Abstract
Background:
How exosomic microRNAs (miRNAs) contribute to the development of drug resistance in the context of the tumor microenvironment has not been previously described in neuroblastoma (NBL).
Methods:
Coculture experiments were performed to assess exosomic transfer of miR-21 from NBL cells to human monocytes and miR-155 from human monocytes to NBL cells. Luciferase reporter assays were performed to assess miR-155 targeting of TERF1 in NBL cells. Tumor growth was measured in NBL xenografts treated with Cisplatin and peritumoral exosomic miR-155 (n = 6 mice per group) CD163, miR-155, and TERF1 levels were assessed in 20 NBL primary tissues by Human Exon Arrays and quantitative real-time polymerase chain reaction. Student’s t test was used to evaluate the differences between treatment groups. All statistical tests were two-sided.
Results:
miR-21 mean fold change (f.c.) was 12.08±0.30 (P < .001) in human monocytes treated with NBL derived exosomes for 48 hours, and miR-155 mean f.c. was 4.51±0.25 (P < .001) in NBL cells cocultured with human monocytes for 48 hours. TERF1 mean luciferase activity in miR-155 transfected NBL cells normalized to scrambled was 0.36 ± 0.05 (P <.001). Mean tumor volumes in Dotap-miR-155 compared with Dotap-scrambled were 322.80±120mm3 and 76.00±39.3mm3, P = .002 at day 24, respectively. Patients with high CD163 infiltrating NBLs had statistically significantly higher intratumoral levels of miR-155 (P = .04) and lower levels of TERF1 mRNA (P = .02).
Conclusions:
These data indicate a unique role of exosomic miR-21 and miR-155 in the cross-talk between NBL cells and human monocytes in the resistance to chemotherapy, through a novel exosomic miR-21/TLR8-NF-кB/exosomic miR-155/TERF1 signaling pathway.
Neuroblastoma (NBL) is the most common solid malignancy in children outside of the skull (1). Amplification of the MYCN oncogene (occurring in about 30% of tumors) defines a group of NBLs with high risk of recurrence (2–6). Unfortunately, despite all current standard treatments the prognosis of patients with high-risk NBL is still poor (7,8). The main reason for failure in treating NBL (and, essentially, every other type of cancer) is the development of resistance to treatments (9). Tumor-associated macrophages (TAMs) promote NBL growth, metastasis (10), and the development of drug resistance (11,12) and represent a negative prognostic factor in NBL and other cancer types (10,13–15). However, the mechanisms responsible for these protumoral functions of TAMs are still poorly understood. MicroRNAs (miRNAs) are small noncoding RNAs (ncRNAs) with gene expression regulatory functions (16) dysregulated in almost all human tumors, including NBL (17). Recently, in lung cancer we showed that miR-21 and miR-29a are secreted by cancer cells within exosomes and can bind to toll-like receptor 8 (TLR8) or its murine ortholog Tlr7 in the surrounding TAMs (18), triggering a protumoral inflammatory reaction (18). The role of miRNAs in the NBL microenvironment is unexplored. In particular, how exosomic miRNAs released within the tumor microenvironment (TME) affect resistance to chemotherapy is currently unknown. We hypothesized that TAMs affect NBL resistance to chemotherapy through the exchange of exosomic miRNAs. The goals of this study are to assess which exosomic miRNAs are involved and through which molecular mechanisms they elicit this function.
Methods
Patient Sample Collection and Microarray Studies
Fresh frozen neuroblastoma primary tissues were collected from patients treated at Children’s Hospital Los Angeles (n = 20). Patients’ characteristics are summarized in Supplementary Table 1 (available online). Informed consent was obtained in accordance with institutional review board policies. RNA was extracted as previously described (19) and analyzed using Affymetrix (Santa Clara, CA) Human Exon Arrays (HuEx), normalized by quantile normalization and summarized using robust multichip average (Affymetrix Power Tools software package version 1.12). CD163 expression levels were obtained by averaging the core unique probe sets for the CD163 transcript (transcript ID: 3442706).
Animal Experiments
All mouse experiments were performed according to protocols approved by the Animal Care and Usage Committee of Children’s Hospital Los Angeles. Female nu/nu mice (Jackson Laboratories, Bar Harbor, ME) at the age of five weeks (n = 6/group) were irradiated with 2 Gy total body irradiation to achieve a more complete immunosuppression, avoid murine macrophage infiltration, and allow better xenograft growth, as previously described (20). The following day mice were injected subcutaneously with 4x106 CHLA-255 and 2x106 human monocytes. All mice were intraperitoneally (i.p) injected with GW4869 (1.25mg/kg/day for 5 days before cell injection). After irradiation, all mice were i.p. injected with GW4869 (1.25mg/kg/day) and Cisplatin (100 nmol/day) three times a week until day 28. All mice received peritumoral injections of Dotap-scrambled or Dotap-miR-155 (100nM) three times a week until day 28. At day 28, mice were killed, necropsy performed, and the tumors were excised, measured, and photographed. Tumor volumes were determined with the equation: V = L x W2/2, where L is the largest diameter and W is the perpendicular diameter. A portion of the excised tumor was used for total RNA isolation, and the other portion was used for identification of TERF1 protein by immunoblotting.
Statistical Analysis
Statistical data are presented as mean ± standard deviation. Statistical significance was calculated by two-tailed Student’s t test. A value of P < .05 was considered statistically significant. The GraphPad software was used for statistical analyses. All statistical tests were two-sided.
All additional experimental methods can be found in the Supplementary Methods (available online).
Results
Effects of NBL-Derived Exosomic miRNAs in Surrounding Human Monocytes
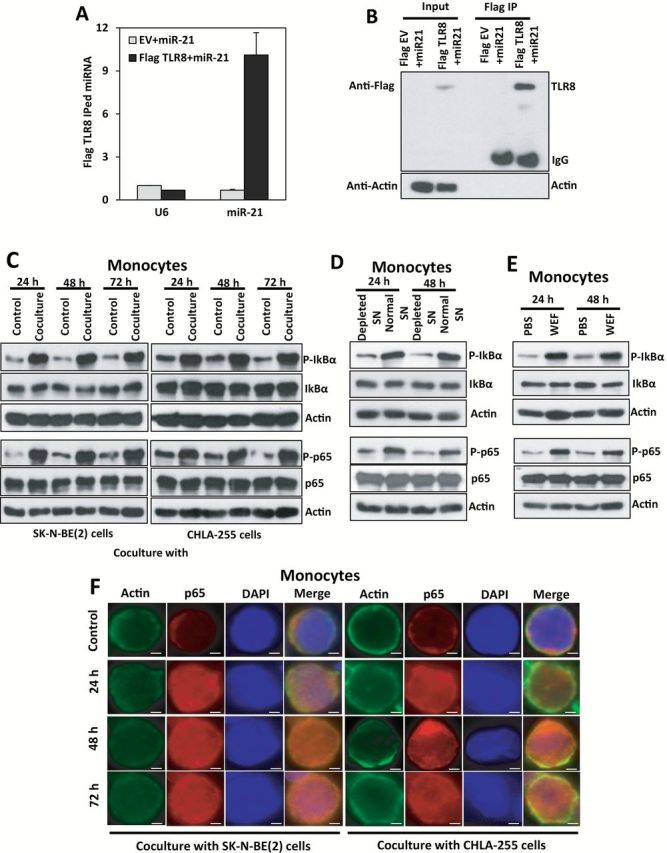
We isolated exosomes from the supernatant (SN) of SK-N-BE(2), CHLA-255, and IMR-32 NBL cell lines (Supplementary Figure 1, A-D, and Supplementary Materials, available online) and assessed their content of miR-21-5p, miR-29a-5p, and miR-155-5p (from now on referred to as miR-21, miR-29a, and miR-155, respectively) by quantitative real-time polymerase chain reaction (qRT-PCR) as indicated in the Supplementary Materials (available online). We focused on these three miRNAs because miR-21 and miR-29a can trigger an NF-кB-mediated pro-inflammatory response in lung cancer (18), and miR-155 is induced during the macrophage inflammatory response (21). Also we have conducted an unbiased miRNA screening of exosomic miRNAs secreted by five different NBL cell lines (CHLA-255, LA-N-1, SK-N-BE(2), KNCR, and IMR-32 cultured as in the Supplementary Materials, available online) by using a noncoding RNA array previously validated in (22). We compared the most secreted exosomic miRNAs common to all NBL cell lines and to the dataset of our previous work (18), and miR-21 was the top represented exosomic miRNA (fold change = 2.46, false discovery rate = 0.052, P = .01). By qRT-PCR we validated that only miR-21 was expressed in exosomes from all three cell lines (Figure 1A). miR-21 (P = .004, .002, .001 with SK-N-BE(2), and .001, .002, .001 with CHLA-255 at 24 hours, 48 hours, and 72 hours, respectively) but not pre-miR-21 levels were statistically significantly increased in human monocytes cocultured with SK-N-BE(2) or CHLA-255 (Figure 1, B and C). When human monocytes were cultured with the SN of NBL cells depleted of exosomes by ultracentrifugation (as in the Supplementary Materials, Supplementary Figure 1, E-J, available online, P < .001 all panels), we observed reduced levels of mature miR-21 compared with monocytes cultured in non-exosome depleted SN (miR-21 mean fold change [f.c.] normalized to non-exosome depleted SN = 0.097±0.10 at 24 hours, P = .001 and 0.079±0.15 at 48 hours, P = .001) (Figure 1D). Conversely, mature miR-21 levels were statistically significantly increased in monocytes grown in media enriched for the whole exosomic fraction (WEF) of NBL cell SN (miR-21 mean f.c. normalized to phosphate buffer saline (PBS) = 6.76±1.36 at 24 hours, P = .002 and 12.08±0.30 at 48 hours, P < .001) (Figure 1E; Supplementary Figure 1, F-J, available online) and in monocytes transfected with the total RNA content extracted from NBL-secreted exosomes (miR-21 mean f.c. normalized to scrambled = 9.21±0.08, P < .001 at 24 hours and 13.12±0.71 at 48 hours, P < .001) (Figure 1F). These data indicate that NBL cells secrete exosomic miR-21 transferred to human monocytes through exosomes. Next, we observed miR-21 binding to TLR8 in human monocytes by co-immunoprecipitation (Figure 2, A and B; Supplementary Materials, available online) and exosome-mediated NF-кB pathway activation in monocytes cocultured with NBL cells (Figure 2, C-F; Supplementary Figure 2 and Supplementary Materials, available online).
Figure 1.
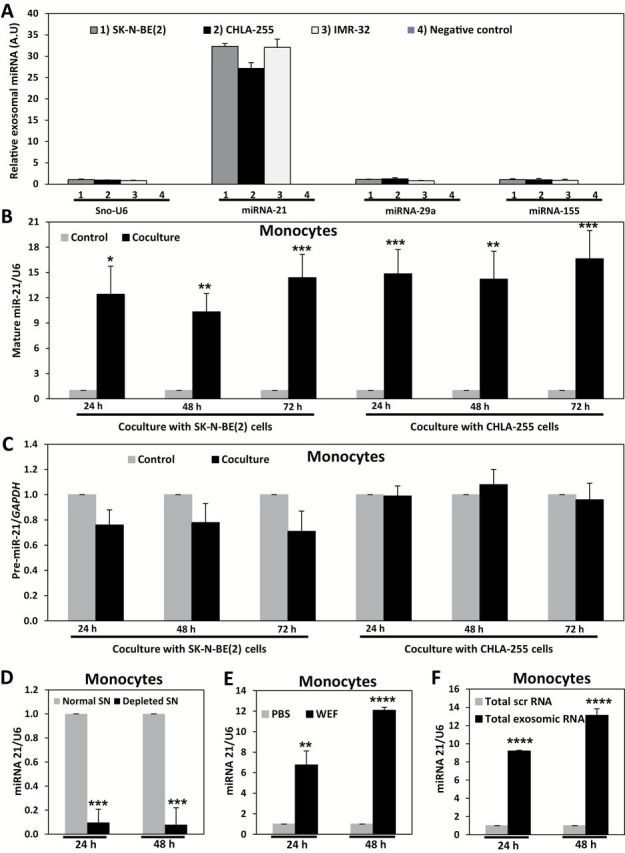
Effects of neuroblastoma (NBL)–derived exosomic microRNAs (miRNAs) in human monocytes. A) Quantitative real-time polymerase chain reaction (qRT-PCR) for miR-21, miR-29a, and miR-155 in exosomes derived from: 1) SK-N-BE(2), 2) CHLA-255, 3) IMR-32 cells, or 4) Remaining supernatant after exosome isolation (negative control). Exosomes were isolated from NBL cells cultured media containing exosome-free fetal bovine serum for a period of 48 hours. B) qRT-PCR for mature miR-21 in monocytes cocultured with SK-N-BE(2) or CHLA-255 cells for the indicated time periods. C) qRT-PCR for pre-miR-21 in monocytes cocultured with SK-N-BE(2) or CHLA-255 cells for the indicated time periods. D) qRT-PCR for mature miR-21 in monocytes cultured for the indicated time periods with the supernatant (SN) of SK-N-BE(2) cells (normal SN) or with exosome-depleted by ultracentrifugation (depleted SN) of SK-N-BE(2) cells. E) qRT-PCR for mature miR-21 in monocytes treated for the indicated time periods with whole exosomes fraction (WEF) isolated from SK-N-BE(2) cells compared with PBS. F) qRT-PCR for mature miR-21 in monocytes transfected with total RNA extracted from exosomes of SK-N-BE(2) cells (compared with a total scrambled RNA) for the indicated time periods. Relative levels of pre-miRNA or mature miRNAs expression were normalized to GAPDH mRNA or U6 snRNA, respectively. Data are presented as mean ± SD of experiments conducted in triplicate. *P = .004, **P = .002, ***P = .001, ****P < .001, Student’s t test, two-sided.
Figure 2.
Exosomic miR-21 and TLR8 interaction in human monocytes. A) Quantitative real-time polymerase chain reaction (qRT-PCR) for miR-21 or U6 in the immunoprecipitate with anti-Flag antibody in human monocytes transfected with empty vector and Dotap-miR-21 (EV+miR-21) or with a Flag-TLR8 vector and Dotap-miR-21 (Flag TLR8+miR-21), after 48 hours from trasfection. Data presented as mean ± SD of experiments conducted in triplicate. B) Immunoblotting with anti-Flag antibody corresponding to the experiment described in (A). C) Immunoblotting for phospho-IкBα, IкBα, phospho-p65, p65, and relative actin proteins in human monocytes cocultured with SK-N-BE(2) or CHLA-255 cells for the indicated time periods. D) Immunoblotting for phospho-IкBα, IкBα, phospho-p65, p65, and relative actin proteins in human monocytes cultured with the supernatant (SN) from SK-N-BE(2) cells that were depleted for exosomes (depleted SN) compared with normal SN for the indicated time periods.
(E) Immunoblotting for phospho-IкBα, IкBα, phospho-p65, p65 and relative actin proteins in human monocytes treated with whole exosomes fraction (WEF) isolated from SK-N-BE(2) cells compared with PBS for the indicated time periods. F) Immunofluorescence (IF) image showing intracellular localization of the p65 NF-кB subunit in human monocytes cocultured with SK-N-BE(2) or CHLA-255 cells. IF staining was performed using monoclonal anti-p65 (red) and polyclonal anti-actin (red) antibodies together with DAPI (blue). Results are a representative of three independent experiments. Scale bar = 2 μm.
Identification of a NBL-TAM Exosomic miRNA Cross-Talk
miR-155 levels progressively increased in human monocytes cocultured with NBL cells (miR-155 mean f.c. normalized to monocytes alone = 4.92±0.26, P < .001, 6.24±0.16, P < .001, 6.94±0.58, P < .001 in SK-N-BE(2), and 4.03±0.49, P < .001, 5.45±1.07, P = .002, 6.41±0.39, P < .001 in CHLA-255 at 24 hours, 48 hours, and 72 hours, respectively) (Figure 3A). This effect was reversed when monocytes were cultured in exosome-depleted NBL-derived media (miR-155 mean f.c. normalized to non-exosome depleted SN = 0.17±0.14 at 24 hours, P = .001 and 0.12±0.16 at 48 hours, P = .001) (Figure 3B), whereas increased miR-155 levels occurred in monocytes treated with NBL-derived WEF (miR-155 mean f.c. normalized to PBS = 4.32±0.32 at 24 hours, P < .001 and 4.51±0.25 at 48 hours, P < .001) (Figure 3C) or transfected with total RNA from SK-N-BE(2)–derived exosomes (miR-155 mean f.c. normalized to scrambled = 3.76±0.15, P < .001 at 24 hours and 6.64±0.52 at 48 hours, P < .001) (Figure 3D). These data suggest that NBL-derived exosomes are able to induce miR-155 upregulation in human monocytes. Next, we observed statistically significant upregulation of miR-155 in human monocytes treated with Dotap-miR-21 (miR-155 mean f.c. normalized to Dotap-Scrambled = 1.52±0.02, P < .001, 3.00±0.03, P < .001, and 3.68±0.22, P < .001 at 24 hours, 48 hours, and 72 hours, respectively) in a TLR8-dependent manner (P < .001 only in cocultures with TLR8-expressing monocytes) (Figure 3, E and F; Supplementary Figure 3, A-C, available online, all P < .001). We observed upregulation of mature miR-155 in the exosomes extracted from the SN of NBL-human monocyte cocultures (miR-155 mean f.c. normalized to SN of monocytes not in coculture with NBL cells = 4.97±0.09, P < .001, 6.20±1.15, P = .001, and 6.99±1.08, P < .001 in coculture with SK-N-BE(2), and 3.14±0.08, P < .001, 3.41±0.50, P = .001, and 5.36±1.08, P = .002 in coculture with CHLA-255 at 24 hours, 48 hours, and 72 hours, respectively) (Figure 3G; Supplementary Figure 3D), as well as in NBL cells (miR-155 mean f.c. normalized to NBL cells not in coculture = 2.63±0.08, P < .001, 3.67±0.72, P = .003, and 7.22±1.27, P = .001 in SK-N-BE(2), and 1.46±0.11, P = .002, 4.83±0.29, P < .001, and 5.45±0.21, P < .001 in CHLA-255 at 24 hours, 48 hours, and 72 hours, respectively) (Figure 3H). Next, we investigated whether exosome transfer or increased endogenous neo-synthesis could explain the increased miR-155 levels in NBL cells cocultured with monocytes. In presence of cancer cells human monocytes secrete higher levels of IL-6 (12,18), which triggers the STAT3 pathway (23), known to induce miR-155 upregulation (24,25). We measured pre-miR-155 levels in NBL cells alone or cocultured with monocytes, however no variations were observed (Supplementary Figure 3, E and F, available online). We also determined the levels of mature miR-155 in monocyte-SK-N-BE(2) cocultures treated with ruxolitinib (a JAK1/2 inhibitor [26,27]), or GW4869 (an inhibitor of exosome secretion and miRNA content in exosomes [18,28]) (see Supplementary Materials, available online). While GW4869 statistically significantly reduced miR-155 levels in NBL cells cocultured with human monocytes (miR-155 mean f.c. normalized to DMSO = 0.31±0.02, P < .001 and 0.41±0.03, P < .001 at 24 hours and 48 hours, respectively), no reduction was observed in cocultures treated with ruxolitinib (Figure 3I; Supplementary Figure 3G, available online). These data suggest that miR-155 is transferred from monocytes to NBL cells through exosomes.
Figure 3.
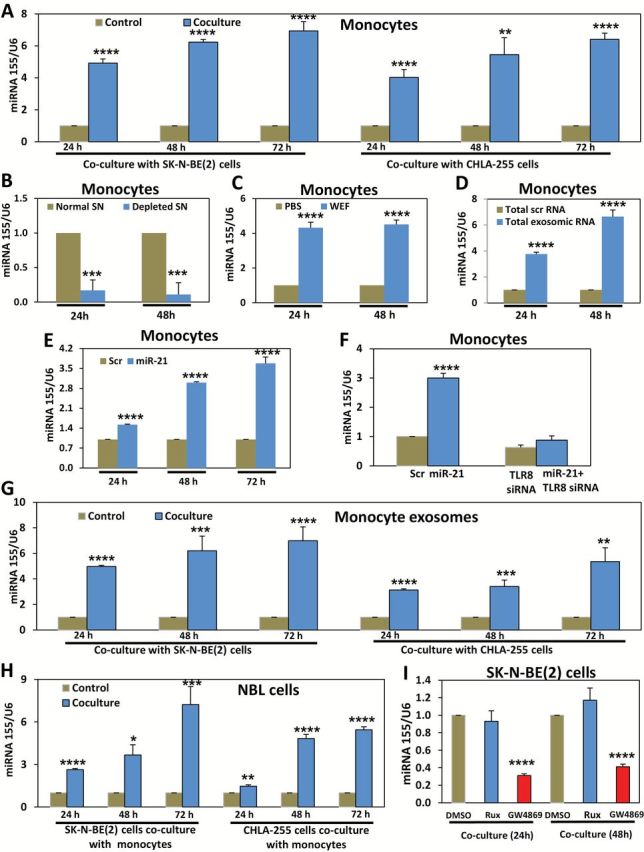
Exosomic miR-155-mediated cross-talk between human monocytes and neuroblastoma (NBL) cells. A) Quantitative real-time polymerase chain reaction (qRT-PCR) for miR-155 in monocytes cocultured with SK-N-BE(2) or CHLA-255 cells for the indicated time periods. B) qRT-PCR for miR-155 in human monocytes cultured for the indicated time periods with the supernatant (SN) of SK-N-BE(2) cells (normal SN) or with exosome-depleted by ultracentrifugation (depleted SN) of SK-N-BE(2) cells. C) qRT-PCR for miR-155 in human monocytes treated for the indicated time periods with whole exosomes fraction (WEF) isolated from SK-N-BE(2) cells compared with PBS. D) qRT-PCR for miR-155 in human monocytes transfected with total RNA extracted from exosomes of SK-N-BE(2) cells (compared with a total scrambled RNA) for the indicated time periods. E) qRT-PCR for miR-155 in human monocytes transfected with Dotap-miR-21 (miR-21) or Dotap-scrambled RNA (scr) for the indicated time periods. F) qRT-PCR for miR-155 in human monocytes transfected for 48 hours with Dotap-Scrambled (scr group) or Dotap-miR-21 (miR-21 group) alone (left histograms) or in human monocytes transfected for 48 hours with anti-TLR8 siRNA and Dotap-Scrambled (TLR8 siRNA group) or with anti-TLR8 siRNA and Dotap-miR-21 (miR-21+TLR8 siRNA group) (right histograms). G) qRT-PCR for miR-155 in exosomes derived from human monocytes alone (control) or cocultured with SK-N-BE(2) or CHLA-255 cells for the indicated time periods. H) qRT-PCR for miR-155 in SK-N-BE(2) or CHLA-255 cells cocultured with human monocytes for the indicated time periods. I) qRT-PCR for miR-155 in SK-N-BE(2) cells cocultured with human monocytes and in presence of Ruxolitinib (Rux), GW4869, or DMSO (as a control) for the indicated time periods. Relative levels of miR-155 expression were normalized to U6 snRNA. Data are presented as mean ± SD of experiments conducted in triplicate. *P = .003, **P = .002, ***P = .001, ****P < .001, Student’s t test, two-sided.
Effects of Human Monocyte–Derived Exosomic miR-155 in NBL Drug Resistance
Increased telomerase activity (T.A.) is associated with increased chemoresistance in NBL (29). One of the predicted targets of miR-155, previously validated in breast cancer (30), is TERF1, an inhibitor of telomerase, whose silencing can induce increased T.A. (31). We transfected SK-N-BE(2) and CHLA-255 cells with miR-155 (or a scrambled oligonucleotide, P < .001 for both cell lines) (Supplementary Figure 4, A and B, available online) and observed downregulation of TERF1 mRNA and protein at 48 hours (TERF1 mRNA mean f.c. normalized to scrambled = 0.58±0.06, P < .001 and 0.51±0.09, P < .001, in SK-N-BE(2) and CHLA-255, respectively) (Figure 4, A and B). Conversely, when miR-155 was silenced with locked nucleic acid (LNA) anti-miR-155, TERF1 mRNA and protein levels were upregulated compared with LNA-scrambled treated NBL cells (TERF1 mRNA mean f.c. normalized to LNA-anti-scrambled = 1.61±0.08, P < .001 and 1.58±0.21, P = .009, in SK-N-BE(2) and CHLA-255, respectively) (Figure 4, C and D). We performed a luciferase reporter assay in three different NBL cell lines and observed direct targeting of TERF1 by miR-155 (TERF1 mean luciferase activity in miR-155 transfected cells normalized to scrambled = 0.36±0.05, P < .001, 0.40±0.05, P < .001 and 0.54±0.14, P = .005, in SK-N-BE(2), IMR-32 and CHLA-255, respectively) (Figure 4, E and F; Supplementary Figure 4, C-E, and Supplementary Materials, available online). We also looked at the R2 database (http://r2.amc.nl) and observed a statistically significant although mild inverse correlation between miR-155 host gene and TERF1 in 649 primary neuroblastomas (r = -0.2, P = .004) (Figure 4G) (32).
Figure 4.
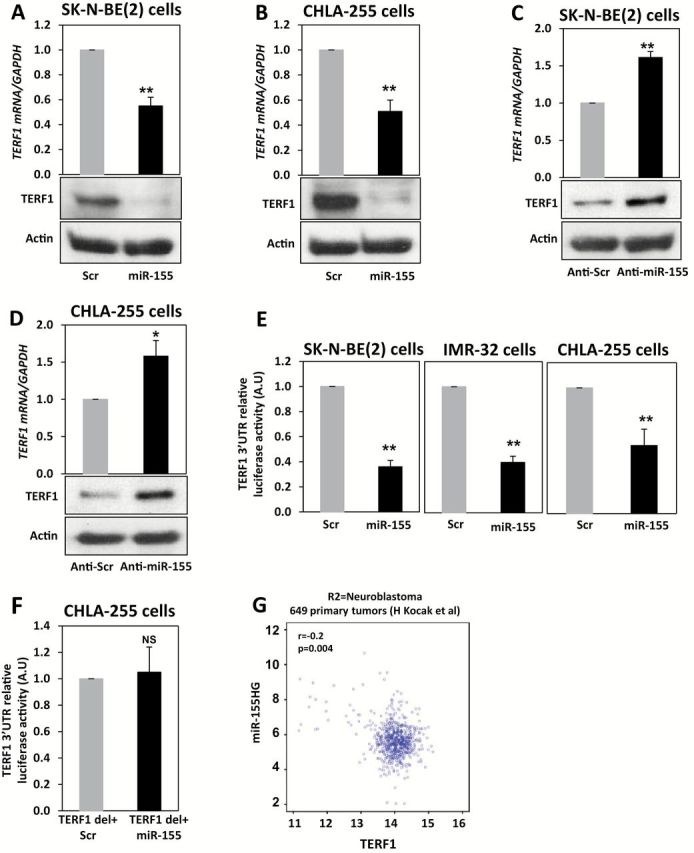
miR-155 and TERF1 regulation in neuroblastoma (NBL). A) Detection of TERF1 mRNA (by quantitative real-time polymerase chain reaction [qRT-PCR]) and protein (by immunoblotting) levels in SK-N-BE(2) cells transfected with a scrambled oligonucleotide (scr group) or with miR-155 (miR-155 group) after 48 hours. B) Same experiment described in (A) but in CHLA-255 NBL cells. C) Detection of TERF1 mRNA (by qRT-PCR) and protein (by immunoblotting) levels in SK-N-BE(2) cells transfected with LNA-anti-miR-155 (anti-miR-155 group) or LNA-anti-scrambled (anti-scr group) oligonucleotide after 48 hours. D) Same experiment described in (C) but in CHLA-255 NBL cells. Relative levels of TERF1 mRNA expression were normalized to GAPDH mRNA whereas β-actin was used as loading control for TERF1 protein. E) Luciferase reporter assay in SK-N-BE(2), IMR-32, and CHLA-255 NBL cells cotransfected with a plasmid expressing the wild-type 3’-UTR of the TERF1 gene downstream of the reporter gene and with miR-155 or a scrambled oligonucleotide and detected after 24 hours. F) Luciferase reporter assay in SK-N-BE(2) cells cotransfected with a plasmid in which the predicted binding site for miR-155 in the 3’-UTR of the TERF1 gene had been deleted (TERF1 del+miR-155) and with miR-155 or a scrambled oligonucleotide and detected after 24 hours from transfection. Data are presented as mean ± SD of experiments conducted in quadruplicate. *P = .009, **P < .001. Student’s t test, two-sided. G) Correlation between miR-155 host gene (miR155HG) and TERF1 mRNA in 649 NBL primary tumors, as detected from a publicly available R2 database. NS = not statistically significant.
Next, we observed a downregulation of TERF1 in NBL cells cocultured with human monocytes (TERF1 mRNA mean f.c. normalized to NBL not in coculture = 0.34±0.03, P < .001, 0.43±0.03, P < .001 and 0.40±0.02, P < .001, in SK-N-BE(2) and 0.40±0.03, P < .001, 0.46±0.01, P < .001 and 0.30±0.01, P < .001 in CHLA-255, at 24 hours, 48 hours, and 72 hours, respectively) (Figure 5, A and B), which was reversed when NBL cells were pretreated with LNA-anti-miR-155, suggesting that TERF1 downregulation in NBL cells cocultured with human monocytes was mediated by miR-155 (P = .004 at 48 hours and <.001 at 72 hours) (Figure 5C; Supplementary Figure 5A, available online). Next, we showed that miR-155 and TERF1 expression were regulated by exosomes in SK-N-BE(2) cells cultured with exosome-depleted SN (TERF1 mRNA and miR-155 mean f.c. normalized to non-exosome depleted SN = 2.8±0.11, P < .001 and 0.2±0.11, P < .001 at 24 hours, and 3.5±0.04, P < .001 and 0.1±0.08, P < .001 at 48 hours, respectively) (Figure 5D) or WEF from SK-N-BE(2)-human monocyte cocultures (TERF1 mRNA and miR-155 mean f.c. normalized to PBS = 0.52±0.03, P < .001 and 4.04±0.04, P < .001 at 24 hours, and 0.39±0.03, P < .001 and 5.35±0.31, P < .001 at 48 hours, respectively) (Figure 5E; Supplementary Figure 5, B and C, available online, P < .001). We also treated SK-N-BE(2)-human monocyte cocultures with ruxolitinib, GW4869, or DMSO as a control. Only GW4869 induced statistically significant upregulation of TERF1 in NBL cells (TERF1 mRNA mean f.c. normalized to DMSO = 1.63±0.11, P < .001 and 1.57±0.07, P < .001 at 24 hours and 48 hours, respectively) (Figure 5, F-G). Moreover, increased T.A. (detected as in the Supplementary Materials, available online) was observed in SK-N-BE(2) and CHLA-255 cells transfected with Dotap-miR-155 (P < .001 in both cell lines, Supplementary Figure 5D, available online), mirroring the increased T.A. in SK-N-BE(2) cells cocultured with human monocytes (P < .001, Supplementary Figure 5E, available online). This effect was reversed when NBL cells were pretreated with LNA-anti-miR-155 (P < .001, Supplementary Figure 5F and Supplementary Materials, available online), suggesting that the increased T.A. in NBL cells cocultured with human monocytes was mediated by miR-155.
Figure 5.
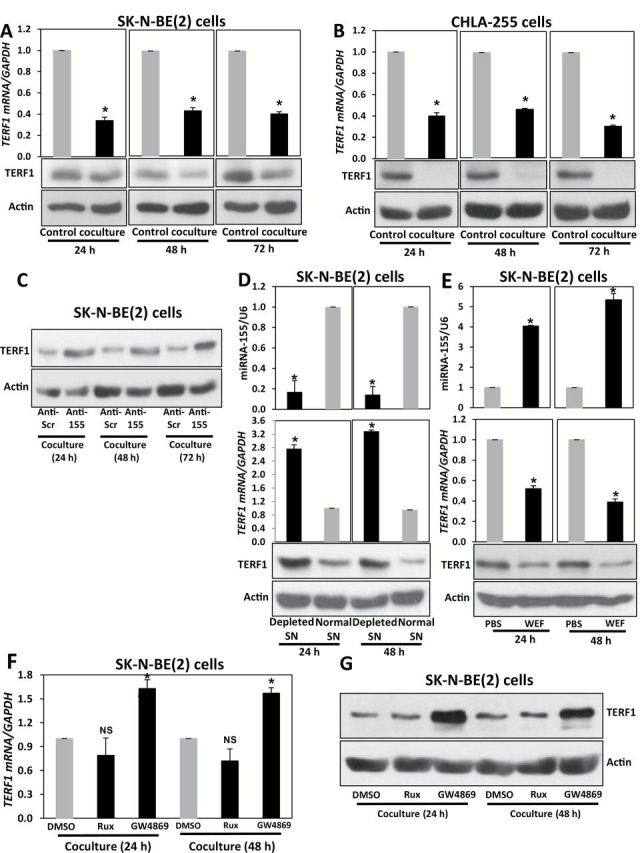
TERF1 regulation in neuroblastoma (NBL)–human monocyte cocultures. A) Detection of TERF1 mRNA (by quantitative real-time polymerase chain reaction [qRT-PCR]) and protein (by immunoblotting) levels in SK-N-BE(2) cells alone (control group) or cocultured with human monocytes (coculture group) for the indicated time periods. B) Same experiment described in (A) but in CHLA-255 NBL cells. C) Immunoblotting for TERF1 in SK-N-BE(2) cells cocultured with human monocytes and pretransfected with LNA-anti-Scrambled (anti-scr group) or with LNA-anti-miR-155 (anti-155 group) for the indicated time periods. D) qRT-PCR for miR-155 (upper panel), TERF1 mRNA (middle panel), and immunoblotting for TERF1 protein (bottom panel) in SK-N-BE(2) cells cultured with the supernatant (SN) from SK-N-BE(2)-human monocytes cocultures (normal SN group) or with exosome-depleted SN of the coculture by ultracentrifugation (depleted SN group) for the indicated time periods. E) qRT-PCR for miR-155 (upper panel), TERF1 mRNA (middle panel), and immunoblotting for TERF1 protein (bottom panel) in SK-N-BE(2) cells treated for the indicated time periods with whole exosomes fraction (WEF) isolated from SK-N-BE(2)- human monocytes cocultures (WEF group) or PBS. F) qRT-PCR for TERF1 mRNA in SK-N-BE(2) cells cocultured with human monocytes and treated with Ruxolitinib (Rux), GW4869, or DMSO (as a control) for the indicated time periods. G) Immunoblotting for TERF1 in SK-N-BE(2) cells treated as described in (F). Relative levels of miR-155 were normalized to U6 snRNA, whereas TERF1 mRNA was normalized to GAPDH mRNA. β-actin was used as a loading control for TERF1 protein. Data are presented as mean ± SD of experiments conducted in triplicate. *P < .001, Student’s t test, two-sided.
Next, we asked whether NBL cells induce M1- or M2-polarization of unpolarized human monocytes. By performing a cytokine profile (Figure 6A) and cytofluorimetry (Supplementary Materials, available online) for the TAM marker CD163 in human monocytes alone or in coculture with SK-N-BE(2) (Figure 6B), we observed upregulation of mixed M2- and M1-markers but with a prevalence of M2 markers, and an increased percentage of CD163+ cells when unpolarized monocytes were cocultured with NBL cells. Also, we artificially polarized human monocytes (see the Supplementary Materials, available online) as M2-, M1-, or dendritic cells with differentiating media and observed statistically significant upregulation of miR-21 and miR-155 in M1- and M2-polarized monocytes (P < .001 for both miRNAs at 24 hours and 48 hours and both M1- and M2-polarized monocytes) and downregulation of TERF1 when SK-N-BE(2) cells were cocultured with M1- and M2-polarized monocytes (P = .001 at 24 hours and =.002 at 48 hours in M2-polarized and P < .001 at 24 hours and =.003 in M1-polarized). No variations in miR-21, miR-155, and TERF1 were observed when human monocytes were differentiated in dendritic cells (Figure 6C). Overall these data indicate that NBL cells induce a mixed (but prevalently M2) polarization of unpolarized human monocytes and the miR-21/155/TERF1 circuitry occurs both in M1- and M2- monocytes but not in dendritic cells.
Figure 6.
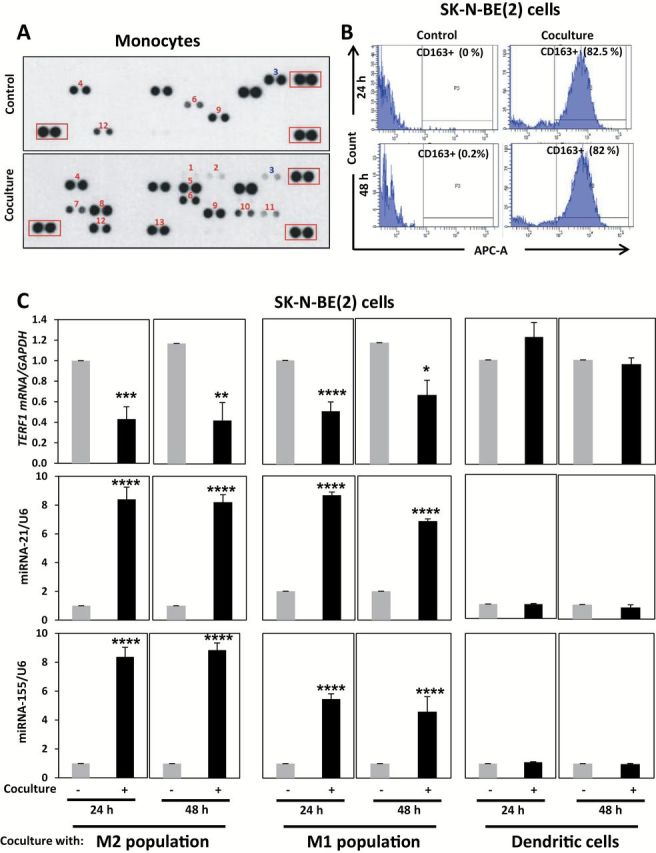
Neuroblastoma (NBL) cells and human monocyte polarization. A) Cytokine Array Panel showing the expression of a panel of cytokines in human monocytes alone (control) or cocultured with SK-N-BE(2) cells for 48 hours (coculture). 1. CXCL1; 2. CCL1; 3. IFN-γ; 4. IL-1α; 5. IL-4; 6. IL-17; 7. IL-32α; 8. CXCL10; 9. CCL3; 10. CCL4; 11. PAI1; 12. CXCL12; 13. sTREM1. Representative image from one of two human monocyte donors. The red boxes refer to reference spots (positive controls). Cytokines shown in red colors and blue colors are upregulated and downregulated in coculture compared with control, respectively. B) Flow cytometry expression of CD163 in human monocytes alone (control, left panels) or cocultured with SK-N-BE(2) cells (coculture, right panels) for 24 hours and 48 hours. C) Quantitative real-time polymerase chain reaction (qRT-PCR) for TERF1 mRNA in SK-N-BE(2) cells alone or cocultured with M1- or M2- or dendritic artificially differentiated human monocytes for 24 hours or 48 hours. qRT-PCR for miR-21 and miR-155 in M1- or M2- or dendritic artificially differentiated human monocytes alone or cocultured with SK-N-BE(2) cells for 24 hours or 48 hours. Data are presented as mean ± SD of experiments conducted in triplicate. *P = .003, **P = .002, ***P = .001, ****P < .001, Student’s t test, two-sided.
To test chemoresistance CDDP was chosen because it is one of the election drugs for NBL treatment. When SK-N-BE(2) and CHLA-255 cells were treated with Dotap-Scrambled or Dotap-miR-155 in presence of CDDP a statistically significantly increased growth (P = .001 at 48 hours, = 0.003 at 72 hours, and <.001 at 96h for SK-N-BE(2), and P = .01 at 48 hours, and <.001 at 72 hours and 96 hours for CHLA-255) and telomere length (P < .001 at 48 hours for both cell lines) in the Dotap-miR-155 treated group was observed (Figure 7, A and B; Supplementary Figure 6, A-C, and Supplementary Materials, available online). Therefore, we transfected SK-N-BE(2) cells with a plasmid expressing full length TERF1 (wt-TERF1 group) or a miR-155-not-regulated TERF1 (no-3’UTR group) (Supplementary Materials, available online), and observed a statistically significant reduction (P < .001 at all time points) in cell number in the no-3’UTR group (Figure 7, C and D), indicating that TERF1 is able to rescue the CDDP-resistant phenotype induced by exosomic miR-155 in SK-N-BE(2) cells.
Figure 7.
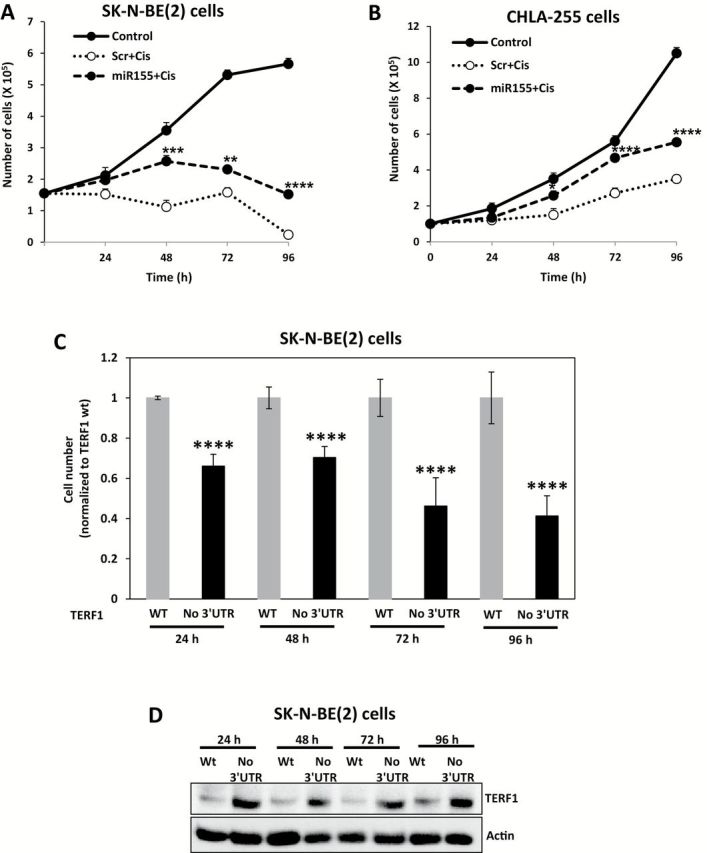
Exosomic miR-155 and cisplatin resistance in neuroblastoma (NBL) cells. A) Growth curves of SK-N-BE(2) cells not Dotap-treated and not treated with cisplatin (control group), or treated with Dotap-Scrambled and cisplatin (scr+cis group), or treated with Dotap-miR-155 and cisplatin (miR-155+cis group). B) Same experiment described in (A) but in CHLA-255 NBL cells. C) Cell count for SK-N-BE(2) cells pretransfected with a plasmid expressing the whole TERF1 gene (WT group) or just TERF1 coding sequence (no 3’-UTR group) and treated with cisplatin and Dotap-miR-155 for the indicated time periods. D) Immunoblotting for TERF1 in the same groups described in (C). Data are presented as mean ± SD of experiments conducted in triplicate. *P = .01, **P = .003, ***P = .001, ****P < .001, Student’s t test, two-sided.
Also, we injected subcutaneously 12 five-week-old nude mice with a coculture of CHLA-255 and human monocytes and treated the animals with CDDP. All mice were pretreated with GW4869 for five days before tumor cell/monocyte injections; however, half of the mice (n = 6) were injected peritumorally with Dotap-scrambled every three days, and the other half (n = 6) with Dotap-miR-155. Xenografts injected with Dotap-miR-155 grew statistically significantly more (mean tumor volumes in Dotap-miR-155 compared with Dotap-scrambled = 322.80±120mm3 vs 76.00±39.3mm3, P = .002 at day 24) and showed higher miR-155 (P = .01) and lower TERF1 expression (P = .008) (Figure 8, A-D; Supplementary Figure 7, A-C, available online, P = .004 for tumor weights and P < .001 for relative band intensity).
Figure 8.
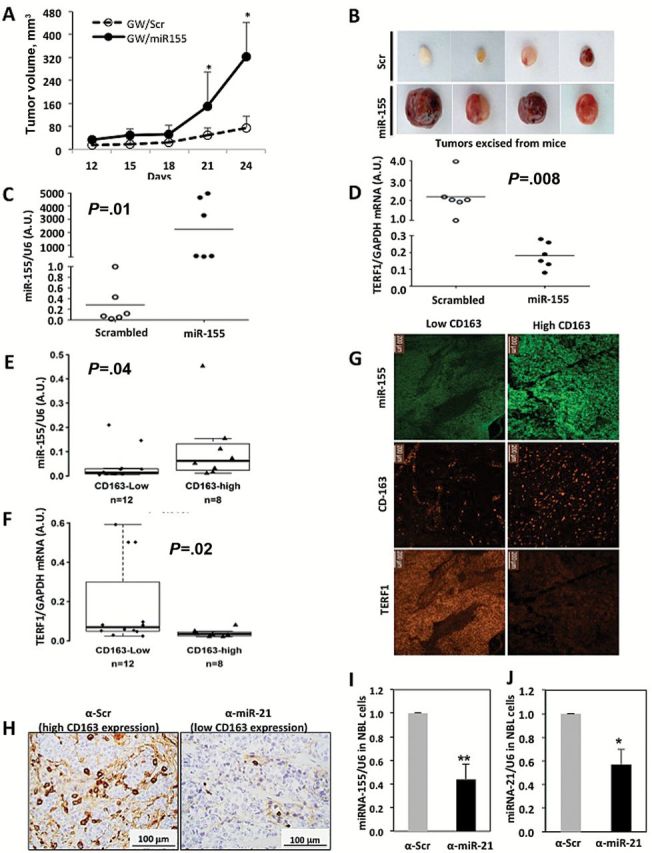
Effects of exosomic miR-155 in cisplatin resistance in vivo and in primary neuroblastoma (NBL) samples. A) Tumor growth curves of xenografts of mice co-injected subcutaneously with CHLA-255-Luc cells (CHLA-255 cells stably expressing a luciferase reporter gene) and human monocytes, pretreated intraperitoneally with GW4869 and treated with cisplatin and injected peritumorally with Dotap-Scrambled (GW/scr group n = 6) or with Dotap-miR-155 (GW/miR-155 group n = 6). Data are presented as the mean tumor volumes ± SD for each animal group. *P < .05. B) Representative images of the excised tumors from the experiment described in (A). C) Quantitative real-time polymerase chain reaction (qRT-PCR) for miR-155 in excised xenografts from experiment (A). Relative levels of miR-155 were normalized to U6. D) qRT-PCR for TERF1 mRNA in excised xenografts from experiment (A). Relative levels of TERF1 mRNA were normalized to GAPDH mRNA. E and F) qRT-PCR for miR-155 (E) and TERF1 (F) in 20 primary NBLs with low and high CD163 infiltrating cells. Data are reported as 2-ΔCT and expressed in arbitrary units (A.U.) as normalized to U6 expression/each patient. G) Representative images of miR-155, CD163, and TERF1 in situ hybridization from two of 14 primary NBL tissues from a patient with high (right) and low (left) CD163 infiltrating cells. Scale bar = 200 μm. H) Representative images of CD163 immunohistochemistry xenograft tumor tissues in mice co-injected with CHLA-255 antiscrambled (left) (n = 3) or CHLA-255 anti-miR-21 (right) (n = 3) + human monocytes + human mesenchymal stem cells. Scale bar = 100 μm. I and J) qRT-PCR for miR-155 (I) and miR-21 (J) in NBL cells isolated from the tumor tissue of xenografts of the experiment as described in H. *P = .005, **P = .002, Student’s t test, two-sided.
Next, we assessed miR-155 and TERF1 expression in 20 primary NBLs in which the level of CD163 expression (representative of TAM infiltration) was determined by HuEx arrays and dichotomized into low (n = 12) and high (n = 8) TAM infiltration. Statistically significantly higher levels of miR-155 (P = .04) and lower levels of TERF1 mRNA (P = .02) were measured in NBLs with higher TAM infiltration (Figure 8, E and F; and additional statistical analysis in the Supplementary Materials, available online). We also performed in situ hybridization for miR-155, CD163, and TERF1 mRNA in 14 of 20 primary NBL tumors (see Supplementary Materials, available online), and we observed that tumors with higher CD163+ cell infiltration also expressed higher levels of miR-155 and lower levels of TERF1 (Figure 8G). Also, we generated CHLA-255 cells with stable lentiviral downregulation of miR-21 (or CHLA-255 antiscrambled as a control) (see Supplementary Materials, available online). Six nude mice were injected subcutaneously with CHLA-255 anti-miR-21 + human monocytes + human mesenchymal stem cells (hMSCs) (n = 3) or with CHLA-255 antiscrambled + human monocytes + hMSCs (n = 3) and after one week tumors were excised and CD163+ cells detected by immunohistochemistry (see Supplementary Materials, available online). A decreased number of CD163+ cells, downregulation of miR-155 (P = .002), and downregulation of miR-21 (P = .005) were observed in the xenografts with CHLA-255 anti-miR-21 cells (Figure 8, H-J). Finally, we assessed the expression of miR-155 and TERF1 mRNA in seven different cancer cell lines alone or in coculture with human monocytes. In all cases we observed upregulation of miR-155 and downregulation of TERF1 mRNA, suggesting that the exosomic miR-155 targeting of TERF1 in cancer cell-monocyte cocultures also occurs in other cancer types (Supplementary Figure 7D, available online, for miR-155 levels: P = .003 for M21 and H1299, =0.002 for FTC and <.001 for all other cell lines; for TERF1 levels: P = .002 for FTC and <.001 for all other cell lines).
Discussion
Cancer is a disease of the TME. Exosomes are involved in intercellular communication (33,34), and in this study we investigated whether the paracrine exchange of exosomic miRNAs between NBL cells and neighboring human monocytes affected drug resistance. We showed that the “educational” process elicited by NBL on human monocytes through the secretion of exosomic miR-21 led to a TLR8 and NF-кB-dependent upregulation of miR-155 in NBL cells. We also showed that NBL cells induced mixed (but prevalently M2-) polarization of unpolarized human monocytes. Interestingly, our model suggests that the high levels of miR-155 frequently observed in homogenates of primary tumors might not be because of high miR-155 expression in cancer cells, but because of the exosomic transfer of miR-155 from the surrounding TAMs. These data revealed a previously unknown positive correlation between two oncogenic miRNAs (miR-21 and miR-155), indirectly mediated by the presence of TLR8-positive cells in the TME. Our data also suggest that in order to interfere with miR-155 upregulation in NBL cells, an anti-IL-6/anti-STAT3 therapy might not be as effective as interfering with exosome secretion.
Once in NBL cells, miR-155 directly targeted TERF1, a component of the shelterin complex and inhibitor of telomerase (35,36). Telomere length is a prognostic factor in NBL (37), and telomerase activity (T.A.) correlates with drug resistance and poor outcome in various malignancies (29,38–43). In this study we showed that exosomic miR-155 transferred by human monocytes was able to directly target TERF1 and affect T.A. and telomere length in NBL, and TERF1 targeting by miR-155 was involved in the acquisition of increased CDDP resistance both in vitro and in in vivo. The use of an exosome inhibitor (GW4869) statistically significantly restored NBL cell sensitivity to CDDP, even in presence of surrounding monocytes, providing the rationale for the use of exosome inhibitors to prevent and/or overcome drug resistance. We conducted the majority of the experiments in two NBL cell lines and this might represent a limitation of this study (44), however we also showed higher TAM infiltration, higher miR-155, and lower TERF1 expression in primary NBL samples. Finally, upregulation of miR-155 and downregulation of TERF1 were observed in seven different types of cancer cell lines cocultured with human monocytes, suggesting a mechanism common to different cancers.
Also, we did not investigate in this study which concentrations of exosomic miR-21 trigger the TLR8-dependent response in TAMs and which other targets might be modulated by TAM-derived exosomic miR-155 in NBL cells. Future studies clarifying these aspects are warranted.
In conclusion our study identifies a new exosomic miR-21/TLR8/NF-кB/exosomic miR-155/TERF1 axis triggered regardless of M1- or M2- polarization, but not in dendritic cells involved in resistance to chemotherapy in NBL, and identifies exosomes within the TME as important molecular targets to restore drug sensitivity.
Funding
Dr. Fabbri is a St. Baldrick Foundation’s Scholar and is supported by the Pablove Foundation, the Concern Foundation, the Saban Research Institute Research Career Development Award, the Southern California Clinical and Translational Science Institute (SC CTSI), the Jean Perkins Foundation, the Nautica Malibu Triathlon Funds, the award number P30CA014089 from the National Cancer Institute, the Hugh and Audy Lou Colvin Foundation, the Concern Foundation, and by a Shirley McKernan stewardship. Drs. Seeger and Asgharzadeh are supported by the award number 5 P01 CA081403-15 from the National Cancer Institute. Dr Calin is The Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. Work in Dr. Calin’s laboratory is supported in part by the National Institutes of Health (NIH)/National Cancer Institute (NCI) grants 1UH2TR00943-01 and 1 R01 CA182905-01, the UT MD Anderson Cancer Center SPORE in Melanoma grant from NCI (P50 CA093459), Aim at Melanoma Foundation and the Miriam and Jim Mulva research funds, the Brain SPORE (2P50CA127001), the Center for radiation Oncology Research Project, the Center for Cancer Epigenetics Pilot project, a 2014 Knowledge GAP MDACC grant, a CLL Moonshot pilot project, the UT MD Anderson Cancer Center Duncan Family Institute for Cancer Prevention and Risk Assessment, an SINF grant in colon cancer, the Laura and John Arnold Foundation, the RGK Foundation and the Estate of C. G. Johnson Jr.
Supplementary Material
The study sponsors had no role in in the design of the study, the collection, analysis, or interpretation of the data, the writing of the manuscript, nor the decision to submit the manuscript for publication.
Author contributions: KBC and MF designed experiments, performed experiments, analyzed data and wrote the manuscript; PMW conducted in vivo experiments; PN, HC, MM, TX, IV, and FF performed experiments; XZ performed in situ hybridization; RK analyzed data and performed experiments; CI analyzed data; MH, HS, and SA analyzed samples and performed experiments; DA, GAC, AJ, and RCS designed experiments and wrote the paper; AG designed experiments, performed experiments, and wrote the manuscript.
References
- 1. Cohn SL, Pearson AD, London WB, et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol. 2009;27(2):289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deyell RJ, Attiyeh EF. Advances in the understanding of constitutional and somatic genomic alterations in neuroblastoma. Cancer Genet. 2011;204(3):113–121. [DOI] [PubMed] [Google Scholar]
- 3. Matthay KK, Reynolds CP, Seeger RC, et al. Long-term results for children with high-risk neuroblastoma treated on a randomized trial of myeloablative therapy followed by 13-cis-retinoic acid: a children’s oncology group study. J Clin Oncol. 2009;27(7):1007–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schmidt ML, Lal A, Seeger RC, et al. Favorable prognosis for patients 12 to 18 months of age with stage 4 nonamplified MYCN neuroblastoma: a Children’s Cancer Group Study. J Clin Oncol. 2005;23(27):6474–6480. [DOI] [PubMed] [Google Scholar]
- 5. George RE, London WB, Cohn SL, et al. Hyperdiploidy plus nonamplified MYCN confers a favorable prognosis in children 12 to 18 months old with disseminated neuroblastoma: a Pediatric Oncology Group study. J Clin Oncol. 2005;23(27):6466–6473. [DOI] [PubMed] [Google Scholar]
- 6. Baker DL, Schmidt ML, Cohn SL, et al. Outcome after reduced chemotherapy for intermediate-risk neuroblastoma. N Engl J Med. 2010;363(14):1313–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3(3):203–216. [DOI] [PubMed] [Google Scholar]
- 8. Pearson AD, Pinkerton CR, Lewis IJ, et al. High-dose rapid and standard induction chemotherapy for patients aged over 1 year with stage 4 neuroblastoma: a randomised trial. Lancet Oncol. 2008;9(3):247–256. [DOI] [PubMed] [Google Scholar]
- 9. Szakacs G, Paterson JK, Ludwig JA, et al. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5(3):219–234. [DOI] [PubMed] [Google Scholar]
- 10. Asgharzadeh S, Salo JA, Ji L, et al. Clinical significance of tumor-associated inflammatory cells in metastatic neuroblastoma. J Clin Oncol. 2012;30(28):3525–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ara T, Song L, Shimada H, et al. Interleukin-6 in the bone marrow microenvironment promotes the growth and survival of neuroblastoma cells. Cancer Res. 2009;69(1):329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ara T, Nakata R, Sheard MA, et al. Critical role of STAT3 in IL-6-mediated drug resistance in human neuroblastoma. Cancer Res. 2013;73(13):3852–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang QW, Liu L, Gong CY, et al. Prognostic significance of tumor-associated macrophages in solid tumor: a meta-analysis of the literature. PLoS One. 2012;7(12):e50946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Colvin EK. Tumor-associated macrophages contribute to tumor progression in ovarian cancer. Front Oncol. 2014;4:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Franklin RA, Liao W, Sarkar A, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344(6186):921–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113(6):673–676. [DOI] [PubMed] [Google Scholar]
- 17. Chen Y, Stallings RL. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 2007;67(3):976–983. [DOI] [PubMed] [Google Scholar]
- 18. Fabbri M, Paone A, Calore F, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109(31):E2110-E2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Asgharzadeh S, Pique-Regi R, Sposto R, et al. Prognostic significance of gene expression profiles of metastatic neuroblastomas lacking MYCN gene amplification. J Natl Cancer Inst. 2006;98(17):1193–1203. [DOI] [PubMed] [Google Scholar]
- 20. Gridley DS, Andres ML, Slater JM. Enhancement of prostate cancer xenograft growth with whole-body radiation and vascular endothelial growth factor. Anticancer Res. 1997;17(2A):923–928. [PubMed] [Google Scholar]
- 21. O’Connell RM, Taganov KD, Boldin MP, et al. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104(5):1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Melo SA, Sugimoto H, O’Connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26(5):707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9(11):798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li P, Grgurevic S, Liu Z, et al. Signal transducer and activator of transcription-3 induces MicroRNA-155 expression in chronic lymphocytic leukemia. PLoS One. 2013;8(6):e64678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Escobar T, Yu CR, Muljo SA, et al. STAT3 activates miR-155 in Th17 cells and acts in concert to promote experimental autoimmune uveitis. Invest Ophthalmol Vis Sci. 2013;54(6):4017–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mesa RA. Ruxolitinib, a selective JAK1 and JAK2 inhibitor for the treatment of myeloproliferative neoplasms and psoriasis. IDrugs. 2010;13(6):394–403. [PubMed] [Google Scholar]
- 27. Pardanani A, Tefferi A. Targeting myeloproliferative neoplasms with JAK inhibitors. Curr Opin Hematol. 2011;18(2):105–110. [DOI] [PubMed] [Google Scholar]
- 28. Kosaka N, Iguchi H, Yoshioka Y, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285(23):17442–17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wesbuer S, Lanvers-Kaminsky C, Duran-Seuberth I, et al. Association of telomerase activity with radio- and chemosensitivity of neuroblastomas. Radiat Oncol. 2010;5:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dinami R, Ercolani C, Petti E, et al. miR-155 drives telomere fragility in human breast cancer by targeting TRF1. Cancer Res. 2014;74(15):4145–4156. [DOI] [PubMed] [Google Scholar]
- 31. Smogorzewska A, van Steensel B, Bianchi A, et al. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol. 2000;20(5):1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kocak H, Ackermann S, Hero B, et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis. 2013;4:e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. [DOI] [PubMed] [Google Scholar]
- 34. Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. [DOI] [PubMed] [Google Scholar]
- 35. Diotti R, Loayza D. Shelterin complex and associated factors at human telomeres. Nucleus. 2011;2(2):119–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. [DOI] [PubMed] [Google Scholar]
- 37. Ohali A, Avigad S, Ash S, et al. Telomere length is a prognostic factor in neuroblastoma. Cancer. 2006;107(6):1391–1399. [DOI] [PubMed] [Google Scholar]
- 38. Deville L, Hillion J, Pendino F, et al. hTERT promotes imatinib resistance in chronic myeloid leukemia cells: therapeutic implications. Mol Cancer Ther. 2011;10(5):711–719. [DOI] [PubMed] [Google Scholar]
- 39. Yamada O, Ozaki K, Furukawa T, et al. Activation of STAT5 confers imatinib resistance on leukemic cells through the transcription of TERT and MDR1. Cell Signal. 2011;23(7):1119–1127. [DOI] [PubMed] [Google Scholar]
- 40. Goldblatt EM, Erickson PA, Gentry ER, et al. Lipid-conjugated telomerase template antagonists sensitize resistant HER2-positive breast cancer cells to trastuzumab. Breast Cancer Res Treat. 2009;118(1):21–32. [DOI] [PubMed] [Google Scholar]
- 41. Guo XL, Ma NN, Zhou FG, et al. Up-regulation of hTERT expression by low-dose cisplatin contributes to chemotherapy resistance in human hepatocellular cancer cells. Oncol Rep. 2009;22(3):549–556. [DOI] [PubMed] [Google Scholar]
- 42. Mukherjee S, Bhattacharya RK, Roy M. Targeting protein kinase C (PKC) and telomerase by phenethyl isothiocyanate (PEITC) sensitizes PC-3 cells towards chemotherapeutic drug-induced apoptosis. J Environ Pathol Toxicol Oncol. 2009;28(4):269–282. [DOI] [PubMed] [Google Scholar]
- 43. Smith V, Dai F, Spitz M, et al. Telomerase activity and telomere length in human tumor cells with acquired resistance to anticancer agents. J Chemother. 2009;21(5):542–549. [DOI] [PubMed] [Google Scholar]
- 44. Domcke S, Sinha R, Levine DA, et al. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


