Abstract
A number of potent broadly neutralizing antibodies against HIV-1 have recently been identified that target epitopes on the viral envelope that contain N-linked glycans. It remains unknown how frequently glycan-dependent neutralizing antibodies generally arise during the course of natural infection or whether particular glycosylation sites are preferentially targeted. We tested sera with a broad range of neutralization activity from individuals infected with HIV-1 clades B or C against panels of HIV-1 Env pseudoviruses that lacked specific glycans in the outer domain glycan cluster (ODGC) or inner domain glycan cluster (IDGC) to determine the presence of glycan-dependent neutralizing antibodies. Overall, 54% of individuals were observed to have neutralizing antibodies targeting these glycan regions. Glycan-specific neutralizing antibodies were readily detected in sera that were selected for having broad, moderate, or weak neutralization potency and breadth. Our results demonstrate that glycan-specific neutralizing antibodies arise with appreciable frequency in individuals chronically infected with HIV-1 clades B and C. Antibody responses that commonly occur during natural infection may be more feasible to induce by vaccination; thus glycan-specific neutralizing antibodies may be desirable responses to elicit with candidate HIV-1 vaccines.
Introduction
The HIV-1 envelope spike is a heterotrimeric glycoprotein composed of gp120 and gp41 subunits that mediates fusion with host cells and serves as the sole target for neutralizing antibodies.1,2 The gp120 surface subunit exhibits an extraordinary degree of glycosylation, the majority of which is N-linked.3,4 These N-linked glycans can act to shield underlying epitopes on the envelope protein backbone from neutralizing antibodies, and can further change conformation in response to antibody-mediated immune pressure to facilitate viral escape.5–7 Over the past several years numerous monoclonal antibodies (mAb) have been isolated from HIV-1-infected individuals that demonstrate broad and potent activity against HIV-1, and a substantial portion of these broadly neutralizing antibodies (bNAbs) targets epitopes that contain N-linked glycans.8–14 It has therefore become increasingly apparent that aside from shielding underlying epitopes, HIV-1 envelope glycans can themselves serve as targets of neutralizing antibodies.
In-depth characterization of recently isolated bNAbs has identified several major epitope targets on HIV-1 Env, some of which include the CD4-binding site (CD4bs), the gp41 membrane proximal external region (MPER), the gp120–gp41 ECTO interface, and glycan-containing epitopes on either the V3 or V1/V2 regions of gp120.15–23 bNAbs targeting the V3 region recognize a high-mannose patch that is centered around the highly conserved N332 glycan. It has recently been demonstrated that these bNAbs can evolve promiscuous glycan recognition that allows binding to proximal glycans in the absence of N332, thus effectively extending their breadth of viral coverage.24
A second class of glycan-dependent bNAbs has been described that targets a conformation-dependent region in V1/V2 that contains the N160 glycan.8–10 Additional glycans have also been found to play a role in epitope formation for bNAbs that target N332 or N160. These include N295, N339, N386, and N392 for mAb 2G1214; N156 for mAbs PG9 and PG16 10; and N301 for mAb PGT128.25 Other antibodies have been recently characterized that target glycans unrelated to the dominant N332-V3 or N160-V1/V2 epitopes. The bNAbs 8ANC195 and 35O22 target novel epitopes spanning both the gp120 and gp41 subunits, and require the glycans at positions N234 and N276, or N88, N241, and N625 for neutralization, respectively.18, 6 HJ16, which recognizes an epitope in the CD4bs region, requires the N276 glycan.27
The recently discovered bNAb PGT151 targets a glycan-dependent epitope that spans both gp120 and gp41.15,17 While the extensive characterization of recently isolated bNAbs has increased our understanding of the key role that glycans can play in forming highly targeted epitopes, these bNAbs were isolated from elite neutralizers, whose sera demonstrated extremely broad and potent neutralizing activity. It remains unclear how frequent glycan-dependent neutralizing antibodies develop in individuals whose sera demonstrate only weak or moderate neutralizing activity. This information may have implications for HIV vaccine design, as neutralizing antibodies that commonly develop in infected individuals may be more readily elicited by vaccination.
A prior study by Lavine et al. characterized the glycan-dependent neutralizing antibodies of nine individuals with chronic HIV-1 infection.28 Those subjects were chosen for investigation as they were elite neutralizers with known broad and potent serum neutralizing antibody activity.29 These individuals were infected with HIV-1 subtypes A or C, and serum samples were tested against glycan-specific deletion mutant viruses derived from the clade B virus YU2. In that cohort, glycans were found to be frequent targets of neutralizing antibodies, with sera from eight of nine subjects containing antibodies with glycan-dependent epitopes. Interestingly, only a few glycosylation sites appeared to be targeted: N332 and N386 in the outer domain glycan cluster (ODGC) of gp120, and N234, N241, and N362 located in the newly described inner domain glycan cluster (IDGC).
Here we sought to expand upon those findings by examining the frequency of neutralizing antibodies targeting ODGC, IDGC, or N160 glycans in a larger cohort of individuals chronically infected with HIV-1 subtypes B or C, with a focus on individuals whose sera demonstrated weak or moderate neutralization potency and breadth. This new cohort displayed a broad range of serum neutralizing activity that may better represent the cross-section of antibody responses found in HIV-1-infected populations.30–32 In addition, we have utilized glycan-deletion mutant viruses generated using both a clade B (YU2) and clade C (ZM135M.PL10a) reference strain to assess whether the clade of virus envelope used in screening assays influenced glycan recognition.
Materials and Methods
Ethics statement
This study utilized deidentified serum samples and was conducted under the approval of the local Institutional Review Boards (IRBs). The following IRBs conducted oversight for their respective sites: Beth Israel Deaconess Medical Center (Boston, MA), Brigham and Women's Hospital (Boston, MA), Duke University Medical Center (Durham, NC), Queen Mary's School of Medicine and Dentistry (United Kingdom), and University of Witwatersrand–Human Research Ethics Committee (Johannesburg, South Africa). The data were analyzed anonymously. Written informed consent was obtained from all subjects.
Serum samples and monoclonal antibodies (mAbs)
The serum samples utilized for this study have been previously described30–32 and were obtained from chronically infected individuals who were antiretroviral drug naive and infected with HIV-1 subtypes B or C. These samples displayed a broad range of neutralization activity when tested against panels of Tier 2 HIV-1 Env pseudoviruses (Supplementary Figs S1 and S2; Supplementary Data are available online at www.liebertpub.com/aid). HIV-1 genetic subtypes were determined by sequencing of a single plasma gp160 gene as described.33 mAbs 2F5 and PG9 were obtained commercially (Polymun Scientific, Vienna, Austria), as was soluble human CD4 protein (sCD4, Progenics, Tarrytown, NY). mAbs 3BNC117, 10-1074, and 8ANC195 were kindly provided by Michel Nussenzweig (Rockefeller University, New York, NY). PGT121 and PGT145 were generously provided by Dennis Burton (The Scripps Research Institute, La Jolla, CA). VRC01 was kindly provided by John Mascola (NIH Vaccine Research Center, Bethesda, MD). HIVIG-B and HIVIG-C were generously provided by David Montefiori (Duke University Medical Center, Durham, NC). mAb 10E8 was produced in the laboratory of Bing Chen (Children's Hospital, Boston, MA).34
Generation of core panel of HIV-1 Env gp160 glycan-deletion mutant viruses
Wild-type (WT) and glycan-deletion mutant YU2 (clade B) and ZM135M.PL10a (clade C) Env gp160 constructs were generated as previously described.28 The YU2 glycan-deletion mutants N234S, N241S, N362K, and N160K were those generated previously, whereas the YU2 N332K and N386T mutants and the panel of ZM135M.PL10a glycan mutants were newly created for this study. Glycan-deletion mutants were created by following the PCR-based QuikChange protocol (Stratagene) using the parental YU2 or ZM135M.PL10a Env, expressed in pcDNA3.1, as the template. WT YU2 gp160 plasmid was a kind gift of Dana Gabuzda (Dana Farber Cancer Institute, Boston, MA), and WT ZM135M.PL10a gp160 plasmid was obtained from the NIH AIDS Research and Reference Reagent Program as donated by C.A. Derdeyn and E. Hunter. Glycan-deletion mutations were designed to replace the asparagine residue in the NXS/T glycosylation motif with either a serine, lysine, or threonine residue as are found in some strains of HIV-1. The integrity of each of the constructs was confirmed by DNA sequencing of the entire env reading frame. The mutants were named via the following formula: the wild-type amino acid residue in single-letter code, the residue number, and the substituted amino acid. Residue numbering is based on that of HXBc2 gp160, according to current conventions.35
Production and titration of glycan-deletion mutant pseudoviruses
To prepare pseudovirus stocks, 293T/17 cells were cotransfected with optimized concentrations of the WT or glycan-deletion mutant env plasmid and SG3ΔEnv backbone plasmid using FuGENE 6 transfection reagent (Promega, Madison, WI) via the manufacturer's instructions. 293T/17 cells were seeded into T75 flasks and allowed to reach 50–80% confluency overnight. The following day, backbone and Env plasmid DNA were combined with the transfection reagent and incubated at room temperature for 30 min to allow for the formation of complexes. The transfection mixture was then added to the flasks and incubated for 6–8 h at 37°C, 5% CO2. After incubation, the medium containing transfection mixture was removed and the cells overlaid with fresh growth media. Twenty-four hours later, cell culture supernatants containing the pseudoviruses were harvested, aliquoted, and stored at −80°C until use.
Titration of pseudovirus stocks to determine the 50% tissue culture infectious dose per ml (TCID50/ml) and dilution for use in neutralization assays were performed using TZM.bl cells as previously described.33 Briefly, a 5-fold serial dilution of virus stock was performed in quadruplicate wells and incubated with TZM.bl cells in growth media containing DEAE-dextran (11 μg/ml final concentration). After 48 h, the cells were measured for luciferase reporter gene expression, indicating the ability of the WT and glycan-deletion mutant pseudoviruses to infect cells. TCID50/ml was calculated using an Excel macro made available on the Las Alamos National Laboratories website (www.hiv.lanl.gov). The dilution of virus stock for use in neutralization assays was based on a read-out of approximately 150,000 relative luminescence unit (RLU) equivalents in TZM.bl cells.
Neutralization assay for phenotyping and detection of glycan-dependent neutralizing antibodies
Neutralizing antibody titers against WT and glycan-deletion mutant pseudoviruses were determined using a luciferase-based TZM.bl cell assay as previously described.36,37 This assay measures a decrease in luciferase reporter gene expression following single-round viral infection of TZM.bl cells. Briefly, 3-fold serial dilutions of mAb reagents or heat-inactivated patient serum samples (56o C, 1 h) were performed in duplicate and incubated with either the WT or glycan-deletion mutant pseudoviruses for 1 h at 37°C. TZM.bl cells were then added in growth media containing DEAE-dextran at a final concentration of 11 μg/ml, and assay plates incubated for 48 h at 37°C, 5% CO2. Luciferase reporter gene expression was measured using Bright-Glo luciferase reagent (Promega) and a Victor 3 luminometer (Perkin Elmer). Neutralization titers (50% inhibitory dose, ID50) were calculated as the serum dilution at which RLU were reduced by 50% compared to RLU in virus control wells after subtraction of background RLU in cell control wells. All assays were performed in a laboratory meeting GCLP standards. For the detection of glycan-dependent neutralizing antibodies against the YU2 and ZM135M.PL10a glycan-deletion mutant panel, a sample was considered to have glycan-dependent neutralizing antibodies if the ID50 titer was at least 2.5-fold lower than that against WT virus.
Results
Generation and phenotyping of YU2 and ZM135M.PL10a glycan-deletion mutant viruses
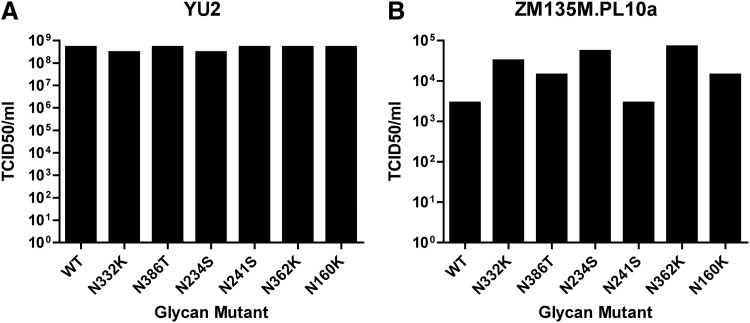
The clade B isolate YU2 was utilized to generate N332K, N386T, N234S, N241S, N362K, and N160K glycan-deletion mutant envelope plasmids. All YU2 glycan-deletion mutant Env plasmids yielded infectious pseudovirus stocks that exhibited equivalent TCID50/ml compared to WT YU2 (Fig. 1A). To study the effect of virus envelope clade on the identification of glycan-dependent NAb, we also created comparable mutants using the clade C virus envelope clone ZM135M.PL10a. This virus belongs to the standard reference panel of clade C Tier 2 isolates,38 and was found to have a similar number of N-linked glycosylation sites as YU2. Using N-GlycoSite (www.hiv.lanl.gov/content/sequence/GLYCOSITE/glycosite.html)39 we determined the N-linked glycosylation motifs that were analogous to the ODGC, IDGC, and N160 glycans identified for YU2. Site-directed mutations were introduced at these sites to remove the glycosylation motif. Pseudoviruses produced with these mutant Envs were found to have infectious titers that were at least equivalent to that of WT ZM135M.PL10a (Fig. 1B).
FIG. 1.
Infectivity of YU2 (clade B) and ZM135M.PL10a (clade C) glycan-deletion pseudoviruses. A panel of glycan-deletion mutant viruses was created in either YU2 (A) or ZM135M.PL10a (B) gp160. Pseudovirus stocks were generated by transfection of 293T cells, and tested for infectivity in a TZM.bl cell-based assay. TZM.bl cells were infected with serial dilutions of each glycan mutant or parental wild-type (WT) virus, and TCID50/ml was calculated.
To ensure that removal of glycans at these positions within the viral envelope did not significantly alter the neutralization phenotype of the virus as compared to WT, viruses were tested in TZM.bl neutralization assays using a panel of neutralizing antibodies against HIV-1 (Table 1). bNAbs were chosen that target known neutralization epitopes on the HIV-1 envelope, including the CD4bs (3BNC117, VRC01, and sCD4), MPER (2F5 and 10E8), glycan-containing epitopes on the V3 loop (PGT121 and 10-1074), the V1/V2 region (PG9 and PGT145), and the glycan at N234 (8ANC195). In addition, we tested the WT and glycan-deletion mutant viruses against a heterologous pool of polyclonal HIV immunoglobulins of patients infected with either clade B (HIVIG-B) or clade C (HIVIG-C) subtypes. Overall, glycan-deletion mutant viruses exhibited similar neutralization sensitivities as WT viruses to mAbs directed against CD4bs and MPER epitopes, with the exception of the ZM-N234S mutant that exhibited a decrease in sensitivity to the CD4bs mAbs 3BNC117 and VRC01, but not sCD4.
Table 1.
Glycan-Deletion Mutant Virus Phenotyping
| IC50 (μg/ml) in TZM.bl cells | |||||||
|---|---|---|---|---|---|---|---|
| YU2-WT | YU2-N332K | YU2-N386T | YU2-N234S | YU2-N241S | YU2-N362K | YU2-160K | |
| sCD4 | 0.117 | 0.047 | 0.148 | 0.203 | 0.117 | 0.180 | 0.075 |
| 3BNC117 | 0.009 | 0.011 | 0.006 | 0.010 | 0.007 | 0.011 | 0.009 |
| VRC01 | 0.125 | 0.087 | 0.042 | 0.080 | 0.192 | 0.192 | 0.177 |
| PGT121 | 0.011 | 0.311 | 0.012 | 0.012 | 0.008 | 0.012 | 0.007 |
| 10-1074 | 0.138 | >25 | 0.201 | 0.141 | 0.134 | 0.096 | 0.079 |
| PGT145 | 0.027 | <0.023 | <0.023 | 0.041 | 0.029 | 0.033 | >50 |
| PG9 | 20.102 | 15.780 | 16.721 | 22.154 | 6.580 | 9.363 | >25 |
| 8ANC195 | 0.284 | 0.321 | 0.287 | >25 | 0.426 | 0.488 | 0.240 |
| 2F5 | 13.632 | 8.716 | 14.385 | 23.579 | 19.275 | 11.430 | 9.728 |
| 10E8 | 0.731 | 0.571 | 0.750 | 0.817 | 1.003 | 0.986 | 0.724 |
| HIVIG-B | 350.960 | 210.589 | 306.248 | 332.909 | 586.594 | 518.778 | 356.832 |
| HIVIG-C | 69.004 | 43.379 | 58.346 | 95.874 | 101.952 | 97.471 | 55.038 |
| ZM135M.PL10a-WT | ZM-N332K | ZM-N386T | ZM-N234S | ZM-N241S | ZM-N362K | ZM-N160K | |
|---|---|---|---|---|---|---|---|
| sCD4 | 12.370 | 8.830 | 21.660 | >25 | >25 | 13.900 | 15.470 |
| 3BNC117 | 0.034 | 0.062 | 0.045 | 0.097 | 0.059 | 0.023 | 0.053 |
| VRC01 | 2.855 | 3.992 | 1.984 | 19.551 | 1.638 | 0.787 | 2.510 |
| PGT121 | 0.186 | >25 | 0.221 | 0.222 | 0.288 | 0.221 | 0.165 |
| 10-1074 | 0.127 | >25 | 0.210 | 0.151 | 0.146 | 0.112 | 0.122 |
| PGT145 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| PG9 | >25 | >25 | 12.770 | >25 | >25 | >25 | >25 |
| 8ANC195 | >25 | >25 | >25 | >25 | >25 | >25 | >25 |
| 2F5 | >25 | >25 | >25 | >25 | >25 | >25 | >25 |
| 10E8 | 0.580 | 1.270 | 1.340 | 0.960 | 0.730 | 1.160 | 1.070 |
| HIVIG-B | 936.439 | 662.977 | 1468.623 | 719.841 | 1157.868 | 1060.778 | 1096.948 |
| HIVIG-C | 304.207 | 391.521 | 355.891 | 237.719 | 514.155 | 308.350 | 288.143 |
YU2 and ZM135M.PL10a glycan-deletion mutant viruses were tested against a panel of bNAbs, HIV immunoglobulin from individuals infected with clades B or C (HIVIG-B and HIVIG-C), and soluble CD4 (sCD4) to determine if neutralization sensitivity was altered compared to parental WT virus. IC50 titers demonstrating resistance have been bolded.
As previously reported, the removal of the glycan at N332 in the YU2 envelope abolished recognition by mAb 10-1074, and significantly increased the IC50 titer of PGT121 to >25-fold that of WT.9,12 The fact that PGT121 still exhibited a degree of neutralizing activity against the YU2-N332K mutant may be due to promiscuous binding to alternative glycans as previously described.24 Similarly, removal of the N332 glycan in the ZM135M.PL10a envelope eliminated neutralizing activity for mAbs PGT121 and 10-1074. While mAb 8ANC195 did not demonstrate neutralizing activity against WT ZM135M.PL10a, removal of the glycan at N234 in the YU2 envelope eliminated recognition as previously reported.40,41 Removal of the glycan at position 160 in YU2 completely abrogated recognition by mAb PGT145 as would be predicted,9 whereas ZM135M.PL10a WT and glycan-deletion mutant viruses were naturally resistant to this mAb. From these data, we conclude that removal of N-linked glycosylation sites at the indicated amino acids did not change the global neutralization sensitivity of mutant viruses compared to the WT parental virus, but did eliminate recognition by certain glycan-dependent bNAbs as would be predicted based on epitope specificities.
Mapping of glycan-specific antibodies using glycan-deletion mutant viruses
In the previous study by Lavine et al., sera from a cohort of elite neutralizers chronically infected with clade A or C viruses were found to target glycans located in the ODGC (N332, N386) and/or IDGC (N234, N241, N362) of clade B virus YU2.28 We sought to further investigate whether separate cohorts of clade C-infected or clade B-infected individuals with a broader range of neutralizing activity would similarly target ODGC, IDGC, and/or the frequent bNAb target N160 glycans.8–10 We selected serum samples from previous studies that had been extensively screened for neutralizing activity against multiclade panels of Tier 2 HIV-1 Env pseudoviruses. As the potency and breadth of neutralizing activity were found to be highly correlated, we categorized samples based on those that neutralized >80% (broad), 40–80% (moderate), or <40% (weak) of viruses in their respective panels (Supplementary Figs S1 and S2).
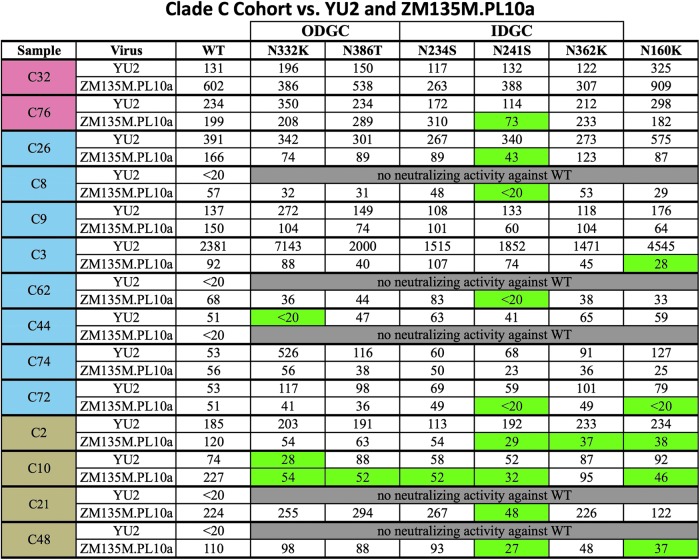
Sera from 35 patients chronically infected with HIV-1 clade C were tested against YU2 and ZM135M.PL10a WT Env pseudoviruses. Of these samples, 14 were able to neutralize either YU2 and/or ZM135M.PL10a WT and were thus subsequently tested against the glycan-deletion mutant panels (Fig. 2). Ten of the clade C serum samples had measurable neutralizing activity against YU2 WT, and the majority of these neutralized the panel of YU2 glycan-deletion mutant viruses with similar ID50 titers. However, samples C44 and C10 exhibited decreased neutralization against the glycan-deletion mutant virus N332K. These data suggest that these individuals had NAbs that targeted epitopes requiring the N332 glycan. When the clade C serum samples were tested against the clade-matched ZM135M.PL10a glycan-deletion mutants, 10 samples in the cohort were determined to have glycan-dependent NAb. Of these, nine were found to have NAbs that targeted glycans located within the IDGC, with the glycan at N241 being the predominant target. Sample C3 recognized the glycan at position N160, but no glycans within the ODGC or IDGC. Within this group of patients, only sample C10 was found to recognize glycans in both the YU2 panel and ZM135M.PL10a panel.
FIG. 2.
Screening of clade C serum samples against YU2 and ZM135M.PL10a WT and panel of glycan-deletion mutant viruses. ID50 titers were calculated as the reciprocal dilution that neutralized 50% of input virus. Samples are ordered based on neutralization breadth, with samples demonstrating >80% neutralization breadth highlighted in red, 40–80% neutralization breadth highlighted in blue, and <40% neutralization breadth highlighted in brown. Glycan-dependent neutralizing antibodies were deemed present if the calculated ID50 titer against a particular glycan-deletion mutant was≥2.5-fold lower than the titer calculated for that same sample against WT virus. Samples containing glycan-dependent neutralizing antibodies against a particular glycan-deletion mutant are highlighted in green. ODGC, outer domain glycan cluster; IDGC, inner domain glycan cluster.
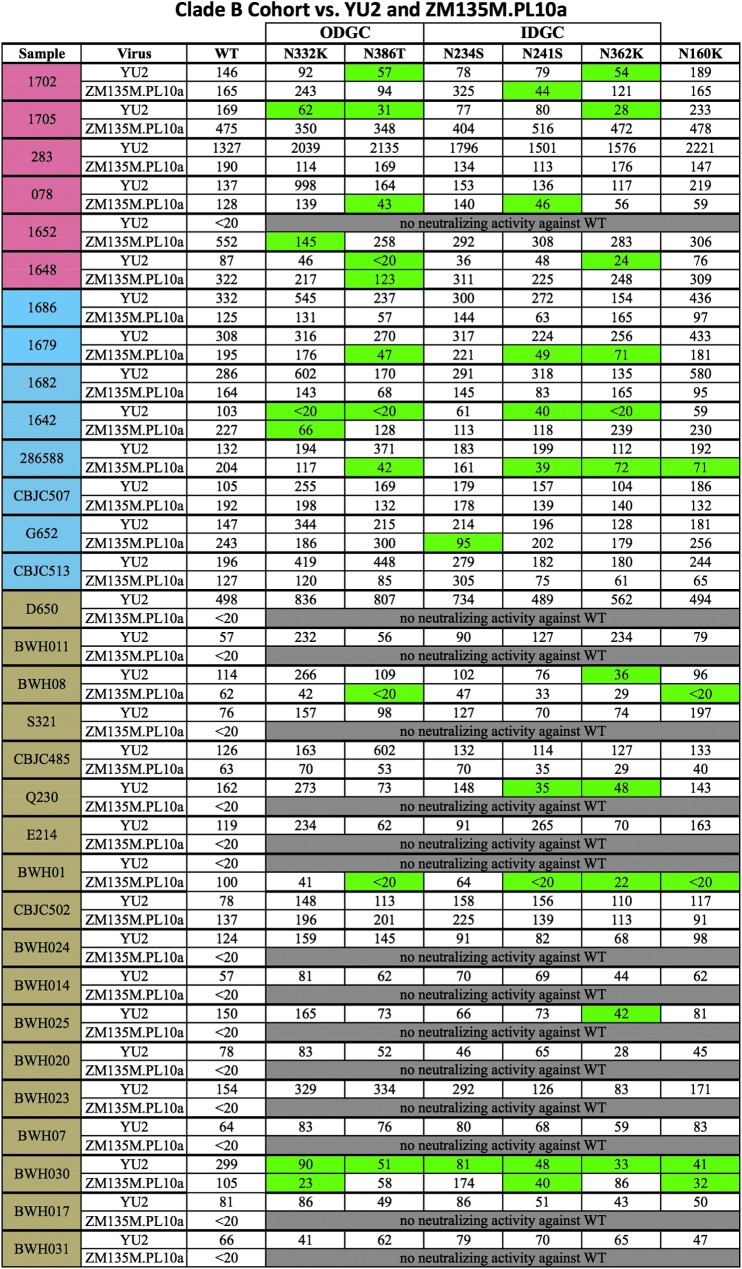
To determine whether sera from individuals infected with HIV-1 clade B viruses would recognize the glycans in the ODGC and IDGC, we tested samples from 39 patients chronically infected with HIV-1 clade B against YU2 and ZM135M.PL10a WT. Of these samples, 32 were able to neutralize either or both WT isolates and were thus subsequently tested against the panels of glycan-deletion mutant viruses (Fig. 3). When screened against the clade-matched YU2 glycan-deletion mutant panel, eight patient samples were found to have glycan-dependent neutralizing antibodies. Five patients exhibited decreased neutralization against both IDGC and ODGC glycans, and three of eight were found to target glycans located in the IDGC only. Sample BWH030 exhibited decreased neutralization against all glycan mutant viruses tested, suggesting a broad glycan-dependent NAb response targeting multiple ODGC, IDGC, and N160 glycan epitopes.
FIG. 3.
Screening of clade B serum samples against YU2 and ZM135M.PL10a WT and panel of glycan-deletion mutant viruses. ID50 titers were calculated as the reciprocal dilution that neutralized 50% of input virus. Samples are ordered based on neutralization breadth, with samples demonstrating >80% neutralization breadth highlighted in red, 40–80% neutralization breadth highlighted in blue, and <40% neutralization breadth highlighted in brown. Glycan-dependent neutralizing antibodies were deemed present if the calculated ID50 titer against a particular glycan-deletion mutant was≥2.5-fold lower than the titer calculated for that same sample against WT virus. Samples containing glycan-dependent neutralizing antibodies against a particular glycan-deletion mutant are highlighted in green. ODGC, outer domain glycan cluster; IDGC, inner domain glycan cluster.
When screened against the clade C virus ZM135M.PL10a mutant panel, 11 clade B samples were found to have glycan-dependent NAbs: five of the samples recognized glycans located in both the ODGC and the IDGC, two of the samples recognized glycans in the IDGC only, and four samples recognized glycans located within the ODGC only. Four samples were found to also contain glycan-dependent NAbs that targeted the glycan located at position N160. Of the 32 samples in the clade B patient group, five were found to cross-recognize glycans in both the YU2 and ZM135M.PL10a glycan-deletion mutant panels (samples 1702, 1648, 1642, BWH08, BWH030).
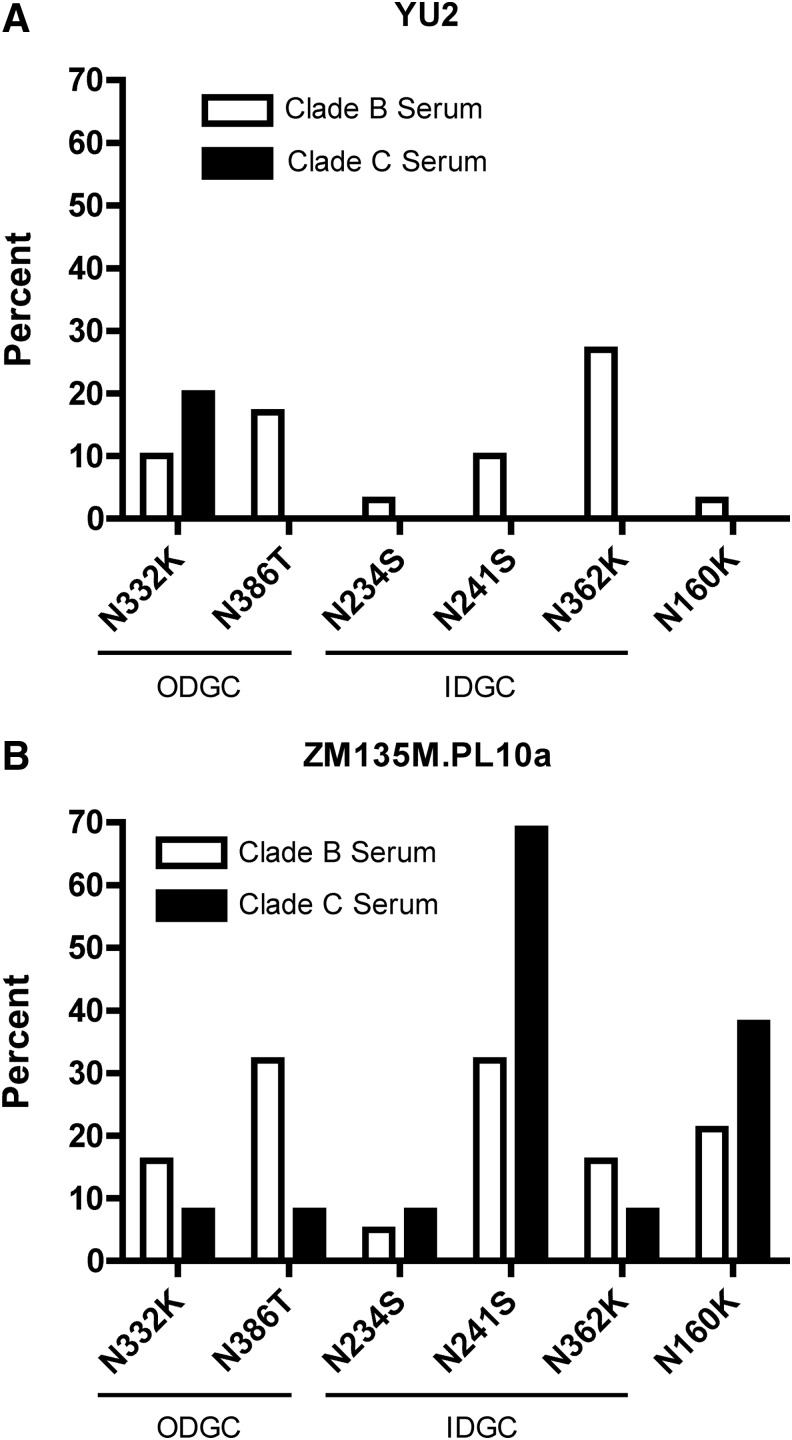
For samples in the clade B and clade C cohorts capable of neutralizing WT virus, the percentage that targeted each glycan in the YU2 and ZM135M.PL10a glycan-deletion mutant panels was calculated. For clade B serum samples screened using the clade-matched YU2 glycan-deletion mutant panel (Fig. 4A), every mutant screened was targeted by at least one serum sample, with the IDGC glycan N362 being the most frequent target of glycan-dependent NAbs (27%). Sera from clade C-infected individuals were found to recognize only the ODGC glycan N332 when screened with this same panel of non-clade-matched viruses. In contrast, when screening the cohort of clade C-infected individuals against the clade-matched ZM135M.PL10a glycan-deletion mutant virus panel (Fig. 4B), the glycan located at position N241 within the IDGC was the most frequently targeted (69%). Importantly, every glycan-deletion mutant virus in the ZM135M.PL10a panel was targeted by at least one serum sample within the clade C patient cohort.
FIG. 4.
Percentage of clade B and clade C serum samples demonstrating decreased neutralizing activity against glycan-deletion mutant viruses. Clade B and clade C serum samples able to neutralize WT viruses were tested against each of the six matched glycan-deletion mutants for both YU2 (A) and ZM135M.PL10a (B). Data are presented as the percentage of samples from each cohort that exhibited a decrease in ID50 titer against each glycan-deletion mutant virus compared to parental WT virus. ODGC, outer domain glycan cluster; IDGC, inner domain glycan cluster.
The most frequent targets of glycan-dependent NAbs found in the clade B cohort were the glycans located at positions N386 (ODGC) and N241 (IDGC). In addition, when screening with ZM135M.PL10a, both clade-matched and non-clade-matched sera were found to target the glycan at position N160 (38% for clade C sera, 21% for clade B sera). This is in contrast to the observation that only a single clade-matched HIV-1 clade B-infected individual contained NAbs to N160 on the YU2 envelope. For both YU2 and ZM135M.PL10a, more glycans were targeted when clade-matched sera were tested. However, greater numbers of glycans were targeted overall in ZM135M.PL10a, independent of sera clade.
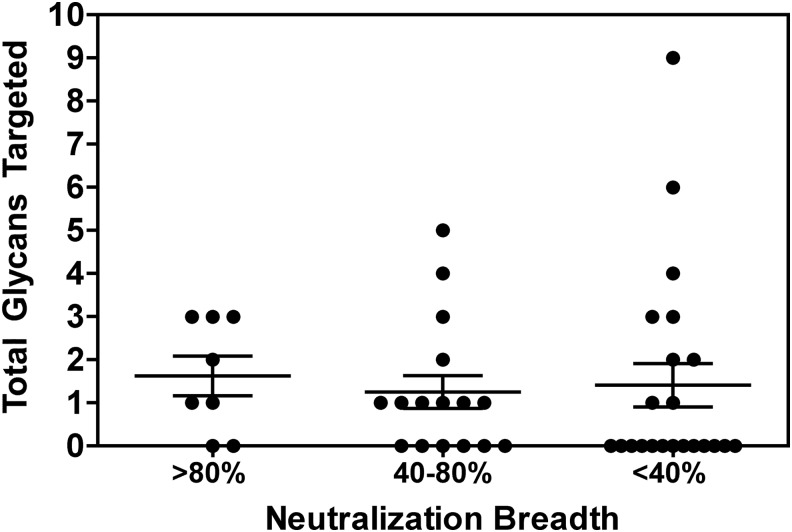
When patient samples were categorized based on overall potency and breadth (Supplementary Figs S1 and S2), we observed that the frequency of detecting glycan-dependent antibodies was 75%, 63%, and 41% for patients exhibiting >80%, 40–80%, or <40% neutralization breadth, respectively. Furthermore, no significant difference was observed when comparing the total number of glycans targeted by neutralizing antibodies in these patient populations (p=0.46, Fig. 5). From these data we conclude that glycan-dependent neutralizing antibodies are readily detected in clade B and C chronically infected subjects that exhibit a wide spectrum of serum neutralizing antibody activity.
FIG. 5.
Glycan targeting by clade B and C serum samples based on sample neutralization breadth. Patient samples are categorized based on neutralization breadth as indicated, and the total number of glycans targeted on both YU2 and ZM135M.PL10a viruses is indicated. Bars indicate the mean number of glycans targeted for each group±SEM.
Discussion
In this study we have assessed the frequency of neutralizing antibody responses targeting specific glycans in the ODGC and IDGC of HIV-1 gp120 in a cohort of individuals chronically infected with HIV-1 subtypes B and C. This study was initiated based on a prior report that identified the glycans N332 and N386 (located within the ODGC) and/or N234, N241, and N362 (located within the newly identified IDGC) as frequent targets of neutralizing antibodies in a cohort of nine individuals selected for having broad and potent serum neutralizing activity.28 Here, we have expanded upon those findings by demonstrating that neutralizing antibodies against ODGC and IDGC glycans, as well as the frequently targeted N160 glycan, are also detected in a larger cohort of HIV-1-infected individuals who exhibit a broad spectrum of serum neutralizing activity. Overall, the frequency of patients in this cohort targeting one or more of these specific glycans in either YU2 or ZM135M.PL10a was 54% (44% in clade B-infected individuals and 79% in clade C-infected individuals).
From these data, we conclude that glycan-specific neutralizing antibodies appear to be readily generated even in chronically infected individuals whose sera demonstrate only low or moderate neutralizing activity, and are not rare responses found in only a subpopulation of individuals exhibiting elite serum neutralizing activity.
We have further investigated the impact of using viral isolates either matched or nonmatched in clade with the patient serum sample to screen for glycan-dependent neutralizing antibodies. HIV-1 clades are known to differ in their glycosylation patterns42; therefore, neutralizing antibodies that develop in vivo against glycan-dependent epitopes on the infecting virus may be more likely to cross-react against similar glycans on a clade-matched virus. Our results demonstrate that clade-matched sera and virus pairings did result in a greater frequency of detecting glycan-specific neutralizing antibodies than clade nonmatched pairings. However, the results were somewhat conflicting, as both clade B and C serum samples targeted more glycans overall when screened against the clade C ZM135M.PL10a virus. It remains unclear why glycan-deletion mutations made in the ZM135M.PL10a envelope were targeted more frequently by the samples in our cohort, as this virus has not previously been studied in a similar setting.
One finding that particularly stood out in this study was that glycans identified as targets of neutralizing antibodies for particular samples often varied depending on the viral envelope that was used for screening. A similar finding was observed in an epitope mapping study examining reactivity29 against the N332 glycan-epitope in the cohort of elite neutralizers. In that report, which employed a multiclade panel of four N332A mutant pseudoviruses, detection of glycan-dependent neutralizing activity from individual serum samples varied depending on the mutant strain tested. Unfortunately due to limited serum sample availability, in the current study we were able to assess glycan-specific neutralizing antibodies only against a single isolate from either clade B or clade C. The specific features of viral envelopes that influence the targeting of N-linked glycan epitopes by neutralizing antibodies have yet to be determined; thus future studies would benefit from utilizing a larger number of cloned virus glycan-mutant panels representing multiple clades.
The recent discovery and characterization of bNAbs isolated from elite neutralizers have defined epitopes on HIV-1 Env that are considered optimal targets for antibodies that may be elicited by vaccination. Multiple bNAbs have been isolated from various individuals that target epitopes containing glycans at position N160 or N332.8–10,12,14,25,43,44 These may be of particular importance, as some studies have suggested that antibodies targeting these epitopes may arise more frequently than bNAbs targeting other major neutralization epitopes on HIV-1 Env, such as the CD4bs or MPER.22,45,46 In our cohort, the number of samples with <80% neutralization breadth that targeted the N160 glycan on YU2 or ZM135MPL10a was 9/38 (24%), and the number of samples targeting the N332 glycan was 4/38 (11%). These data suggest that antibodies against these glycans arise with some frequency even in individuals whose sera demonstrate moderate or low neutralizing activity. How these responses may compare to the frequency of neutralizing antibodies targeting other epitopes, however, is not currently known.
The recently isolated bNAb 35O22 has been found to bind to a novel epitope spanning gp120 and gp41 that includes the N241 glycan.18 Sixty-nine percent of individuals in our cohort infected with clade C viruses that were found to neutralize WT virus harbored antibodies that targeted the N241 glycan on ZM135M.PL10a, which may indicate that this is a prevalent epitope of neutralizing antibodies in clade C chronically infected individuals. Interestingly, of the glycans found to be targets for neutralizing antibodies in the original report by Lavine et al., only ODGC glycans at positions N332 and N386 had been identified at that time to be epitopes for bNAbs.9,14 More recently, glycans at positions N234 and N241 of the IDGC have also been found to be included in the epitopes of newly identified bNAbs.18,26,41 Importantly, of the five specific ODGC and IDGC glycans studied here, all except N362 appear to be highly conserved and are predicted to be glycosylated in greater than 75% of sequences for multiple major subtypes and CRFs.42 These data further support the rationale that glycan epitopes in both the ODGC and IDGC may be desirable targets for vaccine-elicited antibodies.
Our infectivity and phenotypic neutralization data suggest that the YU2 and ZM53M.PL10a glycan-deletion mutant viruses we constructed are similar to the parental WT viruses. However, it remains possible that removal of a particular glycan could induce secondary structural changes in Env that could affect global sensitivity to neutralizing antibodies, and thus influence our estimated frequencies of glycan-dependent neutralization. The glycan deletion mutant ZM-N234S did demonstrate increased resistance to CD4bs antibodies, and it is therefore possible that sera with increased resistance to this particular mutant may have harbored antibodies directed against the CD4bs rather than the N234 glycan. A recent study has demonstrated that bNAbs targeting the N332 glycan can alternately bind other proximal N-linked glycans to neutralize virus in the absence of a glycan at the N332 position.24 The implications of this finding are not clear for polyclonal sera, but considering that the glycan-deletion mutant viruses used in our study only had single deletions, it is possible that our results underestimate the frequency at which the N332 glycan and possibly other glycans are targeted. It should also be noted that the glycan-deletion mutant pseudoviruses utilized in our screening assays were produced via transfection of 293T cells, and thus may express different N-glycan patterns than viruses replicating in vivo.47,48 Further investigation into the contribution of host cell variability on the sensitivity of viruses to glycan-specific neutralizing antibodies is warranted.
The results presented here have implications for future studies that may employ glycan-deletion mutant viruses to evaluate glycan-dependent neutralization epitopes from polyclonal sera. Because different viral envelopes may yield disparate results, the use of at least two viruses, preferably from different clades, is advised. This study also has implications for HIV-1 vaccine design. The induction of bNAbs through traditional vaccination strategies may be extremely difficult, and neutralizing antibodies that arise more frequently during the course of natural infection may be easier to induce with vaccines.49 Our results suggest that candidate vaccine immunogens capable of eliciting neutralizing antibodies targeting ODGC, IDGC, and N160 glycan epitopes may be desirable.
Supplementary Material
Acknowledgments
This work was supported by the Harvard Multidisciplinary AIDS Training Grant (NIH Grant T32AI007387). The authors thank David Montefiori and the CAVD Comprehensive Antibody-Vaccine Immune Monitoring Consortium, Lynn Morris, Michel Nussenzweig, John Mascola, and Dennis Burton for generously providing reagents used in these studies. We thank Raphael Dolin for advice and insightful discussions.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Kwong PD. and Mascola JR: Human antibodies that neutralize HIV-1: Identification, structures, and B cell ontogenies. Immunity 2012;37(3):412–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyatt R. and Sodroski J: The HIV-1 envelope glycoproteins: Fusogens, antigens, and immunogens. Science 1998;280(5371):1884–1888 [DOI] [PubMed] [Google Scholar]
- 3.Huang X, Barchi JJ, Jr, Lung FD, et al. : Glycosylation affects both the three-dimensional structure and antibody binding properties of the HIV-1IIIB GP120 peptide RP135. Biochemistry 1997;36(36):10846–10856 [DOI] [PubMed] [Google Scholar]
- 4.Myers G. and Lenroot R: HIV glycosylation: What does it portend? AIDS Res Hum Retroviruses 1992;8(8):1459–1460 [DOI] [PubMed] [Google Scholar]
- 5.Wei X, Decker JM, Wang S, et al. : Antibody neutralization and escape by HIV-1. Nature 2003;422(6929):307–312 [DOI] [PubMed] [Google Scholar]
- 6.Moore PL, Gray ES, Wibmer CK, et al. : Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med 2012;18(11):1688–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wibmer CK, Bhiman JN, Gray ES, et al. : Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS Pathog 2013;9(10):e1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonsignori M, Hwang KK, Chen X, et al. : Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol 2011;85(19):9998–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker LM, Huber M, Doores KJ, et al. : Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 2011;477(7365):466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker LM, Phogat SK, Chan-Hui PY, et al. : Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 2009;326(5950):285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLellan JS, Pancera M, Carrico C, et al. : Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 2011;480(7377):336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mouquet H, Scharf L, Euler Z, et al. : Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci USA 2012;109(47):E3268–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pancera M, Yang Y, Louder MK, et al. : N332-Directed broadly neutralizing antibodies use diverse modes of HIV-1 recognition: Inferences from heavy-light chain complementation of function. PLoS One 2013;8(2):e55701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scanlan CN, Pantophlet R, Wormald MR, et al. : The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1→2 mannose residues on the outer face of gp120. J Virol 2002;76(14):7306–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blattner C, Lee JH, Sliepen K, et al. : Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity 2014;40(5):669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derking R, Ozorowski G, Sliepen K, et al. : Comprehensive antigenic map of a cleaved soluble HIV-1 envelope trimer. PLoS Pathog 2015;11(3):e1004767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falkowska E, Le KM, Ramos A, et al. : Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 2014;40(5):657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Kang BH, Pancera M, et al. : Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 2014;515(7525):138–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Ofek G, Laub L, et al. : Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 2012;491(7424):406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheid JF, Mouquet H, Feldhahn N, et al. : Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 2009;458(7238):636–640 [DOI] [PubMed] [Google Scholar]
- 21.Scheid JF, Mouquet H, Ueberheide B, et al. : Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 2011;333(6049):1633–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker LM, Simek MD, Priddy F, et al. : A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog 2010;6(8):e1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West AP, Jr, Scharf L, Scheid JF, et al. : Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell 2014;156(4):633–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sok D, Doores KJ, Briney B, et al. : Promiscuous glycan site recognition by antibodies to the high-mannose patch of gp120 broadens neutralization of HIV. Sci Transl Med 2014;6(236):236ra263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pejchal R, Doores KJ, Walker LM, et al. : A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 2011;334(6059):1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scharf L, Scheid JF, Lee JH, et al. : Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Cell Rep 2014;7(3):785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balla-Jhagjhoorsingh SS, Corti D, Heyndrickx L, et al. : The N276 glycosylation site is required for HIV-1 neutralization by the CD4 binding site specific HJ16 monoclonal antibody. PLoS One 2013;8(7):e68863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavine CL, Lao S, Montefiori DC, et al. : High-mannose glycan-dependent epitopes are frequently targeted in broad neutralizing antibody responses during human immunodeficiency virus type 1 infection. J Virol 2012;86(4):2153–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomaras GD, Binley JM, Gray ES, et al. : Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J Virol 2011;85(21):11502–11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Binley JM, Lybarger EA, Crooks ET, et al. : Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol 2008;82(23):11651–11668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.deCamp A, Hraber P, Bailer RT, et al. : Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 2014;88(5):2489–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hraber P, Korber BT, Lapedes AS, et al. : Impact of clade, geography, and age of the epidemic on HIV-1 neutralization by antibodies. J Virol 2014;88(21):12623–12643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li M, Gao F, Mascola JR, et al. : Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 2005;79(16):10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Frey G, Peng H, et al. : Mechanism of HIV-1 neutralization by antibodies targeting a membrane-proximal region of gp41. J Virol 2014;88(2):1249–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korber BFB, Kuiken CL, Pillai SK, and Sodroski J: Numbering positions in HIV relative to HXB2. In: A Compilation and Analysis of Nucleic Acid and Amino Acid Sequences (Korber B KC, Foley B, Hahn B, et al., eds.). Los Alamos, NM, Los Alamos National Laboratory, 1998, pp. 102–111 [Google Scholar]
- 36.Montefiori DC: Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol 2005;Chapter 12:Unit 12 11 [DOI] [PubMed] [Google Scholar]
- 37.Sarzotti-Kelsoe M, Bailer RT, Turk E, et al. : Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods 2014;409:131–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li M, Salazar-Gonzalez JF, Derdeyn CA, et al. : Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol 2006;80(23):11776–11790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang M, Gaschen B, Blay W, et al. : Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 2004;14(12):1229–1246 [DOI] [PubMed] [Google Scholar]
- 40.Chuang GY, Acharya P, Schmidt SD, et al. : Residue-level prediction of HIV-1 antibody epitopes based on neutralization of diverse viral strains. J Virol 2013;87(18):10047–10058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.West AP, Jr, Scharf L, Horwitz J, et al. : Computational analysis of anti-HIV-1 antibody neutralization panel data to identify potential functional epitope residues. Proc Natl Acad Sci USA 2013;110(26):10598–10603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Travers SA: Conservation, compensation, and evolution of N-linked glycans in the HIV-1 group M subtypes and circulating recombinant forms. AIDS 2012;2012:823605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong L, Lee JH, Doores KJ, et al. : Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol 2013;20(7):796–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trkola A, Purtscher M, Muster T, et al. : Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol 1996;70(2):1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gray ES, Madiga MC, Hermanus T, et al. : The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol 2011;85(10):4828–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song H, Chu Y, Zhang H, et al. : Epitope specificity of cross-clade neutralizing sera from Chinese HIV-1-positive individuals. Scand J Immunol 2013;78(4):357–370 [DOI] [PubMed] [Google Scholar]
- 47.Doores KJ, Bonomelli C, Harvey DJ, et al. : Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci USA 2010;107(31):13800–13805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raska M, Takahashi K, Czernekova L, et al. : Glycosylation patterns of HIV-1 gp120 depend on the type of expressing cells and affect antibody recognition. J Biol Chem 2010;285(27):20860–20869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klein F, Mouquet H, Dosenovic P, et al. : Antibodies in HIV-1 vaccine development and therapy. Science 2013; 341: 1199–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.