Abstract
The expression patterns of secreted (MUC2 and MUC5AC) and membrane-tethered (MUC1, MUC4, MUC12 and MUC13) mucins were monitored in healthy pigs and pigs challenged orally with Lawsonia intracellularis. These results showed that the regulation of mucin gene expression is distinctive along the GI tract of the healthy pig, and may reflect an association between the function of the mucin subtypes and different physiological demands at various sites. We identified a specific depletion of secreted MUC2 from goblet cells in infected pigs that correlated with the increased level of intracellular bacteria in crypt cells. We concluded that L. intracellularis may influence MUC2 production, thereby altering the mucus barrier and enabling cellular invasion.
Keywords: Pig, Mucin, Lawsonia intracellularis, Goblet cells
1. Introduction
Lawsonia intracellularis (LI) is a Gram-negative intracellular bacterial pathogen; the aetiological agent that causes proliferative enteropathy (PE) (Jacobson et al., 2010). LI causes both subclinical infection and clinically apparent disease. Both forms affect the well-being of animals, clinically manifesting as poor feed conversion, anorexia, diarrhea and occasionally fatalities due to severe hemorrhage of the intestinal tract. This infection therefore affects health, welfare and productivity of pigs and is also increasingly reported in a wide range of animals, particularly in horses. LI infects the intestinal crypts and invades the immature enterocytes, causing intestinal hyperplasia by stimulating continuous cell division (Smith and Lawson, 2001) and alteration of mucosal cell integrity (Smith et al., 2014, Vannucci et al., 2013, Vannucci and Gebhart, 2014).
The mucosal epithelial tissues have highly specialized functions throughout the gastrointestinal (GI) tract to allow food absorption and processing for excretion. They also serve as a continuous barrier both to the commensal microorganisms and to potential viral, bacterial and eukaryotic pathogens (McGuckin et al., 2011). These epithelial cells constitutively secrete a highly hydrated structural mucus barrier that coats the surfaces of the cells lining the GI tract. This layer of complex fluid consists of a vast repertoire of defense compounds including mucin glycoproteins, antibodies, defensins, protegrins, collectins, cathlecidins, lysozymes, histatins, and nitric oxide. Regulated by underlying innate immune cell defense, the mucus barrier can also respond dynamically to pathogen insults, by altering its production rate, constituents and biophysical properties (Atuma et al., 2001). Mucins are the main component of the viscous layer and are synthesized and secreted by goblet cells which are scattered within the epithelial lining (Kim and Khan, 2013). The mucin gene family comprises 20 different mucin genes ranging from MUC1 to MUC20. They are classified as secreted and membrane bound forms (Bansil and Turner, 2006). Mucins are a family of highly glycosylated macromolecules that are multifunctional and provide the epithelial cells with protection against physical or chemical injures and mechanical stress, lubricating the intestinal tract to ease the passage of food content and preventing mucosal dehydration Bansil and Turner, 2006. Their primary function, however, is to serve as a first line of defence against pathogens such as Yersinia enterocolitica and Shigella flexneri (Kim and Khan, 2013, Mantle and Husar, 1994, Nutten et al., 2002). Mucins have been shown to have direct antimicrobial activity, and the ability to opsonize microbes to aid clearance. However, growing evidence suggests that intestinal bacteria have developed specific pathogenic factors and/or ways of interfering with mucin production in order to enable pathogens to cross the physical mucus barrier. Such a phenomenon has been observed during Helicobacter pylori invasion of the human stomach through its interaction with MUC5AC, enabling the bacteria to colonize and reside in the gastric mucus layer (Van de Bovenkamp et al., 2003). This is followed by alteration of MUC1 and MUC6 production to weaken the mucus gel layer, allowing the bacteria to penetrate through to the epithelial cells below (Byrd et al., 2000, Morgenstern et al., 2001, Van de Bovenkamp et al., 2003). Increased expression of MUC5AC in pig colon has also been observed during salmonellosis (Kim et al., 2009). Alteration of mucin expression has been shown in several diseases such as cystic fibrosis, chronic gastric inflammation caused by H. pylori and certain cancers (Llinares et al., 2004, Roger et al., 2000, Rose and Voynow, 2006). Such observations have highlighted the importance of mucins in relation to diseases and infections, and led us to examine in greater detail the changes in the regulation of mucin gene expression in the pig GI tract, following infection with LI. Recent work conducted by Smith et al. revealed that the gene encoding the glycoprotein MUC2 was down-regulated at the time of peak LI infection burden, suggesting that this pathogen may have a profound impact on mucin production and echoing previous research (Driemeier et al., 2002, Smith et al., 2014).
The objective of this work was to measure the pattern of MUC1, MUC2, MUC4, MUC5AC, MUC12 and MUC13 expression in the GI tract of healthy pigs and in the ileum of LI infected pigs. We found that the accumulation of MUC transcripts were highly regulated during LI infection and that the specific depletion of secreted MUC2 from goblet cells correlated with the increased level of intracellular bacteria in crypt epithelial cells.
2. Materials and methods
2.1. Samples
Tongue, stomach, duodenum, ileum, caecum, colon and rectum tissues were isolated from healthy Duroc pigs (n = 2–4) aged between 5 and 7 months with male and female equally represented. Infected ileum samples originated from a previous challenge study by MacIntyre et al. (2003), in which pigs were randomly selected from a minimal disease herd and fecal samples were culture negative for Brachyspira hyodysenteriae, B. pilosicoli, Yersinia spp, Salmonella spp and LI. Pigs were challenged with a pure culture of LI (isolate LR189/5/83) and euthanized at 3, 7, 14, 21, 28, 35 or 42 days post challenge (dpc). All pigs were then subjected to full necropsy. Infection was confirmed immunohistochemically using a monoclonal antibody VPM53 against LI (MacIntyre et al., 2003). For data normalization, negative control ileum samples were obtained from a separate group of three uninfected age-matched pigs.
2.2. RNA extraction
Total RNA was extracted from the ileum of infected pigs; these samples had been used previously for a microarray analysis (Smith et al., 2014). Briefly, total RNA was extracted from the ileum of infected pigs with Trizol (Invitrogen) using standard methods (Ait-Ali et al., 2007). The RNA was further purified using the Qiagen RNeasy mini-kit following the manufacturer's protocol (Qiagen). Using a Nanodrop spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA) and Agilent 2100 bioanalyser (Agilent Technologies) the quantity and quality of RNAs were assessed.
2.3. Quantification of L. intracellularis-specific genomic DNA
Genomic DNA of ileal samples for the measurement of L. intracellularis-specific genomic DNA originated from previous experiment (Smith et al., 2014). Quantitative measurement was performed as described previously and was expressed as number of 16S rRNA copies per ng of DNA (Smith et al., 2014). Only DNA from pigs at 3, 14 and 28 dpc were selected because they showed statistically significant variations (P < 0.0003) from the peak of infection at 14 dpc. Primers used are described in supplementary material 1.
2.4. Quantitative RT-PCR validations
Expression of MUC transcript levels in healthy and infected tissues was measured using quantitative RT-PCR (qRT-PCR). To reduce the likelihood of amplifying any contaminating genomic DNA that may be present in RNA preparations, MUC specific primer sequences (supplementary material 1) were positioned on exons flanking an intron, and qRT-PCR was run at extension times that favored the amplification of shorter cDNA sequences. RNA samples were analyzed with Brilliant III Ultra-Fast SYBR Green qRT-PCR Master Mix kit (Agilent Technologies) according to manufacturers’ instructions. Quantitative RT-PCR was performed with a Stratagene MX3000P (Stratagene) using the following profile: 48 °C 10 min, 95 °C 3 min; 40 cycles of 95 °C 15 s, 60 °C 20 s, and 95 °C 60 s, 60 °C 30 s, 95 °C 15 s, 25 °C 30 s. Samples were tested in triplicate.
A standard curve was used to quantify mucin transcript in the total RNA samples. Briefly, standard curves were constructed using gBlocks® Gene Fragments (Integrated DNA Technologies) of mucin gene sequences (Gene fragment sequences are listed in supplementary material 2), fused to a T7 promoter. The gBlocks® Gene Fragments were transcribed into cDNA through in vitro transcription, using mMESSAGE mMACHINE T7 transcription kit (Life Technologies, Paisley) according to the manufacturer's instructions. Agarose gel electrophoresis was used to verify transcribed products, followed by lithium chloride precipitation to purify the transcribed gBlocks Gene fragments mixture. Quality and quantity of the DNA was assessed using a Nanodrop spectrophotometer. Serial dilutions of the transcribed gene fragment were run on each plate of samples and analyzed by qRT-PCR. The log of the serial diluted gBlock gene fragment quantities was plotted against Cycle Threshold values to generate a standard curve, which was used to estimate the quantities of MUC transcript copy number in each sample.
2.5. Periodic acid Schiff staining
Sections of paraffin-embedded ileum from pigs euthanized at 3, 14, 28 dpc, and uninfected control pigs were processed, deparaffinized and hydrated according to standard protocols (Smith et al., 2014). Sections were treated for 5 min in 0.5% periodic acid and rinsed in distilled water. Sections were treated with Schiff's reagent at room temperature for 30 min and extensively rinsed in running tap water for 5 min. After mounting, sections were observed using a standard light microscope.
2.6. Immunohistochemistry
Sections of paraffin-embedded ileum from pigs euthanized at 3, 14, 28 dpc, and uninfected control pigs were processed, deparaffinised and hydrated according to standard protocols (Smith et al., 2014). Due to the fact that the detection of LI and MUC antigens required different method of antigen retrieval (see below) a dual staining strategy was not an option. Therefore we opted for single antigen detection on contiguous sections. Sections stained to detect LI were incubated for 10 min in proteinase K (Dako UK Ltd., Ely, UK) to allow antigen retrieval. After 30 min incubation in blocking agent (5% BSA/2% goat serum), the sections were incubated with mouse anti-LI VPM53 (MacIntyre et al., 2003) diluted 1:400 in blocking solution, at 4 °C overnight. After two washes in PBS, the slides were incubated for 30 min at room temperature with a FITC-conjugated goat anti-mouse IgG (Fc specific) F(ab)2 fragment (Sigma–Aldrich, UK) secondary antibody. Sections stained for detection of MUC2 and MUC4 underwent heat-mediated antigen retrieval in citrate buffer following dewaxing of paraffin sections. Sections were incubated with blocking agent for 30 min at room temperature, and were then incubated with either rabbit monoclonal anti-MUC2 (Abcam ab134119) diluted 1:1500 in blocking solution, at 4 °C overnight. After serial washes in PBS, sections were incubated with secondary antibody Alexa-Fluor-647 goat anti-rabbit (Life technologies, Paisley) diluted in 1:1000 in blocking solution. After a final wash as described above sections were incubated with 4′,6-diamidino-2-phenylindole (DAPI) and mounted with Lab Vision PermaFluor aqueous mounting medium (Thermo Fisher Scientific, location). Sections were observed with a LSM 700 confocal laser scanning microscope (Carl Zeiss). Images were captured using Zen Software (Carl Zeiss).
2.7. Quantitative image analysis
For quantitative analysis of PAS staining for Fig. 3, images were acquired at 20 magnification from at least 3 random fields per condition (healthy, 3, 14 and 28 dpc) and processed using Image J 1.49S. Briefly, the green channel was selected and the intensity threshold was adjusted to highlight goblet cells. Crypts were manually outlined and percentage of area with PAS signal was measured for 8–10 crypts per field and per condition.
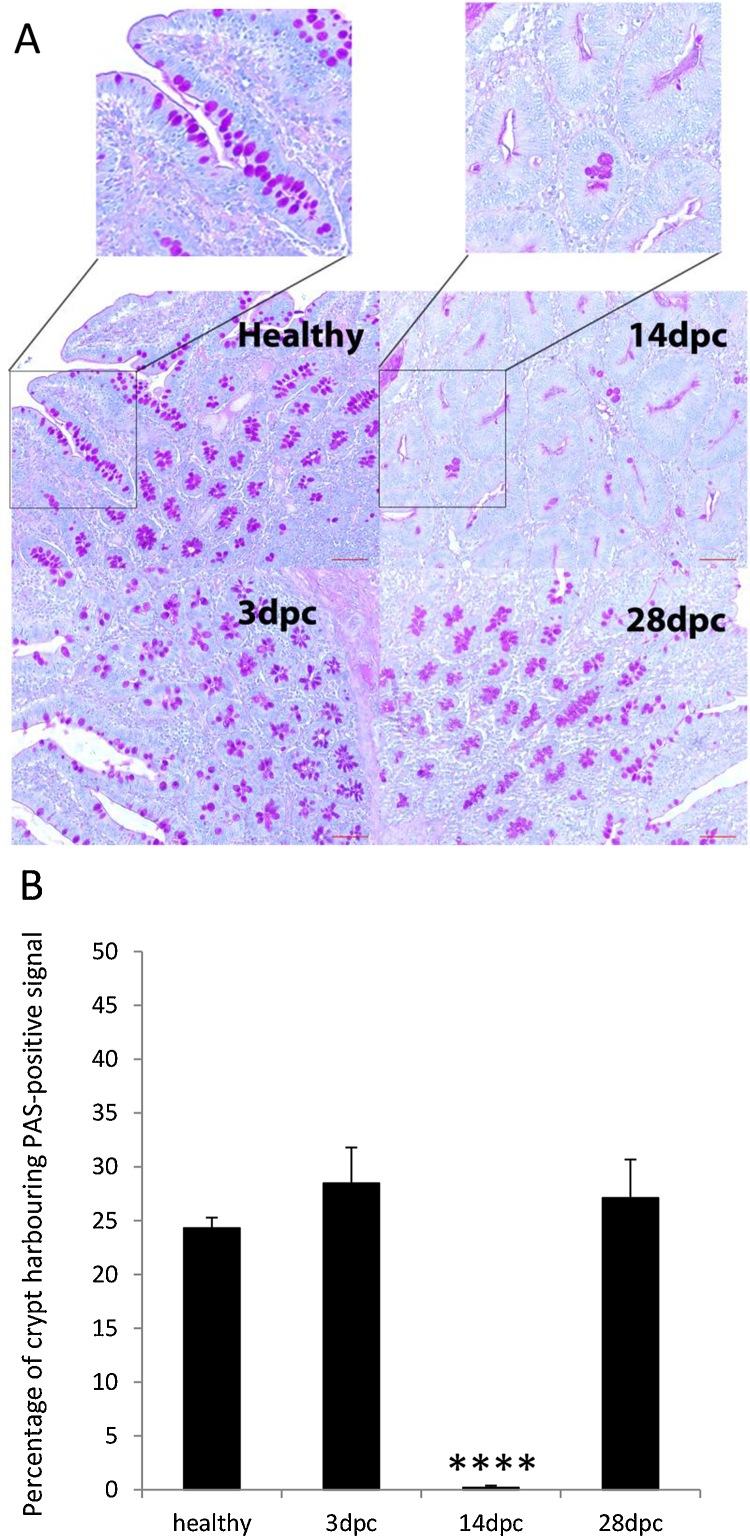
Fig. 3.
Periodic Acid-Schiff's staining of healthy and LI infected Ileum. A, staining was performed in ileum tissues of healthy and infected pigs collected 3, 14 and 28 dpc. Insets denote magnification of crypt region in healthy and in 14 dpc sections. Note the intense pink coloration of crypt which contains goblet cells for the ileum sections of healthy and infected tissues at 3 and 28 dpc. In contrast crypt at 14 dpc sections are lacking mucus-producing cells. Scale bar: 50 mm. B, quantification of PAS signal in crypt of healthy and infected ileum at 3, 14 and 28 dpc. At least 3 random fields and 8–10 crypts per condition (healthy, 3, 14 and 28 dpc) were measured using Image J 1.49S as described in Section 2. Asterisks indicate that the difference between 14 dpc time point and healthy is statistically significant: ****P < 0.0001.
For quantitative assessment of the co-localization of the MUC2 and LI immunofluorescence signals of Fig. 4B, confocal images at 20× magnification were obtained from contiguous sections at 14 dpc that were stained either with anti-MUC2 antibody or an anti-LI antibody followed with recommended secondary fluorescent antibody detection. Two random fields from 2 independent experimental staining were analyzed using Image J 1.49S. Briefly, color channels were selected and intensity thresholds were adjusted to highlight either the MUC2 or the LI signal. In total 25 areas were manually selected and MUC2 and LI signal intensity ratios were measured for the same area on contiguous sections. To model the trend of the MUC2 accumulation with the increase in LI infection within crypt cells at 14 dpc an asymptotic regression was fitted to the data. This was done using the function NLSstAsymptotic from the stats package in R (http://www.r-project.org/), which fits the following equation:
where y is the MUC2 immunofluorescence signal and x the percentage of LI infected crypt; b0 is the estimated intercept on the y-axis, b1 is the estimated difference between the asymptote and the y-intercept, and lrc is the estimated logarithm of the rate constant.
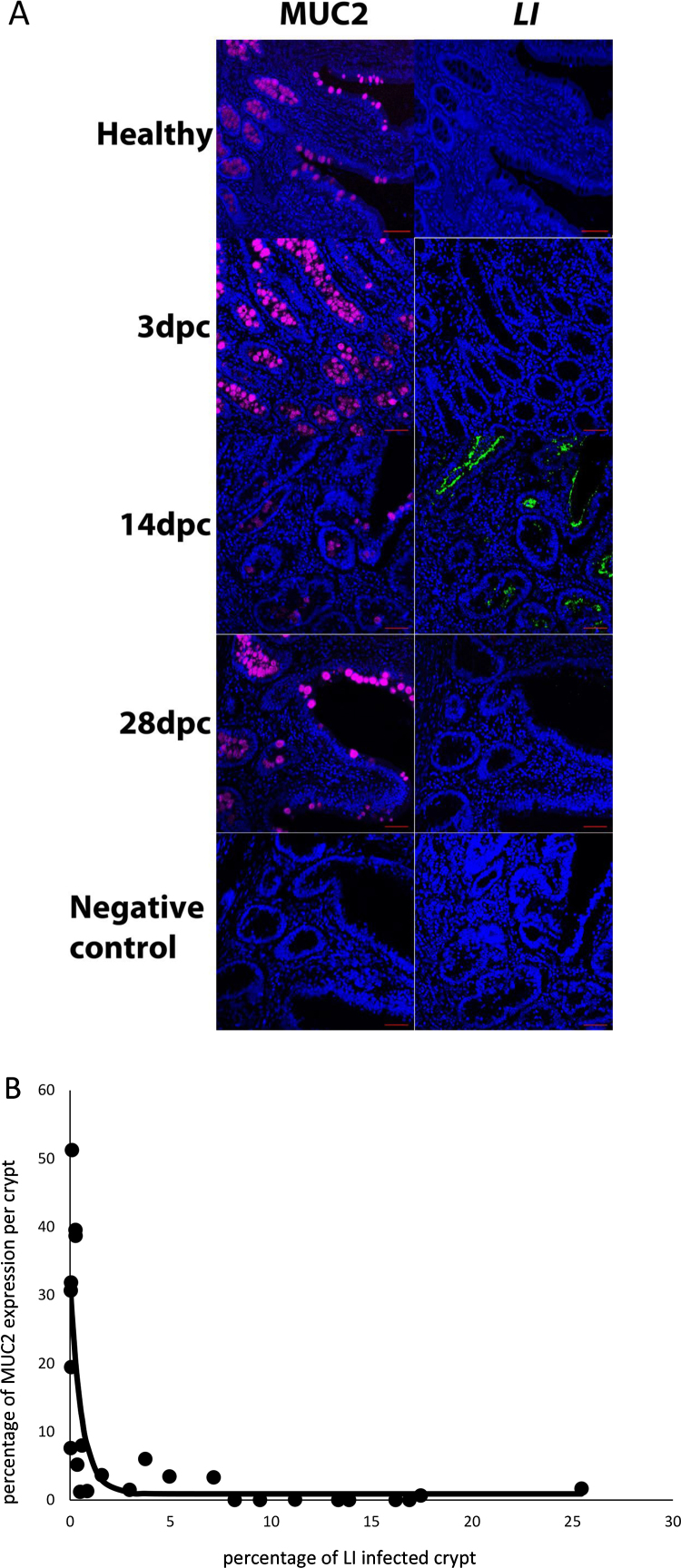
Fig. 4.
Immunohistochemical detection of MUC2 and LI. (A) Immunohistochemical detection in healthy and LI infected ileum collected at 3, 14 and 28 dpc and confocal microscopy as described in material and methods section. Anti MUC2 and anti-LI primary antibodies were detected with Alexa-Fluor 467 (purple) and FITC (green) secondary antibody antibodies, respectively. MUC2 and LI were detected on contiguous sections. Nuclei were revealed with DAPI (blue). For negative controls no primary antibodies were used. Scale bar: 50 μm. B, Quantitative assessment of the co-localization of the MUC2 and LI immunofluorescence signals of A, confocal images at 20× magnification were obtained from contiguous sections at 14 dpc that were stained either with anti-MUC2 antibody or an anti-LI antibody. Two random fields from 2 independent experimental staining were analyzed using Image J 1.49S. Briefly, color channels were selected and intensity thresholds were adjusted to highlight either the MUC2 or the LI signal. In total 25 areas were manually selected and MUC2 and LI signal intensity ratio were measured for the same area on contiguous sections and plotted (white round shape circle). To model the trend of the MUC2 accumulation with the increase in LI infection within crypt cells at 14 dpc an asymptotic regression was fitted to the data (black line). This was done using the function NLSstAsymptotic from the stats package in R (http://www.r-project.org/), see Section 2 in for the description of the equation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.8. Statistical analysis
With the exception of multiple T-tests analysis for the data of Fig. 1 all the statistical analysis of data that involved change of specific parameters over time were performed with a multiple least squares regression using the GLM procedure from SAS. The model accounted for fixed effects of gene, day-after-treatment and their interactions. Contrasts against 14 dpc within gene (and their significant values) were calculated from the analysis.
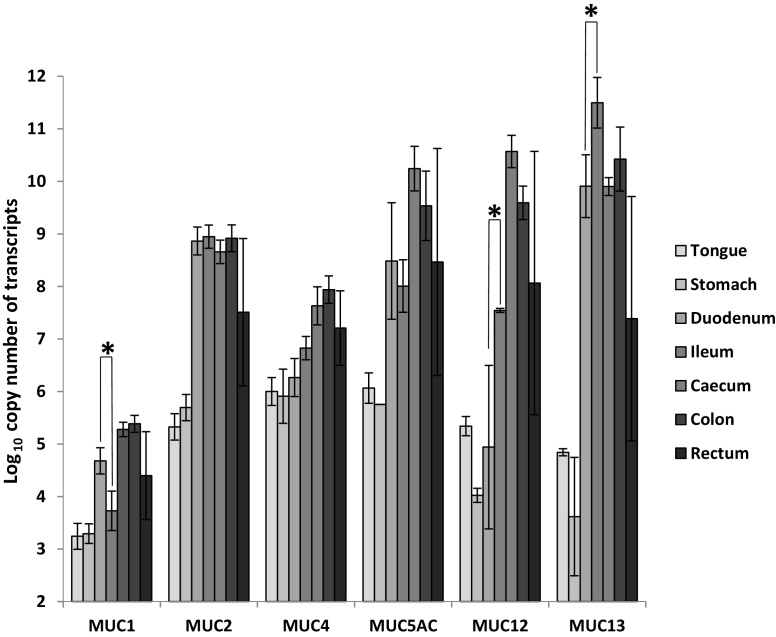
Fig. 1.
Quantification of transcript levels encoding MUC1, MUC2, MUC4, MUC5AC, MUC12, and MUC13 in healthy pig tissues using real time qPCR. Data shows the mean Log10 copy number of RNA/μg of total RNA with error bars representing ± SD for n = 2–4 pigs. One asterisk (*) indicates that the difference between tissues is statistically significant using student's T-test (P < 0.05).
2.9. Pearson correlation
Using the data of the quantitative assessment of the co-localization of the MUC2 and LI signals a Pearson correlation coefficient was calculated in SAS.
3. Results and discussion
3.1. Differential regulation of MUC subtype transcripts in GI tract
Accumulation of transcripts encoding for secreted MUC2 and MUC5AC, and membrane-tethered mucins, MUC1, MUC4, MUC12 and MUC13 in tongue, stomach, duodenum, ileum, caecum and colon, from healthy Duroc pigs were measured using real-time qRT-PCR. Results revealed that transcripts were predominantly expressed in caecum, colon and rectum, i.e. the distal portions of the GI tract, although for MUC2 and MUC13, expression in the duodenum and ileum was similar to the large intestine (Fig. 1). This observation is consistent with the distribution and density of mucin secreting cells (goblet cells), in the mammalian large intestine, since they predominate in the large intestine and increase in frequency aborally, from the caecum to the rectum (Specian, 1991). MUC1 transcript expression was the lowest across most tissues, with levels ranging from 3.3 to 5.3 Log10 copy number/μg of RNA. In the small intestine differences in the accumulation of transcripts encoding for MUC1, MUC12 and MUC13 between the duodenum and ileum were significant (P < 0.05). Interestingly the transcript levels for MUC1, MUC4 and MUC13, with the exception of the high level of MUC1 transcripts in the stomach, were consistent with previous reports (Freeman et al., 2012, Zhang et al., 2008) (supplementary material 3). Collectively, these results show that the regulation of mucin gene expression displays a distinctive pattern along the GI tract and that this pattern may reflect the functions of the mucin subtypes that likely change in relation to the physiological demands at the various sites (Deplancke and Gaskins, 2001).
3.2. Simultaneous modulation of the level of transcripts encoding mucins is associated with high burden of LI infection in ileum
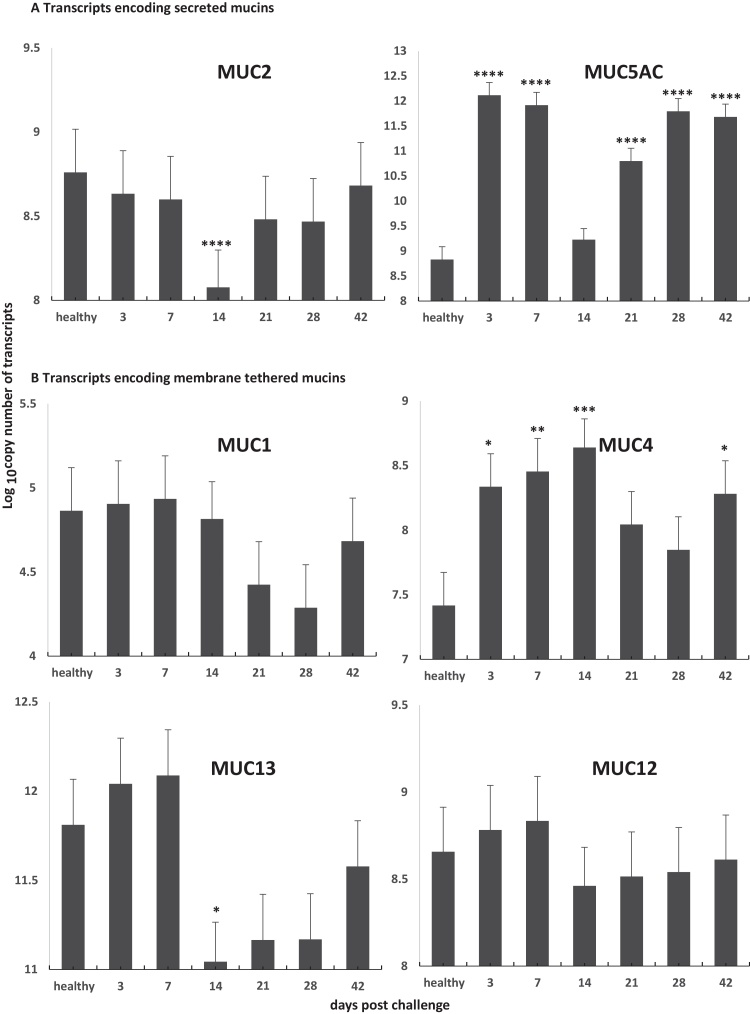
LI infection leads to intestinal crypt epithelial cell invasion and proliferation, but the mechanisms responsible are yet to be defined. To assess whether MUC gene expression changed in pigs with PE we measured transcript levels in ileum of healthy pigs and pigs orally challenged with strain LI LR189/5/83 for 3, 7, 14, 21, 28 and 42 days (Smith et al., 2014). Table 1 shows that LI 16S rRNA gene accumulation was highest at 14 dpc and was statistically significant when compared to time points 3 and 28 dpc (P < 0.003 and P < 0.003), respectively, as previously shown (Smith et al., 2014) and which suggested that LI bacterial load in crypt cells were the highest 14 dpc. When MUC transcripts were measured our results showed evidence of a modulation of the regulation for some of the transcripts, starting as early as 3 dpc in infected ileum (Fig. 2 and supplementary material 4). MUC5AC transcript levels were highest at 3 and 7 dpc (P < 0.001). Interestingly, MUC4 transcript levels remained high up to 14 dpc (P < 0.001). This up-regulation could be due to a host innate immune and/or inflammatory response to the invading pathogen, since inflammatory diseases of the epithelium are often characterized by mucin up-regulation and mucus hypersecretion in response to pathogen aggression. For example in humans, transcripts encoding for MUC5AC, MUC2 and MUC4 are consistently up-regulated by inflammatory and immune processes involving molecules such as TNF-a and/or IFN-a and LPS (Hauber et al., 2006, Li et al., 1997, McNamara and Basbaum, 2001, Perrais et al., 2001). The absence of significant up-regulation of MUC2 transcript levels may be due to (a) differences in regulatory sequences, such as in the promoter region of the gene, or (b) a specific inhibitory effect exerted by L. intracellularis on mucin subtype transcript expression during early infection between 3 and 7 dpc. An apparent coordinated down-regulation of transcripts was observed at 14 dpc for MUC2, MUC5AC and MUC13 (P < 0.05). It is noteworthy that in contrast to MUC2 transcript regulation, MUC5AC and MUC4 transcript levels regulation undergo a reduction of up regulation rather than down-regulation when compared to the levels in the healthy ileum at 14 dpc (no statistical differences when compared to healthy ileum) and 21 dpc (P-value is approaching statistical significance), respectively. While the down-regulation of MUC2 (P < 0.0001), MUC13 (P < 0.05) and possibly MUC5AC transcripts was transient and centered on 14 dpc (supplementary material 4) suggested a general trend of down regulation of these transcripts at that infection time point. In contrast, the changes in MUC1 and MUC12 transcript levels did not reach statistical significance and therefore no conclusion could be reach regarding these transcripts (supplementary material 4). These findings confirm and further enhance the results of Smith et al. (2014), in which a 3–9 fold reduction in MUC2 transcripts was observed at 14 dpc (Smith et al., 2014). It is unclear why MUC2, MUC5AC and MUC13 transcript levels are transiently down-regulated at 14 dpc, but it is noteworthy that at this time of infection bacterial load is at its highest level (Table 1) (MacIntyre et al., 2003, Smith et al., 2014). Hence, the transient down-regulation of secreted MUC2, MUC5AC and MUC13 may either be a consequence of a homeostatic regulation mechanism during hyperplasia or a result of the mechanism of invasion of crypt cells by LI.
Table 1.
Quantification of 16S rRNA Lawsonia intracellularis LR189/5/83 in experimentally challenged pigs.
| Days post challenge | 3 | 14 | 28 |
|---|---|---|---|
| Log10 16S rRNA copies/ng of DNA | 3.83 ± 0.12** | 4.84 ± 0.11 | 3.88 ± 0.11** |
16S rRNA gene copy number per ng of genomic DNA was measured by qPCR as described in Section 2. The mean copy number of the 16S rRNA gene was expressed in Log10 for each time point assessed at 3 (n = 3), 14 (n = 4) and 28 (n = 4) dpc were measured using the same DNA than previously reported (Smith et al., 2014) with the exception of 28 dpc where n = 4. Only DNA from pigs at 3, 14 and 28 dpc were selected because they showed statistical significant variation (P < 0.001) from the peak of infection at 14 dpc. Mean ± standard errors are shown. Statistical analysis of data was performed using a multiple least squares regression versus 14 dpc: ** denotes P < 0.001.
Fig. 2.
Level of transcripts encoding MUC1, MUC2, MUC4, MUC5AC, MUC12, and MUC13 in ileum of healthy pigs and orally challenged pigs with strain LI LR189/5/83 for 3, 7, 14, 21, 28 and 42 days (n = 3–4 pigs). A, transcripts encoding secreted mucins and B, transcripts encoding membrane tethered mucins. The statistical analysis of the data was performed using a multiple least squares regression. Asterisks indicate that the difference between time point and healthy is statistically significant: ****P < 0.0001; **P < 0.01; and *P < 0.05. Statistical analysis of the data is shown in supplementary material 4.
3.3. Depletion of MUC2 protein in goblet cells coincides with the accumulation of LI in crypt cells
To further assess if the modulation of MUC transcripts during the LI infection of ileum tissue at 3, 14, 28 dpc is associated with alterations of protein accumulation in goblet cells, we performed periodic acid Schiff staining (Driemeier et al., 2002, Mcmanus, 1946). Fig. 3A and B shows that at 14 dpc, the peak of LI infection burden, a near complete depletion of goblet cells (P < 0.001) was observed and echoed previous report (Driemeier et al., 2002). Further immunohistochemistry analysis revealed that MUC2 was exclusively and abundantly present in goblet cells of healthy ileum (Fig. 4A). Dense and positive MUC2 staining was localized in the cytoplasm of the cells and within the luminal surface, the location of mucinous secretions. Upon infection with LI, MUC2 staining in the goblet cells declined substantially between 3 and 14 dpc prior to being re-established at 28 dpc (Fig. 4A). This result is consistent with those obtained in Fig. 2 and suggests that MUC2 production may be down-regulated at transcriptional levels. Furthermore, we observed that MUC2 down-regulation may coincide with the concomitant accumulation of LI in the apical region of the crypt cells during early infection. To further investigate the coexistence of MUC2 and LI bacteria in crypt cells we quantified immunofluorescence signals of each entity on confocal microscopy images (Fig. 4B). We found that there was a significant negative correlation between LI and MUC2 of −0.53 (P < 0.007). To model the trend of the MUC2 accumulation with the increase in LI infection within crypt cells at 14 dpc an asymptotic regression was fitted to the data.
These results highlighted that MUC2 protein and LI bacteria may not coexist in the same crypt area. Taken together these data suggest that MUC2 accumulation may be inhibited by LI bacterial infection.
Several factors may explain the regulation of MUC2 in infected ileum at the peak of infection. PE lesions are often associated with enlarged intestinal crypts lined by numerous proliferating immature epithelial cells and a reduction or even absence of goblet cells at both 10 and 21 dpc (Driemeier et al., 2002, McOrist et al., 2006, McOrist and Lawson, 1989, McOrist et al., 1996). It is believed that the basic hyperplastic lesion may develop 2–3 weeks post challenge, which coincides with the time when MUC2 is down-regulated. Since MUC2 is synthesized and secreted by goblet cells, the reduction in goblet cells at time of peak PE lesion may provide an explanation for the down-regulation observed at 14 dpc. Muc2-deficient mice have been shown to have increased permeability in the intestinal mucus layer, a consequence of which is increased bacterial adhesion to the underlying epithelial cells (Johansson et al., 2008).
In conclusion, collectively, these observations suggest that LI may be able to influence the regulation of MUC2 expression in order for it to cross the mucus barrier and invade the mucosal layer. Further investigation would be required to assess if the alteration in MUC2 transcript levels is caused directly by an as yet unidentified LI encoded factor or if it is a consequence of the host immune response to the infection.
Conflict of interest statement
The authors declare no conflict of interest.
Acknowledgement
This work was supported by BBSRC Institute Strategic Program Grant.
Footnotes
Supplementary material related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vetimm.2015.08.005
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Ait-Ali T., Wilson A.D., Westcott D.G., Clapperton M., Waterfall M., Mellencamp M.A., Drew T.W., Bishop S.C., Archibald A.L. Innate immune responses to replication of porcine reproductive and respiratory syndrome virus in isolated Swine alveolar macrophages. Viral Immunol. 2007;20:105–118. doi: 10.1089/vim.2006.0078. [DOI] [PubMed] [Google Scholar]
- Atuma C., Strugala V., Allen A., Holm L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am. J. Physiol. Gastrointest. Liver Physiol. 2001;280:G922–G929. doi: 10.1152/ajpgi.2001.280.5.G922. [DOI] [PubMed] [Google Scholar]
- Bansil R.T., Turner B.S. Mucin structure, aggregation, physiological functions and biomedical applications. Curr. Opin. Colloid Interface Sci. 2006;11:164–170. [Google Scholar]
- Byrd J.C., Yunker C.K., Xu Q.S., Sternberg L.R., Bresalier R.S. Inhibition of gastric mucin synthesis by Helicobacter pylori. Gastroenterology. 2000;118:1072–1079. doi: 10.1016/s0016-5085(00)70360-x. [DOI] [PubMed] [Google Scholar]
- Deplancke B., Gaskins H.R. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 2001;73:1131S–1141S. doi: 10.1093/ajcn/73.6.1131S. [DOI] [PubMed] [Google Scholar]
- Driemeier D., Faccini G.S., de Oliveira R.T., Colodel E.M., Traverso S.D., Cattani C. Silver staining combined with alcian blue and hematoxylin–eosin for the detection of Lawsonia intracellularis in swine proliferative enteropathy. Acta Histochem. 2002;104:285–287. doi: 10.1078/0065-1281-00647. [DOI] [PubMed] [Google Scholar]
- Freeman T.C., Ivens A., Baillie J.K., Beraldi D., Barnett M.W., Dorward D., Downing A., Fairbairn L., Kapetanovic R., Raza S., Tomoiu A., Alberio R., Wu C., Su A.I., Summers K.M., Tuggle C.K., Archibald A.L., Hume D.A. A gene expression atlas of the domestic pig. BMC Biol. 2012;10:90. doi: 10.1186/1741-7007-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber H.P., Foley S.C., Hamid Q. Mucin overproduction in chronic inflammatory lung disease. Can. Respir. J. Can. Thorac. Soc. 2006;13:327–335. doi: 10.1155/2006/901417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M., Aspan A., Nordengrahn A., Lindberg M., Wallgren P. Monitoring of Lawsonia intracellularis in breeding herd gilts. Vet. Microbiol. 2010;142:317–322. doi: 10.1016/j.vetmic.2009.09.034. [DOI] [PubMed] [Google Scholar]
- Johansson M.E., Phillipson M., Petersson J., Velcich A., Holm L., Hansson G.C. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.H., Kim D., Ha Y., Cho K.D., Lee B.H., Seo I.W., Kim S.H., Chae C. Expression of mucins and trefoil factor family protein-1 in the colon of pigs naturally infected with Salmonella typhimurium. J. Comp. Pathol. 2009;140:38–42. doi: 10.1016/j.jcpa.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Kim J.J., Khan W.I. Goblet cells and mucins: role in innate defense in enteric infections. Pathogens. 2013;2:55–70. doi: 10.3390/pathogens2010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.D., Dohrman A.F., Gallup M., Miyata S., Gum J.R., Kim Y.S., Nadel J.A., Prince A., Basbaum C.B. Transcriptional activation of mucin by Pseudomonas aeruginosa lipopolysaccharide in the pathogenesis of cystic fibrosis lung disease. Proc. Natl. Acad. Sci. U. S. A. 1997;94:967–972. doi: 10.1073/pnas.94.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llinares K., Escande F., Aubert S., Buisine M.P., de Bolos C., Batra S.K., Gosselin B., Aubert J.P., Porchet N., Copin M.C. Diagnostic value of MUC4 immunostaining in distinguishing epithelial mesothelioma and lung adenocarcinoma. Mod. Pathol. 2004;17:150–157. doi: 10.1038/modpathol.3800027. [DOI] [PubMed] [Google Scholar]
- MacIntyre N., Smith D.G., Shaw D.J., Thomson J.R., Rhind S.M. Immunopathogenesis of experimentally induced proliferative enteropathy in pigs. Vet. Pathol. 2003;40:421–432. doi: 10.1354/vp.40-4-421. [DOI] [PubMed] [Google Scholar]
- Mantle M., Husar S.D. Binding of Yersinia enterocolitica to purified, native small intestinal mucins from rabbits and humans involves interactions with the mucin carbohydrate moiety. Infect. Immun. 1994;62:1219–1227. doi: 10.1128/iai.62.4.1219-1227.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuckin M.A., Linden S.K., Sutton P., Florin T.H. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 2011;9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- Mcmanus J.F.A. Histological demonstration of mucin after periodic acid. Nature. 1946;158:202. doi: 10.1038/158202a0. [DOI] [PubMed] [Google Scholar]
- McNamara N., Basbaum C. Signaling networks controlling mucin production in response to Gram-positive and Gram-negative bacteria. Glycoconjug. J. 2001;18:715–722. doi: 10.1023/a:1020875423678. [DOI] [PubMed] [Google Scholar]
- McOrist S., Gebhart C.J., Bosworth B.T. Evaluation of porcine ileum models of enterocyte infection by Lawsonia intracellularis. Can. J. Vet. Res. = Rev. can. rech. vet. 2006;70:155–159. [PMC free article] [PubMed] [Google Scholar]
- McOrist S., Lawson G.H. Reproduction of proliferative enteritis in gnotobiotic pigs. Res. Vet. Sci. 1989;46:27–33. [PubMed] [Google Scholar]
- McOrist S., Roberts L., Jasni S., Rowland A.C., Lawson G.H., Gebhart C.J., Bosworth B. Developed and resolving lesions in porcine proliferative enteropathy: possible pathogenetic mechanisms. J. Comp. Pathol. 1996;115:35–45. doi: 10.1016/s0021-9975(96)80026-0. [DOI] [PubMed] [Google Scholar]
- Morgenstern S., Koren R., Moss S.F., Fraser G., Okon E., Niv Y. Does Helicobacter pylori affect gastric mucin expression? Relationship between gastric antral mucin expression and H. pylori colonization. Eur. J. Gastroenterol. Hepatol. 2001;13:19–23. doi: 10.1097/00042737-200101000-00004. [DOI] [PubMed] [Google Scholar]
- Nutten S., Sansonetti P., Huet G., Bourdon-Bisiaux C., Meresse B., Colombel J.F., Desreumaux P. Epithelial inflammation response induced by Shigella flexneri depends on mucin gene expression. Microbes Infect./Inst. Pasteur. 2002;4:1121–1124. doi: 10.1016/s1286-4579(02)01636-2. [DOI] [PubMed] [Google Scholar]
- Perrais M., Pigny P., Ducourouble M.P., Petitprez D., Porchet N., Aubert J.P., Van Seuningen I. Characterization of human mucin gene MUC4 promoter: importance of growth factors and proinflammatory cytokines for its regulation in pancreatic cancer cells. J. Biol. Chem. 2001;276:30923–30933. doi: 10.1074/jbc.M104204200. [DOI] [PubMed] [Google Scholar]
- Roger P., Gascard J.P., Bara J., Brink C. Development of a functional human bronchial model of mucin secretion. Therapie. 2000;55:51–54. [PubMed] [Google Scholar]
- Rose M.C., Voynow J.A. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol. Rev. 2006;86:245–278. doi: 10.1152/physrev.00010.2005. [DOI] [PubMed] [Google Scholar]
- SAS Institute Inc., SAS 9.3 Help and Documentation, Cary, NC: SAS Institute Inc., 2002-2011.
- Smith D.G., Lawson G.H. Lawsonia intracellularis: getting inside the pathogenesis of proliferative enteropathy. Vet. Microbiol. 2001;82:331–345. doi: 10.1016/s0378-1135(01)00397-2. [DOI] [PubMed] [Google Scholar]
- Smith S.H., Wilson A.D., Van Ettinger I., MacIntyre N., Archibald A.L., Ait-Ali T. Down-regulation of mechanisms involved in cell transport and maintenance of mucosal integrity in pigs infected with Lawsonia intracellularis. Vet. Res. 2014;45:55. doi: 10.1186/1297-9716-45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specian R.G., Oliver M.G. Functional biology of intestinal goblet cells. Am J Physiol. 1991;260:183–193. doi: 10.1152/ajpcell.1991.260.2.C183. [DOI] [PubMed] [Google Scholar]
- Van de Bovenkamp J.H., Mahdavi J., Korteland-Van Male A.M., Buller H.A., Einerhand A.W., Boren T., Dekker J. The MUC5AC glycoprotein is the primary receptor for Helicobacter pylori in the human stomach. Helicobacter. 2003;8:521–532. doi: 10.1046/j.1523-5378.2003.00173.x. [DOI] [PubMed] [Google Scholar]
- Vannucci F.A., Foster D.N., Gebhart C.J. Laser microdissection coupled with RNA-seq analysis of porcine enterocytes infected with an obligate intracellular pathogen (Lawsonia intracellularis) BMC Genomics. 2013;14:421. doi: 10.1186/1471-2164-14-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannucci F.A., Gebhart C.J. Recent advances in understanding the pathogenesis of Lawsonia intracellularis infections. Vet. Pathol. 2014;51:465–477. doi: 10.1177/0300985813520249. [DOI] [PubMed] [Google Scholar]
- Zhang B., Ren J., Yan X., Huang X., Ji H., Peng Q., Zhang Z., Huang L. Investigation of the porcine MUC13 gene: isolation, expression, polymorphisms and strong association with susceptibility to enterotoxigenic Escherichia coli F4ab/ac. Anim. Genet. 2008;39:258–266. doi: 10.1111/j.1365-2052.2008.01721.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.