Abstract
Aim:
Fatty acid-binding protein 4 (FABP4) plays an important role in maintaining glucose and lipid homeostasis. The aim of this study was to find new inhibitors of FABP4 for the treatment of type 2 diabetes.
Methods:
Human FABP4 protein was expressed, and its inhibitors were detected in 1,8-ANS displacement assay. The effect of the inhibitor on lipolysis activity was examined in mouse 3T3-L1 preadipocytes. The db/db mice were used to evaluate the anti-diabetic activity of the inhibitor. Molecular docking and site-directed mutagenesis studies were carried out to explore the binding mode between the inhibitor and FABP4.
Results:
From 232 compounds tested, benzbromarone (BBR), an old uricosuric drug, was discovered to be the best inhibitor of FABP4 with an IC50 value of 14.8 μmol/L. Furthermore, BBR (25 μmol/L) significantly inhibited forskolin-stimulated lipolysis in 3T3-L1 cells. Oral administration of BBR (25 or 50 mg/kg, for 4 weeks) dose-dependently reduced the blood glucose level and improved glucose tolerance and insulin resistance in db/db mice. Molecular docking revealed that the residues Ser55, Asp76, and Arg126 of FABP4 formed important interactions with BBR, which was confirmed by site-directed mutagenesis studies.
Conclusion:
BBR is an inhibitor of FABP4 and a potential drug candidate for the treatment of type 2 diabetes and atherosclerosis.
Keywords: benzbromarone, uricosuric drug, fatty acid-binding protein 4, adipocyte, diabetes, db/db mouse, molecular docking
Introduction
Fatty acid-binding protein 4 (FABP4), also known as A-FABP or aP2, is highly expressed in mature adipocytes and activated macrophages. It plays an important role in maintaining glucose and lipid homeostasis1,2. Disruption of FABP4 in mice prevented them from developing diet-induced insulin resistance3,4. Population genetic studies showed that a genetic variant of FABP4 reduced FABP4 expression in the adipose tissue of human beings, resulting in lower serum triglyceride levels and significantly reducing the risk of developing type 2 diabetes5. Total or macrophage-specific gene deletion of FABP4 was shown to suppress the process of atherosclerosis in apolipoprotein E-deficient mice6. Thus, inhibitors of FABP4 have been considered potential drug candidates for diabetes and atherosclerosis7.
Benzbromarone (BBR), an old uricosuric drug, has been widely used as a therapeutic agent for hyperuricemia for 30 years8. In this study, we discovered that BBR was an inhibitor of FABP4. Because FABP4 inhibitors confer beneficial effects against diabetes7, both in vitro and in vivo, the anti-diabetic activities of BBR were evaluated. Furthermore, to reveal the binding mechanism of BBR in the active site of FABP4, molecular docking and point mutation studies were carried out.
Materials and methods
The expression and purification of FABP4
The human FABP4 (wild type or mutant) gene was subcloned into the pET28a vector (synthesized by Shanghai Generay Biotech Co, Ltd). The pET28a-FABP4 plasmid contains a C-terminal His6 tag. The synthesized genes were verified by DNA sequence analysis. Rosetta bacterial cells (purchased from Transgene) that had been transformed with a pET28a plasmid encoding a His-tagged human FABP4 were grown in Luria-Bertani (LB) medium. To induce protein expression, the cells were incubated with 0.8 mmol/L isopropyl-β-D-thiogalactoside (IPTG) in the medium for 4 h. The cells were then harvested by centrifugation and lysed in ice-cold lysis buffer [20 mmol/L HEPES (pH 7.5), 500 mmol/L NaCl and protease inhibitor cocktail solution (Sigma-Aldrich)]. The debris was pelleted by centrifugation at 12 000×g for 45 min at 4 °C. Finally, His-tagged FABPs were isolated by nickel-affinity chromatography; the purity of the isolated proteins was verified by SDS-PAGE.
In vitro FABP4 inhibition assay
The inhibitory activities of the compounds (BBR and linoleic acid) against wild-type and mutated FABP4 were evaluated using the 1,8-ANS displacement assay developed by Kane and Ber with some modification as described previously9. Briefly, 10 μmol/L of 1,8-ANS in phosphate buffered solution (PBS, pH 7.4) was mixed with FABP4 protein (10 μmol/L, final concentration). Compounds at varying concentrations were added and incubated for 3 min at room temperature. The fluorescence signal at 370 nm (excitation)/470 nm (emission) of the assay system was then determined with a Flexstation III instrument (Molecular Devices, CA, USA).
Analysis of adipocyte lipolysis
Mouse 3T3-L1 preadipocytes were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% newborn calf serum (NCS). Cells were seeded in 48-well plates for differentiation. Two days after confluence, a mixture containing 0.1 mmol/L 3-isobutyl-L-methylxanthine, 1 μmol/L dexamethasone and 2 μg/mL insulin was added to the DMEM with 10% fetal bovine serum (FBS) to create an induction medium. After 48 h, cells were switched to differentiation medium containing DMEM with 10% FBS plus insulin (2 μg/mL). Thereafter, the differentiation medium was replaced every 2 d. After 8 d, cells were treated with different concentrations of BBR or with DMSO (final concentration 0.2%) for 24 h. On the day of lipolysis experimentation, cells were incubated in Krebs Ringer HEPES buffer for 2 h in the presence or absence of 20 μmol/L forskolin. To determine lipolysis, glycerol levels in supernatants were measured using a commercial glycerol assay kit (Applygen Technologies Inc, China).
Treatment of db/db diabetic mice with BBR
Male C57BL/KsJ-Lepdb (db/db) mice were obtained from Jackson Laboratories (Bar Harbor, ME, USA). Animals were maintained in a light-dark (12–12 h) cycle at a temperature of 23 °C with free access to regular chow and drinking water. To investigate the anti-diabetic effect of BBR in db/db mice, 8-week-old male db/db mice were divided into three groups (n=8/group) and were given vehicle (0.5% sodium carboxyl methyl cellulose, CMC-Na) or BBR (25 or 50 mg/kg body weight) by gastric lavage for 4 weeks. The body weight and food intake of the mice were regularly recorded. After fasting for 6 h, the blood of each animal was obtained via the tail vein, and the glucose concentration was measured by a glucometer (One-Tough Ultra, Lifescan). At the end of the experiment, mice were fasted for 12 h for the oral glucose tolerance test (OGTT) and 6 h for the insulin tolerance test (ITT) as described10.
Molecular docking
Molecular docking was carried out using AutoDock4.2 software11. The X-ray structure of FABP4 bound with the inhibitor BMS309403 (PDB ID: 2NNQ12) was retrieved from the Protein Data Bank (http://www.rcsb.org/pdb) for docking calculation. Water, buffer molecules and ions were all removed. To prepare both the protein and BBR, all hydrogens were added, Gasteiger charges were computed, and then non-polar hydrogens were merged. The active site was defined by a grid box as large as 60×60×60 points with a grid spacing of 0.375 Å using AutoGrid4. The box was centered on the center of BMS309403 in the crystal structure. The protein was considered rigid for the docking study. The docking parameters were set as follows: ga_pop_size=150 (number of individuals in population), ga_run=200 (the number of dockings that were performed); other parameters were set to the software's default values. The Lamarckian genetic algorithm was used to account for protein-ligand interactions. Finally, the conformation was selected taking into account both the predicted binding free energy and binding modes in the FABP4 active pocket.
Statistical analysis
The IC50 values of BBR and LA for FABP4 were determined by nonlinear regression using GraphPad Prism software (San Diego, CA, USA). Statistical significance of the data were assessed using a one-way analysis of variance (ANOVA) followed by Dunnett's test. P<0.05 and P<0.01 indicate significant differences compared with the control.
Results
The discovery of BBR
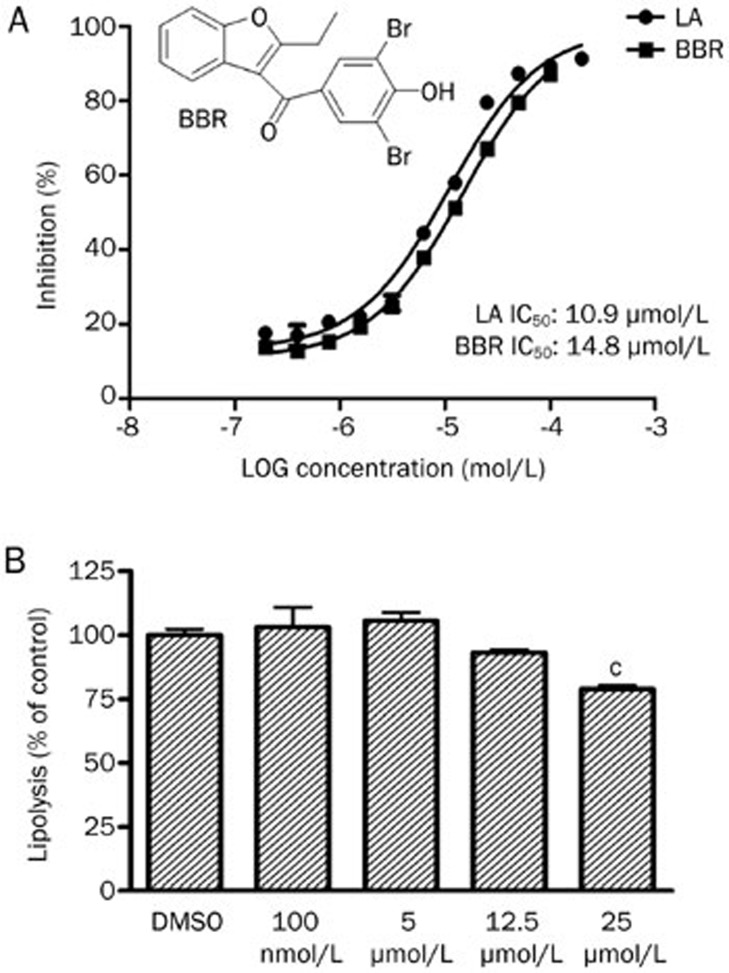
To discover inhibitors of FABP4, the inhibitory activity against FABP4 of our in-house chemical library, which is composed of 232 compounds, was determined. Preliminary screening was performed at the concentration of 100 μmol/L for each compound. Compounds that exhibited an inhibition rate greater than 50% at 100 μmol/L were selected for further IC50 value determination. The results showed that four compounds had IC50 values lower than 100 μmol/L. The best inhibitor of BBR was an old uricosuric drug with an IC50 value of 14.8 μmol/L (Figure 1A). The inhibitory activity of BBR is similar to that of linoleic acid (an endogenous ligand of FABP4) (Figure 1A).
Figure 1.
(A) Effects of BBR and LA on FABP4. (B) Effect of BBR on forskolin-stimulated lipolysis in 3T3-L1 adipocytes. cP<0.01 indicate significant differences compared with DMSO.
BBR inhibited lipolysis in 3T3-L1 adipocytes
Targeted deletion or chemical inhibition of FABP4 was reported to decrease lipolysis in adipocytes13,14. Thus, we assessed whether BBR could modulate the levels of lipolysis. The results showed BBR significantly reduced forskolin-stimulated lipolysis at a dose level of 25 μmol/L (Figure 1B). This result is consistent with BBR functioning as an inhibitor of FABP4 in adipocytes.
In vivo anti-diabetic activity of BBR on db/db mice
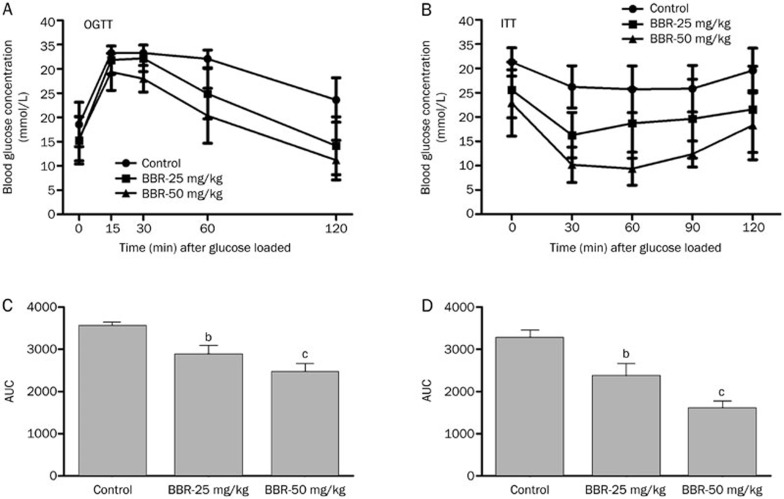
The in vivo anti-diabetic activity of BBR was evaluated in db/db mice. After 4 weeks of treatment with BBR at a dose of 25 or 50 mg/kg, the fasting-blood glucose was reduced in the BBR-treated db/db mice compared with the vehicle group (Figure 2B). In addition, BBR-treated mice showed an obvious decrease in blood glucose levels during OGTT and ITT in a dose-dependent manner (Figure 2). The above results indicated that BBR could lower blood glucose levels and improve glucose tolerance and insulin resistance in the db/db mouse model.
Figure 2.
Effects of 4 weeks treatment of BBR on OGTT and ITT in db/db mice. (A) After po loaded with 2 g dextrose per kg body weight at 0 min, blood glucose concentrations of the mice were measured at the indicated times in the graph. (B) Blood glucose concentrations at the indicated time were measured after ip injection of 1 unit insulin per kg body weight in db/db mice. (C, D) Area under the curve (AUC) for OGTT (C) and ITT (D). Values are mean±SEM (n=8). bP<0.05, cP<0.01 indicate significant differences compared with vehicle group.
Interaction mechanisms between FABP4 and BBR
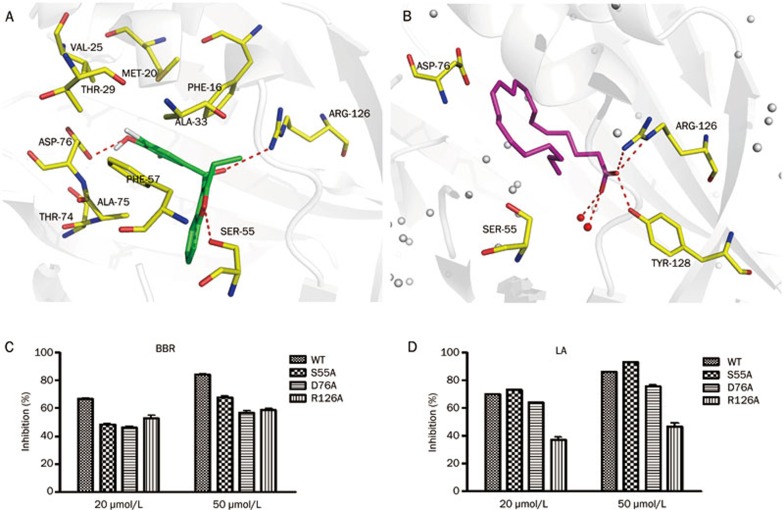
To investigate the interaction mechanism between BBR and FABP4, molecular docking (via software AutoDock4.211) was carried out. The best conformation of BBR was selected mainly considering the free binding energy (the lowest free binding energy value for BBR is -8.31 kcal/mol). The binding mode demonstrated that the 3,5-dibromo-4-hydroxyphenyl moiety of BBR is nicely accommodated in a lipophilic pocket defined by Phe16, Tyr19, Met20, Val25, Thr29, Ala33, Phe57, and Thr74. The two Br atoms on the benzene ring may engage in the key hydrophobic interactions, and the hydroxyl group is hydrogen bonded to the carbonyl oxygen of residue Asp76 (Figure 3A). Moreover, the carbonyl group of BBR forms stable polar interaction with Arg126, a critical residue for the binding of inhibitors12,14,15,16,17. In addition, the oxygen atom in the benzofuran of BBR forms a hydrogen bond with Ser55 (Figure 3A). It is also noted that BBR occupies a corner of the large binding pocket of FABP4. Thus, introducing a large group to the benzofuran moiety might enhance the binding affinity of the compounds to FABP4 (Figure 3A).
Figure 3.
(A) Predicted conformations of BBR in the binding pocket of FABP4. Residues provided for hydrophobic interactions of FABP4 are indicated in yellow sticks. (B) The complex of FABP4 and arachidonic acid from crystal structure (PDB ID: 1ADL). Arachidonic acid and BBR are shown in green and magenta sticks. The dashed lines in red represent hydrogen bonds. (C, D) Inhibition rate of BBR and LA on wild type and mutants: S55A, D76A, and R126A at the concentration of 20 μmol/L and 50 μmol/L, respectively. LA, linoleic acid.
Point mutation
To verify our predicted binding mode of BBR and FABP4, residues Ser55, Asp76, and Arg126 were experimentally substituted with an Alanine (Ala). The inhibitory activities on mutants by the endogenous ligand of FABP4 and BBR were evaluated. The inhibitory activities were determined as described in the Materials and methods section. Because linoleic acid (one of the endogenous ligands of FABP4) has the most stable activity against FABP4 in our study, we used linoleic acid as the control compound in our experimental system. However, no complex structure of FABP4 and linoleic acid has been reported. Fortunately, we obtained the complex of FABP4-PA (palmitic acid) and FABP4-ACD (arachidonic acid) from the PDB databank. Moreover, fatty acids bind in similar conformations in the FABP4 active site18,19. The structure shows that the carboxylic acid group of the fatty acid forms stable polar interactions with the key residues Arg126 and Tyr128. Additionally, the aliphatic chain is located in the hydrophobic site of the pocket. Residue Asp76 is close enough to the carbon chain of the fatty acid to form Van der Waals interactions, while Ser55 is more distant (Figure 3B).
Our mutation studies demonstrated that for linoleic acid, the inhibition rate of the R126A mutants markedly decreased at both 20 μmol/L and 50 μmol/L (Table 1, Figure 3D). For the D76A mutants, the inhibition rate was reduced by approximately 10% at a concentration of 50 μmol/L. However, little change was observed in the ability of linoleic acid to inhibit both the wild type and the S55A mutant (Table 1, Figure 3D). These results indicated that Ser55 in FABP4 is not important for linoleic acid binding. Asp76 might form Van der Waals interactions with linoleic acid while Arg126 is essential for linoleic acid binding. Therefore, our results are consistent with the reported binding modes between FABP4 and fatty acids. The results also confirmed the power of mutagenesis for verifying the binding between proteins and ligands. For BBR, inhibition of D76A decreased by more than 20% at both 20 μmol/L and 50 μmol/L (Table 1, Figure 3C), and the inhibitory rate was also obviously reduced for Ser55A and R126A (Table 1, Figure 3C). These data demonstrated that residues Ser55, Asp76, and Arg126 of FABP4 are important for binding with BBR.
Table 1. Comparison of the inhibitory activity of the compounds on wild-type and mutated FABP4.
| Compound | Inhibition at 20 μmol/La (%) |
Inhibition at 50 μmol/La (%) |
||||||
|---|---|---|---|---|---|---|---|---|
| Wild type | S55A | D76A | R126A | Wild type | S55A | D76A | R126A | |
| Lab | 69.7±0.6 | 72.8±0.7 | 63.8±0.6 | 36.7±4.0 | 86.0±0.5 | 92.9±0.7 | 75.3±2.2 | 46.5±4.4 |
| BBR | 66.9±0.6 | 48.5±1.0 | 46.2±0.6 | 52.5±3.5 | 84.2±0.5 | 67.7±2.0 | 56.8±2.0 | 58.6±2.3 |
a Values are means of triplicate experiments with standard deviation values.
b LA: linoleic acid.
Discussion
Inhibitors of FABP4 are potential anti-diabetic drugs. Of a series of chemical inhibitors of FABP4 that was discovered over the last decade12,14,15,16,17, the selective inhibitor BMS309403 was proven to be effective for improving insulin resistance in the ob/ob mouse model and can ameliorate the symptoms of atherosclerosis7. However, none of these compounds are in clinical development. Our in vitro data showed that the well-known uricosuric drug BBR inhibits FABP4 with an IC50 value of 14.8 μmol/L, which was similar to the activity of the endogenous ligand, linoleic acid, against FABP4 (Figure 1A). Finding new applications for old drugs represents a very promising strategy for drug discovery with obvious advantages, including optimal ADME/T properties. The inhibitory activity of BBR on FABP4 made us wonder if BBR could have new applications in treating type 2 diabetes. Therefore, the cellular and in vivo activities of BBR were characterized in our study. Our results demonstrated that BBR could significantly reduce forskolin-stimulated lipolysis in 3T3-L1 cells (Figure 1B) and improve blood glucose levels, glucose tolerance and insulin resistance in db/db diabetic mice (Figure 2). These results are consistent with the reported results of small molecular inhibitors of FABP47, indicating that BBR is an inhibitor of FABP4 and that it could be effective against diabetes in db/db mice.
However, it was reported that BBR was also an activator of PPARγ (peroxisome proliferator-activated receptor γ), which is a well-known target of anti-diabetic drugs. Thus, it may be assumed that the observed anti-diabetic effects of BBR are attributable to a multi-target mechanism. The study showed that BBR activates PPARγ at a concentration of 100 μmol/L. However, the EC50 value is much higher than 10 μmol/L, which means that BBR is a very weak activator of PPARγ (the EC50 value is 40 nmol/L for the well-known PPARγ activator rosiglitazone)20,21,22. In addition, BBR could inhibit the lipolysis of mouse 3T3-L1 adipocytes while agonists of PPARγ were reported to increase rat adipose tissue lipolysis23. Moreover, during treatment with rosiglitazone, the daily food intake increased and adipose tissue weight increased for ob/ob mice24. In our study, no significant change was observed in adipose tissue weight and daily food intake (Supplemental Figure S1). Therefore, we deduced that inhibition of FABP4 is the primary effector of the observed in vivo anti-diabetic effect of BBR.
To investigate the mechanism of interaction between FABP4 and BBR, a molecular docking strategy was applied. Three important residues (Ser55, Asp76, and Arg126) were observed to form essential polar interactions between FABP4 and BBR. To verify our predictions, we carried out site-directed mutagenesis studies. Ser55, Asp76, and Arg126 were mutated to Alanine. We first tested the effects of linoleic acid on the mutants. The results were consistent with the reported studies, which confirmed the efficacy of using mutagenesis for studying binding between proteins and ligands. For BBR, the inhibitory activities were reduced for all the mutants (Table 1), indicating the importance of these residues (Ser55, Asp76, and Arg126) in binding with BBR. Therefore, the above results confirmed our predicted mechanism of interaction between FABP4 and BBR. Furthermore, we also performed molecular docking studies between BBR and the mutated proteins (the mutated proteins were obtained using the software Pymol). The trend of the estimated binding free energies (Supplemental Table S1) was in agreement with that of the inhibitory rates from experimental data (Table 1). These results again confirmed the binding mode of FABP4-BBR. Moreover, these data also implied that docking (rigid docking) is a reliable method for studying the interactions between FABP4 and BBR.
The hepatotoxicity of BBR was reported in 2003, leading to its withdrawal from the markets of some European countries25,26, which should be taken into account if BBR is developed as a new anti-diabetic drug. Therefore, structural optimization of BBR to reduce its hepatotoxicity will be a major research direction in the future. The binding mode that we presented here should be helpful for the structural optimization.
In summary, we demonstrated that BBR, a uricosuric drug, is an inhibitor of FABP4. Its anti-diabetic activity on db/db diabetic mice might be mediated through its multi-target binding mechanisms, and FABP4 should play a main role. Molecular docking and point mutations studies revealed that Ser55, Asp76, and Arg126 are the key residues of FABP4 that bind with BBR. This work indicated that BBR might be a new potential therapeutic agent for type 2 diabetes, and our study also should be an important reference for further design of novel inhibitors of FABP4.
Author contribution
Prof He-yao WANG and Wei-liang ZHU designed and supervised the research and revised the manuscript; Hai-yan CAI and Ting WANG performed the research, analyzed data and wrote the manuscript; Prof Kai-xian Chen and Ying-xia LI helped develop part of the research idea and helped revise the manuscript; and Jian-chun ZHAO, Peng SUN, Hai-peng DING, and Gui-rui YAN helped with part of the research.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (20721003 and 81072681), the International S&T Cooperation (2010DFB73280) and the National 863 High Performance Computing Project (2012AA01A305), China.
Footnotes
Supplemental Table S1 and Figure S1 are available at the website of Acta Pharmacologica Sinica.
Supplementary Information
Comparison of the predicted binding free energy of BBR and FABP4 (wild type/mutants)
References
- 1Spiegelman BM, Frank M, Green H. Molecular cloning of mRNA from 3T3 adipocytes. Regulation of mRNA content for glycerophosphate dehydrogenase and other differentiation-dependent proteins during adipocyte development. J Biol Chem 1983; 258: 10083–9. [PubMed] [Google Scholar]
- 2Hunt CR, Ro JH, Dobson DE, Min HY, Spiegelman BM. Adipocyte P2 gene: developmental expression and homology of 5′-flanking sequences among fat cell-specific genes. Proc Natl Acad Sci U S A 1986; 83: 3786–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Hotamisligil GS, Johnson RS, Distel RJ, Ellis R, Papaioannou VE, Spiegelman BM. Uncoupling of obesity from insulin resistance through a targeted mutation in aP2, the adipocyte fatty acid binding protein. Science 1996; 274: 1377–9. [DOI] [PubMed] [Google Scholar]
- 4Uysal KT, Scheja L, Wiesbrock SM, Bonner-Weir S, Hotamisligil GS. Improved glucose and lipid metabolism in genetically obese mice lacking aP2. Endocrinology 2000; 141: 3388–96. [DOI] [PubMed] [Google Scholar]
- 5Tuncman G, Erbay E, Hom X, De Vivo I, Campos H, Rimm EB, et al. A genetic variant at the fatty acid-binding protein aP2 locus reduces the risk for hypertriglyceridemia, type 2 diabetes, and cardiovascular disease. Proc Natl Acad Sci U S A 2006; 103: 6970–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Makowski L, Boord JB, Maeda K, Babaev VR, Uysal KT, Morgan MA, et al. Lack of macrophage fatty-acid-binding protein aP2 protects mice deficient in apolipoprotein E against atherosclerosis. Nat Med 2001; 7: 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Furuhashi M, Tuncman G, Gorgun CZ, Makowski L, Atsumi G, Vaillancourt E, et al. Treatment of diabetes and atherosclerosis by inhibiting fatty-acid-binding protein aP2. Nature 2007; 447: 959–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Heel RC, Brogden RN, Speight TM, Avery GS. Benzbromarone: a review of its pharmacological properties and therapeutic use in gout and hyperuricaemia. Drugs 1977; 14: 349–66. [DOI] [PubMed] [Google Scholar]
- 9Cai HY, Yan GR, Zhang XD, Gorbenko O, Wang HY, Zhu WL. Discovery of highly selective inhibitors of human fatty acid binding protein 4 (FABP4) by virtual screening. Bioorg Med Chem Lett 2010; 20: 3675–9. [DOI] [PubMed] [Google Scholar]
- 10Zhang XD, Yan JW, Yan GR, Sun XY, Ji J, Li YM, et al. Pharmacological inhibition of diacylglycerol acyltransferase 1 reduces body weight gain, hyperlipidemia, and hepatic steatosis in db/db mice. Acta Pharmacol Sin 2010; 31: 1470–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 1998; 19: 1639–62. [Google Scholar]
- 12Sulsky R, Magnin DR, Huang Y, Simpkins L, Taunk P, Patel M, et al. Potent and selective biphenyl azole inhibitors of adipocyte fatty acid binding protein (aFABP). Bioorg Med Chem Lett 2007; 17: 3511–5. [DOI] [PubMed] [Google Scholar]
- 13Coe NR, Simpson MA, Bernlohr DA. Targeted disruption of the adipocyte lipid-binding protein (aP2 protein) gene impairs fat cell lipolysis and increases cellular fatty acid levels. J Lipid Res 1999; 40: 967–72. [PubMed] [Google Scholar]
- 14Hertzel AV, Hellberg K, Reynolds JM, Kruse AC, Juhlmann BE, Smith AJ, et al. Identification and characterization of a small molecule inhibitor of Fatty Acid binding proteins. J Med Chem 2009; 52: 6024–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Lehmann F, Haile S, Axen E, Medina C, Uppenberg J, Svensson S, et al. Discovery of inhibitors of human adipocyte fatty acid-binding protein, a potential type 2 diabetes target. Bioorg Med Chem Lett 2004; 14: 4445–8. [DOI] [PubMed] [Google Scholar]
- 16Ringom R, Axen E, Uppenberg J, Lundback T, Rondahl L, Barf T. Substituted benzylamino-6-(trifluoromethyl)pyrimidin-4(1H)-ones: a novel class of selective human A-FABP inhibitors. Bioorg Med Chem Lett 2004; 14: 4449–52. [DOI] [PubMed] [Google Scholar]
- 17Barf T, Lehmann F, Hammer K, Haile S, Axen E, Medina C, et al. N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte fatty-acid binding protein (A-FABP) inhibitors. Bioorg Med Chem Lett 2009; 19: 1745–8. [DOI] [PubMed] [Google Scholar]
- 18LaLonde JM, Levenson MA, Roe JJ, Bernlohr DA, Banaszak LJ. Adipocyte lipid-binding protein complexed with arachidonic acid. Titration calorimetry and X-ray crystallographic studies. J Biol Chem 1994; 269: 25339–47. [DOI] [PubMed] [Google Scholar]
- 19Marr E, Tardie M, Carty M, Brown Phillips T, Wang IK, Soeller W, et al. Expression, purification, crystallization and structure of human adipocyte lipid-binding protein (aP2). Acta Crystallogr Sect F Struct Biol Cryst Commun 2006; 62: 1058–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Butler EG, Ichida T, Maruyama H, Schulte-Hermann R, Williams GM. Toxicological studies on a benzofurane derivative. II. Demonstration of peroxisome proliferation in rat liver. Toxicol Appl Pharmacol 1990; 106: 500–8. [DOI] [PubMed] [Google Scholar]
- 21Kunishima C, Inoue I, Oikawa T, Nakajima H, Komoda T, Katayama S. Activating effect of benzbromarone, a uricosuric drug, on peroxisome proliferator-activated receptors. PPAR Res 2007; 2007: 36092–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Montanari R, Saccoccia F, Scotti E, Crestani M, Godio C, Gilardi F, et al. Crystal structure of the peroxisome proliferator-activated receptor gamma (PPARgamma) ligand binding domain complexed with a novel partial agonist: a new region of the hydrophobic pocket could be exploited for drug design. J Med Chem 2008; 51: 7768–76. [DOI] [PubMed] [Google Scholar]
- 23Festuccia WT, Laplante M, Berthiaume M, Gelinas Y, Deshaies Y. PPARgamma agonism increases rat adipose tissue lipolysis, expression of glyceride lipases, and the response of lipolysis to hormonal control. Diabetologia 2006; 49: 2427–36. [DOI] [PubMed] [Google Scholar]
- 24Carmona MC, Louche K, Nibbelink M, Prunet B, Bross A, Desbazeille M, et al. Fenofibrate prevents Rosiglitazone-induced body weight gain in ob/ob mice. Int J Obes (Lond) 2005; 29: 864–71. [DOI] [PubMed] [Google Scholar]
- 25WHO Drug Information: Benzbromarone and hepatitis. WHO Drug Information 2000; 14.
- 26Lee MH, Graham GG, Williams KM, Day RO. A benefit-risk assessment of benzbromarone in the treatment of gout. Was its withdrawal from the market in the best interest of patients? Drug Saf 2008; 31; 643–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of the predicted binding free energy of BBR and FABP4 (wild type/mutants)