SUMMARY
Why some viruses are enveloped while others lack an outer lipid bilayer is a major question in viral evolution but one that has received relatively little attention. The viral envelope serves several functions, including protecting the RNA or DNA molecule(s), evading recognition by the immune system, and facilitating virus entry. Despite these commonalities, viral envelopes come in a wide variety of shapes and configurations. The evolution of the viral envelope is made more puzzling by the fact that nonenveloped viruses are able to infect a diverse range of hosts across the tree of life. We reviewed the entry, transmission, and exit pathways of all (101) viral families on the 2013 International Committee on Taxonomy of Viruses (ICTV) list. By doing this, we revealed a strong association between the lack of a viral envelope and the presence of a cell wall in the hosts these viruses infect. We were able to propose a new hypothesis for the existence of enveloped and nonenveloped viruses, in which the latter represent an adaptation to cells surrounded by a cell wall, while the former are an adaptation to animal cells where cell walls are absent. In particular, cell walls inhibit viral entry and exit, as well as viral transport within an organism, all of which are critical waypoints for successful infection and spread. Finally, we discuss how this new model for the origin of the viral envelope impacts our overall understanding of virus evolution.
INTRODUCTION
The majority of organisms that act as hosts for viruses possess a cell wall. Cell walls are robust layers that surround the cell membrane and are best known in plants, fungi, protists, algae, and bacteria. Cell walls are clearly ancient, and while the similarity of cell wall components indicates a shared ancestry among algae and plants (1), studies of brown algae and Archeaplastida (i.e., green and red algae and land plants) suggest that cell walls have evolved convergently (2). The cell wall has a variety of functions from protection to the maintenance of cell shape, although its most important role is to provide structural support to counteract high internal osmotic pressure. The cell wall is also a selective filter, allowing free diffusion of small molecules and ions. Experiments with cell walls in plants and bacteria have determined an exclusion size of approximately 50 to 60 kDa (3–5). This allows the diffusion of important signaling molecules, such as phytohormones in plants, but not virus particles.
Cell walls differ in number and composition, depending on the organism. Several plants have a secondary cell wall (6), while bacteria and Archaea possess only a single cell wall. The diversity of cell wall components has led to several classification systems based on their complexity and composition, such as the classification systems for algae (7) and flagellates (8), and these systems can be used to assess the rigidity of a cell wall. While the majority of bacteria possess a rigid cell wall due to the presence of peptidoglycan, in some cases, such as Mycoplasma, there is no such rigid “shell,” and the cell walls consist of a plasma membrane reinforced with glycocalyx, a glycoprotein polysaccharide (9, 10). Similarly, most members of the Archaea domain have a crystalline protein layer, called the surface layer (S-layer), as their cell wall lacks peptidoglycans (10–12). As a consequence, the cell walls of most Archaea are less rigid than those of bacteria.
In marked contrast, animal cells lack cell walls and are surrounded by a flexible lipid bilayer, the cell membrane, that can contain numerous important functional modifications such as receptors or other membrane-bound structures. These structures are responsible for molecule uptake and excretion, are involved in cell signaling, and maintain a stable osmotic pressure and pH (13). Hence, the cell walls found in plants, fungi, protists, algae, and bacteria provide a rigid and strong barrier for viral entry and exit not seen in animal cells. Critically, viruses cannot enter cells that possess cell walls by endocytosis or exit these cells by budding, and instead they rely on a number of different approaches.
While viral genomes encode the structural proteins they require, enveloped viruses acquire a major component of their envelope from the host cell through budding and are able to modify it by inserting their own proteins (14). The envelope may be acquired from the host cell membrane or intracellular compartment, such as the endoplasmic reticulum or Golgi compartment (15). Upon virus entry, each layer of a virus serves to overcome a specific host cell barrier. After each successful breach, the corresponding layer of the virus is lost, eventually delivering the unpacked genomic payload to its origin of replication. Inversely, successful virus exit involves the acquisition of these layers. However, the pathways for virus entry and exit differ substantially, especially among viruses infecting cells surrounded by a cell wall.
To understand the evolution of the viral envelope, we reviewed and compared the mechanisms of virus entry, spread, and exit among all known virus families. Strikingly, this revealed that enveloped viruses predominantly infect organisms without cell walls, while viruses without an envelope can infect hosts with and without cell wells, although the majority of their hosts possess cell walls. From this analysis, we hypothesize that the lack of an envelope is a specific viral adaptation to the presence of cell walls, while the viral envelope is an adaptation to hosts that lack cell walls. Although there are a number of exceptions to this simple evolutionary rule, closer inspection reveals that these individual adaptations support the general distinction noted above. Indeed, we show that viruses from organisms possessing cell walls have evolved a variety of ways to ensure successful infection and spread. While entry pathways of known viruses have been compared and analyzed extensively in previous publications (16–21), this is, to our knowledge, the first synthesis that links viral evolution to the structure of host cells.
VIRUS ENTRY, TRANSMISSION, AND EXIT
We selected 101 virus families from the 2013 release of the International Committee on Taxonomy of Viruses (ICTV) (22). We excluded the viroid families Avsunviroidae and Pospiviroidae, virus satellites, and the family Metaviridae, since they contain eukaryotic retrotransposons. Of the 101 virus families analyzed, 65 were nonenveloped virus families, while 37 were enveloped (the Iridoviridae can be both enveloped and nonenveloped and hence were included in both groups [23, 24]). To identify the host range of these virus families, we created seven broad classes of host organisms based on their identified hosts (see Data Sets S1 and S2 in the supplemental material) and their taxonomic position in the tree of life (D. R. Maddison and K.-S. Schultz, Tree of Life Web Project [http://tolweb.org]). In total, we identified 123 host types, of which 64 were animal cells with no cell walls, while 59 had cells surrounded by a cell wall. All bacteria were grouped in the class (simplified taxonomic class) “Eubacteria” and hence distinct from the Archaea. The eukaryotes were split into five classes (simplified taxonomic classes): “Plants” (which contains all plants and algae), “Protozoa,” “Fungi,” “Invertebrates,” and “Vertebrates” (Fig. 1). “Fungi” contains all Eumycota, while animals were subdivided into “Vertebrates” (Chordata) and “Invertebrates” (all non-Chordata). The remaining members of the animal clade were classified as “Protozoa.” Importantly, this classification was developed only as a general guide for data analysis and did not impact any of the major conclusions drawn.
FIG 1.
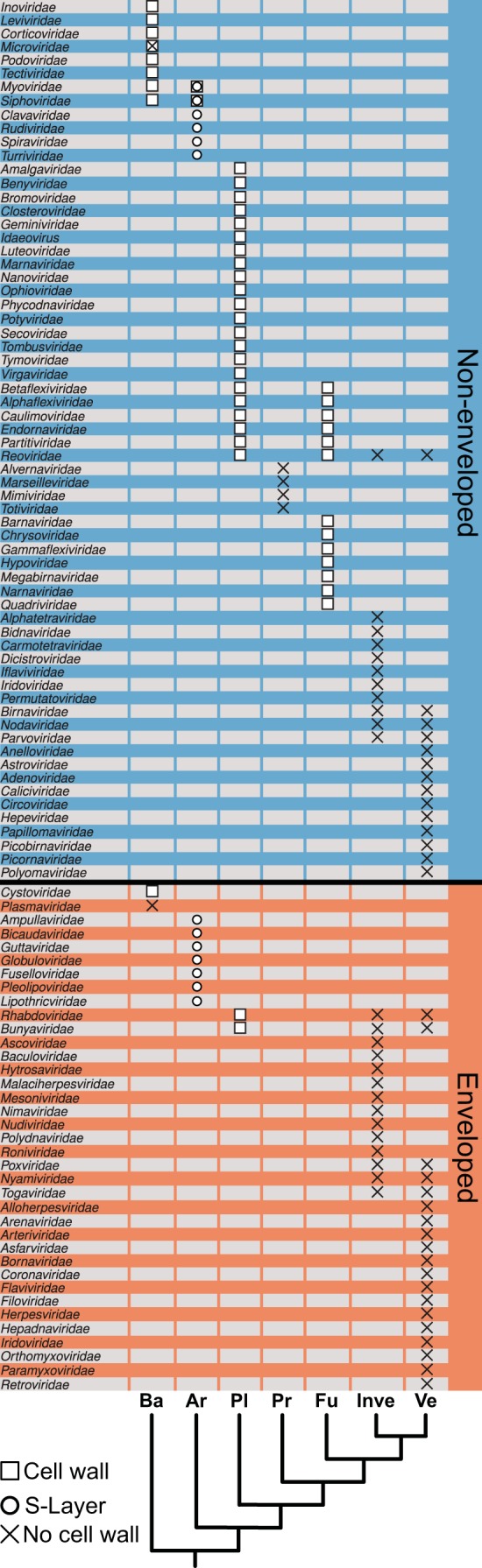
Association between known virus families and the presence of a cell wall, surface layer (S-layer), or absence in the hosts they infect. The schematic phylogenetic tree represents our simplified taxonomic classes as defined in the text. The abbreviations for the different host classes are as follows: Ba, Bacteria; Ar, Archaea; Pl, Plants; Pr, Protozoa; Fu, Fungi; Inve, Invertebrates; Ve, Vertebrates.
We then analyzed the 101 virus families to determine the taxonomic distribution of the presence/absence of envelopes among viruses. This revealed a strong association between the presence of the viral envelope and the absence of a cell wall in the host organism. Specifically, the 65 nonenveloped virus families infected 79 host types, of which 49 had cells with a cell wall while 30 did not (Table 1 and Fig. 1). In contrast, of the 37 enveloped virus families, only 10 infected host types with cell walls compared to 34 host types without cell walls. Hence, the majority of host types with cell walls are infected by nonenveloped viruses, while the majority of enveloped viruses infect animal cells. Only a few enveloped viruses are known to infect cells with cell walls, representing unique cases that are likely to be highly specialized adaptations (see below).
TABLE 1.
Summary of the pattern of association between virus envelopes (presence or absence) and hosts (with and without cell wall) among 101 virus familiesa
| Host | No. of virus families |
||
|---|---|---|---|
| Nonenveloped | Enveloped | Total | |
| With cell wall | 49 | 10 | 59 |
| No cell wall | 30 | 34 | 64 |
| Total | 79 | 44 | |
The S-layer of Archaea has been treated as cell wall. Note that some virus families can infect hosts with and without cell wall and are therefore present in more than one category.
We also analyzed the pathways for virus entry, transmission, and exit (Tables 2 and 3; see below). Viral entry into animal cells relies on endocytosis pathways for both enveloped and nonenveloped viruses. However, endocytosis is not possible in organisms that possess a cell wall, since it creates an important physical barrier. Virus release by excretion pathways or budding is similarly hindered. Of the 65 nonenveloped virus families analyzed, 21 are released by lysis, while 10 are released in a nonlytic pathway (Table 3). In contrast, only five enveloped virus families exit the host cell by lysis, while 21 utilize a nonlytic pathway, mostly budding or the endosomal sorting complex required for transport (ESCRT). ESCRT is a conserved molecular complex that modulates membrane scission into the cytoplasm. However, several viruses have managed to use parts of the ESCRT complex for budding and subsequent release into the cytoplasm (26). In addition, some plant and fungal viruses spread vertically, never leaving the cell (16). Finally, our analysis of pathways of viral transmission within hosts showed that, among multicellular organisms with cell walls like plants, the capsid or ribonucleoprotein (RNP) is the key factor, such that an envelope is not required (see below). Accordingly, we propose that nonenveloped viruses are an adaptation to the evolution of the cell wall, while the viral envelope constitutes an adaptation to cells without cell walls (i.e., animal cells). We now discuss, in more detail, how these observations relate to aspects of the virus life cycle.
TABLE 2.
Cell entry pathways of the virus families analyzeda
| Cell entry pathway | Virus family [reference(s)]b |
|---|---|
| Endocytosis | Caliciviridae (102) |
| Hepeviridae (111) | |
| Parvoviridae (123, 124) | |
| Phycodnaviridae (46) | |
| Hepadnaviridae* (135) | |
| Macropinocytosis | Adenoviridae (103) |
| Birnaviridae (108) | |
| Papillomaviridae (117) | |
| Mimiviridae (125) | |
| Totiviridae (130) | |
| Filoviridae* (136–138) | |
| Herpesviridae* (141) | |
| Nodaviridae* (145) | |
| Paramyxoviridae* (148, 149) | |
| Poxviridae* (27) | |
| Clathrin mediated | Adenoviridae (104–106) |
| Astroviridae (112) | |
| Circoviridae (118) | |
| Luteoviridae (126) | |
| Papillomaviridae (131, 132) | |
| Pestiviridae (139) | |
| Picornaviridae (142, 143) | |
| Polyomaviridae (146) | |
| Reoviridae (150, 151) | |
| Iridoviridae(*) (152) | |
| Coronaviridae* (154) | |
| Arenaviridae* (156) | |
| Arteriviridae* (158–160) | |
| Asfarviridae* (162) | |
| Baculoviridae* (166, 167) | |
| Bornaviridae* (169) | |
| Bunyaviridae* (170) | |
| Filoviridae* (171) | |
| Flaviviridae* (172, 173) | |
| Orthomyxoviridae* (174) | |
| Paramyxoviridae* (175) | |
| Retroviridae* (176, 177) | |
| Rhabdoviridae* (178) | |
| Togaviridae* (179–181) | |
| Caveolae | Papillomaviridae (107) |
| Picornaviridae (113) | |
| Polyomaviridae (119, 120) | |
| Hepadnaviridae* (127) | |
| Retroviridae* (133) | |
| Lipid raft | Birnaviridae (108) |
| Caliciviridae (114) | |
| Orthomyxoviridae* (128) | |
| Fusion | Corticoviridae (109) |
| Phycodnaviridae (45) | |
| Picornaviridae (121) | |
| Tectiviridae (109) | |
| Iridoviridae(*) (134) | |
| Arenaviridae* (140) | |
| Baculoviridae* (144) | |
| Coronaviridae* (147) | |
| Cystoviridae* (50) | |
| Herpesviridae* (153) | |
| Malacoherpesviridae* (155) | |
| Paramyxoviridae* (157) | |
| Plasmaviridae* (59, 161) | |
| Polydnaviridae* (163–165) | |
| Retroviridae* (168) | |
| Ejectionc | Microviridae (47) |
| Myoviridae (115, 116) | |
| Podoviridae (122) | |
| Siphoviridae (129) | |
| Pilus retraction | Inoviridae (61) |
| Leviviridae? (43) | |
| Membrane penetration | Picobirnaviridae? (110) |
Families where no entry pathways have been published are not listed.
Enveloped virus families are indicated by a * symbol, while (*) indicates virus families containing enveloped and nonenveloped forms. A? symbol indicates putative exit pathways. The corresponding source publication(s) or reference(s) is shown in parentheses at the end of an entry.
Ejection indicates membrane penetration, cell wall digestion, and genome ejection.
TABLE 3.
Cell exit pathways of the virus families analyzeda
| Cell exit pathway | Virus family [reference(s)]b |
|---|---|
| Unknown/nonlyticc | Hepeviridae (182, 183) |
| Inoviridae (61) | |
| Luteoviridae (189) | |
| Mesoniviridae (194) | |
| Nodaviridae (198) | |
| Papillomaviridae (202) | |
| Rudiviridae (207) | |
| Totiviridae (212) | |
| Bornaviridae* (216) | |
| Bunyaviridae* (221) | |
| Fuselloviridae* (225) | |
| Malacoherpesviridae* (155) | |
| ESCRT | Picornaviridae (93) |
| Arenaviridae* (186) | |
| Filoviridae* (190) | |
| Flaviviridae* (195) | |
| Rhabdoviridae* (199) | |
| Hepadnaviridae* (203) | |
| Herpesviridae* (208) | |
| Paramyxoviridae* (213) | |
| Poxviridae* (217) | |
| Retroviridae* (222) | |
| Budding | Phycodnaviridae (46) |
| Reoviridae (187) | |
| Asfarviridae* (191) | |
| Baculoviridae* (144, 196) | |
| Coronaviridae* (200) | |
| Iridoviridae(*) (204) | |
| Nyamiviridae* (209) | |
| Orthomyxoviridae* (214) | |
| Plasmaviridae* (218, 219) | |
| Togaviridae* (223) | |
| Lysis | Annelloviridae (184, 185) |
| Astroviridae (188) | |
| Birnaviridae (192, 193) | |
| Caliciviridae (197) | |
| Corticoviridae (201) | |
| Leviviridae (205, 206) | |
| Marnaviridae (210, 211) | |
| Marseilleviridae (215) | |
| Microviridae (220) | |
| Mimiviridae (224) | |
| Myoviridae (226) | |
| Parvoviridae (227) | |
| Phycodnaviridae (228) | |
| Picornaviridae (229) | |
| Podoviridae (230, 231) | |
| Polyomaviridae (232) | |
| Reoviridae (233) | |
| Rudiviridae (234) | |
| Siphoviridae (82, 235) | |
| Tectiviridae (236) | |
| Turriviridae (237) | |
| Adenoviridae* (238) | |
| Ascoviridae*? (239) | |
| Circoviridae* (240, 241) | |
| Cystoviridae* (242) | |
| Polydnaviridae*? (243) |
Virus families without (published) exit pathways are not listed.
Enveloped virus families are indicated by a * symbol, while (*) indicates families containing enveloped and nonenveloped forms. A? symbol indicates putative exit pathways. The corresponding source publication(s) or reference(s) is shown in parentheses.
Unknown/nonlytic indicates release pathways where no lytic pathway exists but viral release has been observed.
Although our review of the literature covers all those virus families for which data are available—entry and exit pathways for 71 and 57 virus families, respectively—it is important to note that it does not include all known viruses (Tables 2 and 3). Although we are able to describe pathways from all known host kingdoms, most data are necessarily from the better-known viruses. Clearly, it will be important to determine whether the generalities noted here can be extended to all known virus groups, including those only recently described, and it is striking that there is relatively little data from most archaeal and insect viruses.
Virus Entry
The major role of membranes in animal cells is to create distinct compartments and to receive and send signals from outside the cell. Therefore, viruses have to enter and exit animal cells in a systemic infection or to reach their target tissue. Viruses have overcome this barrier in animals by hijacking endo- and exocytosis pathways.
Animal viruses have evolved several ways to enter animal cells, although these pathways are always based on the flexibility of the cell membrane (17). This flexibility allows different pathways for virus uptake for both enveloped and nonenveloped viruses. Viruses are adapted to endocytosis pathways, as they offer entry points usually used for nonspecific uptake of fluids, solutes, or particles. As an example, vaccinia virus enters cells by mimicking an apoptotic body, thereby triggering macropinocytosis (27, 28). Virus uptake through endocytosis is induced upon binding of the virus to cell surface receptors (20). For enveloped viruses, uptake into animal cells involves the fusion and subsequent release of the capsid (29), while nonenveloped viruses can create pores in the cell membrane to deliver their viral genome (30, 31). A single virus can induce several endocytosis pathways as observed for dengue virus and HIV-1. While both can enter cells by triggering macropinocytosis (32, 33), additional entry pathways for dengue via the clathrin-mediated pathway (34) and HIV-1 through fusion have been observed (35).
Such entry pathways are blocked in plants and bacteria due to the presence of the cell wall. While the plant cell wall allows diffusion of water and ions, the diffusion of macromolecules is restricted. However, endocytosis-like pathways have been observed in plants (36) and bacteria (37). Lonhienne et al. (37) used green fluorescent protein (GFP) to highlight endocytosis in Gemmata obscuriglobus, a budding bacterium with Gram-negative cell wall structure (38), and showed that GFP was able to diffuse through the cell wall. The maximum exclusion size for cell walls of plants and bacteria is approximately 60 kDa (3–5). We estimated the diameter of a spherical protein that can diffuse freely through the cell wall to be ∼5.126 nm, which approximately corresponds to the width of two DNA double helices (Appendix). Consequently, while the GFP, with a molecular mass of 26.9 kDa and a diameter of 2.4 nm (39, 40), is able to diffuse through cell walls, viruses cannot. Critically, therefore, the intrinsic rigidity of cell walls in plants means that plant pathogens have evolved a variety of ways to penetrate and infect their hosts (41). We now discuss some of these adaptations.
In plants and fungi, viruses do not actively breach the cell wall. Plant viruses are obligate intracellular parasites in that they remain with their host indefinitely but can be transmitted by vectors, fungi (42), mechanical injuries, or vertically (16). Fungal viruses have adapted to cell walls by using hyphal anastomosis (fusion of encountering vegetative hyphae) for horizontal transmission and a persistent lifestyle for vertical transmission. Vertical transmission allows fungal viruses to stay in the host (43). Similarly, some plant viruses remain asymptomatic inside the host, relying in vertical transmission through seeds (16, 21, 44).
The situation is complex in algae. While algae share similarities with plants with respect to cell architecture, notable exceptions exist. Chlorella, a single-cell green algae, is infected by Paramecium bursaria chlorella virus 1 (PBCV-1) (45). PBCV-1 has an internal membrane (that is, the membrane is surrounded by the capsid). To enter its host, PBCV-1 degrades the Chlorella cell wall and fuses its inner membrane with the cell membrane (45). Another algal virus, Emiliana huxleyi virus 86, belongs to the Coccolthovirus genus and infects a wide range of eukaryotic algae in marine and freshwater environments. Emiliana huxleyi is a marine calcifying unicellular phytoplankton. Rather than a typical cell wall, these phytoplanktons possess a characteristic calcite covering that surrounds the cell membrane. Although it belongs to the Phycodnaviridae family, like PBCV-1, Emiliana huxleyi virus 86 has an additional outer membrane that covers the capsid, and to infect its host, the virus fuses its outer membrane with the host membrane or enters via an endocytic process (46). Since budding of Emiliana huxleyi virus 86 particles from infected Emiliana huxleyi has been demonstrated (46), we assume that the cell covering is not tight enough to exclude viral particles. However, it has been proposed that the calcified shell offers a certain degree of viral defense (46). It should be noted that its capsid may possess cell wall-degrading enzymes, although they are not required in this case. This example of an “animal virus-like” entry mechanism shows that viruses infecting unicellular algae have evolved several approaches to enter their hosts.
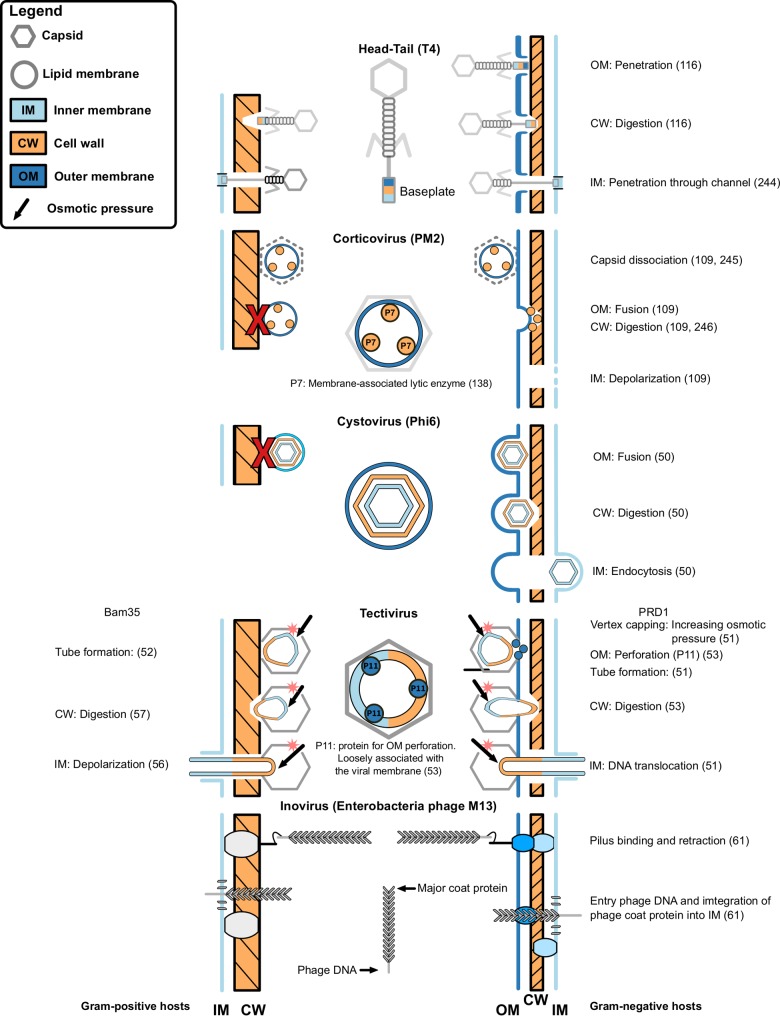
Similar to plant pathogens, most bacteriophage have evolved diverse entry pathways (Fig. 2). All known bacteriophage use lytic enzymes to penetrate the cell wall, while different mechanisms have been described to overcome the bacterial membranes. Most bacteriophage follow a three-step program: (i) puncture the outer cell wall, if present; (ii) digest the cell wall; (iii) insert the phage genome into the host cell. Tail-less, nonenveloped bacteriophage, such as ϕX174, form a tube to deliver their genome into the host (47). However, enveloped bacteriophage have also been observed. Interestingly, these envelopes can surround the capsid, as in the case of Cystovirus, or the envelope can be encapsulated by a capsid, as in the case of Corticovirus or Tectivirus (48). To infect Gram-negative bacteria, enveloped bacteriophage found in the Cystoviridae and Corticoviridae families fuse their envelope with the outer membrane of their hosts (48–50). Phages PRD1 and Bam35 belong to the Tectiviridae. Both are nonenveloped, but the capsid encloses an internal membrane containing the genome. Despite their similarity, PRD1 infects Gram-negative bacteria, while Bam35 infects Gram-positive bacteria. The entry pathway from Bam35 differs in some steps form PRD1 (Fig. 2). Both phages form a tube for DNA delivery which is initiated by capping vertices from the capsid. The osmotic difference between the capsid and cytosol pushes the internal membrane through a special vertex in the capsid. The emerging membrane has lytic properties and digests the cell wall, thereby forming a tube for subsequent DNA delivery (51, 52). PRD1 possess proteins that are loosely associated with the internal membrane and are able to puncture the outer membrane (53, 54). In contrast, as Bam35 infects Gram-positive bacteria, it does not need to perforate an outer membrane, although the genes for outer membrane perforation are present in its genome (55). In addition, these phages differ in how they pass the internal membrane (56–58). Bam35 depolarizes the internal membrane, while PRD1 does not, although mechanisms by which it functions are not fully understood. Bacteriophage infecting Gram-positive bacteria do not need to pass an outer membrane and can attack the cell wall directly. In the case of bacteriophage that have an envelope covered by a protein capsid, such as Bam35, the envelope facilitates the fusion with the inner membrane (57). Notably, Plasmavirus, an enveloped bacteriophage, exclusively infects Mycoplasma, one of the few bacteria without a cell wall (59).
FIG 2.
Schematic overview of different bacteriophage entry mechanisms. Several different entry mechanism for nonenveloped (Head-Tail, Corticovirus, and Tectivirus), enveloped (Cystovirus), and filamentous (Enterobacteria phage M13) bacteriophage are shown. Structures are not drawn to scale, and only key structures for viral entry are shown and color coded according to the part they breach during entry, e.g., components responsible for cell wall degradation have the same color as the cell wall indicated in the legend. Associated membrane proteins are indicated as circles. Mechanisms for Gram-positive bacterial hosts are shown on the left, while those Gram-negative hosts are shown on the right. Numbers in parentheses indicate references for the corresponding step (steps without references are putative and inferred by the authors) (see references 50 to 53, 56, 57, 61, 109, 116, and 244 to 246). No Gram-positive hosts are known for corticovirus and cystovirus, and a red X indicates possible interference of the cell wall onto the entry mechanism.
Another bacteriophage family has evolved a very different approach. Members of the Inoviridae attach to the pili of Gram-negative bacteria (60). The retraction of the pili brings the capsid into contact with the inner membrane where it disassembles and is released into the cytoplasm (61). This approach circumvents the outer membrane and cell wall altogether, abolishing the need for an envelope and cell wall-digesting properties (Fig. 2).
The host range for enveloped bacteriophage does not include Gram-positive bacteria, since the envelope cannot fuse and the cell wall is not digested, as in the case of Cystovirus. The Inoviridae similarly do not possess an envelope, since it is not required for infection, as they bypass the outer membrane and cell wall by using the pili of their host. The presence of the cell wall requires cell wall-degrading enzymes for successful infection, which are largely associated with base plates and capsids of bacteriophages.
Overall, the analysis of viral entry pathways strongly supports our hypothesis that the presence of a virus envelope is associated with the absence of cell walls and vice versa, such that these two traits have an intimate evolutionary relationship (Fig. 1). In particular, the presence or absence of a viral envelope is clearly better associated with cell structure, especially the presence or absence of a cell wall, than to a specific type of host species.
Intrahost Virus Spread
We now examine how the presence of the cell wall, which influences cell-to-cell communications, impacts viral spread within an individual host. Once plant viruses enter epidermal or mesophyll cells, systemic transport is possible by taking advantage of the plant cell architecture. It is known that plant viruses move from cell to cell by plasmodesmata and across whole vascular plants by phloem (62). Multicellular fungi are either coenocytic (large cells with several nuclei) or the cells are separated by septa, i.e., end walls that can be perforated and therefore connect neighboring hyphae. The movement of viral capsids within or between fungi is not restricted and can occur horizontally by hyphal anastomosis, a naturally occurring process in which two hyphal cells create a fusion aperture to allow the migration and exchange of nuclei and cytoplasm (63, 64).
Due to a general inability to infect new hosts by penetrating the cell walls, plant and fungal viruses rely on different mechanisms to gain entry into new hosts, with arthropod vectors a key element. Using vectors to infect new hosts is possible, since the cell wall is breached upon feeding, which we therefore propose to be a secondary adaptation in plant and fungal viruses (see below). Viruses in insects can be classified into two groups based on their mode of transmission—noncirculative and circulative (18)—which reflect how long a virus is viable in the vector during transmission to a new host. Noncirculative transmission is essential for viruses that remain within the vector at the mouthparts or foregut and need to be immediately inoculated into a new host after acquisition by the vector (65). In contrast, circulative transmission allows longer times between acquisition and transmission of the virus into the new host by circulating across the gut, hemolymph, and salivary gland before being inoculated into a new host. Circulative plant and insect viruses can undergo this process with or without replication.
Transport across the plasmodesmata requires a virus-encoded movement protein which interacts with the plasmodesmata to allow the passage of the virus particles (66). The transport of viruses within plants occurs either as a RNP or viral capsid (67, 68) but, importantly, not as enveloped viruses. Experiments in tomatoes infected with Tomato leaf curly virus (69, 70) and Tomato bushy stunt virus (TBSV) (71–73) showed that viruses without the ability to form capsids were transported from cell to cell but with a lower efficiency. Interestingly, only four plant-infecting virus genera possess an envelope: Cytorhabdovirus, Nucleorhabdovirus (both of which are members of the family Rhabdoviridae), Emaravirus, and Tospovirus. Since the envelope is not required for cell entry and subsequent cell-to-cell movement, we argue that its limited presence in these genera is because it facilitates vector-borne viral transmission.
Rhadboviruses are unusual in that they are able to infect both plants and animals, with Cytorhabdovirus and Nucleorhabdovirus able to bud in the plant and insect host (74). In plants, budding virions are found in the perinuclear space and at the cell membrane (74). Since the enveloped form of plant viruses is not transported to neighboring cells (67, 75), it has to be assumed that enveloped Rhabdoviridae in plants are transmitted solely by vectors. This scenario has also been reported for Tospovirus, the only genus of the Bunyaviridae infecting plants. Mature Tosposvirus virions accumulate in the plant cells, waiting to be transmitted by feeding thrips (68). The enveloped, vector-borne emaviruses have been recently discovered in several plant species (76), and their capability for cell-to-cell movement is likely based on the capsid, rather than the envelope (77).
In the enveloped Tospovirus, two transmembrane glycoproteins GN and GC, are required for vector transmission, as repeated passages through plants led to accumulated mutations in those proteins that subsequently impaired insect transmission (78). In addition, targeted point mutations in GN and GC inhibited transmission through thrips (79), although plant infection was not impaired. Cytorhabdovirus and Toposvirus are all circulative and persistent within the vector. In addition, Rhabdoviruses show a wider array of vectors, while Toposvirus is associated only with thrips (18, 80). This strongly suggests that the envelopes of enveloped plant viruses are an adaptation to the vector, not the host.
Cell walls impair cell-to-cell communications, and structures like the plasmodesmata serve as communication channels between plant cells. Viruses have adapted them for viral movement within the plant hosts. While plant viruses can acquire an envelope in plant cells, the envelope is not required for viral cell-to-cell movement, which is facilitated by the capsid or RNP. That all enveloped plant viruses are vector-borne strengthens our theory that nonenveloped viruses are an adaptation to the cell wall, and envelopes are needed only upon vector-aided translocation due to the fact that viral transport is possible as capsid, RNP, or naked DNA/RNA, such that the viral envelope is not required.
Virus Exit
The absence of a cell wall in animal cells favors endocytosis for cell entry and budding for cell exit. Budding pathways have been successfully adopted by viruses. Several enveloped viruses hijack the ESCRT pathway (19, 81) that is responsible for a variety of functions in a cell, including endosomal sorting, receptor signaling, and cytokinesis (26). Only a few enveloped viruses lyse the host cell to be released, while virtually all nonenveloped viruses exit the host cell through lysis (Table 2). Interestingly, nonenveloped viruses infecting animals do not use excretion pathways and lyse their host cell (Table 2).
With the exception of the Inoviridae, all bacteriophage escape the host cell through lysis. Inoviridae encode three proteins that create a secretion channel through the cell wall and bacterial membranes (61). Recent research with Gram-negative bacteria indicates that both the cell wall and outer membrane are actively disrupted through a spanin complex (82). Permeabilization of the inner membrane is the first step, whereby holins and pinholins, small viral membrane proteins, are secreted into the inner membrane of the host and upon activation allow cell wall-degrading enzymes to leave the cytoplasm (83–87). The subsequent release of endolysins into the periplasm degrades the peptidoglycan. While the spanin complexes are required to disrupt the outer membrane, its mechanics are unknown (88). Similarly, the release pathway of the enveloped bacteriophage Cystovirus is currently unclear. Bacteriophage that do not possess an envelope can induce lysis by holins without being permeabilized themselves. In contrast, virus envelopes can be targeted by holins, especially as the envelope is acquired from the host.
Lysis of a bacterial cell involves membrane-disrupting proteins. Therefore, viruses that acquire an envelope from the inner membrane of the host turn themselves into a putative target for membrane permeabilization. This, in turn, would release capsids that are capable of digesting cell walls but not getting past the outer or inner membranes of bacteria. Hence, we propose that members of the Tectiviridae and Corticoviridae evolved the outer capsid to protect their envelope during host cell lysis. Since virus particles cannot diffuse through the cell wall, exocytosis pathways in plants and bacteria are not used for viral release.
EVOLUTIONARY IMPACT OF CELL WALLS ON VIRAL ENVELOPES
Our association study of 101 viral families and their hosts revealed a strong relationship between enveloped viruses and animal host cells and nonenveloped viruses and host cells with cell walls. An extensive literature review of viral entry, transmission, and exit strategies of these viral families supports our main hypothesis that cell walls were central to the evolution of nonenveloped viruses, while the lack of a cell wall provides an adaptive advantage to viruses with envelopes. The cell wall constitutes an important physical barrier that cannot be breached by endocytosis for entry or exocytosis for exit. In bacteria, where membranes are present, viral envelopes are used to get past either the outer or inner membrane but lack the sophisticated arsenal of receptors found on enveloped viruses that infect animal cells.
The Viral Envelope Is a Result of Convergent Evolution
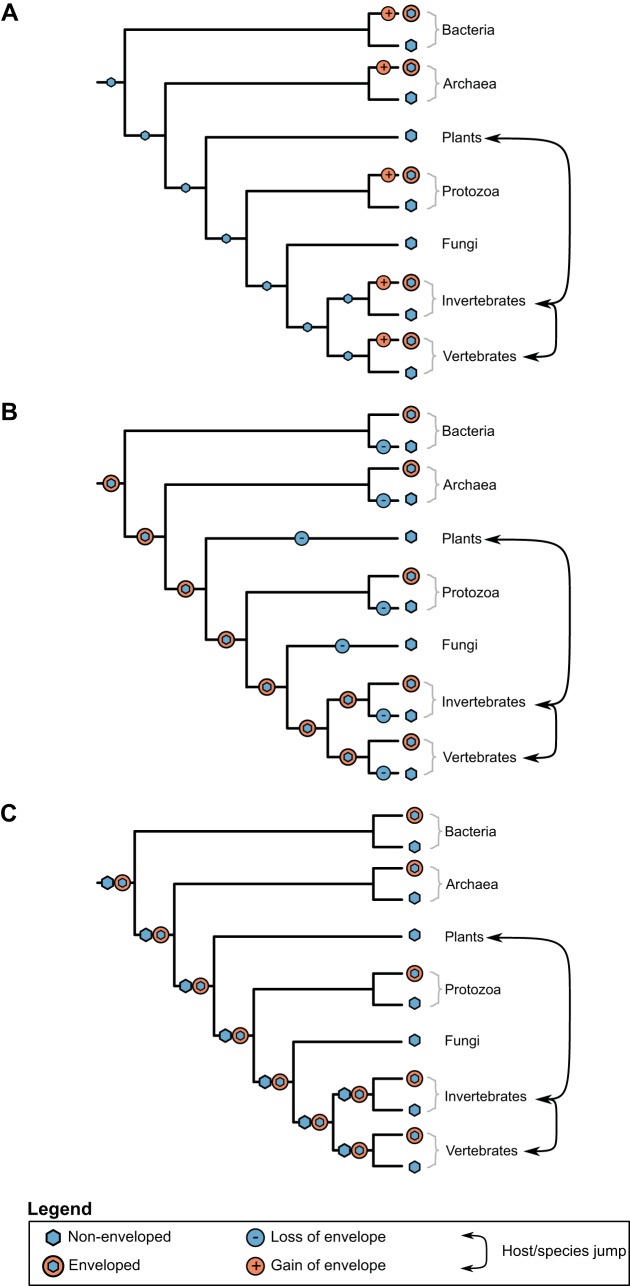
A variety of models can be proposed to explain the evolution of the viral envelope. If we assume that early viruses were enveloped, then they must have lost their envelope several times (Fig. 3A). Conversely, if early viruses were not enveloped, as seems more likely, then they have gained their envelope several times (Fig. 3B). A third possibility is the initial coexistence of enveloped and nonenveloped viruses and subsequent selection in the corresponding hosts leading to either gain or loss of the envelope (Fig. 3C). The scattered presence of envelopes among viral taxa strongly suggests that they have evolved convergently, which we propose reflects the presence or absence of cell walls in phylogenetically diverse host species.
FIG 3.
Three models for the loss and/or gain of the viral envelope during evolutionary history, as well as putative host jump events. The phylogenetic tree is the same as that used in Fig. 1. (A) Early, nonenveloped viruses with subsequent gain (multiple times independently) of the viral envelope. (B) Early, enveloped viruses with its subsequent loss in multiple host lineages. (C) Early, coexisting nonenveloped and enveloped viruses.
It is also possible that host jumps allowed nonenveloped viruses to infect animals and enveloped viruses to infect hosts with cell walls. For example, a large number of new RNA viruses have recently been identified in arthropods, constituting a potentially huge viral reservoir (89). Since arthropods have a close ecological relationship to both plants and vertebrates, host jumps from plants to animals via arthropods are not unlikely. As mentioned above, animal cells show less discrimination between enveloped and nonenveloped viruses than organisms that possess a cell wall, and the ability of plant virus capsids to release genes into mammalian cells has been demonstrated (89). Hence, the pivotal position of arthropods between plants and vertebrates could have facilitated the adaptation of nonenveloped viruses to vertebrates.
The only enveloped viruses in plants are Emaravirus, Bunyavirus, and Rhabdovirus. As noted above, the envelopes of plant viruses appear to be an adaptation to the vector, rather than to the plant, and hence could be the result of a host jump. Since all other plant viruses are not enveloped, they have obviously lost the envelope or were never enveloped. However, the former scenario seems highly unlikely, since plants evolved before insects (90, 91). Entering the plant through mechanical injuries after being transported by environmental factors like wind or rain would still be possible, although likely inefficient. As a consequence, early enveloped plant viruses appear to have few ways to be transmitted.
Plant viruses can move within their host by plasmodesmata and phloem, while fungal viruses can transverse their hosts due to perforated septa. These specialized cell-to-cell links evolved to facilitate cell communication, overcoming the rigidness and impermeability of cell walls. Crucially, we argue that this development also led to preferential infection by nonenveloped viruses. Hence, most plant and fungal viruses are not enveloped, since fusion or budding from a plant or fungal cell is not feasible due to the presence of a cell wall and because transport inside the host is possible only via the RNP or capsid. The adaptation of viral capsids or RNPs for transport by plasmodesmata and the later emergence of arthropods means that early plant viruses were very likely nonenveloped. In turn, this means that Emaravirus, bunyaviruses, and rhabdoviruses infected plants subsequent to the emergence of arthropods.
Cystoviridae and Plasmaviridae are the only known enveloped bacteriophage families, and both have a very limited known host range, the former infecting only Pseudomonas, while the latter infect only Mycoplasma, suggesting that the envelope is a highly specialized adaptation. Although several bacteriophage with internal membranes exist, such membranes lack the receptors required for cell entry. Therefore, viruses infecting cells with a cell wall do not need an envelope per se, and if it is present, it serves as a tool to gain access to the cell wall by fusion with an outer membrane or fusion with the inner membrane after cell wall digestion. As mentioned earlier, numerous bacteriophage encode their own membrane proteins but gain the lipids required for their membrane from their hosts. Therefore, a scenario of coexisting nonenveloped and enveloped early viruses (Fig. 3C) is unlikely. Assuming early bacteriophage were able to synthesize their own lipids and lost this ability over time in favor of using host lipids, we speculate that bacteriophage will have a wider host range than currently seen, as in the case of the cystoviruses where a mutation in a coding region would allow them to infect Gram-positive hosts (Fig. 2).
In sum, we argue that early viruses were likely nonenveloped with the viral envelope a later adaptation (Fig. 3A). In support of this, nonenveloped bacteriophage show the simplest adaptation for bacterial infection, since they are able to enter and exit their hosts with the least interference. In contrast, enveloped bacteriophage need to deal with the lytic pathway and limited entry possibilities. Without a cell wall, endocytosis of enveloped and nonenveloped viruses would most likely occur, as seen in animal viruses. However, the cell wall renders endocytosis and exocytosis not feasible. The use of lytic enzymes to exit the host requires the permeabilization of the cell membrane, thereby potentially threatening the virus itself. Without an envelope, membrane permeabilization is not a concern. This, in turn, influences virus entry, since membranes are required for several bacteriophage to enter the host cell.
The enormous diversity among virus families greatly complicates phylogenetic analysis, including whether virus envelopes have been gained or lost through evolutionary history. However, previous studies have revealed clear evolutionary relationships between the so-called alphavirus-like (nonenveloped) and flavivirus-like (enveloped) positive-sense RNA viruses (92) and among the Mononegavirales group of negative-sense RNA viruses (89). In addition, it has also been shown that nonenveloped picornaviruses can acquire an envelope from the cellular membrane (93). Together, these data offer support to the idea that the viral envelope evolved convergently.
The Viral Envelope as an Adaptation to Animal Cells
Entering animal cells requires the correct signals to trigger endocytosis. Animal cells use membrane-bound receptors for cell signaling, which viruses use to gain entry into the cell. The viral envelope is advantageous in such cases, since different viral receptors can be expressed, providing the virus with the ability to trigger more than one endocytosis pathway. In contrast, capsids (in the absence of envelopes) offer less flexibility to attach different receptors. Acquiring the host's membrane not only offers less visibility to the immune system but allows a flexible way to mount receptors. For example, Ebola virus uses glycoproteins to mask its epitopes, a strategy not applicable to viral capsids due to its rigidity. Experiments with the nonenveloped plant viruses Luteovirus and Begomovirus revealed that they interact with GroEL, a chaperone of a symbiotic bacterium in aphid vectors (94, 95). This interaction is required for circulative transmission and protects against degradation in the vector (96, 97). Chaperones are not only involved in protein folding but also in membrane translocation. Luteovirus and Begomovirus enter the primary salivary glands in the vector via endocytosis before infecting the host via the saliva. We assume that GroEL functions as an envelope substitute, since the receptors on the viral capsids do not trigger endocytosis, indicating that capsids have a limited flexibility to attach different receptors. However, cases where nonenveloped viruses can attach to several receptors are also known. For example, foot-and-mouth-disease virus is known to attach to two different receptors in vivo, integrin (98) and heparan sulfate proteoglycans (99).
A common denominator among organisms with cell walls is the lack of an adaptive immune system. While innate immunity recognizes pathogens in a generic way, the adaptive immune system has virtually unlimited possibilities to recognize pathogens. Viral membranes offer the possibility to adapt to different cell types by expressing or including different varieties of membrane-bound entry receptors than on a single capsid. Such complexity is not required to evade innate immune systems. In addition, viral transport from the entry site to different organs increases the exposure of the viruses to the adaptive immune system. In such a scenario, the envelope may serve as a decoy, as the virus appears to be a host cell.
In sum, our extensive review has revealed a close association between cell walls and nonenveloped viruses that was not bound to particular types of host organism. The cell wall provides a physical barrier that hinders the interaction of receptors on the viral envelope with receptors in the cell membrane, an interaction that is central to the infection of animal cells. Although there are exceptions to this important evolutionary generality, we show that they can be considered to be individual adaptations. We also propose that early viruses were nonenveloped and that the viral envelope has evolved several times independently, reflecting the diversity of hosts encountered; this provides a new perspective on our understanding of virus origins and evolution.
APPENDIX
Calculating the Radius of a Spherical Protein of 60 kDa To Estimate the Particle Exclusion Size for Cell Walls
We calculated the volume of the protein (V) and used this to calculate its diameter. The average density of a protein of 60 kDa can be calculated as described previously (100, 101), resulting in 1.4114 g/cm3. The volume for a protein of this size is then calculated as follows:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
where V is the volume of the protein, p is the density of the protein (in grams/cubic centimeter), M is the mass of the protein (in daltons), and Na is Avogadro constant.
Assuming a sphere with volume V, the diameter (d) is calculated as follows:
| (7) |
| (8) |
| (9) |
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a National Health and Medical Research (NHMRC) Australia Fellowship awarded to E.C.H.
We declare that we have no conflicts of interest.
Biographies
Jan P. Buchmann obtained his Ph.D. in plant biology from the University of Zurich under the supervision of Professors Beat Keller and Thomas Wicker. The main focus of his thesis was the analysis of transposable elements in plant genomes and the impact of their activity on the genomic landscape and genome evolution. With a postdoctoral fellowship from the Swiss National Science Foundation, he continued the analysis of transposable elements in grasses in the group of Professor Alan H. Schulman at the University of Helsinki, Finland. In 2014, he joined the group of Professor Edward C. Holmes (University of Sydney, Australia) as Postdoctoral Researcher, where he is now studying the molecular evolution of viruses.
Edward C. Holmes is an NHMRC Australia Fellow and a Professor in the School of Biological Sciences and Sydney Medical School, joining the University of Sydney in October 2012. He received his undergraduate degree from the University of London (1986) and his Ph.D. from the University of Cambridge (1990). Following that, he performed postdoctoral research at the Universities of California (Davis), Edinburgh, and Oxford. Between 1993 and 2004, he held various positions at the University of Oxford, including University Lecturer in Evolutionary Biology and Fellow of New College, before moving to The Pennsylvania State University in early 2005. His current research focuses on the emergence, evolution, and spread of RNA viruses, with special emphasis on revealing the genetic and epidemiological processes that underpin viral emergence, the molecular epidemiology of important human pathogens, and the major mechanisms of virus evolution.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MMBR.00017-15.
REFERENCES
- 1.Popper ZA, Tuohy MG. 2010. Beyond the green: understanding the evolutionary puzzle of plant and algal cell walls. Plant Physiol 153:373–383. doi: 10.1104/pp.110.158055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niklas KJ. 2004. The cell walls that bind the tree of life. BioScience 54:831–841. doi: 10.1641/0006-3568(2004)054[0831:TCWTBT]2.0.CO;2. [DOI] [Google Scholar]
- 3.Bidnenko E, Mercier C, Tremblay J, Tailliez P, Kulakauskas S. 1998. Estimation of the state of the bacterial cell wall by fluorescent in situ hybridization. Appl Environ Microbiol 64:3059–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambert PA. 2002. Cellular impermeability and uptake of biocides and antibiotics in Gram-positive bacteria and mycobacteria. J Appl Microbiol 92(Suppl):46S–54S. doi: 10.1046/j.1365-2672.92.5s1.7.x. [DOI] [PubMed] [Google Scholar]
- 5.Tepfer M, Taylor IE. 1981. The permeability of plant cell walls as measured by gel filtration chromatography. Science 213:761–763. doi: 10.1126/science.213.4509.761. [DOI] [PubMed] [Google Scholar]
- 6.Buchanan BB, Gruissem W, Jones RL. 2000. Biochemistry & molecular biology of plants. American Society of Plant Physiologists, Rockville, MD. [Google Scholar]
- 7.Leadbeater BSC, Green JC. 1993. Cell coverings of microalgae, p 71–98. In Berner T. (ed), Ultrastructure of microalgae. CRC Press, Boca Raton, FL. [Google Scholar]
- 8.Becker B. 2000. Flagellates: unity, diversity and evolution, p 110–123. Taylor & Francis Group, New York, NY. [Google Scholar]
- 9.Kandler O. 1994. Cell wall biochemistry and three-domain concept of life. Syst Appl Microbiol 16:501–509. [Google Scholar]
- 10.Kandler O, König H. 1998. Cell wall polymers in Archaea (Archaebacteria). Cell Mol Life Sci 54:305–308. doi: 10.1007/s000180050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howland JL. 2000. The surprising archaea: discovering another domain of life. Oxford University, Oxford, United Kingdom. [Google Scholar]
- 12.Albers S-V, Meyer BH. 2011. The archaeal cell envelope. Nat Rev Microbiol 9:414–426. doi: 10.1038/nrmicro2576. [DOI] [PubMed] [Google Scholar]
- 13.Uzman A. 1999. Biochemistry and molecular biology education, p 126–128. In Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J, Molecular cell biology, 4th ed WH Freeman & Co., New York, NY. [Google Scholar]
- 14.Laurinavicius S, Käkelä R, Bamford DH, Somerharju P. 2004. The origin of phospholipids of the enveloped bacteriophage ϕ6. Virology 326:182–190. doi: 10.1016/j.virol.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 15.Welsch S, Müller B, Kräusslich H-G. 2007. More than one door – budding of enveloped viruses through cellular membranes. FEBS Lett 581:2089–2097. doi: 10.1016/j.febslet.2007.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanc S. 2007. Virus transmission—getting out and in, p 1–28. In Waigmann E, Heinlein M (ed), Viral transport in plants, vol 7 Springer, Berlin, Germany. [Google Scholar]
- 17.Yamauchi Y, Helenius A. 2013. Virus entry at a glance. J Cell Sci 126:1289–1295. doi: 10.1242/jcs.119685. [DOI] [PubMed] [Google Scholar]
- 18.Blanc S, Drucker M, Uzest M. 2014. Localizing viruses in their insect vectors. Annu Rev Phytopathol 52:403–425. doi: 10.1146/annurev-phyto-102313-045920. [DOI] [PubMed] [Google Scholar]
- 19.McDonald B, Martin-Serrano J. 2009. No strings attached: the ESCRT machinery in viral budding and cytokinesis. J Cell Sci 122:2167–2177. doi: 10.1242/jcs.028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mercer J, Schelhaas M, Helenius A. 2010. Virus entry by endocytosis. Annu Rev Biochem 79:803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- 21.Roossinck MJ. 2010. Lifestyles of plant viruses. Philos Trans R Soc Lond B Biol Sci 365:1899–1905. doi: 10.1098/rstb.2010.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International Committee on Taxonomy of Viruses. 2013. ICTV Master Species List v2. International Committee on Taxonomy of Viruses. [Google Scholar]
- 23.Fukaya M, Nasu S. 1966. A chilo iridescent virus (CIV) from the rice stem borer, Chilo suppressalis Walker (Lepidoptera: Pyralidae). Appl Entomol Zool 1:69–72. [Google Scholar]
- 24.Tidona CA, Schnitzler P, Kehm R, Darai G. 1998. Is the major capsid protein of iridoviruses a suitable target for the study of viral evolution? Virus Genes 16:59–66. doi: 10.1023/A:1007949710031. [DOI] [PubMed] [Google Scholar]
- 25.Reference deleted.
- 26.Rusten TE, Vaccari T, Stenmark H. 2012. Shaping development with ESCRTs. Nat Cell Biol 14:38–45. doi: 10.1038/nrm3495. [DOI] [PubMed] [Google Scholar]
- 27.Mercer J, Helenius A. 2008. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science 320:531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- 28.Conner SD, Schmid SL. 2003. Regulated portals of entry into the cell. Nature 422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 29.Helenius A, Kartenbeck J, Simons K, Fries E. 1980. On the entry of Semliki forest virus into BHK-21 cells. J Cell Biol 84:404–420. doi: 10.1083/jcb.84.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prchla E, Plank C, Wagner E, Blaas D, Fuchs R. 1995. Virus-mediated release of endosomal content in vitro: different behavior of adenovirus and rhinovirus serotype 2. J Cell Biol 131:111–123. doi: 10.1083/jcb.131.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schober D, Kronenberger P, Prchla E, Blaas D, Fuchs R. 1998. Major and minor receptor group human rhinoviruses penetrate from endosomes by different mechanisms. J Virol 72:1354–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, Lew E, Lemke G, Schwartz O, Amara A. 2012. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe 12:544–557. doi: 10.1016/j.chom.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morizono K, Xie Y, Olafsen T, Lee B, Dasgupta A, Wu AM, Chen ISY. 2011. The soluble serum protein Gas6 bridges virion envelope phosphatidylserine to the TAM receptor tyrosine kinase Axl to mediate viral entry. Cell Host Microbe 9:286–298. doi: 10.1016/j.chom.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Schaar HM, Rust MJ, Chen C, van der Ende-Metselaar H, Wilschut J, Zhuang X, Smit JM. 2008. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog 4:e1000244. doi: 10.1371/journal.ppat.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 36.Kitakura S, Vanneste S, Robert S, Löfke C, Teichmann T, Tanaka H, Friml J. 2011. Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23:1920–1931. doi: 10.1105/tpc.111.083030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lonhienne TGA, Sagulenko E, Webb RI, Lee K-C, Franke J, Devos DP, Nouwens A, Carroll BJ, Fuerst JA. 2010. Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci U S A 107:12883–12888. doi: 10.1073/pnas.1001085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franzmann PD, Skerman VB. 1984. Gemmata obscuriglobus, a new genus and species of the budding bacteria. Antonie Van Leeuwenhoek 50:261–268. doi: 10.1007/BF02342136. [DOI] [PubMed] [Google Scholar]
- 39.Ormö M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science 273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 40.Yang F, Moss LG, Phillips JGN. 1996. The molecular structure of green fluorescent protein. Nat Biotechnol 14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 41.Giraldo MC, Valent B. 2013. Filamentous plant pathogen effectors in action. Nat Rev Microbiol 11:800–814. doi: 10.1038/nrmicro3119. [DOI] [PubMed] [Google Scholar]
- 42.Lot H, Campbell RN, Souche S, Milne RG, Roggero P. 2002. Transmission by Olpidium brassicae of Mirafiori lettuce virus and Lettuce big-vein virus, and their roles in lettuce big-vein etiology. Phytopathology 92:288–293. doi: 10.1094/PHYTO.2002.92.3.288. [DOI] [PubMed] [Google Scholar]
- 43.Fields BN, Knipe DM, Howley PM (ed). 2013. Fields virology, 6th ed. Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 44.Lister RM. 1960. Transmission of soil-borne viruses through seed. Virology 10:547–549. doi: 10.1016/0042-6822(60)90138-0. [DOI] [PubMed] [Google Scholar]
- 45.Van Etten JL. 2003. Unusual life style of giant chlorella viruses. Annu Rev Genet 37:153–195. doi: 10.1146/annurev.genet.37.110801.143915. [DOI] [PubMed] [Google Scholar]
- 46.Mackinder LCM, Worthy CA, Biggi G, Hall M, Ryan KP, Varsani A, Harper GM, Wilson WH, Brownlee C, Schroeder DC. 2009. A unicellular algal virus, Emiliania huxleyi virus 86, exploits an animal-like infection strategy. J Gen Virol 90:2306–2316. doi: 10.1099/vir.0.011635-0. [DOI] [PubMed] [Google Scholar]
- 47.Sun L, Young LN, Zhang X, Boudko SP, Fokine A, Zbornik E, Roznowski AP, Molineux IJ, Rossmann MG, Fane BA. 2014. Icosahedral bacteriophage ϕX174 forms a tail for DNA transport during infection. Nature 505:432–435. [DOI] [PubMed] [Google Scholar]
- 48.Abrescia NGA, Grimes JM, Kivelä HM, Assenberg R, Sutton GC, Butcher SJ, Bamford JKH, Bamford DH, Stuart DI. 2008. Insights into virus evolution and membrane biogenesis from the structure of the marine lipid-containing bacteriophage PM2. Mol Cell 31:749–761. doi: 10.1016/j.molcel.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 49.Bamford DH, Romantschuk M, Somerharju PJ. 1987. Membrane fusion in prokaryotes: bacteriophage ϕ6 membrane fuses with the Pseudomonas syringae outer membrane. EMBO J 6:1467–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daugelavicius R, Cvirkaite V, Gaidelyte A, Bakiene E, Gabrenaite-Verkhovskaya R, Bamford DH. 2005. Penetration of enveloped double-stranded RNA bacteriophages ϕ13 and ϕ6 into Pseudomonas syringae cells. J Virol 79:5017–5026. doi: 10.1128/JVI.79.8.5017-5026.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peralta B, Gil-Carton D, Castaño-Dez D, Bertin A, Boulogne C, Oksanen HM, Bamford DH, Abrescia NGA. 2013. Mechanism of membranous tunnelling nanotube formation in viral genome delivery. PLoS Biol 11:e1001667. doi: 10.1371/journal.pbio.1001667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laurinmäki PA, Huiskonen JT, Bamford DH, Butcher SJ. 2005. Membrane proteins modulate the bilayer curvature in the bacterial virus Bam35. Structure 13:1819–1828. doi: 10.1016/j.str.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 53.Grahn AM, Daugelavicius R, Bamford DH. 2002. Sequential model of phage PRD1 DNA delivery: active involvement of the viral membrane. Mol Microbiol 46:1199–1209. doi: 10.1046/j.1365-2958.2002.03250.x. [DOI] [PubMed] [Google Scholar]
- 54.Mattila S, Oksanen HM, Bamford JKH. 2015. Probing protein interactions in the membrane-containing virus PRD1. J Gen Virol 96:453–462. doi: 10.1099/vir.0.069187-0. [DOI] [PubMed] [Google Scholar]
- 55.Strömsten NJ, Benson SD, Burnett RM, Bamford DH, Bamford JKH. 2003. The Bacillus thuringiensis linear double-stranded DNA phage Bam35, which is highly similar to the Bacillus cereus linear plasmid pBClin15, has a prophage state. J Bacteriol 185:6985–6989. doi: 10.1128/JB.185.23.6985-6989.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaidelyte A, Jaatinen ST, Daugelavicius R, Bamford JKH, Bamford DH. 2005. The linear double-stranded DNA of phage Bam35 enters lysogenic host cells, but the late phage functions are suppressed. J Bacteriol 187:3521–3527. doi: 10.1128/JB.187.10.3521-3527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaidelyte A, Cvirkaite-Krupovic V, Daugelavicius R, Bamford JKH, Bamford DH. 2006. The entry mechanism of membrane-containing phage Bam35 infecting Bacillus thuringiensis. J Bacteriol 188:5925–5934. doi: 10.1128/JB.00107-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daugelavicius R, Bamford JK, Bamford DH. 1997. Changes in host cell energetics in response to bacteriophage PRD1 DNA entry. J Bacteriol 179:5203–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maniloff J. 2011. Plasmavirus, p 1341–1345. In Tidona C, Darai G (ed), The Springer index of viruses. Springer, New York, NY. [Google Scholar]
- 60.Holland SJ, Sanz C, Perham RN. 2006. Identification and specificity of pilus adsorption proteins of filamentous bacteriophages infecting Pseudomonas aeruginosa. Virology 345:540–548. doi: 10.1016/j.virol.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 61.Rakonjac J, Bennett NJ, Spagnuolo J, Gagic D, Russel M. 2011. Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr Issues Mol Biol 13:51–76. [PubMed] [Google Scholar]
- 62.Carrington JC, Kasschau KD, Mahajan SK, Schaad MC. 1996. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8:1669–1681. doi: 10.1105/tpc.8.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen JT, Wu HK. 1977. Hyphal anastomosis in Pyricularia oryzae cav. Protoplasma 92:281–287. doi: 10.1007/BF01279465. [DOI] [Google Scholar]
- 64.Glass NL, Dementhon K. 2006. Non-self recognition and programmed cell death in filamentous fungi. Curr Opin Microbiol 9:553–558. doi: 10.1016/j.mib.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 65.Watson MA, Roberts FM. 1939. A comparative study of the transmission of Hyoscyamus virus 3, potato virus Y and cucumber virus 1 by the vectors Myzus persicae (Sulz), M. circumflexus (Buckton), and Macrosiphum gei (Koch). Proc R Soc Lond B Biol Sci 127:543–576. doi: 10.1098/rspb.1939.0039. [DOI] [Google Scholar]
- 66.Benitez-Alfonso Y, Faulkner C, Ritzenthaler C, Maule AJ. 2010. Plasmodesmata: gateways to local and systemic virus infection. Mol Plant Microbe Interact 23:1403–1412. doi: 10.1094/MPMI-05-10-0116. [DOI] [PubMed] [Google Scholar]
- 67.Hipper C, Brault V, Ziegler-Graff V, Revers F. 2013. Viral and cellular factors involved in phloem transport of plant viruses. Front Plant Sci 4:154. doi: 10.3389/fpls.2013.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kikkert M, Van Lent J, Storms M, Bodegom P, Kormelink R, Goldbach R. 1999. Tomato spotted wilt virus particle morphogenesis in plant cells. J Virol 73:2288–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Padidam M, Beachy RN, Fauquet CM. 1995. Tomato leaf curl geminivirus from India has a bipartite genome and coat protein is not essential for infectivity. J Gen Virol 76:25–35. doi: 10.1099/0022-1317-76-1-25. [DOI] [PubMed] [Google Scholar]
- 70.Padidam M, Beachy RN, Fauquet CM. 1996. The role of AV2 (“precoat”) and coat protein in viral replication and movement in tomato leaf curl geminivirus. Virology 224:390–404. doi: 10.1006/viro.1996.0546. [DOI] [PubMed] [Google Scholar]
- 71.Scholthof HB, Morirs TJ, Jackson AO. 1993. The capsid protein gene of tomato bushy stunt virus is dispensable for systemic movement and can be replaced for localized expression of foreign genes. Mol Plant Microbe Interact 6:309–322. doi: 10.1094/MPMI-6-309. [DOI] [Google Scholar]
- 72.Desvoyes B, Scholthof HB. 2002. Host-dependent recombination of a Tomato bushy stunt virus coat protein mutant yields truncated capsid subunits that form virus-like complexes which benefit systemic spread. Virology 304:434–442. doi: 10.1006/viro.2002.1714. [DOI] [PubMed] [Google Scholar]
- 73.Qu F, Morris TJ. 2002. Efficient infection of Nicotiana benthamiana by Tomato bushy stunt virus is facilitated by the coat protein and maintained by p19 through suppression of gene silencing. Mol Plant Microbe Interact 15:193–202. doi: 10.1094/MPMI.2002.15.3.193. [DOI] [PubMed] [Google Scholar]
- 74.Ammar E-D, Tsai C-W, Whitfield AE, Redinbaugh MG, Hogenhout SA. 2009. Cellular and molecular aspects of rhabdovirus interactions with insect and plant hosts. Annu Rev Entomol 54:447–468. doi: 10.1146/annurev.ento.54.110807.090454. [DOI] [PubMed] [Google Scholar]
- 75.Tilsner J, Taliansky ME, Torrance L. 2001. Plant virus movement. John Wiley & Sons, Ltd, London, United Kingdom. [Google Scholar]
- 76.Mielke-Ehret N, Mühlbach H-P. 2012. Emaravirus: a novel genus of multipartite, negative strand RNA plant viruses. Viruses 4:1515–1536. doi: 10.3390/v4091515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ishikawa K, Maejima K, Komatsu K, Netsu O, Keima T, Shiraishi T, Okano Y, Hashimoto M, Yamaji Y, Namba S. 2013. Fig mosaic emaravirus p4 protein is involved in cell-to-cell movement. J Gen Virol 94:682–686. doi: 10.1099/vir.0.047860-0. [DOI] [PubMed] [Google Scholar]
- 78.Nagata T, Inoue-Nagata AK, Prins M, Goldbach R, Peters D. 2000. Impeded thrips transmission of defective Tomato spotted wilt virus isolates. Phytopathology 90:454–459. doi: 10.1094/PHYTO.2000.90.5.454. [DOI] [PubMed] [Google Scholar]
- 79.Whitfield AE, Ullman DE, German TL. 2004. Expression and characterization of a soluble form of tomato spotted wilt virus glycoprotein GN. J Virol 78:13197–13206. doi: 10.1128/JVI.78.23.13197-13206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hogenhout SA, Ammar E-D, Whitfield AE, Redinbaugh MG. 2008. Insect vector interactions with persistently transmitted viruses. Annu Rev Phytopathol 46:327–359. doi: 10.1146/annurev.phyto.022508.092135. [DOI] [PubMed] [Google Scholar]
- 81.Chen BJ, Lamb RA. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Berry J, Rajaure M, Pang T, Young R. 2012. The spanin complex is essential for lambda lysis. J Bacteriol 194:5667–5674. doi: 10.1128/JB.01245-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang I-N, Deaton J, Young R. 2003. Sizing the holin lesion with an endolysin-beta-galactosidase fusion. J Bacteriol 185:779–787. doi: 10.1128/JB.185.3.779-787.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dewey JS, Savva CG, White RL, Vitha S, Holzenburg A, Young R. 2010. Micron-scale holes terminate the phage infection cycle. Proc Natl Acad Sci U S A 107:2219–2223. doi: 10.1073/pnas.0914030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.White R, Chiba S, Pang T, Dewey JS, Savva CG, Holzenburg A, Pogliano K, Young R. 2011. Holin triggering in real time. Proc Natl Acad Sci U S A 108:798–803. doi: 10.1073/pnas.1011921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Park T, Struck DK, Dankenbring CA, Young R. 2007. The pinholin of lambdoid phage 21: control of lysis by membrane depolarization. J Bacteriol 189:9135–9139. doi: 10.1128/JB.00847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pang T, Fleming TC, Pogliano K, Young R. 2013. Visualization of pinholin lesions in vivo. Proc Natl Acad Sci U S A 110:E2054–E2063. doi: 10.1073/pnas.1222283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Young R. 2014. Phage lysis: three steps, three choices, one outcome. J Microbiol 52:243–258. doi: 10.1007/s12275-014-4087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li C-X, Shi M, Tian J-H, Lin X-D, Kang Y-J, Chen L-J, Qin X-C, Xu J, Holmes EC, Zhang Y-Z. 2015. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife 4:e05378. doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peterson KJ, Lyons JB, Nowak KS, Takacs CM, Wargo MJ, McPeek MA. 2004. Estimating metazoan divergence times with a molecular clock. Proc Natl Acad Sci U S A 101:6536–6541. doi: 10.1073/pnas.0401670101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Labandeira CC. 2005. Invasion of the continents: cyanobacterial crusts to tree-inhabiting arthropods. Trends Ecol Evol 20:253–262. doi: 10.1016/j.tree.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 92.Goldbach R, de Haan P. 1994. RNA viral supergroups and the evolution of RNA viruses, p 105–119. In Morse SS. (ed), The evolutionary biology of viruses. Raven Press, New York, NY. [Google Scholar]
- 93.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong S-H, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morin S, Ghanim M, Sobol I, Czosnek H. 2000. The GroEL protein of the whitefly Bemisia tabaci interacts with the coat protein of transmissible and nontransmissible begomoviruses in the yeast two-hybrid system. Virology 276:404–416. doi: 10.1006/viro.2000.0549. [DOI] [PubMed] [Google Scholar]
- 95.Bouvaine S, Boonham N, Douglas AE. 2011. Interactions between a luteovirus and the GroEL chaperonin protein of the symbiotic bacterium Buchnera aphidicola of aphids. J Gen Virol 92:1467–1474. doi: 10.1099/vir.0.029355-0. [DOI] [PubMed] [Google Scholar]
- 96.Morin S, Ghanim M, Zeidan M, Czosnek H, Verbeek M, van den Heuvel JF. 1999. A GroEL homologue from endosymbiotic bacteria of the whitefly Bemisia tabaci is implicated in the circulative transmission of tomato yellow leaf curl virus. Virology 256:75–84. doi: 10.1006/viro.1999.9631. [DOI] [PubMed] [Google Scholar]
- 97.van den Heuvel JF, Bruyère A, Hogenhout SA, Ziegler-Graff V, Brault V, Verbeek M, van der Wilk F, Richards K. 1997. The N-terminal region of the luteovirus readthrough domain determines virus binding to Buchnera GroEL and is essential for virus persistence in the aphid. J Virol 71:7258–7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jackson T, Sheppard D, Denyer M, Blakemore W, King AM. 2000. The epithelial integrin αvβ6 is a receptor for foot-and-mouth disease virus. J Virol 74:4949–4956. doi: 10.1128/JVI.74.11.4949-4956.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.O'Donnell V, Larocco M, Baxt B. 2008. Heparan sulfate-binding foot-and-mouth disease virus enters cells via caveola-mediated endocytosis. J Virol 82:9075–9085. doi: 10.1128/JVI.00732-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Quillin ML, Matthews BW. 2000. Accurate calculation of the density of proteins. Acta Crystallogr D Biol Crystallogr 56:791–794. doi: 10.1107/S090744490000679X. [DOI] [PubMed] [Google Scholar]
- 101.Fischer H, Polikarpov I, Craievich AF. 2004. Average protein density is a molecular-weight-dependent function. Protein Sci 13:2825–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Perry JW, Wobus CE. 2010. Endocytosis of murine norovirus 1 into murine macrophages is dependent on dynamin II and cholesterol. J Virol 84:6163–6176. doi: 10.1128/JVI.00331-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Amstutz B, Gastaldelli M, Kälin S, Imelli N, Boucke K, Wandeler E, Mercer J, Hemmi S, Greber UF. 2008. Subversion of CtBP1-controlled macropinocytosis by human adenovirus serotype 3. EMBO J 27:956–969. doi: 10.1038/emboj.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Leopold PL, Crystal RG. 2007. Intracellular trafficking of adenovirus: many means to many ends. Adv Drug Deliv Rev 59:810–821. doi: 10.1016/j.addr.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 105.Salinas S, Bilsland LG, Henaff D, Weston AE, Keriel A, Schiavo G, Kremer EJ. 2009. CAR-associated vesicular transport of an adenovirus in motor neuron axons. PLoS Pathog 5:e1000442. doi: 10.1371/journal.ppat.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bartlett JS, Wilcher R, Samulski RJ. 2000. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J Virol 74:2777–2785. doi: 10.1128/JVI.74.6.2777-2785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sapp M, Bienkowska-Haba M. 2009. Viral entry mechanisms: human papillomavirus and a long journey from extracellular matrix to the nucleus. FEBS J 276:7206–7216. doi: 10.1111/j.1742-4658.2009.07400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yip CW, Hon CC, Zeng F, Leung FCC. 2012. Cell culture-adapted IBDV uses endocytosis for entry in DF-1 chicken embryonic fibroblasts. Virus Res 165:9–16. doi: 10.1016/j.virusres.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 109.Kivelä HM, Daugelavicius R, Hankkio RH, Bamford JKH, Bamford DH. 2004. Penetration of membrane-containing double-stranded-DNA bacteriophage PM2 into Pseudoalteromonas hosts. J Bacteriol 186:5342–5354. doi: 10.1128/JB.186.16.5342-5354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Duquerroy S, Da Costa B, Henry C, Vigouroux A, Libersou S, Lepault J, Navaza J, Delmas B, Rey FA. 2009. The picobirnavirus crystal structure provides functional insights into virion assembly and cell entry. EMBO J 28:1655–1665. doi: 10.1038/emboj.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kalia M, Chandra V, Rahman SA, Sehgal D, Jameel S. 2009. Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J Virol 83:12714–12724. doi: 10.1128/JVI.00717-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Méndez E, Muñoz-Yañez C, Sánchez-San Martín C, Aguirre-Crespo G, del Rocio Baños-Lara M, Gutierrez M, Espinosa R, Acevedo Y, Arias CF, López S. 2014. Characterization of human astrovirus cell entry. J Virol 88:2452–2460. doi: 10.1128/JVI.02908-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bergelson JM. 2008. New (fluorescent) light on poliovirus entry. Trends Microbiol 16:44–47. doi: 10.1016/j.tim.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gerondopoulos A, Jackson T, Monaghan P, Doyle N, Roberts LO. 2010. Murine norovirus-1 cell entry is mediated through a non-clathrin-, non-caveolae-, dynamin- and cholesterol-dependent pathway. J Gen Virol 91:1428–1438. doi: 10.1099/vir.0.016717-0. [DOI] [PubMed] [Google Scholar]
- 115.Leiman PG, Shneider MM. 2012. Contractile tail machines of bacteriophages. Adv Exp Med Biol 726:93–114. doi: 10.1007/978-1-4614-0980-9_5. [DOI] [PubMed] [Google Scholar]
- 116.Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, Rossmann MG. 2002. Structure of the cell-puncturing device of bacteriophage T4. Nature 415:553–557. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- 117.Schelhaas M, Shah B, Holzer M, Blattmann P, Kühling L, Day PM, Schiller JT, Helenius A. 2012. Entry of human papillomavirus type 16 by actin-dependent, clathrin- and lipid raft-independent endocytosis. PLoS Pathog 8:e1002657. doi: 10.1371/journal.ppat.1002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Misinzo G, Meerts P, Bublot M, Mast J, Weingartl HM, Nauwynck HJ. 2005. Binding and entry characteristics of porcine circovirus 2 in cells of the porcine monocytic line 3D4/31. J Gen Virol 86:2057–2068. doi: 10.1099/vir.0.80652-0. [DOI] [PubMed] [Google Scholar]
- 119.Neu U, Stehle T, Atwood WJ. 2009. The Polyomaviridae: contributions of virus structure to our understanding of virus receptors and infectious entry. Virology 384:389–399. doi: 10.1016/j.virol.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Eash S, Querbes W, Atwood WJ. 2004. Infection of Vero cells by BK virus is dependent on caveolae. J Virol 78:11583–11590. doi: 10.1128/JVI.78.21.11583-11590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nash TC, Buchmeier MJ. 1997. Entry of mouse hepatitis virus into cells by endosomal and nonendosomal pathways. Virology 233:1–8. doi: 10.1006/viro.1997.8609. [DOI] [PubMed] [Google Scholar]
- 122.Aksyuk AA, Bowman VD, Kaufmann B, Fields C, Klose T, Holdaway HA, Fischetti VA, Rossmann MG. 2012. Structural investigations of a Podoviridae streptococcus phage C1, implications for the mechanism of viral entry. Proc Natl Acad Sci U S A 109:14001–14006. doi: 10.1073/pnas.1207730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Canaan S, Zádori Z, Ghomashchi F, Bollinger J, Sadilek M, Moreau ME, Tijssen P, Gelb MH. 2004. Interfacial enzymology of parvovirus phospholipases A2. J Biol Chem 279:14502–14508. doi: 10.1074/jbc.M312630200. [DOI] [PubMed] [Google Scholar]
- 124.Girod A, Wobus CE, Zádori Z, Ried M, Leike K, Tijssen P, Kleinschmidt JA, Hallek M. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J Gen Virol 83:973–978. [DOI] [PubMed] [Google Scholar]
- 125.Ghigo E, Kartenbeck J, Lien P, Pelkmans L, Capo C, Mege J-L, Raoult D. 2008. Ameobal pathogen mimivirus infects macrophages through phagocytosis. PLoS Pathog 4:e1000087. doi: 10.1371/journal.ppat.1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Reinbold C, Herrbach E, Brault V. 2003. Posterior midgut and hindgut are both sites of acquisition of Cucurbit aphid-borne yellows virus in Myzus persicae and Aphis gossypii. J Gen Virol 84:3473–3484. doi: 10.1099/vir.0.19415-0. [DOI] [PubMed] [Google Scholar]
- 127.Macovei A, Radulescu C, Lazar C, Petrescu S, Durantel D, Dwek RA, Zitzmann N, Nichita NB. 2010. Hepatitis B virus requires intact caveolin-1 function for productive infection in HepaRG cells. J Virol 84:243–253. doi: 10.1128/JVI.01207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Takeda M, Leser GP, Russell CJ, Lamb RA. 2003. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc Natl Acad Sci U S A 100:14610–14617. doi: 10.1073/pnas.2235620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mackay DJ, Bode VC. 1976. Events in lambda injection between phage adsorption and DNA entry. Virology 72:154–166. doi: 10.1016/0042-6822(76)90320-2. [DOI] [PubMed] [Google Scholar]
- 130.Rivero MR, Jausoro I, Bisbal M, Feliziani C, Lanfredi-Rangel A, Touz MC. 2013. Receptor-mediated endocytosis and trafficking between endosomal-lysosomal vacuoles in Giardia lamblia. Parasitol Res 112:1813–1818. doi: 10.1007/s00436-012-3253-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Horvath CAJ, Boulet GAV, Renoux VM, Delvenne PO, Bogers J-PJ. 2010. Mechanisms of cell entry by human papillomaviruses: an overview. Virol J 7:11. doi: 10.1186/1743-422X-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sapp M, Day PM. 2009. Structure, attachment and entry of polyoma- and papillomaviruses. Virology 384:400–409. doi: 10.1016/j.virol.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 133.Beer C, Andersen DS, Rojek A, Pedersen L. 2005. Caveola-dependent endocytic entry of amphotropic murine leukemia virus. J Virol 79:10776–10787. doi: 10.1128/JVI.79.16.10776-10787.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gendrault JL, Steffan AM, Bingen A, Kirn A. 1981. Penetration and uncoating of frog virus 3 (FV3) in cultured rat Kupffer cells. Virology 112:375–384. doi: 10.1016/0042-6822(81)90284-1. [DOI] [PubMed] [Google Scholar]
- 135.Stoeckl L, Funk A, Kopitzki A, Brandenburg B, Oess S, Will H, Sirma H, Hildt E. 2006. Identification of a structural motif crucial for infectivity of hepatitis B viruses. Proc Natl Acad Sci U S A 103:6730–6734. doi: 10.1073/pnas.0509765103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, Halfmann P, Kawaoka Y. 2010. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog 6:e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. 2010. Cellular entry of Ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog 6:e1001110. doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hunt CL, Kolokoltsov AA, Davey RA, Maury W. 2011. The Tyro3 receptor kinase Axl enhances macropinocytosis of Zaire ebolavirus. J Virol 85:334–347. doi: 10.1128/JVI.01278-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mathapati BS, Mishra N, Rajukumar K, Nema RK, Behera SP, Dubey SC. 2010. Entry of bovine viral diarrhea virus into ovine cells occurs through clathrin-dependent endocytosis and low pH-dependent fusion. In Vitro Cell Dev Biol Anim 46:403–407. doi: 10.1007/s11626-009-9263-9. [DOI] [PubMed] [Google Scholar]
- 140.Castilla V, Mersich SE. 1996. Low-pH-induced fusion of Vero cells infected with Junin virus. Arch Virol 141:1307–1317. doi: 10.1007/BF01718832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Raghu H, Sharma-Walia N, Veettil MV, Sadagopan S, Chandran B. 2009. Kaposi's sarcoma-associated herpesvirus utilizes an actin polymerization-dependent macropinocytic pathway to enter human dermal microvascular endothelial and human umbilical vein endothelial cells. J Virol 83:4895–4911. doi: 10.1128/JVI.02498-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kapur N, Thakral D, Durgapal H, Panda SK. 2012. Hepatitis E virus enters liver cells through receptor-dependent clathrin-mediated endocytosis. J Viral Hepat 19:436–448. doi: 10.1111/j.1365-2893.2011.01559.x. [DOI] [PubMed] [Google Scholar]
- 143.Fuchs R, Blaas D. 2010. Uncoating of human rhinoviruses. Rev Med Virol 20:281–297. doi: 10.1002/rmv.654. [DOI] [PubMed] [Google Scholar]
- 144.Granados RR, Lawler KA. 1981. In vivo pathway of Autographa californica baculovirus invasion and infection. Virology 108:297–308. doi: 10.1016/0042-6822(81)90438-4. [DOI] [PubMed] [Google Scholar]
- 145.Odegard AL, Kwan MH, Walukiewicz HE, Banerjee M, Schneemann A, Johnson JE. 2009. Low endocytic pH and capsid protein autocleavage are critical components of Flock House virus cell entry. J Virol 83:8628–8637. doi: 10.1128/JVI.00873-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pho MT, Ashok A, Atwood WJ. 2000. JC virus enters human glial cells by clathrin-dependent receptor-mediated endocytosis. J Virol 74:2288–2292. doi: 10.1128/JVI.74.5.2288-2292.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Belouzard S, Millet JK, Licitra BN, Whittaker GR. 2012. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pernet O, Pohl C, Ainouze M, Kweder H, Buckland R. 2009. Nipah virus entry can occur by macropinocytosis. Virology 395:298–311. doi: 10.1016/j.virol.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 149.Krzyzaniak MA, Zumstein MT, Gerez JA, Picotti P, Helenius A. 2013. Host cell entry of respiratory syncytial virus involves macropinocytosis followed by proteolytic activation of the F protein. PLoS Pathog 9:e1003309. doi: 10.1371/journal.ppat.1003309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Danthi P, Guglielmi KM, Kirchner E, Mainou B, Stehle T, Dermody TS. 2010. From touchdown to transcription: the reovirus cell entry pathway. Curr Top Microbiol Immunol 343:91–119. doi: 10.1007/82_2010_32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Forzan M, Marsh M, Roy P. 2007. Bluetongue virus entry into cells. J Virol 81:4819–4827. doi: 10.1128/JVI.02284-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Braunwald J, Nonnenmacher H, Tripier-Darcy F. 1985. Ultrastructural and biochemical study of frog virus 3 uptake by BHK-21 cells. J Gen Virol 66:283–293. doi: 10.1099/0022-1317-66-2-283. [DOI] [PubMed] [Google Scholar]
- 153.Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH. 2012. Herpes virus fusion and entry: a story with many characters. Viruses 4:800–832. doi: 10.3390/v4050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Burkard C, Verheije MH, Wicht O, van Kasteren SI, van Kuppeveld FJ, Haagmans BL, Pelkmans L, Rottier PJM, Bosch BJ, de Haan CAM. 2014. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog 10:e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jouaux A, Lafont M, Blin J-L, Houssin M, Mathieu M, Lelong C. 2013. Physiological change under OsHV-1 contamination in pacific oyster Crassostrea gigas through massive mortality events on fields. BMC Genomics 14:590. doi: 10.1186/1471-2164-14-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Martinez MG, Forlenza MB, Candurra NA. 2009. Involvement of cellular proteins in Junin arenavirus entry. Biotechnol J 4:866–870. doi: 10.1002/biot.200800357. [DOI] [PubMed] [Google Scholar]
- 157.Smith EC, Popa A, Chang A, Masante C, Dutch RE. 2009. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. FEBS J 276:7217–7227. doi: 10.1111/j.1742-4658.2009.07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nitschke M, Korte T, Tielesch C, Ter-Avetisyan G, Tünnemann G, Cardoso MC, Veit M, Herrmann A. 2008. Equine arteritis virus is delivered to an acidic compartment of host cells via clathrin-dependent endocytosis. Virology 377:248–254. doi: 10.1016/j.virol.2008.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nauwynck HJ, Duan X, Favoreel HW, Van Oostveldt P, Pensaert MB. 1999. Entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophages via receptor-mediated endocytosis. J Gen Virol 80:297–305. [DOI] [PubMed] [Google Scholar]
- 160.Van Breedam W, Delputte PL, Van Gorp H, Misinzo G, Vanderheijden N, Duan X, Nauwynck HJ. 2010. Porcine reproductive and respiratory syndrome virus entry into the porcine macrophage. J Gen Virol 91:1659–1667. doi: 10.1099/vir.0.020503-0. [DOI] [PubMed] [Google Scholar]
- 161.Maniloff J. 1988. Mycoplasma viruses. Crit Rev Microbiol 15:339–389. doi: 10.3109/10408418809104462. [DOI] [PubMed] [Google Scholar]
- 162.Hernaez B, Alonso C. 2010. Dynamin- and clathrin-dependent endocytosis in African swine fever virus entry. J Virol 84:2100–2109. doi: 10.1128/JVI.01557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Stoltz DB, Vinson SB. 1979. Penetration into caterpillar cells of virus-like particles injected during oviposition by parasitoid ichneumonid wasps. Can J Microbiol 25:207–216. doi: 10.1139/m79-032. [DOI] [PubMed] [Google Scholar]
- 164.Stoltz DB, Vinson SB, MacKinnon EA. 1976. Baculovirus-like particles in the reproductive tracts of female parasitoid wasps. Can J Microbiol 22:1013–1023. doi: 10.1139/m76-148. [DOI] [PubMed] [Google Scholar]
- 165.Stoltz D, Lapointe R, Makkay A, Cusson M. 2007. Exposure of ichnovirus particles to digitonin leads to enhanced infectivity and induces fusion from without in an in vitro model system. J Gen Virol 88:2977–2984. doi: 10.1099/vir.0.83118-0. [DOI] [PubMed] [Google Scholar]
- 166.Long G, Pan X, Kormelink R, Vlak JM. 2006. Functional entry of baculovirus into insect and mammalian cells is dependent on clathrin-mediated endocytosis. J Virol 80:8830–8833. doi: 10.1128/JVI.00880-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Volkman LE, Goldsmith PA. 1985. Mechanism of neutralization of budded Autographa californica nuclear polyhedrosis virus by a monoclonal antibody: inhibition of entry by adsorptive endocytosis. Virology 143:185–195. doi: 10.1016/0042-6822(85)90107-2. [DOI] [PubMed] [Google Scholar]
- 168.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 169.Clemente R, de la Torre JC. 2009. Cell entry of Borna disease virus follows a clathrin-mediated endocytosis pathway that requires Rab5 and microtubules. J Virol 83:10406–10416. doi: 10.1128/JVI.00990-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Simon M, Johansson C, Mirazimi A. 2009. Crimean-Congo hemorrhagic fever virus entry and replication is clathrin-, pH- and cholesterol-dependent. J Gen Virol 90:210–215. doi: 10.1099/vir.0.006387-0. [DOI] [PubMed] [Google Scholar]
- 171.Bhattacharyya S, Warfield KL, Ruthel G, Bavari S, Aman MJ, Hope TJ. 2010. Ebola virus uses clathrin-mediated endocytosis as an entry pathway. Virology 401:18–28. doi: 10.1016/j.virol.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Suksanpaisan L, Susantad T, Smith DR. 2009. Characterization of dengue virus entry into HepG2 cells. J Biomed Sci 16:17. doi: 10.1186/1423-0127-16-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chu JJH, Ng ML. 2004. Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J Virol 78:10543–10555. doi: 10.1128/JVI.78.19.10543-10555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chen C, Zhuang X. 2008. Epsin 1 is a cargo-specific adaptor for the clathrin-mediated endocytosis of the influenza virus. Proc Natl Acad Sci U S A 105:11790–11795. doi: 10.1073/pnas.0803711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Diederich S, Moll M, Klenk H-D, Maisner A. 2005. The Nipah virus fusion protein is cleaved within the endosomal compartment. J Biol Chem 280:29899–29903. doi: 10.1074/jbc.M504598200. [DOI] [PubMed] [Google Scholar]
- 176.Diaz-Griffero F, Jackson AP, Brojatsch J. 2005. Cellular uptake of avian leukosis virus subgroup B is mediated by clathrin. Virology 337:45–54. doi: 10.1016/j.virol.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 177.Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. 2009. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137:433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cureton DK, Massol RH, Whelan SPJ, Kirchhausen T. 2010. The length of vesicular stomatitis virus particles dictates a need for actin assembly during clathrin-dependent endocytosis. PLoS Pathog 6:e1001127. doi: 10.1371/journal.ppat.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.DeTulleo L, Kirchhausen T. 1998. The clathrin endocytic pathway in viral infection. EMBO J 17:4585–4593. doi: 10.1093/emboj/17.16.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kee S-H, Cho E-J, Song J-W, Park KS, Baek LJ, Song K-J. 2004. Effects of endocytosis inhibitory drugs on rubella virus entry into VeroE6 cells. Microbiol Immunol 48:823–829. doi: 10.1111/j.1348-0421.2004.tb03614.x. [DOI] [PubMed] [Google Scholar]
- 181.Vonderheit A, Helenius A. 2005. Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLoS Biol 3:e233. doi: 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Blank CA, Anderson DA, Beard M, Lemon SM. 2000. Infection of polarized cultures of human intestinal epithelial cells with hepatitis A virus: vectorial release of progeny virions through apical cellular membranes. J Virol 74:6476–6484. doi: 10.1128/JVI.74.14.6476-6484.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Emerson SU, Nguyen HT, Torian U, Burke D, Engle R, Purcell RH. 2010. Release of genotype 1 hepatitis E virus from cultured hepatoma and polarized intestinal cells depends on open reading frame 3 protein and requires an intact PXXP motif. J Virol 84:9059–9069. doi: 10.1128/JVI.00593-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Desai M, Pal R, Deshmukh R, Banker D. 2005. Replication of TT virus in hepatocyte and leucocyte cell lines. J Med Virol 77:136–143. doi: 10.1002/jmv.20426. [DOI] [PubMed] [Google Scholar]
- 185.Maggi F, Bendinelli M. 2010. Human anelloviruses and the central nervous system. Rev Med Virol 20:392–407. doi: 10.1002/rmv.668. [DOI] [PubMed] [Google Scholar]
- 186.Perez M, Craven RC, de la Torre JC. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc Natl Acad Sci U S A 100:12978–12983. doi: 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wirblich C, Bhattacharya B, Roy P. 2006. Nonstructural protein 3 of bluetongue virus assists virus release by recruiting ESCRT-I protein Tsg101. J Virol 80:460–473. doi: 10.1128/JVI.80.1.460-473.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Méndez E, Salas-Ocampo E, Arias CF. 2004. Caspases mediate processing of the capsid precursor and cell release of human astroviruses. J Virol 78:8601–8608. doi: 10.1128/JVI.78.16.8601-8608.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Garret A, Kerlan C, Thomas D. 1993. The intestine is a site of passage for potato leafroll virus from the gut lumen into the haemocoel in the aphid vector, Myzus persicae Sulz. Arch Virol 131:377–392. doi: 10.1007/BF01378639. [DOI] [PubMed] [Google Scholar]
- 190.Dolnik O, Kolesnikova L, Stevermann L, Becker S. 2010. Tsg101 is recruited by a late domain of the nucleocapsid protein to support budding of Marburg virus-like particles. J Virol 84:7847–7856. doi: 10.1128/JVI.00476-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Viñuela E. 1985. African swine fever virus. Curr Top Microbiol Immunol 116:151–170. [DOI] [PubMed] [Google Scholar]
- 192.Wu Y, Hong L, Ye J, Huang Z, Zhou J. 2009. The VP5 protein of infectious bursal disease virus promotes virion release from infected cells and is not involved in cell death. Arch Virol 154:1873–1882. doi: 10.1007/s00705-009-0524-4. [DOI] [PubMed] [Google Scholar]
- 193.Galloux M, Libersou S, Morellet N, Bouaziz S, Da Costa B, Ouldali M, Lepault J, Delmas B. 2007. Infectious bursal disease virus, a non-enveloped virus, possesses a capsid-associated peptide that deforms and perforates biological membranes. J Biol Chem 282:20774–20784. doi: 10.1074/jbc.M701048200. [DOI] [PubMed] [Google Scholar]
- 194.Zirkel F, Kurth A, Quan P-L, Briese T, Ellerbrok H, Pauli G, Leendertz FH, Lipkin WI, Ziebuhr J, Drosten C, Junglen S. 2011. An insect nidovirus emerging from a primary tropical rainforest. mBio 2(3):e00077-11. doi: 10.1128/mBio.00077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ariumi Y, Kuroki M, Maki M, Ikeda M, Dansako H, Wakita T, Kato N. 2011. The ESCRT system is required for hepatitis C virus production. PLoS One 6:e14517. doi: 10.1371/journal.pone.0014517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Okano K, Vanarsdall AL, Mikhailov VS, Rohrmann GF. 2006. Conserved molecular systems of the Baculoviridae. Virology 344:77–87. doi: 10.1016/j.virol.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 197.Kaufman SS, Chatterjee NK, Fuschino ME, Morse DL, Morotti RA, Magid MS, Gondolesi GE, Florman SS, Fishbein TM. 2005. Characteristics of human calicivirus enteritis in intestinal transplant recipients. J Pediatr Gastroenterol Nutr 40:328–333. doi: 10.1097/01.MPG.0000155182.54001.48. [DOI] [PubMed] [Google Scholar]
- 198.Félix M-A, Ashe A, Piffaretti J, Wu G, Nuez I, Bélicard T, Jiang Y, Zhao G, Franz CJ, Goldstein LD, Sanroman M, Miska EA, Wang D. 2011. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol 9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wirblich C, Tan GS, Papaneri A, Godlewski PJ, Orenstein JM, Harty RN, Schnell MJ. 2008. PPEY motif within the rabies virus (RV) matrix protein is essential for efficient virion release and RV pathogenicity. J Virol 82:9730–9738. doi: 10.1128/JVI.00889-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Klumperman J, Locker JK, Meijer A, Horzinek MC, Geuze HJ, Rottier PJ. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J Virol 68:6523–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Krupovic M, Daugelavicius R, Bamford DH. 2007. A novel lysis system in PM2, a lipid-containing marine double-stranded DNA bacteriophage. Mol Microbiol 64:1635–1648. doi: 10.1111/j.1365-2958.2007.05769.x. [DOI] [PubMed] [Google Scholar]
- 202.Rautava J, Syrjänen S. 2012. Biology of human papillomavirus infections in head and neck carcinogenesis. Head Neck Pathol 6(Suppl 1):S3–S15. doi: 10.1007/s12105-012-0367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lambert C, Döring T, Prange R. 2007. Hepatitis B virus maturation is sensitive to functional inhibition of ESCRT-III, Vps4, and gamma 2-adaptin. J Virol 81:9050–9060. doi: 10.1128/JVI.00479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chinchar VG, Yu KH, Jancovich JK. 2011. The molecular biology of frog virus 3 and other iridoviruses infecting cold-blooded vertebrates. Viruses 3:1959–1985. doi: 10.3390/v3101959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Karnik S, Billeter M. 1983. The lysis function of RNA bacteriophage Qbeta is mediated by the maturation (A2) protein. EMBO J 2:1521–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Vinjé J, Oudejans SJG, Stewart JR, Sobsey MD, Long SC. 2004. Molecular detection and genotyping of male-specific coliphages by reverse transcription-PCR and reverse line blot hybridization. Appl Environ Microbiol 70:5996–6004. doi: 10.1128/AEM.70.10.5996-6004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Vestergaard G, Häring M, Peng X, Rachel R, Garrett RA, Prangishvili D. 2005. A novel rudivirus, ARV1, of the hyperthermophilic archaeal genus Acidianus. Virology 336:83–92. doi: 10.1016/j.virol.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 208.Ruiss R, Jochum S, Wanner G, Reisbach G, Hammerschmidt W, Zeidler R. 2011. A virus-like particle-based Epstein-Barr virus vaccine. J Virol 85:13105–13113. doi: 10.1128/JVI.05598-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Mihindukulasuriya KA, Nguyen NL, Wu G, Huang HV, Travassos da Rosa APA, Popov VL, Tesh RB, Wang D. 2009. Nyamanini and Midway viruses define a novel taxon of RNA viruses in the order Mononegavirales. J Virol 83:5109–5116. doi: 10.1128/JVI.02667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Tai V, Lawrence JE, Lang AS, Chan AM, Culley AI, Suttle CA. 2003. Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae). J Phycol 39:343–352. doi: 10.1046/j.1529-8817.2003.01162.x. [DOI] [Google Scholar]
- 211.Lawrence JE, Brussaard CPD, Suttle CA. 2006. Virus-specific responses of Heterosigma akashiwo to infection. Appl Environ Microbiol 72:7829–7834. doi: 10.1128/AEM.01207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wang AL, Wang CC. 1986. Discovery of a specific double-stranded RNA virus in Giardia lamblia. Mol Biochem Parasitol 21:269–276. doi: 10.1016/0166-6851(86)90132-5. [DOI] [PubMed] [Google Scholar]
- 213.Ciancanelli MJ, Basler CF. 2006. Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J Virol 80:12070–12078. doi: 10.1128/JVI.01743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Leser GP, Lamb RA. 2005. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 342:215–227. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 215.Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L, Robert C, Azza S, Sun S, Rossmann MG, Suzan-Monti M, La Scola B, Koonin EV, Raoult D. 2009. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci U S A 106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Watanabe Y, Ibrahim MS, Hagiwara K, Okamoto M, Kamitani W, Yanai H, Ohtaki N, Hayashi Y, Taniyama H, Ikuta K, Tomonaga K. 2007. Characterization of a Borna disease virus field isolate which shows efficient viral propagation and transmissibility. Microbes Infect 9:417–427. doi: 10.1016/j.micinf.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 217.Honeychurch KM, Yang G, Jordan R, Hruby DE. 2007. The vaccinia virus F13L YPPL motif is required for efficient release of extracellular enveloped virus. J Virol 81:7310–7315. doi: 10.1128/JVI.00034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Poddar SK, Cadden SP, Das J, Maniloff J. 1985. Heterogeneous progeny viruses are produced by a budding enveloped phage. Intervirology 23:208–221. doi: 10.1159/000149607. [DOI] [PubMed] [Google Scholar]
- 219.Putzrath RM, Cadden SP, Maniloff J. 1980. Effect of cell membrane composition on the growth and composition of a nonlytic enveloped mycoplasmavirus. Virology 106:162–167. doi: 10.1016/0042-6822(80)90235-4. [DOI] [PubMed] [Google Scholar]
- 220.Zheng Y, Struck DK, Young R. 2009. Purification and functional characterization of ϕX174 lysis protein E. Biochemistry 48:4999–5006. doi: 10.1021/bi900469g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Elliott RM. 1990. Molecular biology of the Bunyaviridae. J Gen Virol 71:501–522. doi: 10.1099/0022-1317-71-3-501. [DOI] [PubMed] [Google Scholar]
- 222.Weiss ER, Göttlinger H. 2011. The role of cellular factors in promoting HIV budding. J Mol Biol 410:525–533. doi: 10.1016/j.jmb.2011.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Jose J, Przybyla L, Edwards TJ, Perera R, Burgner JW II, Kuhn RJ. 2012. Interactions of the cytoplasmic domain of Sindbis virus E2 with nucleocapsid cores promote alphavirus budding. J Virol 86:2585–2599. doi: 10.1128/JVI.05860-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Suzan-Monti M, La Scola B, Barrassi L, Espinosa L, Raoult D. 2007. Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus. PLoS One 2:e328. doi: 10.1371/journal.pone.0000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Xiang X, Chen L, Huang X, Luo Y, She Q, Huang L. 2005. Sulfolobus tengchongensis spindle-shaped virus STSV1: virus-host interactions and genomic features. J Virol 79:8677–8686. doi: 10.1128/JVI.79.14.8677-8686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Josslin R. 1970. The lysis mechanism of phage T4: mutants affecting lysis. Virology 40:719–726. doi: 10.1016/0042-6822(70)90216-3. [DOI] [PubMed] [Google Scholar]
- 227.Chen AY, Qiu J. 2010. Parvovirus infection-induced cell death and cell cycle arrest. Future Virol 5:731–743. doi: 10.2217/fvl.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Vardi A, Van Mooy BAS, Fredricks HF, Popendorf KJ, Ossolinski JE, Haramaty L, Bidle KD. 2009. Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton. Science 326:861–865. doi: 10.1126/science.1177322. [DOI] [PubMed] [Google Scholar]
- 229.Sánchez-Martínez S, Huarte N, Maeso R, Madan V, Carrasco L, Nieva JL. 2008. Functional and structural characterization of 2B viroporin membranolytic domains. Biochemistry 47:10731–10739. doi: 10.1021/bi800997a. [DOI] [PubMed] [Google Scholar]
- 230.Rennell D, Poteete AR. 1985. Phage P22 lysis genes: nucleotide sequences and functional relationships with T4 and lambda genes. Virology 143:280–289. doi: 10.1016/0042-6822(85)90115-1. [DOI] [PubMed] [Google Scholar]
- 231.Lynch KH, Abdu AH, Schobert M, Dennis JJ. 2013. Genomic characterization of JG068, a novel virulent podovirus active against Burkholderia cenocepacia. BMC Genomics 14:574. doi: 10.1186/1471-2164-14-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Raghava S, Giorda KM, Romano FB, Heuck AP, Hebert DN. 2011. The SV40 late protein VP4 is a viroporin that forms pores to disrupt membranes for viral release. PLoS Pathog 7:e1002116. doi: 10.1371/journal.ppat.1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Han Z, Harty RN. 2004. The NS3 protein of bluetongue virus exhibits viroporin-like properties. J Biol Chem 279:43092–43097. doi: 10.1074/jbc.M403663200. [DOI] [PubMed] [Google Scholar]
- 234.Bize A, Karlsson EA, Ekefjärd K, Quax TEF, Pina M, Prevost M-C, Forterre P, Tenaillon O, Bernander R, Prangishvili D. 2009. A unique virus release mechanism in the Archaea. Proc Natl Acad Sci U S A 106:11306–11311. doi: 10.1073/pnas.0901238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Chang CY, Nam K, Young R. 1995. S gene expression and the timing of lysis by bacteriophage lambda. J Bacteriol 177:3283–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Krupovic M, Bamford DH. 2008. Virus evolution: how far does the double beta-barrel viral lineage extend? Nat Rev Microbiol 6:941–948. doi: 10.1038/nrmicro2033. [DOI] [PubMed] [Google Scholar]
- 237.Snyder JC, Brumfield SK, Peng N, She Q, Young MJ. 2011. Sulfolobus turreted icosahedral virus c92 protein responsible for the formation of pyramid-like cellular lysis structures. J Virol 85:6287–6292. doi: 10.1128/JVI.00379-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Tollefson AE, Scaria A, Hermiston TW, Ryerse JS, Wold LJ, Wold WS. 1996. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J Virol 70:2296–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Federici BA. 1983. Enveloped double-stranded DNA insect virus with novel structure and cytopathology. Proc Natl Acad Sci U S A 80:7664–7668. doi: 10.1073/pnas.80.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Lv Y, Dai L, Han H, Zhang S. 2012. PCV2 induces apoptosis and modulates calcium homeostasis in piglet lymphocytes in vitro. Res Vet Sci 93:1525–1530. doi: 10.1016/j.rvsc.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 241.Karuppannan AK, Kwang J. 2011. ORF3 of porcine circovirus 2 enhances the in vitro and in vivo spread of the virus. Virology 410:248–256. doi: 10.1016/j.virol.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 242.Mindich L, Lehman J. 1979. Cell wall lysin as a component of the bacteriophage ϕ6 virion. J Virol 30:489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Wyler T, Lanzrein B. 2003. Ovary development and polydnavirus morphogenesis in the parasitic wasp Chelonus inanitus. II. Ultrastructural analysis of calyx cell development, virion formation and release. J Gen Virol 84:1151–1163. [DOI] [PubMed] [Google Scholar]
- 244.Boulanger P, Letellier L. 1988. Characterization of ion channels involved in the penetration of phage T4 DNA into Escherichia coli cells. J Biol Chem 263:9767–9775. [PubMed] [Google Scholar]
- 245.Huiskonen JT, Kivelä HM, Bamford DH, Butcher SJ. 2004. The PM2 virion has a novel organization with an internal membrane and pentameric receptor binding spikes. Nat Struct Mol Biol 11:850–856. doi: 10.1038/nsmb807. [DOI] [PubMed] [Google Scholar]
- 246.Tsukagoshi N, Schäfer R, Franklin RM. 1977. Structure and synthesis of a lipid-containing bacteriophage. An endolysin activity associated with bacteriophage PM2. Eur J Biochem 77:585–588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.