help in designing the circular construct and general molecular biology help. Thanks to Dr. Blanca Barquera for use of equipment and laboratory space and for helpful suggestions. Thanks to Philippa Reeder for helpful suggestions.
Footnotes
This work was supported by Career Award DBI-0448072 from the National Science Foundation.
Abbreviations: GFP, green fluorescent protein; OPT, optimized superfolder GFP, from ref 38; PDB, Protein Data Bank; BCA, bicinchoninic acid assay; NMR, nuclear magnetic resonance; PAGE, polyacrylamide gel electrophoresis; PCR, polymerase chain reaction; UV, ultraviolet light; F0, initial fluorescence intensity; Kd, dissociation constant; OD, optical density; SD, standard deviation; A, amplitude; k, rate constant; t1/2, half-life; λ, wavelength; CHAPS, 3-[(3-cholami-dopropyl)dimethylammonio]-1-propanesulfonate; DTT, dithiothreitol; IPTG, isopropyl d-1-thiogalactopyranoside; LB, Luria-Bertani broth; SDS, sodium dodecyl sulfate.
SUPPORTING INFORMATION AVAILABLE
Oligonucleotide sequences and PCR primers used, diagrams indicating locations of omitted segments in the primary sequence and the three-dimensional structure of superfolder GFP OPT, diagrams showing arrangements of β-strands in primary sequences of GFP variants, examples of in vivo complementation, and examples of s7 concentration-dependent kinetics of in vitro complementation. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Rossi F, Charlton CA, Blau HM. Monitoring protein-protein interactions in intact eukaryotic cells by β-galactosidase complementation. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8405–8410. doi: 10.1073/pnas.94.16.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Remy I, Michnick SW. Clonal selection and in vivo quantitation of protein interactions with protein-fragment complementation assays. Proc. Natl. Acad. Sci. U.S.A. 1999;96:5394–5399. doi: 10.1073/pnas.96.10.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghosh I, Hamilton AD, Regan L. Antiparallel Leucine Zipper-Directed Protein Reassembly: Application to the Green Fluorescent Protein. J. Am. Chem. Soc. 2000;122:5658–5659. [Google Scholar]
- 5.Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozawa T, Nogami S, Sato M, Ohya Y, Umezawa Y. A fluorescent indicator for detecting protein-protein interactions in vivo based on protein splicing. Anal. Chem. 2000;72:5151–5157. doi: 10.1021/ac000617z. [DOI] [PubMed] [Google Scholar]
- 7.Ozawa T, Takeuchi TM, Kaihara A, Sato M, Umezawa Y. Protein splicing-based reconstitution of split green fluorescent protein for monitoring protein-protein interactions in bacteria: Improved sensitivity and reduced screening time. Anal. Chem. 2001;73:5866–5874. doi: 10.1021/ac010717k. [DOI] [PubMed] [Google Scholar]
- 8.Ozawa T, Umezawa Y. Detection of protein-protein interactions in vivo based on protein splicing. Curr. Opin. Chem. Biol. 2001;5:578–583. doi: 10.1016/s1367-5931(00)00244-1. [DOI] [PubMed] [Google Scholar]
- 9.Ozawa T, Umezawa Y. Inteins for split-protein reconstitutions and their applications. In: Belfort M, Wood DW, Stoddard BL, Derbyshire V, editors. Homing Endonucleases and Inteins. Berlin: Springer; 2005. pp. 307–323. [Google Scholar]
- 10.Shimomura O, Johnson FH, Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan. Aequorea. J. Cell. Comp. Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 11.Bokman SH, Ward WW. Renaturation of Aequorea gree-fluorescent protein. Biochem. Biophys. Res. Commun. 1981;101:1372–1380. doi: 10.1016/0006-291x(81)91599-0. [DOI] [PubMed] [Google Scholar]
- 12.Ward WW, Bokman SH. Reversible denaturation of Aequorea green-fluorescent protein: Physical separation and characterization of the renatured protein. Biochemistry. 1982;21:4535–4540. doi: 10.1021/bi00262a003. [DOI] [PubMed] [Google Scholar]
- 13.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 14.Yang F, Moss LG, Phillips GN., Jr The molecular structure of green fluorescent protein. Nat. Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 15.Cody CW, Prasher DC, Westler WM, Prendergast FG, Ward WW. Chemical structure of the hexapeptide chromophore of the Aequorea green-fluorescent protein. Biochemistry. 1993;32:1212–1218. doi: 10.1021/bi00056a003. [DOI] [PubMed] [Google Scholar]
- 16.Palm GJ, Zdanov A, Gaitanaris GA, Stauber R, Pavlakis GN, Wlodawer A. The structural basis for spectral variations in green fluorescent protein. Nat. Struct. Biol. 1997;4:361–365. doi: 10.1038/nsb0597-361. [DOI] [PubMed] [Google Scholar]
- 17.Wachter RM, King BA, Heim R, Kallio K, Tsien RY, Boxer SG, Remington SJ. Crystal structure and photodynamic behavior of the blue emission variant Y66H/Y145F of green fluorescent protein. Biochemistry. 1997;36:9759–9765. doi: 10.1021/bi970563w. [DOI] [PubMed] [Google Scholar]
- 18.Wachter RM, Yarbrough D, Kallio K, Remington SJ. Crystallographic and energetic analysis of binding of selected anions to the yellow variants of green fluorescent protein. J. Mol. Biol. 2000;301:157–171. doi: 10.1006/jmbi.2000.3905. [DOI] [PubMed] [Google Scholar]
- 19.Wall MA, Socolich M, Ranganathan R. The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nat. Struct. Biol. 2000;7:1133–1138. doi: 10.1038/81992. [DOI] [PubMed] [Google Scholar]
- 20.Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 2001;98:462–467. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 22.Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 23.Bastiaens PI, Pepperkok R. Observing proteins in their natural habitat: The living cell. Trends Biochem. Sci. 2000;25:631–637. doi: 10.1016/s0968-0004(00)01714-x. [DOI] [PubMed] [Google Scholar]
- 24.Hanson GT, McAnaney TB, Park ES, Rendell ME, Yarbrough DK, Chu S, Xi L, Boxer SG, Montrose MH, Remington SJ. Green fluorescent protein variants as ratiometric dual emission pH sensors. 1. Structural characterization and preliminary application. Biochemistry. 2002;41:15477–15488. doi: 10.1021/bi026609p. [DOI] [PubMed] [Google Scholar]
- 25.Kneen M, Farinas J, Li Y, Verkman AS. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys. J. 1998;74:1591–1599. doi: 10.1016/S0006-3495(98)77870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 27.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc. Natl. Acad. Sci. U.S.A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goh YY, Frecer V, Ho B, Ding JL. Rational design of green fluorescent protein mutants as biosensor for bacterial endotoxin. Protein Eng. 2002;15:493–502. doi: 10.1093/protein/15.6.493. [DOI] [PubMed] [Google Scholar]
- 30.Goh YY, Ho B, Ding JL. A novel fluorescent protein-based biosensor for Gram-negative bacteria. Appl. Environ. Microbiol. 2002;68:6343–6352. doi: 10.1128/AEM.68.12.6343-6352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richmond TA, Takahashi TT, Shimkhada R, Bernsdorf J. Engineered metal binding sites on green fluorescence protein. Biochem. Biophys. Res. Commun. 2000;268:462–465. doi: 10.1006/bbrc.1999.1244. [DOI] [PubMed] [Google Scholar]
- 32.Fukuda H, Arai M, Kuwajima K. Folding of green fluorescent protein and the cycle3 mutant. Biochemistry. 2000;39:12025–12032. doi: 10.1021/bi000543l. [DOI] [PubMed] [Google Scholar]
- 33.Kimata Y, Iwaki M, Lim CR, Kohno K. A novel mutation which enhances the fluorescence of green fluorescent protein at high temperatures. Biochem. Biophys. Res. Commun. 1997;232:69–73. doi: 10.1006/bbrc.1997.6235. [DOI] [PubMed] [Google Scholar]
- 34.Siemering KR, Golbik R, Sever R, Haseloff J. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr. Biol. 1996;6:1653–1663. doi: 10.1016/s0960-9822(02)70789-6. [DOI] [PubMed] [Google Scholar]
- 35.Battistutta R, Negro A, Zanotti G. Crystal structure and refolding properties of the mutant F99S/M153T/V163A of the green fluorescent protein. Proteins. 2000;41:429–437. doi: 10.1002/1097-0134(20001201)41:4<429::aid-prot10>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 36.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 37.Stemmer WP. DNA shuffling by random fragmentation and reassembly: In vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cabantous S, Terwilliger TC, Waldo GS. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 2005;23:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- 39.Graf R, Schachman HK. Random circular permutation of genes and expressed polypeptide chains: Application of the method to the catalytic chains of aspartate transcarbamoylase. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11591–11596. doi: 10.1073/pnas.93.21.11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwakura M. In search of circular permuted variants of Escherichia coli dihydrofolate reductase. Biosci., Biotechnol., Biochem. 1998;62:778–781. doi: 10.1271/bbb.62.778. [DOI] [PubMed] [Google Scholar]
- 41.Deleted at proof [Google Scholar]
- 42.Deleted at proof [Google Scholar]
- 43.Deleted at proof [Google Scholar]
- 44.Ruiz-Sanz J, de Prat Gay G, Otzen DE, Fersht AR. Protein fragments as models for events in protein folding pathways: Protein engineering analysis of the association of two complementary fragments of the barley chymotrypsin inhibitor 2 (CI-2) Biochemistry. 1995;34:1695–1701. doi: 10.1021/bi00005a026. [DOI] [PubMed] [Google Scholar]
- 45.Seifert MH, Georgescu J, Ksiazek D, Smialowski P, Rehm T, Steipe B, Holak TA. Backbone dynamics of green fluorescent protein and the effect of histidine 148 substitution. Biochemistry. 2003;42:2500–2512. doi: 10.1021/bi026481b. [DOI] [PubMed] [Google Scholar]
- 46.Helms V, Straatsma TP, McCammon JA. Internal dynamics of green fluorescent protein. J. Phys. Chem. B. 1999;103:3263–3269. [Google Scholar]
- 47.Akemann W, Raj CD, Knopfel T. Functional characterization of permuted enhanced green fluorescent proteins comprising varying linker peptides. Photochem. Photobiol. 2001;74:356–363. doi: 10.1562/0031-8655(2001)074<0356:fcopeg>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 48.Hoover DM, Lubkowski J. DNA Works: An automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 50.Tolia NH, Joshua-Tor L. Strategies for protein coexpression in Escherichia coli. Nat. Methods. 2006;3:55–64. doi: 10.1038/nmeth0106-55. [DOI] [PubMed] [Google Scholar]
- 51.Oganesyan N, Kim SH, Kim R. On-column protein refolding for crystallization. J. Struct. Funct. Genomics. 2005;6:177–182. doi: 10.1007/s10969-005-2827-3. [DOI] [PubMed] [Google Scholar]
- 52.Ward WW, Cody CW, Hard RC, Cormier MJ. Spectrophotometric identity of the energy transfer chromophores in Renilla and Aequorea green-fluorescent proteins. Photochem. Photobiol. 1980;31:611–615. [Google Scholar]
- 53.Demidov VV, Dokholyan NV, Witte-Hoffmann C, Chalasani P, Yiu HW, Ding F, Yu Y, Cantor CR, Broude NE. Fast complementation of split fluorescent protein triggered by DNA hybridization. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2052–2056. doi: 10.1073/pnas.0511078103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chattoraj M, King BA, Bublitz GU, Boxer SG. Ultra-fast excited state dynamics in green fluorescent protein: Multiple states and proton transfer. Proc. Natl. Acad. Sci. U.S.A. 1996;93:8362–8367. doi: 10.1073/pnas.93.16.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barondeau DP, Kassmann CJ, Tainer JA, Getzoff ED. Understanding GFP posttranslational chemistry: Structures of designed variants that achieve backbone fragmentation, hydrolysis, and decarboxylation. J. Am. Chem. Soc. 2006;128:4685–4693. doi: 10.1021/ja056635l. [DOI] [PubMed] [Google Scholar]
- 56.Lim WA, Fox RO, Richards FM. Stability and peptide binding affinity of an SH3 domain from the Caenorhabditis elegans signaling protein Sem-5. Protein Sci. 1994;3:1261–1266. doi: 10.1002/pro.5560030812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roche PA, Cresswell P. High-affinity binding of an influenza hemagglutinin-derived peptide to purified HLA-DR. J. Immunol. 1990;144:1849–1856. [PubMed] [Google Scholar]
- 58.Enoki S, Saeki K, Maki K, Kuwajima K. Acid denaturation and refolding of green fluorescent protein. Biochemistry. 2004;43:14238–14248. doi: 10.1021/bi048733+. [DOI] [PubMed] [Google Scholar]
- 59.Liao JC, Roider J, Jay DG. Chromophore-assisted laser inactivation of proteins is mediated by the photogeneration of free radicals. Proc. Natl. Acad. Sci. U.S.A. 1994;91:2659–2663. doi: 10.1073/pnas.91.7.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beck S, Sakurai T, Eustace BK, Beste G, Schier R, Rudert F, Jay DG. Fluorophore-assisted light inactivation: A high-throughput tool for direct target validation of proteins. Proteomics. 2002;2:247–255. doi: 10.1002/1615-9861(200203)2:3<247::aid-prot247>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 61.Jay DG, Sakurai T. Chromophore-assisted laser inactivation (CALI) to elucidate cellular mechanisms of cancer. Biochim. Biophys. Acta. 1999;1424:M39–M48. doi: 10.1016/s0304-419x(99)00022-0. [DOI] [PubMed] [Google Scholar]
- 62.Rajfur Z, Roy P, Otey C, Romer L, Jacobson K. Dissecting the link between stress fibres and focal adhesions by CALI with EGFP fusion proteins. Nat. Cell Biol. 2002;4:286–293. doi: 10.1038/ncb772. [DOI] [PubMed] [Google Scholar]
- 63.Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, Merzlyak EM, Shkrob MA, Lukyanov S, Lukyanov KA. A genetically encoded photosensitizer. Nat. Biotechnol. 2006;24:95–99. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- 64.Reid BG, Flynn GC. Chromophore formation in green fluorescent protein. Biochemistry. 1997;36:6786–6791. doi: 10.1021/bi970281w. [DOI] [PubMed] [Google Scholar]
- 65.Dietz H, Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16192–16197. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jackson SE, Craggs TD, Huang JR. Understanding the folding of GFP using biophysical techniques. Expert Rev. Proteomics. 2006;3:545–559. doi: 10.1586/14789450.3.5.545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.