Abstract
To develop applicable and susceptible models to evaluate the toxicity of nanoparticles, the antimicrobial effects of CuO nanoparticles (CuO-NPs) on various Saccharomyces cerevisiae (S. cerevisiae) strains (wild type, single-gene-deleted mutants, and multiple-gene-deleted mutants) were determined and compared. Further experiments were also conducted to analyze the mechanisms associated with toxicity using copper salt, bulk CuO (bCuO), carbon-shelled copper nanoparticles (C/Cu-NPs), and carbon nanoparticles (C-NPs) for comparisons. The results indicated that the growth inhibition rates of CuO-NPs for the wild-type and the single-gene-deleted strains were comparable, while for the multiple-gene deletion mutant, significantly higher toxicity was observed (P < 0.05). When the toxicity of the CuO-NPs to yeast cells was compared with the toxicities of copper salt and bCuO, we concluded that the toxicity of CuO-NPs should be attributed to soluble copper rather than to the nanoparticles. The striking difference in adverse effects of C-NPs and C/Cu-NPs with equivalent surface areas also proved this. A toxicity assay revealed that the multiple-gene-deleted mutant was significantly more sensitive to CuO-NPs than the wild type. Specifically, compared with the wild-type strain, copper was readily taken up by mutant strains when cell permeability genes were knocked out, and the mutants with deletions of genes regulated under oxidative stress (OS) were likely producing more reactive oxygen species (ROS). Hence, as mechanism-based gene inactivation could increase the susceptibility of yeast, the multiple-gene-deleted mutants should be improved model organisms to investigate the toxicity of nanoparticles.
INTRODUCTION
With recent developments in nanotechnology around the world, there are mounting concerns regarding the potential environmental and human health risks caused by exposure to engineered nanomaterials (ENMs). Relative to the rapid development of nanotechnology, ecotoxicology and environmental hazard assessments have obviously fallen behind and still represent huge knowledge gaps (1). It is thus urgent to establish approaches to speed up safety analysis, and the insights gained from these approaches could give us a better understanding of the hazardous effects ENMs at the biological level and assist the development of safe-design approaches.
As one of the increasingly used metal oxide nanoparticles (NPs), CuO-NPs have been applied to superconducting materials, sensing materials, glass, and ceramics and in other primarily antimicrobial applications (2). The extensive production and usage of CuO-NPs increase the harmful effects to organisms. Investigators have demonstrated that CuO-NPs are toxic to Escherichia coli HB101 (3, 4), prokaryotic alga Microcystis aeruginosa (5), airway epithelial (HEp-2) cells (6), the crustacean Thamnocephalus platyurus (7), Lemna gibba (8), mice (9), and other organisms. A considerable amount of research has been done about the toxicity of CuO-NPs, and comparative interspecies sensitivity was found to vary greatly; however, the mechanism of CuO-NP toxicity at the cellular level is still unclear (10–12).
The yeast Saccharomyces cerevisiae as a unicellular eukaryotic model organism in studying toxicity of chemicals is typical and widely used because its structure and functional organization are similar to those of higher organisms and because the genetic background of yeast is clear (13, 14). S. cerevisiae is increasingly used in the toxicity assessment of chemicals such as heavy metals (15, 16) and ENMs (17–19). Kasemets et al. (19) first used S. cerevisiae to evaluate the biocidal effect of ZnO-NPs, CuO-NPs, and TiO2-NPs and demonstrated that ZnO-NPs and bulk ZnO had comparable toxicities, which might be due to significant release of Zn ions from ZnO particles, while soluble Cu ions only partly explained the toxicity of CuO-NPs. Study of the antimicrobial activity of Ag-NPs of a different primary size than yeast revealed that the toxicity of 20- to 80-nm Ag-NPs could be fully explained by soluble Ag ions, whereas for 10-nm Ag-NPs, particle-related effects might also be involved (20). Although controversial, the mechanisms of nanoparticle toxicity to unicellular organisms can be traced to oxidative stress (OS), soluble ions, or particle-related effects (3, 19–22). Kasemets et al. (23) once used a suite of yeast single-gene deletion mutants (related to OS response) and one copper-vulnerable mutant to elucidate the role of oxidative stress and soluble copper in leading to toxicity to S. cerevisiae and indicated that the copper-vulnerable mutants, but not the OS-vulnerable mutants, were 16-fold more sensitive to CuO-NPs than the wild type. Indeed, it has been reported that the deficiency of a single gene may not necessarily result in the desired phenotype because of the existence of compensatory mechanisms in cells (24–26). Also, since metal oxide NP-induced toxicity results from multiple factors, a multiple-gene deletion mutant should be more susceptible and suitable for cytotoxicity estimation than a single-gene deletion mutant. However, current studies on nanoparticle toxicities to unicellular-organism mutants were limited to single-gene deletion mutants (23, 27); little is known about their toxic effect to multiple-gene deletion mutants, and there is doubt about the susceptibility profile of the multiple-gene-deleted mutant.
In order to evaluate the sensitivity of multiple-gene deletion mutants exposed to CuO-NPs, we focused on the toxicity of CuO-NPs to S. cerevisiae wild-type, single-gene deletion mutant, and multiple-gene deletion mutant strains. Based on the potential role of OS in inducing toxicity to yeast cells, for the single-gene deletion mutant, we used the mutant yap1Δ strain (yap1 mutant). YAP1 is a posttranscription factor and leads to expression of genes encoding protective enzymes under oxidative stress conditions (28), and it has been shown that the yap1 mutant is more hypersensitive to oxides such as hydrogen peroxide (H2O2) (29, 30) than the wild type. In addition, concerning the significance of toxicant accumulation in producing toxicity, for the multiple-gene deletion mutant, we chose the cwp1Δ cwp2Δ snq2Δ pdr5Δ mutant strain (quadruple mutant). CWP1 and CWP2 are two major mannoproteins of the outer cell wall. Since disruption of CWP1 and CWP2 had profound effects on the cell wall structure and permeability, the exogenous substances would be more easily taken up inside cells (31, 32). SNQ2 encodes an ATP-binding cassette transporter and is highly homologous to PDR5; inactivation of these proteins would increase intracellular concentration of foreign matter (33). It has been reported that yeast cells regulate the accumulation of exogenous substances at three different levels: cell wall, cell membrane, and the pleiotropic drug resistance (PDR) pathway (24, 34). Therefore, we hypothesized that after deleting genes related to cell wall permeability (i.e., cwp1 and cwp2) and the PDR pathway (i.e., snq2 and pdr5), the yeast cells should take up more nanoparticles/ions and, furthermore, show higher susceptibility. Our previously published study confirmed that deletion of the cwp1, cwp2, snq2, and pdr5 genes has a profound effect on cell permeability (24). The permeability-vulnerable mutant will also help us clarify the role of the yeast cell wall in defense against CuO-NPs. Furthermore, the cwp1Δ cwp2Δ snq2Δ pdr5Δ yap1Δ mutant strain (quintuple mutant) was also used to elucidate whether the combined-gene deletion mutant would be more sensitive to CuO-NPs.
The aim of this study was to investigate the feasibility of using a mechanism-based multiple-gene-deleted mutant to assess the toxicity of CuO-NPs and to gain a better understanding of the mechanism of CuO-NP toxicity to yeast cells. In our study, the total amount of copper element taken up by yeast cells was first quantified, and a well-characterized C/Cu-NP was also used as a control. We concluded that the multiple-gene-deleted mutant was significantly more sensitive to CuO-NPs than the wild type and demonstrated that the toxicity of CuO-NPs was associated with the amount of copper taken up. We expect that these results will provide some useful information for a better understanding of the application of a multiple-gene-deleted mutant in assessing the toxicity of ENMs.
MATERIALS AND METHODS
Materials.
Strains of the yeast S. cerevisiae used in this study are listed in Table S1 in the supplemental material. Yeast cells were grown at 30°C in Sabouraud dextrose (SD) medium (35). SD medium contains 2% glucose, 0.17% yeast nitrogen base, 0.5% ammonium sulfate, 20 mg/liter each of adenine, histidine, methionine, tryptophan, and uracil, and 30 mg/liter each of valine, leucine and lysine. Frozen permanent copies of yeast strains were stored at −80°C. Plates of the strains were incubated on SD agar (sterile petri dishes; Nest Biotechnology) at 30°C for 48 h and further maintained at 4°C. Growth curves of wild-type and mutant strains can be found in the supplemental material; different strains had comparable biomasses during the exposure process (see Fig. S1 in the supplemental material for details).
CuO-NPs (20 nm) and C-NPs (40 nm) were purchased from Beijing Nachen S&T, Ltd., and had a purity of 99.9%. Carbon shell-protected copper nanoparticles (C/Cu-NPs; 25 nm) were obtained from Shenzhen Junye Nano Materials, Ltd.; the carbon comprised 4% (wt/wt) of the total particles, and the purity of C/Cu-NPs was 99.9%. CuSO4·5H2O, bulk CuO (bCuO), and H2O2 were purchased from Sinopharm Group Co., Ltd. 2′,7′-Dichlorodihydrofluorescein diacetate (H2DCFDA) was purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA). All other chemicals were reagent grade or better.
Nanoparticle dispersion protocols and characterization.
The nanoparticle suspensions were prepared in deionized (DI) water (18.2 MΩ/cm; Sartorius Stedim) and ultrasonicated for 30 min (JY92-IIN; Ningbo Scientz Biotechnology Co., Ltd., China). A stock solution of CuSO4 was also prepared in DI water, and all of the solutions were stored in the dark at 4°C. Before the toxicity test, stock solutions were vigorously mixed. The shapes of the nanoparticles and bCuO particles dispersed in DI water and SD medium were determined using a transmission electron microscope (TEM) (HT-7700; Hitachi, Ltd., Japan) operated at 180 kV. The hydrodynamic sizes of particles in suspension after incubation for 4 h in the DI water and SD medium were characterized by a dynamic light-scattering (DLS) system (Nano ZS; Malvern, Instruments, Ltd., United Kingdom). Surface areas of particles were measured by the Brunauer-Emmer-Teller (BET) method (ASAP 2020; Micromeritics Instrument Corp., USA), based on the adsorption-desorption isotherm of N2 at 77.5 K.
Cytotoxicity tests.
To compare the growth curves of yeast wild-type and mutant strains and to select the appropriate exposure time, the yeast strains were cultivated in SD medium at 30°C for 28 h (see the supplemental material for details). The procedures for cytotoxicity tests were adopted from a method described previously (36). Yeast cells were inoculated in the SD medium by overnight (16 to 18 h) culturing at 30°C at 200 rpm under dark conditions from a single agar plate colony. The initial culture optical density (OD) at 600 nm was diluted to 0.110 ± 0.001 (0.5-cm path length) with SD medium, and then aliquots of 1 ml of cell culture were pipetted into 24-well microplates (Corning), and toxicants were added to the nominal concentrations. The nominal concentrations of chemicals in toxicity experiments were as follows: CuO-NPs, 10 to 100 mg/liter; bCuO, 10 to 100 mg/liter; CuSO4, 20 to 200 mg/liter; C/Cu-NPs, 8 to 80 mg/liter Cu; C-NPs, 0.18 to 1.76 mg/liter; H2O2, 1.5 to 6.0 mmol/liter. The copper-containing materials had equivalent doses of copper, and the specific surface areas were identical for CuO-NPs and C-NPs. Then the yeast strains were cultivated in SD medium at 30°C at 200 rpm for 4 h. Unexposed yeast cells were used as controls. The biomass was measured by the viable cell counts on SD agar plates. CFU counts were determined after 2 days at 30°C in the dark for the growth inhibition analysis.
Measurement of soluble copper.
Copper-containing materials may release cupric ions when added into liquid. After the process of centrifugation and filtration (through a 0.22-μm-pore-size cellulose membrane; Millipore), copper in the filtrate was estimated as the amount of soluble copper (3, 37, 38). In our study, copper ion released from CuO-NPs in SD medium of various pH values (the initial pHs for SD medium were 6.00, 4.96, 4.23, and 3.60) was evaluated; CuO-NPs, C/Cu-NPs, and bCuO were added into a volume of 5 ml of SD medium, and the copper concentrations were adjusted to 16 mg/liter. After shaking at 200 rpm for 4 h under dark conditions, the mixtures were centrifuged at 10,000 × g (centrifuge 5810R; Eppendorf, Hamburg, Germany) for 30 min and then filtered with a 0.22-μm-pore-size cellulose membrane (3, 37, 38). The particles trapped on the filter membrane were washed continuously three times to get rid of the residual Cu as much as possible. In addition, removal of the nonsoluble fraction of CuO was also confirmed by a dynamic light-scattering system (Nano ZS; Malvern, Instruments Ltd., United Kingdom). The concentration of soluble copper was quantified by atomic absorption spectrometry (AAS) (Avanta M; GBS Scientific Equipment, Ltd., Australia). Furthermore, amounts of soluble copper released from various quantities of CuO-NPs in the SD medium (with yeast cells or without yeast cells) were measured. After the chemicals were adjusted to nominal concentrations with shaking at 200 rpm for 4 h, the soluble copper was measured following the same experimental setup as described above.
Intracellular ROS assays.
The ability of CuO-NPs and its counterparts to induce intracellular reactive oxygen species (ROS) formation was determined using a 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) assay. Once H2DCFDA was internalized, it was hydrolyzed by intracellular esterase to active 2′,7′-dichlorodihydrofluorescein (DCFH). In the presence of ROS, DCFH was oxidized to the highly fluorescent compound, 2′,7′-dichlorofluorescein (DCF). Hence, its fluorescence intensity was proportional to the amount of ROS produced by the cells (39). The procedures of exposure were the same with the toxicity test. A 1-ml sample of cells was collected and rinsed twice with phosphate-buffered saline (PBS; pH 7.2) to remove the uninternalized toxicants (40) and incubated with 10 μM H2DCFDA at 37°C for 30 min in the dark. Subsequently, samples were washed twice in PBS to remove excess H2DCFDA and then suspended in PBS to analyze the fluorescence. The DCF fluorescence was recorded by using a microplate reader (SpectraMax M5; Molecular Devices, Sunnyvale, CA, USA) at an excitation wavelength of 488 nm and an emission wavelength of 530 nm. Unexposed yeast cells were used as the blank control. Relative ROS levels were calculated by the relative fluorescence ratio of the treatments to that of the blank control. All the procedures were conducted without exposure to light, and the data were normalized to the same numbers of cells.
Quantification of intracellular copper.
For the nanoparticle cellular uptake experiments, the exposure procedure was the same as that for the cytotoxicity tests. After being cultivated with CuO-NPs, copper salt, and bCuO in 5 ml of SD medium for 4 h, the yeast cells were harvested by centrifugation at 8,000 × g for 10 min and washed with PBS (pH 7.2) two times to remove copper adsorbed to the surface of the yeast (41, 42). The process of separating nanoparticles from cells followed a method described previously (43). Briefly, 2 ml of ferric nitrate (1 mol/liter) was added into the sample as etchant solution; after 5 min of etching, the solution was removed by centrifugation, and samples were washed two times again with PBS. Afterwards, the cells were digested by a wet method (44, 45). The residue also was filtered with a 0.22-μm-pore-size cellulose membrane, and the concentration of Cu was determined by AAS. The data were normalized to the same numbers of cells and then converted to the total copper internalized by the yeast cells.
Statistical analysis.
Data were analyzed using the statistical package SPSS (version 13.0; SPSS, Inc., Chicago, IL, USA) and are presented as means ± standard deviations (SD) from at least two independent experiments. The figures were generated by using GraphPad Prism, version 6 (GraphPad Software, Inc., La Jolla, CA, USA). A one-way analysis of variance (ANOVA) and a paired-sample t test were run to measure difference.
RESULTS
Characteristics and properties of particles.
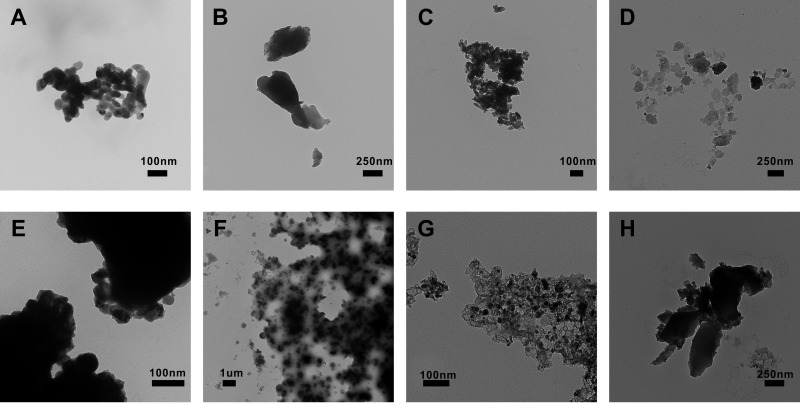
Particle observation by TEM confirmed CuO-NPs to be noticeably smaller than bCuO particles (Fig. 1A and E versus B and F). Additionally, both CuO-NPs and C/Cu-NPs were mostly spherical in shape; in contrast, bCuO particles appeared irregularly shaped, and an irregular shape was also observed for C-NPs. The initial diameters of nanoparticles were in accordance with the information provided by the suppliers. As shown in Fig. 1A to D, particle aggregations were observed when particles were suspended in DI water. In addition, we further characterized the sizes of particles in the culture medium used for the cytotoxicity test (Fig. 1E to H). TEM micrographs revealed that the particles in the SD medium showed a relatively higher degree of aggregation than those in DI water.
FIG 1.
Representative TEM images of the particles. CuO-NPs (A and E), bCuO (B and F), C/Cu-NPs (C and G), and C-NPs (D and H), are shown in DI water (A to D) and in SD medium (E to H).
Particles used in our study were thoroughly characterized for their size distributions, specific surface areas, and key physical parameters. The data are summarized in Table 1. In this study, dynamic light scattering (DLS) was used to determine the particle size in DI water and the SD medium after 4 h of incubation. The data in Table 1 showed that the average hydrodynamic diameter of CuO-NPs in the DI water was 332.3 nm and increased to 750.4 nm in the SD medium. Similarly, the particle sizes of bCuO, C/Cu-NP, and C-NP dispersed in solution also increased from DI water to SD medium. The results indicated stronger aggregation for the particles dispersed in the culture medium than in the DI water; this result could be ascribed to the fact that the SD medium was rich in organic matter and inorganic ions since abundant organic matter and ions in SD medium could cause pronounced effects on the charge balance and then on CuO-NP behavior with respect to aggregation. Specially, in the SD medium, the average hydrodynamic sizes of CuO-NPs and bCuO particles were comparable. The phenomenon has also been reported elsewhere (20, 46, 47), and the explanation maybe that the nutrient-rich medium promotes the equilibrium of nanoparticles in solution (forming particle-nutrient complexes) and, as a result, the formation of bigger aggregations. However, if aggregation exceeds a certain size, sedimentation would happen, which could ensure comparable hydrodynamic sizes in solution. In addition, to compare the surface areas of nanoparticles and bulk counterparts used in our work and further to make sure that the surface areas of C-NPs were equivalent to those of C/Cu-NPs in the exposure experiments, we also determined the specific surface areas of the materials used in our study. The data are shown in Table 1, and they suggest that CuO-NPs exhibited obviously bigger surface areas than bCuO particles.
TABLE 1.
Characterization of particles in this study
| Material | Purity (%) | Diam (nm)a | Profile in suspension in: |
Surface area (m2/g)d | Carbon content (%)e | |||
|---|---|---|---|---|---|---|---|---|
| DI water |
SD medium |
|||||||
| Hydrodynamic size (nm)b | PDIc | Hydrodynamic size (nm) | PDI | |||||
| CuO-NP | 99.9 | 20 | 332.3 (185.6–668.8) | 0.164 | 750.4 (470.1–2,034.7) | 0.304 | 9.1858 | 0 |
| bCuO | 99 | >100 | NAf | NA | 782.0 (269.6–2,268.5) | 0.521 | 0.2005 | 0 |
| C/Cu-NP | 99.8 | 25 | 244.4 (178.6–597.3) | 0.201 | 1,810 (598.5–3,376.4) | 0.328 | 4.1769 | 4 |
| C-NP | 99.9 | 40 | 1,612 (467.8–2,969.6) | 0.237 | 1,655 (409.7–3,187.6) | 0.285 | 198.2058 | 100 |
Diameter of nanoparticles as provided by the suppliers.
Hydrodynamic sizes of particles suspended in DI water and SD medium were determined after incubation for 4 h. CuO-NPs and bCuO were analyzed at concentrations of 50 mg/liter. C/Cu-NPs and C-NPs were analyzed at concentrations of 40 and 0.5 mg/liter, respectively.
Polydispersity index.
Specific surface area, as determined by ASAP 2020 (Micromeritics Instrument, Ltd., USA).
Nominal content.
NA, not available (because of unstable suspension).
Toxicity to yeast strains of different chemicals.
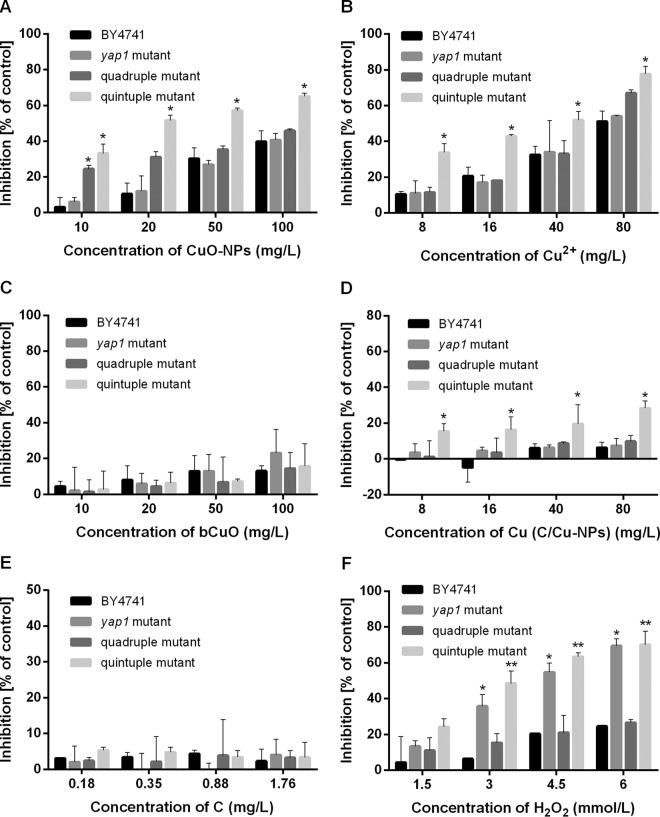
To investigate the toxic effects of CuO-NPs, bCuO, and Cu2+ on yeast, various amounts of CuO-NPs (10 to 100 mg/liter), bCuO (10 to 100 mg/liter), and Cu2+ (8 to 80 mg Cu/liter) were added to the yeast cell culture. Data shown in Fig. 2A and C indicate that the relatively higher growth inhibitions of yeast strains were in response to the increasing concentrations of chemicals. In our study, the yap1 mutant and the wild-type strain exhibited comparable levels of resistance to CuO-NPs, whereas both quadruple and quintuple mutants showed higher sensitivity to CuO-NPs than the wild-type BY4741, and the quintuple mutant was significantly more sensitive (P < 0.05) under identical CuO-NP concentrations (Fig. 2A). Remarkably, low-dose (10 mg/liter) CuO-NP exposure yielded 3.1% ± 5.3% growth inhibition of BY4741 but 24.4% ± 2.0% inhibition of the quadruple mutant (P < 0.05) (Fig. 2A), indicating that the toxicity of CuO-NPs to yeast cells was related to cell permeability to some extent. The toxicity of copper salt to the quadruple mutant was comparable to that to BY4741 but remarkably greater to the quintuple mutant (P < 0.05) (Fig. 2B). In contrast, bCuO showed no serious growth inhibition of wild-type yeast and its mutants. Moreover, CuO-NPs and Cu2+ showed higher toxicities to the wild type and the tested mutants than bCuO (Fig. 2).
FIG 2.
Dose-response of CuO-NPs, Cu2+, bCuO, C/Cu-NPs, C-NPs, and H2O2 to wild-type and mutant strains after exposure for 4 h. Inhibition rates were evaluated by CFU counts on SD agar, and results are expressed as a percentage of the CFU count relative to the CFU count in unexposed cells. The inhibition ratio (percent) was calculated as (1 − Ecfu/CKcfu) × 100%, where CKcfu is the number of CFU of unexposed cells and Ecfu represents the number of CFU of exposed cells. Data are means ± SD of three replicates of independent experiments. Statistically significant differences from results for the wild-type group were determined by a Student's t test (*, P < 0.05; **, P < 0.01).
Similar to CuO-NPs, C/Cu-NPs also showed strikingly higher adverse effects to the quintuple mutant than to the wild type (P < 0.05), and a higher exposure concentration resulted in higher inhibition for the quintuple mutant, with the inhibition rate as high as 28.4%. For C-NPs, no clear dose-effect relationship was displayed, and the inhibition rate was much lower (up to 5.2%).
Analogously to CuO-NPs and Cu2+, H2O2 showed dose-dependent toxic effects on the growth of yeast strains. At an exposure concentration of 1.5 mmol/liter, the inhibition rates for BY4741, the yap1 mutant, the quadruple mutant, and the quintuple mutant were 4.3%, 13.2%, 10.9%, and 24.2%, respectively, and increased to 24.6%, 69.4%, 26.5%, and 70.1%, respectively, when the exposure concentration shifted to 6.0 mmol/liter. As the data in Fig. 2F show, the quadruple mutant was slightly more susceptible to H2O2 than the BY4741 strain, while a significant difference was observed for the yap1 mutant (P < 0.05) and an extremely remarkable difference was observed for the quintuple mutant (P < 0.01) at relatively high concentrations of H2O2 (3 to 6 mmol/liter).
Soluble copper in SD medium.
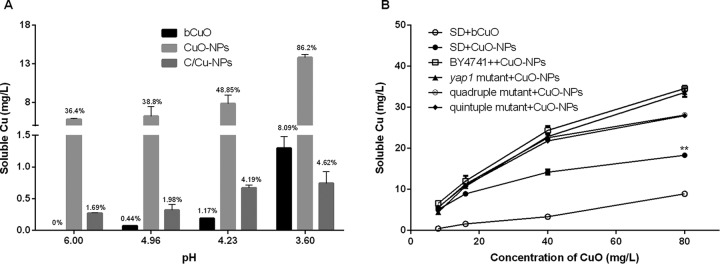
A previous investigation indicated that pH has a significant impact on the dissolution of metal oxide nanoparticles in the exposure medium (21). Based on the literature, the influence of pH on the amount of copper leaching from CuO-NPs, C/Cu-NPs, and bCuO was measured and evaluated. The pH of the culture medium was decreased during the cultivation process for all the studied strains, and the pH, which varied from 6.00 to 3.60, covered the range of all the yeast strains (see Fig. S2 in the supplemental material). Hence, in our research, SD media with pHs of 6.00, 4.96, 4.23, and 3.60 were used to investigate the effects of pH values on copper leaching. The data shown in Fig. 3A indicate that copper release was affected by the solution pH, with a higher leaching ratio at a lower pH, suggesting an increasing release of copper-containing particles during incubation in this study. After 4 h of incubation, about 35%, 40%, 50%, and 85% of the CuO-NPs were dissolved when the pH values were 6.00, 4.96, 4.23, and 3.60, respectively. At an intermediate pH (pH 6.0), almost 35% of copper leachate could be detected for CuO-NPs, but no copper ions for bCuO and almost none for C/Cu-NPs were detected. If the pH was adjusted to 3.60, the nanoparticles of copper oxide rapidly dissolved, and the more stable bCuO gradually leached copper into the surrounding solution; moreover, the copper dissolved from C/Cu-NPs achieved a level of 4.62% (Fig. 3A).
FIG 3.
(A) Soluble copper released from copper-containing materials in SD medium at different pH values after 4 h of incubation at 30°C. CuO-NPs, C/Cu-NPs, and bCuO had constant copper concentrations of 16 mg/liter. The percentage numbers above the bars give the amount of copper released from particles. (B) Soluble copper in SD culture in the absence and presence of yeast strains after cells were treated with various doses of CuO for 4 h. Data are means ± SD of three replicates of independent experiments. **, P < 0.01 compared to the results using SD medium treated with bCuO (SD+bCuO).
As depicted in Fig. 3B, about 4.2 to 8.9% of dissolved copper relative to total copper was observed in SD medium containing 10 to 100 μg/ml bCuO, and a significantly higher ratio of soluble copper (5 to 17%) was detected for CuO-NPs (P < 0.01). This outcome was consistent with the results we mentioned earlier. The results from AAS showed that CuO-NPs dissolved slightly more in the presence of yeast strains than in abiotic conditions (SD medium only). The results also showed that more soluble copper appeared in the test medium when CuO-NPs were incubated with wild-type cells than in mutant cultures, but this effect was not statistically significant (P > 0.05).
Intracellular ROS generation.
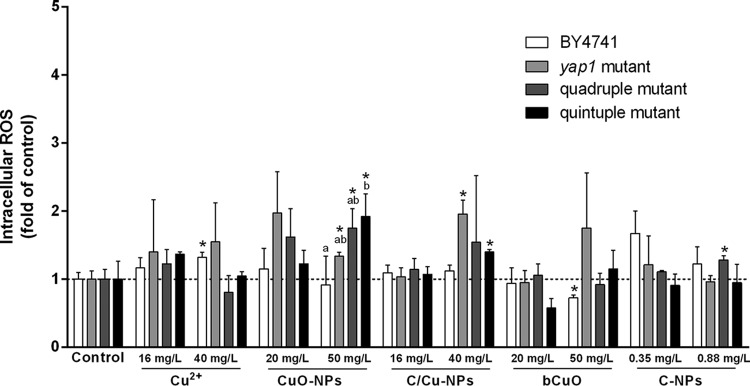
Previous research indicated that oxidative damage is a common mechanism to explain the cytotoxicity of nanoparticles (40, 48, 49). Herein, we measured the generation of intracellular ROS to determine if it was involved with the toxic mechanism of CuO-NPs. Tracking of intracellular ROS generation (with the H2DCFDA assay) was carried out during yeast growth in the presence of various forms of copper, i.e., CuO-NPs, bCuO, Cu2+, and C/Cu-NPs. The data in Fig. 4 show the relative ROS contents of yeast cells under different treatments compared to the amount in the untreated control. After incubation with 40 mg/liter ionic copper for 4 h, a significant difference in ROS generation was detected for the wild type (P < 0.05); notably higher ROS formation was induced for all strains except for the wild-type strain at an exposure concentration of 50 mg/liter CuO-NPs while there was no evident dose-response effect on ROS formation for the studied materials. It is noteworthy that for the quintuple mutant the ROS level induced was markedly higher than that of the wild type at a concentration of 50 mg/liter CuO-NPs (P < 0.05).
FIG 4.
Effects of copper-containing materials and C-NPs on ROS generation. The intracellular ROS level was detected after exposure for 4 h. Results are presented as fold of increase over control values. Data are means ± SD of three independent replicates. The asterisk denotes a significant difference from the control value, and letters (a and b) indicate results statistically different from each other (P < 0.05).
Copper internalization.
To investigate whether the differences in cytotoxicity levels could be explained by the differences in cellular uptake of nanoparticles, i.e., a Trojan horse mechanism, we followed the method described by Xiong et al. (43) to measure the copper content inside yeast cells. By carefully etching with ferric nitrate and washing samples with PBS, free particles and copper ions in medium as well as those absorbed on the cells were removed. The AAS analysis showed that all the yeast strains take up a higher content of copper element at higher copper exposures and that when they were treated with equal copper concentrations of CuO-NPs and copper salts, the specific yeast strains internalized comparable amounts of copper. However, when strains were cultivated with bCuO, lower levels of accumulation were observed (Fig. 5). In addition, after 4 h of exposure, the quadruple mutant readily takes up more copper than the wild type at an exposure concentration of 50 mg/liter CuO-NPs (P < 0.01); at concentrations of 16 and 40 mg/liter copper ions, the quadruple mutant also takes up more copper (P < 0.05). There was no major difference between the yap1 mutant and the wild type, and the quintuple mutant is similar to the quadruple mutant in its assimilation of copper.
FIG 5.
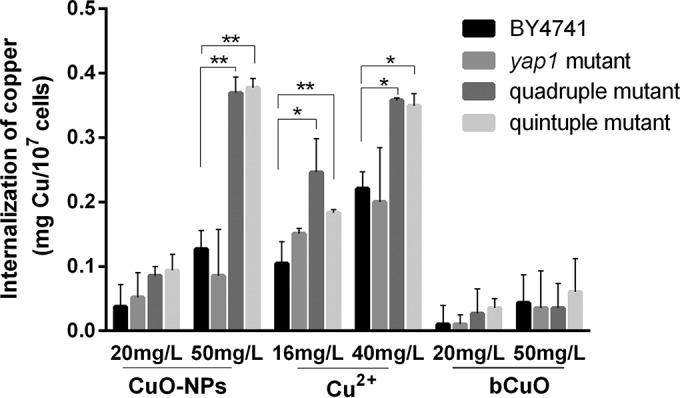
Cellular uptake of copper by yeast cells. The metal content of cells was determined by atomic absorption spectrometry after digestion. Internalization is presented as the total amount of copper element inside cells. The tested copper concentrations of CuO-NPs and bCuO were equivalent to the Cu2+ concentration. Data are means ± SD of three independent replicates. Statistically significant differences were determined by a Student's t test (*, P < 0.05; **, P < 0.01).
DISCUSSION
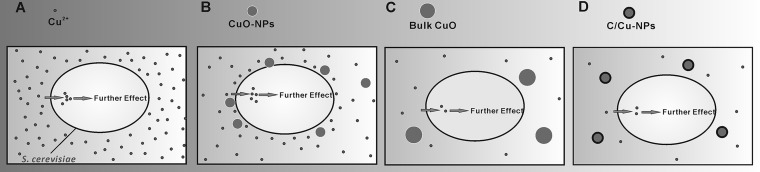
Recent study indicated that the toxicity of CuO-NPs and its counterpart bCuO was due to the copper leachate rather than to the particles themselves. The dissolved copper from CuO-NPs and bCuO induces the same level of cytotoxicity to E. coli as that in the presence of particles (3). In addition, Mortimer et al. (50) reported that the toxicity of CuO-NPs to T. thermophila was traced to dissolved copper, and they also demonstrated that the high toxicity of CuO-NPs compared to that of its bulk control was attributed to increased amounts of dissolved copper (50). In our research, we discovered similar results with previous reports that CuO-NPs showed much higher toxicity than bCuO for all yeast strains; for example, CuO-NPs were remarkably 3- to 4-fold more toxic than bCuO particles. As indicated above, CuO-NPs more easily leached copper than bCuO, and significant differences existed between CuO-NPs and bCuO in terms of their ability to leach copper (P < 0.05) (Fig. 3B). On the basis that S. cerevisiae strains were more sensitive to CuO-NPs than to bCuO and that CuO-NPs have a higher tendency to dissolve copper, the toxicity of CuO-NPs was assumed to be mainly caused by soluble copper. Furthermore, as C/Cu-NPs are well characterized and representative carbon-coated copper nanoparticles, when the copper exposure dose is kept constant, C/Cu-NPs should be less harmful to cells than cupric ions or CuO-NPs. Herein we detected the toxicity of C/Cu-NPs and verified that C/Cu-NPs were less toxic to yeast strains than CuO-NPs and ionic copper; this might be due to the low capability of C/Cu-NPs to leach copper. This result was analogous to that of previous work (21).
As CuO-NPs were undergoing a slow, dynamic dissolving process (3, 51), the available mass of copper was absolutely less than that of its counterpart copper salt. However, the same doses of CuO-NPs and Cu2+ showed no detectable differences in inhibiting the growth of yeast strains in our study. Jiang et al. (38) mentioned that the toxicity of nanoparticles resulted not only from the dissolved metal ions but also from their greater tendency to attach to the cell walls (38). Kasemets et al. (23) indicated that when the toxicity tests were done in organic matter-containing medium, CuO-NPs enhanced the toxic effect, probably due to the stronger sorption of protein-coated nanoparticles onto the cell surface, which facilitates the dissolving of CuO-NPs in the close vicinity of the yeast cell walls (23). Figure 6 depicts possible schematic representations of different copper-containing materials for soluble copper distribution. The release of ions was supposed to be the main mechanism that accounts for the toxicity observed in S. cerevisiae treated with CuO-NPs. Specifically, CuO-NPs were leaching more copper in the presence of the BY4741 strain than in the presence of the quintuple mutant (Fig. 3B); paradoxically, the CuO-NPs were more toxic to the quintuple mutant than to the wild type, indicating that the toxicity of CuO-NPs may not be mediated only by soluble copper.
FIG 6.
Diagrams of different copper-containing materials showing soluble copper distribution and internalization. Soluble copper from copper salt was distributed evenly in the medium. However, for the soluble copper distribution in CuO-NP suspension, as CuO-NPs tend to adsorb on the cell wall, the sorption may lead to enhanced dissolving of CuO-NPs around the cells, and consequently higher ionic stress takes place in the close vicinity of the cell walls. For the hardly soluble bCuO and C/Cu-NPs, only a very limited amount of particles dissolved. Consequently, soluble copper was taken up by the cells, and then a series of toxicological responses were activated.
Oxidative stress occurs when ROS affect the balance between oxidative pressure and antioxidant defense (52, 53). Soluble copper leached from CuO-NPs would induce the generation of ROS (3, 54), and nanoparticles themselves, like graphite and graphene oxide, could cause oxidative stress to bacteria through direct interaction with the cell membrane (55). As ROS could induce oxidative stress in the proteins, nucleic acids, and macromolecules, it may have a profound effect on cell structure as well as on physiological activity. Unlike TiO2-NPs and ZnO-NPs, CuO-NP suspensions did not generate measurable extracellular ROS even under UV irradiation (56); hence, the intracellular ROS generation initiated by CuO-NPs directly and/or by dissolved copper was considered to be one of the factors leading to toxic effects. Inactivation of the yap1 gene could increase cell sensitivity to oxidative stress caused by H2O2 (Fig. 2F) and tert-butyl hydroperoxide (tBHP) (29). Conversely, these chemicals could induce activation of the yap1 gene (57–59). In the present experiments, the yap1 mutant and the quintuple mutant were more sensitive to H2O2 than the wild-type strain and the quadruple mutant, and the differences were significant (P < 0.05) (Fig. 2F). Data in Fig. 2 also indicate that the susceptibility patterns of the yeast strains upon exposure to CuO-NPs were different from those upon exposure to H2O2. Additionally, the absence of a dose-response effect in ROS formation was not coherent with a dose-response effect in toxicity for CuO-NPs (Fig. 2 and 4) and, combined with the relatively low increase in ROS (up to 1.97-fold), indicated that the ROS-mediated toxicity could not be the main reason accounting for the toxicity of CuO-NPs to yeast cells. Furthermore, comparable ROS levels were detected between micro- and nano-CuO treated groups, while a significantly higher toxic effect observed for CuO-NPs also demonstrated this point, and this conclusion was in line with the published literature (5, 21).
As yeast cell walls were rigid, it could be supposed that under typical conditions nanoparticles cannot enter the cell (23, 43, 60). TEM observation also confirmed that no particles were inside yeast cells when cells were cultivated with CuO-NPs in our study (data not shown). However, studies determined that ionic copper will penetrate into the cells under copper stress and eventually lead to cell injury and cytolysis, and microscopic examination of copper-stressed cells revealed cell wall damage (61). Hence, CuO particle (both micro- and nanoparticles) and copper salt uptake experiments were performed and compared to investigate the role of copper internalization in inducing toxicity in yeast cells. Zhang et al. (34) mentioned that after deletion of cwp1 and cwp2, genotoxic compounds and uncharacterized chemicals would cause intense toxicity (34), and disruption of snq2 and pdr5 would lead to about 30-fold greater estradiol accumulation in yeast, thereby increasing estradiol toxicity (62). To explore the effect of deleting genes cwp1, cwp2, snq2, and pdr5 on the mutant's sensitivity to CuO-NPs and to understand the role of cellular copper in conferring cytotoxicity in yeast cells, we evaluated the uptake dose of copper in cells. Once extensive copper is taken up by the cells, Cu would interact with cellular nucleic acids and enzyme active sites, which would further inevitably lead to loss of cell viability (63, 64). As noted above, the lack of cwp1, cwp2, snq2, and pdr5 had a profound effect on cell permeability, and this change would probably accelerate the penetration of copper ions or copper complexes into yeast cells. In the present study, the cwp1 cwp2 snq2 pdr5 deletion mutant accumulated more copper than the wild type and yap1 mutant (Fig. 5), and increases in internalization were observed in the quadruple mutant and quintuple mutant compared to the wild-type level when cells were cultivated with copper salt or CuO-NPs. However, as mentioned above, the mass of copper leached from CuO-NPs in the presence of the quadruple mutant was comparable to that of wild-type BY4741. While the toxic effect was higher for the quintuple mutant (P < 0.05) (Fig. 2A), the reverse could be explained by the relatively higher ability of the quintuple mutant to assimilate copper. Furthermore, the comparable copper uptake from CuO-NPs and copper salt, with much less copper internalization from bCuO, ruled out the possibility that nanoparticle internalization was mediated by the soluble copper and confirmed the greater tendency of CuO-NPs to attach to the cell surface, resulting in copper stress around the cells comparable to that with copper salt as discussed above. Hence, we concluded that the cell walls and ABC transporters play an important role in preventing copper from being taken up by yeast cells; i.e., inactivation of cwp1, cwp2, snq2, and pdr5 would enhance the internalization of copper by yeast and result in higher toxicity to cells.
Conclusions.
In summary, it is worth noting that the cytotoxicity of CuO-NPs was mainly associated with soluble copper and the amount of copper uptake and that ROS-mediated toxicity may also have a role but could not be the dominant mechanism for CuO-NP toxicity. The present work provides the possibility of using single- or multiple-gene deletion mutants to study the toxicity mechanism of nanoparticles and/or chemicals. Furthermore, on the basis of facts discussed above, we concluded that inactivation of a mechanism-based gene would increase cell susceptibility, especially for the multiple-gene deletion mutants. In these cases, less exposure time and a smaller dose were needed to assess the toxicity of ENMs and, as a result, accelerated the nanomaterial safety assessment.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Natural Science Foundation of China grants (numbers 21477159 and 21037004).
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02035-15.
REFERENCES
- 1.von Moos N, Slaveykova VI. 2014. Oxidative stress induced by inorganic nanoparticles in bacteria and aquatic microalgae–state of the art and knowledge gaps. Nanotoxicology 8:605–630. doi: 10.3109/17435390.2013.809810. [DOI] [PubMed] [Google Scholar]
- 2.Jiang XC, Herricks T, Xia YN. 2002. CuO nanowires can be synthesized by heating copper substrates in air. Nano Lett 2:1333–1338. doi: 10.1021/nl0257519. [DOI] [Google Scholar]
- 3.Gunawan C, Teoh WY, Marquis CP, Amal R. 2011. Cytotoxic origin of copper(II) oxide nanoparticles: comparative studies with micron-sized particles, leachate, and metal salts. ACS Nano 5:7214–7225. doi: 10.1021/nn2020248. [DOI] [PubMed] [Google Scholar]
- 4.Baek YW, An YJ. 2011. Microbial toxicity of metal oxide nanoparticles (CuO, NiO, ZnO, and Sb2O3) to Escherichia coli, Bacillus subtilis, and Streptococcus aureus. Sci Total Environ 409:1603–1608. doi: 10.1016/j.scitotenv.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Li J, Zhao J, Xing B. 2011. Toxicity and internalization of CuO nanoparticles to prokaryotic alga Microcystis aeruginosa as affected by dissolved organic matter. Environ Sci Technol 45:6032–6040. doi: 10.1021/es2010573. [DOI] [PubMed] [Google Scholar]
- 6.Fahmy B, Cormier SA. 2009. Copper oxide nanoparticles induce oxidative stress and cytotoxicity in airway epithelial cells. Toxicol In Vitro 23:1365–1371. doi: 10.1016/j.tiv.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blinova I, Ivask A, Heinlaan M, Mortimer M, Kahru A. 2010. Ecotoxicity of nanoparticles of CuO and ZnO in natural water. Environ Pollut 158:41–47. doi: 10.1016/j.envpol.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 8.Perreault F, Oukarroum A, Pirastru L, Sirois L, Gerson Matias W, Popovic R. 2010. Evaluation of copper oxide nanoparticles toxicity using chlorophyll a fluorescence imaging in Lemna gibba. J Bot 2010:763142. doi: 10.1155/2010/763142. [DOI] [Google Scholar]
- 9.Chen Z, Meng H, Xing G, Chen C, Zhao Y, Jia G, Wang T, Yuan H, Ye C, Zhao F, Chai Z, Zhu C, Fang X, Ma B, Wan L. 2006. Acute toxicological effects of copper nanoparticles in vivo. Toxicol Lett 163:109–120. doi: 10.1016/j.toxlet.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Regier N, Cosio C, von Moos N, Slaveykova VI. 2015. Effects of copper-oxide nanoparticles, dissolved copper and ultraviolet radiation on copper bioaccumulation, photosynthesis and oxidative stress in the aquatic macrophyte Elodea nuttallii. Chemosphere 128:56–61. doi: 10.1016/j.chemosphere.2014.12.078. [DOI] [PubMed] [Google Scholar]
- 11.Ivask A, Juganson K, Bondarenko O, Mortimer M, Aruoja V, Kasemets K, Blinova I, Heinlaan M, Slaveykova V, Kahru A. 2014. Mechanisms of toxic action of Ag, ZnO and CuO nanoparticles to selected ecotoxicological test organisms and mammalian cells in vitro: a comparative review. Nanotoxicology 8:57–71. doi: 10.3109/17435390.2013.855831. [DOI] [PubMed] [Google Scholar]
- 12.Bondarenko O, Juganson K, Ivask A, Kasemets K, Mortimer M, Kahru A. 2013. Toxicity of Ag, CuO and ZnO nanoparticles to selected environmentally relevant test organisms and mammalian cells in vitro: a critical review. Arch Toxicol 87:1181–1200. doi: 10.1007/s00204-013-1079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. 1996. Life with 6000 genes. Science 274:546, 563–547. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 14.Gromozova EN, Voychuk SI. 2007. Influence of radiofrequency Emf on the yeast Saccharomyces cerevisiae as model eukaryotic system, p 167–175. In Beloussov LV, Voeikov VL, Martynyuk VS (ed), Biophotonics and coherent systems in biology. Springer, New York, NY. [Google Scholar]
- 15.De Freitas J, Wintz H, Kim JH, Poynton H, Fox T, Vulpe C. 2003. Yeast, a model organism for iron and copper metabolism studies. Biometals 16:185–197. doi: 10.1023/A:1020771000746. [DOI] [PubMed] [Google Scholar]
- 16.Kungolos A, Aoyama I, Muramoto S. 1999. Toxicity of organic and inorganic mercury to Saccharomyces cerevisiae. Ecotoxicol Environ Saf 43:149–155. doi: 10.1006/eesa.1999.1767. [DOI] [PubMed] [Google Scholar]
- 17.Kim JS, Kuk E, Yu KN, Kim JH, Park SJ, Lee HJ, Kim SH, Park YK, Park YH, Hwang CY, Kim YK, Lee YS, Jeong DH, Cho MH. 2007. Antimicrobial effects of silver nanoparticles. Nanomed Nanotechnol Biol Med 3:95–101. doi: 10.1016/j.nano.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Lee S, Lee J, Kim K, Sim SJ, Gu MB, Yi J, Lee J. 2009. Eco-toxicity of commercial silver nanopowders to bacterial and yeast strains. Biotechnol Bioprocess Eng 14:490–495. doi: 10.1007/s12257-008-0254-6. [DOI] [Google Scholar]
- 19.Kasemets K, Ivask A, Dubourguier HC, Kahru A. 2009. Toxicity of nanoparticles of ZnO, CuO and TiO2 to yeast Saccharomyces cerevisiae. Toxicol In Vitro 23:1116–1122. doi: 10.1016/j.tiv.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Ivask A, Kurvet I, Kasemets K, Blinova I, Aruoja V, Suppi S, Vija H, Kakinen A, Titma T, Heinlaan M, Visnapuu M, Koller D, Kisand V, Kahru A. 2014. Size-dependent toxicity of silver nanoparticles to bacteria, yeast, algae, crustaceans and mammalian cells in vitro. PLoS One 9:e102108. doi: 10.1371/journal.pone.0102108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Studer AM, Limbach LK, Van Duc L, Krumeich F, Athanassiou EK, Gerber LC, Moch H, Stark WJ. 2010. Nanoparticle cytotoxicity depends on intracellular solubility: comparison of stabilized copper metal and degradable copper oxide nanoparticles. Toxicol Lett 197:169–174. doi: 10.1016/j.toxlet.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 22.Dasari TP, Pathakoti K, Hwang HM. 2013. Determination of the mechanism of photoinduced toxicity of selected metal oxide nanoparticles (ZnO, CuO, Co3O4 and TiO2) to E. coli bacteria. J Environ Sci (China) 25:882–888. doi: 10.1016/S1001-0742(12)60152-1. [DOI] [PubMed] [Google Scholar]
- 23.Kasemets K, Suppi S, Kunnis-Beres K, Kahru A. 2013. Toxicity of CuO nanoparticles to yeast Saccharomyces cerevisiae BY4741 wild-type and its nine isogenic single-gene deletion mutants. Chem Res Toxicol 26:356–367. doi: 10.1021/tx300467d. [DOI] [PubMed] [Google Scholar]
- 24.Zhang M, Hanna M, Li J, Butcher S, Dai HP, Xiao W. 2010. Creation of a hyperpermeable yeast strain to genotoxic agents through combined inactivation of PDR and CWP genes. Toxicol Sci 113:401–411. doi: 10.1093/toxsci/kfp267. [DOI] [PubMed] [Google Scholar]
- 25.Jamieson DJ. 1998. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14:1511–1527. [DOI] [PubMed] [Google Scholar]
- 26.Jo WJ, Loguinov A, Chang M, Wintz H, Nislow C, Arkin AP, Giaever G, Vulpe CD. 2008. Identification of genes involved in the toxic response of Saccharomyces cerevisiae against iron and copper overload by parallel analysis of deletion mutants. Toxicol Sci 101:140–151. [DOI] [PubMed] [Google Scholar]
- 27.Xiu Z, Liu Y, Mathieu J, Wang J, Zhu D, Alvarez PJ. 2014. Elucidating the genetic basis for Escherichia coli defense against silver toxicity using mutant arrays. Environ Toxicol Chem 33:993–997. doi: 10.1002/etc.2514. [DOI] [PubMed] [Google Scholar]
- 28.Kuge S, Jones N, Nomoto A. 1997. Regulation of yap1 nuclear localization in response to oxidative stress. EMBO J 16:1710–1720. doi: 10.1093/emboj/16.7.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang M, Zhang C, Li J, Hanna M, Zhang X, Dai H, Xiao W. 2011. Inactivation of YAP1 enhances sensitivity of the yeast RNR3-lacZ genotoxicity testing system to a broad range of DNA-damaging agents. Toxicol Sci 120:310–321. doi: 10.1093/toxsci/kfq391. [DOI] [PubMed] [Google Scholar]
- 30.Dumond H, Danielou N, Pinto M, Bolotin-Fukuhara M. 2000. A large-scale study of Yap1p-dependent genes in normal aerobic and H2O2-stress conditions: the role of Yap1p in cell proliferation control in yeast. Mol Microbiol 36:830–845. doi: 10.1046/j.1365-2958.2000.01845.x. [DOI] [PubMed] [Google Scholar]
- 31.Dielbandhoesing SK, Zhang H, Caro LHP, Van Der Vaart JM, Klis FM, Verrips CT, Brul S. 1998. Specific cell wall proteins confer resistance to nisin upon yeast cells. Appl Environ Microbiol 64:4047–4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Vaart JM, Caro LH, Chapman JW, Klis FM, Verrips CT. 1995. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J Bacteriol 177:3104–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsujimoto Y, Shimizu Y, Otake K, Nakamura T, Okada R, Miyazaki T, Watanabe K. 2015. Multidrug resistance transporters Snq2p and Pdr5p mediate caffeine efflux in Saccharomyces cerevisiae. Biosci Biotechnol Biochem 79:1103–1110. doi: 10.1080/09168451.2015.1010476. [DOI] [PubMed] [Google Scholar]
- 34.Zhang M, Liang Y, Zhang X, Xu Y, Dai H, Xiao W. 2008. Deletion of yeast CWP genes enhances cell permeability to genotoxic agents. Toxicol Sci 103:68–76. doi: 10.1093/toxsci/kfn034. [DOI] [PubMed] [Google Scholar]
- 35.Sherman F, Fink GR, Hicks J (ed). 1983. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 36.Jia X, Zhu Y, Xiao W. 2002. A stable and sensitive genotoxic testing system based on DNA damage induced gene expression in Saccharomyces cerevisiae. Mutat Res 519:83–92. doi: 10.1016/S1383-5718(02)00129-8. [DOI] [PubMed] [Google Scholar]
- 37.Li M, Zhu L, Lin D. 2011. Toxicity of ZnO nanoparticles to Escherichia coli: mechanism and the influence of medium components. Environ Sci Technol 45:1977–1983. doi: 10.1021/es102624t. [DOI] [PubMed] [Google Scholar]
- 38.Jiang W, Mashayekhi H, Xing B. 2009. Bacterial toxicity comparison between nano- and micro-scaled oxide particles. Environ Pollut 157:1619–1625. doi: 10.1016/j.envpol.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 39.Hajare SN, Subramanian M, Gautam S, Sharma A. 2013. Induction of apoptosis in human cancer cells by a Bacillus lipopeptide bacillomycin D. Biochimie 95:1722–1731. doi: 10.1016/j.biochi.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 40.Zhang XY, Hu WB, Li J, Tao L, Wei Y. 2012. A comparative study of cellular uptake and cytotoxicity of multi-walled carbon nanotubes, graphene oxide, and nanodiamond. Toxicol Res 1:62–68. doi: 10.1039/c2tx20006f. [DOI] [Google Scholar]
- 41.Kotrba P, Doleckova L, De Lorenzo V, Ruml T. 1999. Enhanced bioaccumulation of heavy metal ions by bacterial cells due to surface display of short metal binding peptides. Appl Environ Microbiol 65:1092–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooksey DA, Azad HR. 1992. Accumulation of copper and other metals by copper-resistant plant-pathogenic and saprophytic pseudomonads. Appl Environ Microbiol 58:274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong YJ, Brunson M, Huh J, Huang AR, Coster A, Wendt K, Fay J, Qin D. 2013. The role of surface chemistry on the toxicity of Ag nanoparticles. Small 9:2628–2638. doi: 10.1002/smll.201202476. [DOI] [PubMed] [Google Scholar]
- 44.Altundag H, Tuzen M. 2011. Comparison of dry, wet and microwave digestion methods for the multi element determination in some dried fruit samples by ICP-OES. Food Chem Toxicol 49:2800–2807. doi: 10.1016/j.fct.2011.07.064. [DOI] [PubMed] [Google Scholar]
- 45.Bakircioglu D, Kurtulus YB, Ucar G. 2011. Determination of some traces metal levels in cheese samples packaged in plastic and tin containers by ICP-OES after dry, wet and microwave digestion. Food Chem Toxicol 49:202–207. doi: 10.1016/j.fct.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 46.Xiong DW, Fang T, Yu LP, Sima XF, Zhu WT. 2011. Effects of nano-scale TiO2, ZnO and their bulk counterparts on zebrafish: acute toxicity, oxidative stress and oxidative damage. Sci Total Environ 409:1444–1452. doi: 10.1016/j.scitotenv.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 47.Wang HH, Wick RL, Xing BS. 2009. Toxicity of nanoparticulate and bulk ZnO, Al2O3 and TiO2 to the nematode Caenorhabditis elegans. Environ Pollut 157:1171–1177. doi: 10.1016/j.envpol.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 48.Nel A, Xia T, Madler L, Li N. 2006. Toxic potential of materials at the nanolevel. Science 311:622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 49.Pulskamp K, Diabate S, Krug HF. 2007. Carbon nanotubes show no sign of acute toxicity but induce intracellular reactive oxygen species in dependence on contaminants. Toxicol Lett 168:58–74. doi: 10.1016/j.toxlet.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 50.Mortimer M, Kasemets K, Kahru A. 2010. Toxicity of ZnO and CuO nanoparticles to ciliated protozoa Tetrahymena thermophila. Toxicology 269:182–189. doi: 10.1016/j.tox.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 51.Zhao J, Wang Z, Dai Y, Xing B. 2013. Mitigation of CuO nanoparticle-induced bacterial membrane damage by dissolved organic matter. Water Res 47:4169–4178. doi: 10.1016/j.watres.2012.11.058. [DOI] [PubMed] [Google Scholar]
- 52.Birben E, Sahiner UM, Sackesen C, Erzurum S, Kalayci O. 2012. Oxidative stress and antioxidant defense. World Allergy Organ J 5:9–19. doi: 10.1097/WOX.0b013e3182439613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu J, Li ZG, Xu PJ, Xiao L, Yang Z. 2013. Nanosized copper oxide induces apoptosis through oxidative stress in podocytes. Arch Toxicol 87:1067–1073. doi: 10.1007/s00204-012-0925-0. [DOI] [PubMed] [Google Scholar]
- 54.Ivask A, Bondarenko O, Jepihhina N, Kahru A. 2010. Profiling of the reactive oxygen species-related ecotoxicity of CuO, ZnO, TiO2, silver and fullerene nanoparticles using a set of recombinant luminescent Escherichia coli strains: differentiating the impact of particles and solubilised metals. Anal Bioanal Chem 398:701–716. doi: 10.1007/s00216-010-3962-7. [DOI] [PubMed] [Google Scholar]
- 55.Liu S, Zeng TH, Hofmann M, Burcombe E, Wei J, Jiang R, Kong J, Chen Y. 2011. Antibacterial activity of graphite, graphite oxide, graphene oxide, and reduced graphene oxide: membrane and oxidative stress. ACS Nano 5:6971–6980. doi: 10.1021/nn202451x. [DOI] [PubMed] [Google Scholar]
- 56.Li Y, Zhang W, Niu J, Chen Y. 2012. Mechanism of photogenerated reactive oxygen species and correlation with the antibacterial properties of engineered metal-oxide nanoparticles. ACS Nano 6:5164–5173. doi: 10.1021/nn300934k. [DOI] [PubMed] [Google Scholar]
- 57.Schnell N, Krems B, Entian KD. 1992. The Par1 (Yap1/Snq3) gene of Saccharomyces cerevisiae, a c-Jun homolog, is involved in oxygen metabolism. Curr Genet 21:269–273. doi: 10.1007/BF00351681. [DOI] [PubMed] [Google Scholar]
- 58.Hirata D, Yano K, Miyakawa T. 1994. Stress-induced transcriptional activation mediated by Yap1 and Yap2 genes that encode the Jun family of transcriptional activators in Saccharomyces cerevisiae. Mol Gen Genet 242:250–256. doi: 10.1007/BF00280413. [DOI] [PubMed] [Google Scholar]
- 59.Kuge S, Jones N. 1994. Yap1 dependent activation of Trx2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J 13:655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stark WJ. 2011. Nanoparticles in biological systems. Angew Chem Int Ed Engl 50:1242–1258. doi: 10.1002/anie.200906684. [DOI] [PubMed] [Google Scholar]
- 61.Zevenhuizen LP, Dolfing J, Eshuis EJ, Scholten-Koerselman IJ. 1979. Inhibitory effects of copper on bacteria related to the free ion concentration. Microb Ecol 5:139–146. doi: 10.1007/BF02010505. [DOI] [PubMed] [Google Scholar]
- 62.Mahe Y, Lemoine Y, Kuchler K. 1996. The ATP binding cassette transporters Pdr5 and Snq5 of Saccharomyces cerevisiae can mediate transport of steroids in vivo. J Biol Chem 271:25167–25172. doi: 10.1074/jbc.271.41.25167. [DOI] [PubMed] [Google Scholar]
- 63.Ohsumi Y, Kitamoto K, Anraku Y. 1988. Changes induced in the permeability barrier of the yeast plasma membrane by cupric ion. J Bacteriol 170:2676–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Avery SV, Howlett NG, Radice S. 1996. Copper toxicity towards Saccharomyces cerevisiae: dependence on plasma membrane fatty acid composition. Appl Environ Microbiol 62:3960–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.