Abstract
The role that neutrophilic iron-oxidizing bacteria play in the Arctic tundra is unknown. This study surveyed chemosynthetic iron-oxidizing communities at the North Slope of Alaska near Toolik Field Station (TFS) at Toolik Lake (lat 68.63, long −149.60). Microbial iron mats were common in submerged habitats with stationary or slowly flowing water, and their greatest areal extent is in coating plant stems and sediments in wet sedge meadows. Some Fe-oxidizing bacteria (FeOB) produce easily recognized sheath or stalk morphotypes that were present and dominant in all the mats we observed. The cool water temperatures (9 to 11°C) and reduced pH (5.0 to 6.6) at all sites kinetically favor microbial iron oxidation. A microbial survey of five sites based on 16S rRNA genes found a predominance of Proteobacteria, with Betaproteobacteria and members of the family Comamonadaceae being the most prevalent operational taxonomic units (OTUs). In relative abundance, clades of lithotrophic FeOB composed 5 to 10% of the communities. OTUs related to cyanobacteria and chloroplasts accounted for 3 to 25% of the communities. Oxygen profiles showed evidence for oxygenic photosynthesis at the surface of some mats, indicating the coexistence of photosynthetic and FeOB populations. The relative abundance of OTUs belonging to putative Fe-reducing bacteria (FeRB) averaged around 11% in the sampled iron mats. Mats incubated anaerobically with 10 mM acetate rapidly initiated Fe reduction, indicating that active iron cycling is likely. The prevalence of iron mats on the tundra might impact the carbon cycle through lithoautotrophic chemosynthesis, anaerobic respiration of organic carbon coupled to iron reduction, and the suppression of methanogenesis, and it potentially influences phosphorus dynamics through the adsorption of phosphorus to iron oxides.
INTRODUCTION
The Arctic tundra biome is fascinating in its own right and has the potential to be heavily affected by changes in climate associated with increased atmospheric CO2 concentrations and global warming. One of the most dramatic impacts is likely to be a change in the dynamics of permanently frozen soils (permafrost) as overall temperatures rise and the shoulder seasons of thaw and freeze-up expand (1–3). Understanding the biogeochemical implications of climate change in the Arctic is important, in part because relative to its total landmass area, permafrost stores an outsized fraction of organic carbon (4). The fate of that carbon, especially the portion that is mineralized to CO2 and/or methane, has the potential to impact further climate change through the release of greenhouse gases. Understanding the range of biogeochemical processes in the Arctic and how they impact the carbon cycle, either directly or indirectly, is thus of vital importance.
In general, the microbial iron cycle in the Arctic tundra is poorly understood. Only in the past few years have studies started to investigate the reductive aspects of the iron cycle, which have shown that Fe-reducing bacteria can account for a large fraction of the respiration in anoxic Arctic soils (5). There are no published reports on the role of bacteria in iron oxidation in the Arctic, nor is there much information, beyond anecdotal reports, about the occurrence or abundance of biogenically produced iron oxides associated with tundra wetlands or streams. In contrast, in temperate ecosystems, it is now well established that specific communities of bacteria inhabit a variety of aqueous habitats where there are persistent gradients of Fe(II) and O2 that result in visible precipitation of rust-colored iron oxyhydroxides (6).
Fe-oxidizing bacteria (FeOB) that utilize Fe(II) as their primary energy source are dominant members of these communities (6). These organisms precipitate large quantities of Fe oxides through the production of morphologically unique extracellular structures that form the primary fabric of the microbial mat. For example, two iconic FeOB are sheath-forming Leptothrix ochracea and stalk-forming Gallionella ferruginea, which produce iron-encrusted sheaths and stalks, respectively, both of which are easily recognized by light microscopy. Iron mats impact the local environment by increasing the tortuosity of water flow, and the oxides provide a large reactive surface area for the sorption of other metals, phosphates, and dissolved organic matter (7, 8). As a result, iron mats have the capacity to influence the local water chemistry, extending the influence of these microbes beyond their immediate environment.
Under aerobic conditions, FeOB must compete with abiotic iron oxidation, according to the following reaction (9): −d[Fe(II)]/dt = k × [Fe(II)] × [O2] × [OH−]2, where k is a rate constant. The pH of natural waters exerts the greatest control over the kinetics of abiotic iron oxidation; however, the rate constant k is also temperature dependent, and a 10°C reduction in temperature can lower abiotic oxidation rates by severalfold (10). Finally, the presence of organic ligands can also stabilize Fe(II) and result in its being less prone to oxidation (11). These kinetic properties, together with the knowledge that submerged and partially submerged moderately acidic (pH 5 to 6) soils (often referred to as moist acidic soils) are common in the tundra (4), led to a hypothesis that permafrost regions with mineral-containing soils might be good habitats for FeOB and result in a biologically driven iron cycle. These conditions are quite common on the North Slope of the Brooks Range in Alaska, which led to this investigation for microbial iron mats around the Toolik Field Station (TFS). As it turns out, iron mat communities were found to be very abundant in the area around Toolik. An initial characterization of several iron-oxidizing communities is presented here, and some possible implications of their presence for other biogeochemical processes in the tundra are discussed.
MATERIALS AND METHODS
Study overview, site description, and sampling.
All sampling was done between 13 and 20 July 2014 at the long-term ecological research (LTER) site at the TFS, located at Toolik Lake on the North Slope of the Brooks Range in Alaska. Most of the sampling was done within a 10-km radius of TFS by visiting different rivers and catchment basins accessible by road and/or foot. On two occasions, a helicopter was used to fly approximately 50 km north to the site of the Anaktuvuk River tundra fire that occurred in 2007 (12). More details about specific sample sites are provided in the Results. Microbial mats were sampled in volumes ranging from 10 to 45 ml using either a 5- or 10-ml pipette, with care given to acquire only material within the top centimeter of the mat, unless otherwise stated. Mat samples were placed in either 15- or 50-ml plastic conical tubes, put in a cooler containing blue ice, and returned to the laboratory for processing. Samples for DNA extraction were frozen and returned to Bigelow Laboratory. Samples for microscopy were either viewed live within a few hours of collection or fixed with 2% glutaraldehyde for later analysis. These fixed samples were used for doing direct total cell counts from mat samples using a previously described method (13), with the following modifications: the nucleic acid dye SYTO 13 (Invitrogen) was substituted for acridine orange, and fluorescent antibody slides (Gold Seal; Thermo Scientific) with circumscribed 1-cm circles were used in place of regular microscope slides.
The ferrozine assay was used to determine the iron concentrations in waters associated with the surface of iron mats (14). Approximately 0.5 ml of sample water was collected in a 1-ml syringe and passed through a 0.2-μm-pore-size filter, and 0.1 ml of filtrate was added to 0.9 ml of ferrozine reagent in the field. To determine iron concentrations deeper within microbial iron mats, a prefilter step was included to eliminate thick mat precipitates that can rapidly clog syringes and filters. For this purpose, the barrel end of a 10-ml plastic syringe was cut off, such that about 2.5 cm of the barrel remained; this was filled loosely with glass wool, and a 100-μm Nitex mesh was placed over the open end of the syringe barrel and held in place with a zip tie. The prefilter was connected to a regular 10-ml syringe with a small length (<1 cm) of plastic tubing. For use, the prefilter was placed to the selected depth in the mat and a sample was drawn up, the prefilter was removed, and the sample was filtered through a 0.2-μm-pore-size filter into the ferrozine reagent in a 1:20 dilution. Ferrozine samples were returned to the lab for analysis, as described previously (15).
The pH of field waters was determined either with pH paper (0.5-pH-unit increments) or in the field using a handheld SG23 SevenGo Duo pH meter (Mettler Toledo). Temperature was recorded with a standard thermometer. At some sites, bulk water samples were collected from water overlying the microbial mats in acid-washed bottles for analyses of dissolved organic carbon (DOC), total dissolved nitrogen (TDN), and total dissolved phosphorus (TDP). Upon return to the laboratory, the water samples were filtered through a 0.2-μm-pore-size filter and stored at −20°C prior to analysis. DOC and TDN were analyzed at the Rubenstein Ecosystem Science Laboratory at the University of Vermont using a Shimadzu TOC-TN chemiluminescent analyzer. TDP was analyzed at the same laboratory by doing a persulfate digestion, followed by analysis on a Shimadzu UV-2600 spectrophotometer at 885 nm using the ascorbic acid method (13) (see the Arctic LTER streams protocol for details [http://ecosystems.mbl.edu/ARC/streams/protocol2.html]). Samples for methane were also collected at a subset of sites by adding 10 ml of water to 5 ml of 0.1 N NaOH in 20-ml serum vials capped with butyl rubber stoppers. These samples were returned to the laboratory, frozen inverted at −20°C, and returned to Bigelow Laboratory, where they were analyzed for methane content on an SRI model 310 gas chromatograph equipped with a flame ionization detector, as previously described (16).
Oxygen profiling.
Field profiles for O2 were done using a battery-powered FireSting optode (Pyro Science GmbH) connected to a laptop computer loaded with the FireSting software package. The optode was held with a manually operated micromanipulator mounted on a ring stand weighed down with a rock in such a way as to minimize disturbance to the microbial mat. The tip of the optode was placed at the water surface, and then the tip was moved to the mat surface and into the mat at vertical increments of as small as 50 μm. The O2 data (obtained in μM) were recorded at 3-s intervals on a laptop computer, with a minimum of 20 s of data collected at each depth. In cases in which the water overlying the iron mat was too deep (>5 cm) for the micromanipulator, hand measurements were made by holding the optode at the approximate desired depth, as determined with a ruler, while data were recorded. The oxygen data were transferred to Microsoft Excel for analysis and graphed using the DataGraph software package.
Iron reduction assays.
Testing freshly collected mat material for the potential to support iron reduction was done in anoxic medium inoculated with iron mat, incubated up to 8 days, and the production of Fe(II) was regularly measured using ferrozine, as previously described (15). Briefly, serum bottles (total volume, 110 ml) capped with butyl rubber stoppers containing modified Wolfe's mineral medium (40 ml) with 20 mM bicarbonate buffer (pH 6.7) and 10 mM sodium acetate were prepared anoxically, and the bottles were sterilized by autoclaving prior to traveling to TFS. Mat samples were added directly to duplicate serum bottles for each incubation via syringe in volumes ranging from 5 to 10 ml, and the bottles were incubated in the dark at room temperature (18 to 21°C). Each day, approximately 0.3 ml of the liquid was removed in duplicate to quantitate the release of Fe(II) with the ferrozine assay. Samples collected during the later period of field work had iron measurements done for the first 3 days at TFS and then were placed in a cooler with blue ice and shipped to Bigelow Laboratory, where incubations were continued for an additional 4 days.
Microscopy of samples was done on fresh samples using a Zeiss Axio imager equipped with phase-contrast and epifluorescence capabilities and housed at TFS, or on glutaraldehyde-fixed samples at Bigelow Laboratory using an Olympus BX 60 microscope. For epifluorescence, the nucleic acid dye SYTO 13 (Invitrogen) was used to stain cells associated with iron oxides.
DNA extraction and 16S rRNA gene 454 pyrosequencing.
Approximately 250 mg (wet weight) of mat material was extracted from each sample and sequenced using previously published methods (17).
Sequence processing.
All sequence processing was performed using mothur version 1.34.0, in accordance with a previously published methodology (17) (mothur.org/wiki/Schloss_SOP). Primer and barcode sequences were removed, followed by the elimination of any short reads (<300 bp), reads containing more than six homopolymers, and/or any ambiguities. Alignments were generated against a SILVA-based reference alignment (mothur.org/wiki/Silva_reference_files), in accordance with previously published methods (17). Putative chimeras were eliminated from the filtered alignments using UCHIME as implemented in mothur (template, self; settings, default), and a Phylip-formatted distance matrix (calc, one gap; count end penalty, true) was clustered using the average-neighbor algorithm. For taxonomic classification of pyrotag reads, we used a mothur-modified Greengenes reference taxonomy, in accordance with previously published methods (17). Based on the resultant taxonomic summaries, we also calculated the consensus taxonomy for individual OTUs at 97% sequence identity. For a comparison of eight samples collected as part of a previous study of an iron mat in Maine (18), the pyrotag sequences that used the same V4 region from that study were processed together with the TFS samples to produce a single data set for analysis.
Estimates of community diversity.
Alpha diversity was estimated for each sample (based on the number of reads in the smallest sample; Table 1) using the following indices: the inverse Simpson index (1/D, nonparametric index, sensitive to abundant OTUs), Berger-Parker index (d, proportional abundance of dominant OTU); the Q-statistic (Q, parametric index not skewed by very rare/abundant OTUs), and Good's coverage (C, an estimate of sample coverage based on the proportion of OTUs to reads).
TABLE 1.
Characteristics of nine separate iron mat sitesa
| Site | Temp (°C) | pH | Mean level (μM) of: |
Description | ||||
|---|---|---|---|---|---|---|---|---|
| Fe(II) | TDN | DOC | TDP | Methane (SD) | ||||
| South River | 9 | 5.5 | 270 | Flocculant mat, seep adjacent to river | ||||
| Ref 6 site | 11 | 5.5 | 480 | Wet sedge meadow | ||||
| Toolik River TK 1 | 8.2 | 5.2 | 60 | Pool near edge of stream | ||||
| Toolik River TK 3 | 9 | NDb | 20 | Wet sedge meadow | ||||
| Kuparuk River Fe seepc | 11 | 6.5 | 170 | 33.7 | 861 | 0.91 | 33.14 (25.6) | Flocculant mat, seep at river bank |
| Kuparuk River potholec | 9 | 5 | 320 | 64 | 1,578 | 0.5 | 0.56 (0.07) | Small pool |
| Toolik BW1c | 10 | 5 | 60 | 16.4 | 616 | 0.07 | 5.26 (7) | Wet sedge meadow |
| Toolik BW2c | 10 | 5 | 20 | Wet sedge meadow | ||||
| Toolik base Fe seepc | 11 | 6 | 170 | 8.2 | 237 | 0.14 | 2.13 (2.64) | Flocculant mat adjacent to lake |
South River and the Ref 6 site are located near the burn site on the Anaktuvuk River. The other sites are in the vicinity (≤20 km) of TFS. Blank spots in the table signify that no data were collected.
ND, not determined.
Samples collected for microbial community analysis.
Oligotyping and MED.
Minimum entropy decomposition (MED) analysis was performed on 20,354 aligned pyrotag reads from 13 samples (Alaska, 5; Maine, 8) with the MED pipeline (version 1.7; available at http://oligotyping.org/MED) (19). After quality filtering using default parameters, 20,060 reads remained. Dendrograms and a heatmap (columns represent samples, rows represent MED nodes) were generated using the o-heatmap.R script available as part of the MED pipeline, with default parameters. Oligotyping was performed on 3,001 aligned pyrotag reads corresponding to OTU1 (97%) with the oligotyping pipeline (version 1.7; available from http://oligotyping.org) using five components following the initial entropy analysis. To reduce noise, each oligotype had to (i) appear in ≥3 samples, (ii) occur in >10.0% of the reads for at least one sample, (iii) represent a minimum of 100 reads in all samples combined, and (iv) have a most-abundant unique sequence with a minimum abundance of 100. Oligotypes that did not meet these criteria were removed from the analysis. The final number of quality-controlled oligotypes revealed by the analysis was 5, representing 2,484 reads, equivalent to 82.77% of all reads analyzed from OTU1.
Nucleotide sequence accession numbers.
All pyrosequencing libraries were deposited at the European Nucleotide Archive under the study accession numbers PRJEB10276 and ERP011506.
RESULTS
Evidence for microbial iron oxidation.
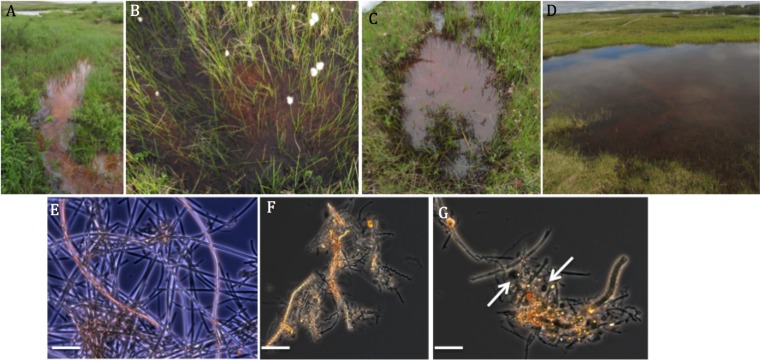
At nearly all the sites (9 of 10) visited at TFS in July 2014, evidence of microbial iron oxidation was observed in the form of rust-colored precipitates that were often striking in their prevalence. Iron mats were either associated with submerged plant stems or found on sediment surfaces, and in many cases, the overlying water bore a metallic sheen. To help make sense of the distribution of iron-oxidizing communities, four different types of iron mats were classified based on associated landscape features (Fig. 1). These were (i) stream/river bank iron seeps, typically 5 to 30 cm deep, with slow flowing water, and centimeters-thick light orange-yellow-colored iron mats; (ii) tundra potholes, small (<1.5 m in diameter) and shallow (<0.5 m deep) open pools of either standing or slow flowing water with iron oxides associated with either sediment layers, or with plant stems in the water; (iii) partially submerged wet sedge meadows dominated by cottongrass with flocculent iron mats, 0.5- to 2-cm thick, which coated the plant stems and/or sediments; and (iv) larger pools or small ponds typically 5 to 30 m across and 1 to 2 m deep, with clearly visible bottom sediments coated with flocculent iron oxides. For logistical reasons, these larger pools were not sampled during this study. The microbial iron mats associated with wet sedge meadows had the largest areal extent and could cover hundreds of square meters.
FIG 1.
Examples of microbial iron mats in the vicinity of TFS (A to D) and photomicrographs of iron mat communities (E to G). (A) Iron microbial mat associated with a seep along the bank of the Kuparuk River. (B) Wet sedge meadow dominated by the cottongrass Eriophorum angustifolium. (C) Small pool or pothole. (D) Small pond with iron oxides on sediment. (E) Mat dominated by sheaths of L. ochracea. (F) Mat dominated by helical stalks reminiscent of Gallionella species. (G) Mixed morphotypes, with arrows denoting capsular oxides, indicating the possible presence of Siderocapsa species. Scale bars = 5 μm.
The temperature and pH of the different sites were quite consistent, with temperatures between 9 and 11°C (mid-July) and a pH range of 5.0 to 6.5 (Table 1). The DOC and TDN levels measured at a subset of sites were consistent with concentrations found in nearby streams, while TDP levels were slightly higher than concentrations found in stream and river waters in the Toolik region. Phase-contrast or bright-field microscopy of oxide samples from the different microbial mat types immediately confirmed the presence of morphotypes that are diagnostic for the presence of FeOB. These were either stalks, characteristic of Gallionella spp., sheaths, characteristic of Leptothrix ochracea, or a combination thereof (Fig. 1). Microscope slides placed in a wet sedge meadow site adjacent to Toolik Lake and close to the BW1 and BW2 sites were colonized within 24 h by stalked and sheathed morphotypes of FeOB (see Fig. S1 in the supplemental material), confirming active growth.
Molecular analysis of microbial communities.
Iron mats from five different sites representing three of the different landforms, as described in the section above, were collected for DNA extraction and community analysis using 16S rRNA gene surveys. Consistent with other freshwater iron-oxidizing communities, Betaproteobacteria had the greatest relative abundance (between 18 and 40%) of any higher (class or above) taxonomic group (Table 2). A comparison of the most abundant orders found from the different TFS sites showed that Burkholderiales were universally abundant in all TFS samples (Fig. 2), while the Sphingobacteriales (Bacteroidetes) and Rhizobiales (Alphaproteobacteria) showed the most variability. The Nitrosomonadales, dominated by the family Gallionellaceae, were present at all sites, but the relative abundance of this group varied substantially between sites. Other higher taxonomic groups that were abundant included Bacteroidetes (15%), Deltaproteobacteria (11%), Alphaproteobacteria (9%), and Cyanobacteria (7%) (Table 2).
TABLE 2.
Phylogenetic breakdown of different 16S rRNA gene OTUs as percentages of the total numbers of OTUs from different sites
| Phylogenetic groupa | % of total OTUs by site |
|||||
|---|---|---|---|---|---|---|
| BW1 | BW2 | Kuparuk River seep | Toolik River seep | Kuparuk River pothole | All sites (mean) | |
| Acidobacteria | 1.6 | 1.9 | 2.4 | 3 | 0.5 | 1.4 |
| Bacteroidetes | 8.7 | 10.2 | 2.9 | 12 | 25 | 14.8 |
| Cyanobacteria | 13 | 6.3 | 25 | 3.3 | 4 | 7.3 |
| Proteobacteria | 48.1 | 60.3 | 52 | 49 | 61 | 56.5 |
| Gammaproteobacteria | 6.2 | 4.5 | 2.7 | 4.1 | 2.8 | 4 |
| Alphaproteobacteria | 6.8 | 14.8 | 4.6 | 17.2 | 2.7 | 9.3 |
| Deltaproteobacteria | 16 | 8.4 | 7.7 | 5.7 | 14.7 | 10.9 |
| Betaproteobacteria | 17.5 | 32.2 | 33.5 | 21 | 40.8 | 31.5 |
| Comamonadaceae | 9.0 | 21.4 | 19.3 | 15 | 31.3 | 22 |
| Rhodoferax spp. | 3.6 | 12.5 | 11.2 | 11 | 17.5 | 12.8 |
| Gallionellaceae | 2.6 | 1 | 7 | 0.3 | 3.2 | 2.3 |
| Leptothrix spp. | 0.2 | 3 | 4 | 1.3 | 4.7 | 3 |
| Gal + Leptob | 2.8 | 4 | 11 | 1.6 | 7.9 | 5.3 |
The first group comprises prevalent phyla, the second group Proteobacteria, the third group members of the Comamonadaceae, and the last group putative FeOB.
Gallionellaceae and Leptothrix spp. combined.
FIG 2.
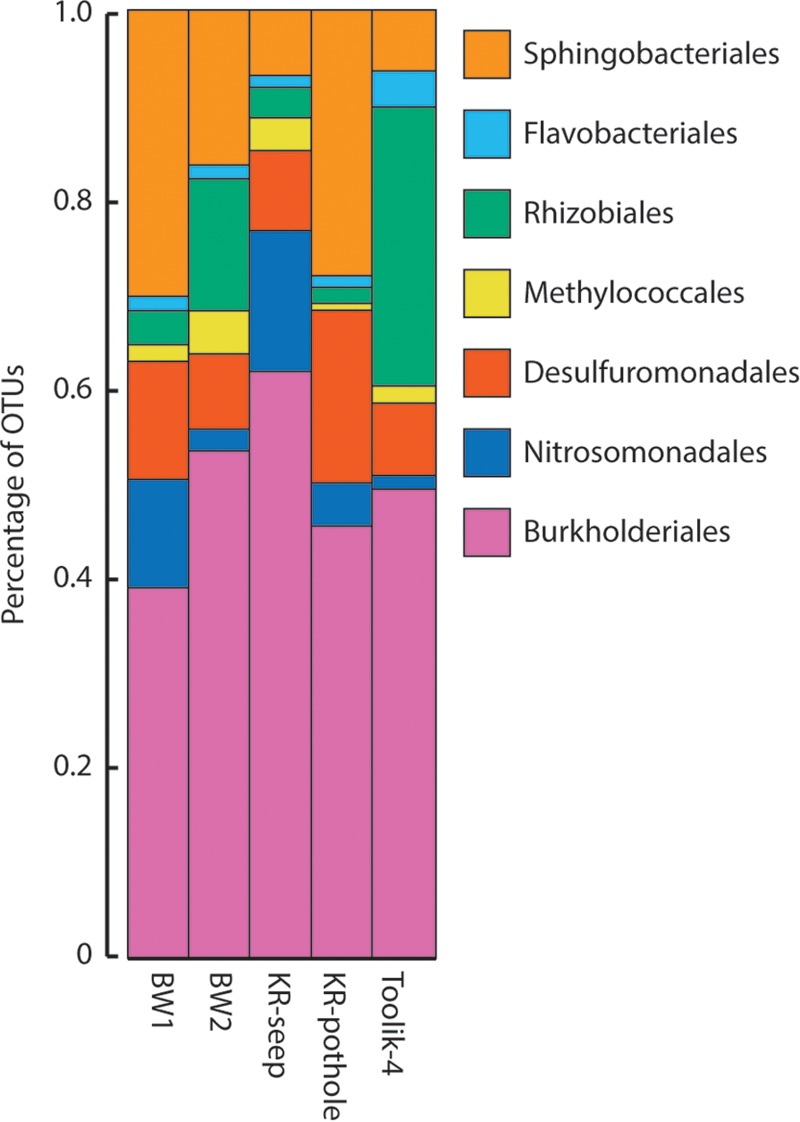
Comparison of the relative abundances of predominant class-level OTUs from the five TFS sites.
At a finer scale of taxonomic resolution, the family Gallionellaceae and genus Leptothrix (both Betaproteobacteria) accounted for about 5.3% of the total reads (range, 1.6 to 11.2%) (Table 2). These two taxa have the strongest association with known FeOB, since all isolates of the Gallionellaceae are microaerophilic lithoautotrophic FeOB, e.g., Gallionella ferruginea, while the Leptothrix reads were closely related to L. ochracea, an uncultured but long recognized sheath-forming FeOB (20). Comamonadaceae had the greatest abundance of any lower (family or below) taxonomic level, accounting for 22% of the total reads at the five sites (range, 9.0 to 31.3%) (Table 2). The majority of reads most closely associated with the genus Rhodoferax accounted for the majority, 13% on average, of the total OTU reads within this family. The overall diversity indices for the different sites are shown in Table S1 in the supplemental material.
Oxygen profiles.
High-spatial-resolution oxygen measurements showed interesting variations in O2 profiles among the profiled mats (Fig. 3). An oxygen profile from a mat at the Kuparuk River seep that had visual evidence for the presence of photosynthetic organisms, based on a slight greenish tinge and microscopic evidence for algal cells, showed an increase in O2 concentration at the surface. This confirmed that an active community of photosynthetic microbes was present. The overall O2 concentrations in this mat were relatively high (50 to 170 μM), and advective flow was visible, likely also contributing to O2 replenishment at deeper depths within the mat. In another mat at the Toolik Lake seep (Fig. 3, blue circles), covered by about 0.5 cm of water and with no visible flow, a steady O2 consumption profile was observed initially; inexplicably, however, the O2 concentration increased deeper within the mat and then decreased below detection at the sediment interface at a total depth of 3 cm. Dissection of this mat after profiling revealed it was actually about 1 cm thick and floated 1 cm above the sediment, resulting in a water channel beneath the mat that was a source of O2 from beneath. The third profile done at BW1 (Fig. 3, red squares) was from a mat that formed on a sediment surface where advective flow was not observed, resulting in a relatively steep O2 gradient. In this case, the Fe(II) concentration in the surface waters above the mat was 135 μM, while an Fe(II) concentration of 615 μM was measured in the pore water sample taken from within the mat near the sediment interface, indicating relatively steep opposing gradients of Fe(II) and O2 that promote the growth of FeOB.
FIG 3.
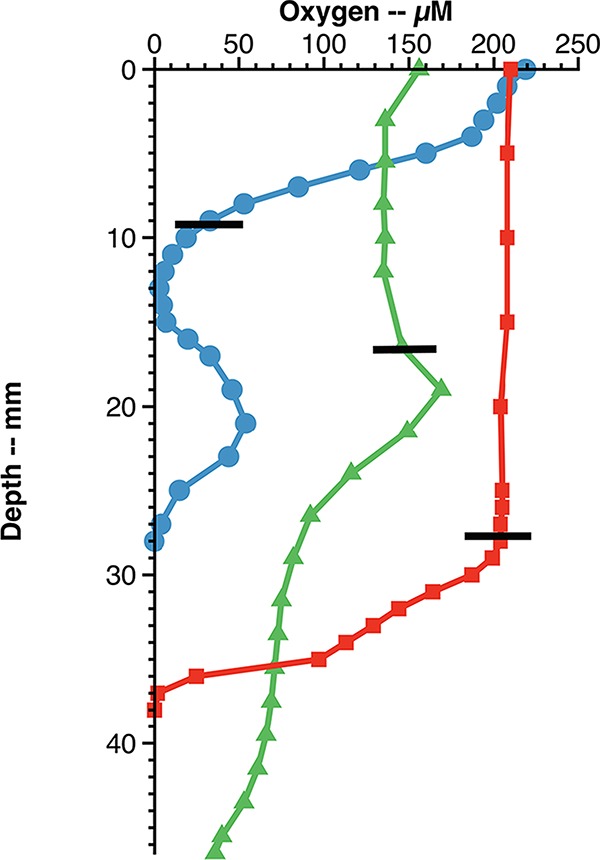
Oxygen profiles made at three different iron mats at TFS. The zero depth is the air-water interface, and the black bars represent the approximate surfaces of the iron mat beneath the water surface. The blue profile is from the Toolik Lake seep station, the green profile is from the Kuparuk River seep, and the red profile is from a mat close to the sediment layer in a wet sedge meadow near site BW1. See the text for details.
Iron reduction.
The potential rates of iron reduction in the presence of 10 mM acetate, normalized to the mat volume, were comparable for all sites, although the total amount of Fe(III) that was reduced varied between sites (Table 3). This might partially be a function of the total iron available in the mats. If mat samples were not supplemented with acetate, little reduction occurred (data not shown). For all the sites, there was a net release of Fe(II) due to reduction within ≤48 h, and the Toolik Lake and Kuparuk River samples showed detectable Fe reduction within ≤24 h (Table 3). These two sites had relatively high abundances of Deltaproteobacteria that include known FeRB belonging to the Geobacteraceae; community analysis was not done for the other sites. There was no evidence for magnetite formation following iron reduction; however, in at least one case, some iron sulfide precipitated 2 weeks after the initiation of reduction, suggesting that sulfate reduction had also taken place.
TABLE 3.
Iron reduction rates, as calculated from Fe reduction experiments from four different sites
| Site | Mean Fe reduction rate (μM h−1 · ml of mat) | Mean total Fe reduced (SD) (μM) | Lag time (h)a |
|---|---|---|---|
| South River | 0.94 | 580 (138) | <48 |
| Ref 6 site | 2.5 | 847 (182) | <48 |
| Kuparuk River seep | 1.7 | 2,930 (237) | <24 |
| Toolik River seep | 1.68 | 2,200 | <24 |
Time until detectable Fe2+.
Structure within a mat.
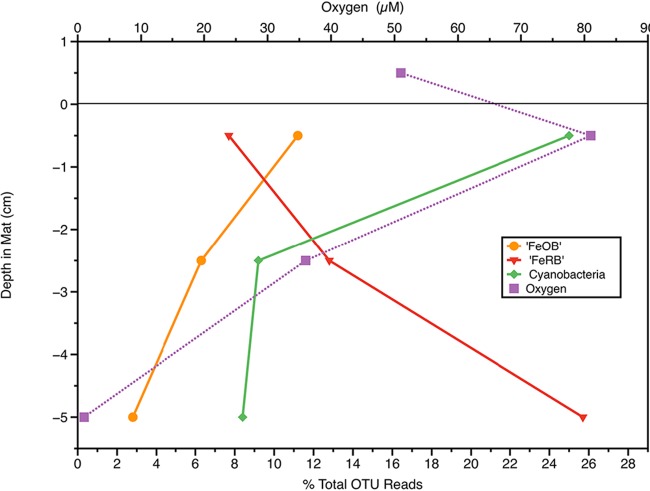
The iron mat associated with the Kuparuk River iron seep was 6 to 7 cm thick and covered by approximately 8 cm of slowly flowing water (see Fig. S2 in the supplemental material). A low-resolution (1- to 2-cm scale) profile of physicochemical and microbial diversity revealed interesting distribution patterns within the mat. The O2 concentration was higher in the surface mat layer (0 to 1 cm) than it was in the overlying water (Fig. 4), but then at a depth of 3 cm, it fell below detection. The 60% increase in O2 concentration in the mat surface compared to that of the overlying water is most likely explained by the high relative abundance of cyanobacterial (and chloroplast) OTU reads in the surface layer, indicating that an active photosynthetic population was producing O2. The relative abundance of FeOB decreased with increasing depth, while the abundance of Deltaproteobacteria reads increased with increasing depth (Fig. 4), consistent with the majority of OTUs of the Deltaproteobacteria being related to known lineages of anaerobes, including the Geobacteraceae. The Fe(II) concentration near the mat surface was 250 μM, while values of 650 and 880 μM were found at depths of 2.5 and 5 cm, respectively. The total cell number in the surface mat layer was 1.1 × 107 ml−1 and increased to 3.1 × 107 ml−1 at a depth of 2.5 cm and 6.7 × 107 ml−1at a depth of 5 cm, while total iron increased from 4.2 mM ml−1 at the surface to around 20 mM ml−1 at the two deeper depths. Together, these results indicate a community structure in which iron oxidation is likely occurring primarily in the surface of the mat, with the potential for iron reduction in deeper regions.
FIG 4.
Low-resolution depth profile in an iron mat from the Kuparuk River iron seep showing the relative percentage of reads for representative groups of bacteria and the O2 concentrations at different depths. The y axis shows depth relative to the mat surface; the air-water interface was about 8 cm above the mat surface. The FeOB and FeRB are inferred based on phylogenetic relationships to groups associated with the respective function. The black horizontal line represents the surface of the mat.
DISCUSSION
The results presented here systematically document the presence of FeOB and their associated communities in the Arctic tundra around TFS. We are not aware of any other published reports of iron mats in the Arctic, and one of us (D.E.) in >20 years of studying communities of FeOB has never witnessed such extensive development of iron mats in freshwater habitats. This abundance suggests that chemosynthetic iron-oxidizing communities can potentially contribute to biogeochemical processes in tundra ecosystems. It is likely a combination of factors that lead to robust iron-cycling communities in these mineral-containing soils. The cold (8 to 11°C) and relatively low-pH (5 to 6.0) waters slow the chemical oxidation of Fe(II) (10). This results in an increased half-life for Fe(II), even in moderately well-oxygenated waters, allowing more opportunity for FeOB to couple iron oxidation to growth. Perhaps most important, the permafrost layer beneath the soil impedes the downward movement of anoxic Fe(II)-rich waters into deeper soil layers or aquifers, resulting in more contact with oxygenated surface waters. This creates a habitat well suited to the close coupling of iron oxidation and reduction and the development of iron mat communities.
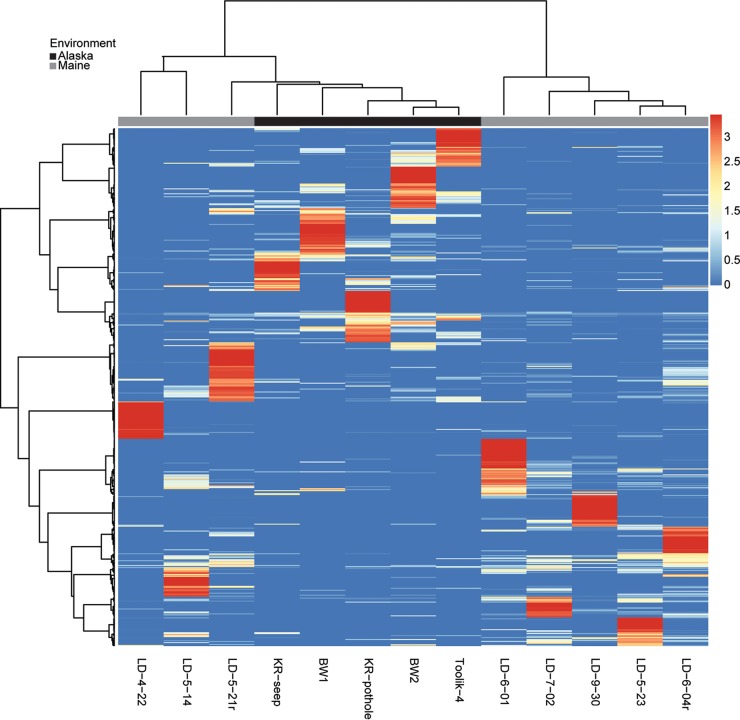
The community composition of tundra iron mats bears an overall similarity to that of temperate iron mats. A comparison of a microbial mat community in Maine that was extensively sampled for community composition over 6 months (18) with TFS samples by minimum entropy decomposition (19) showed that TFS communities clustered together and were more similar to early season communities from the iron mat in Maine (Fig. 5). The specific reasons for clustering of the TFS samples with the early successional community in Maine are not understood; however, the average temperature was 11°C for the early season community in Maine, similar to the temperature for TFS sites, while later in the season, the temperature in Maine rose to 15 to 18°C (18). A more complete analysis of the microbiome (13 sites and 40 samples) of microbial iron mats from Maine, Virginia, Texas, and Wyoming and in Denmark found that TFS iron mats cluster much more closely to these other circumneutral terrestrial sites than they do to iron-oxidizing communities from acidic (pH <4) or marine environments (J. J. Scott and D. Emerson, unpublished data).
FIG 5.
Dendrogram/heatmap based on minimum entropy decomposition (MED) comparing different TFS sites with sites from the Lakeside Drive iron mat in Maine; the data from Maine samples are from reference 18. The values are scaled by taxon relative abundance across all samples. Red indicates a taxon with skewed distribution, in which presence is concentrated in one or two samples; white indicates an even distribution among samples; and blue represents a lower relative abundance or absence.
Morphologically, the iron mats were composed of the sheath and stalk morphotypes typical of the classically described FeOB L. ochracea and G. ferruginea, respectively. Somewhat surprisingly, the overall relative abundance of Gallionellales and Leptothrix spp. ranged from 3 to 10%, whereas in temperate iron seeps, these groups may account for between 25 and 50% of the total population (18, 21, 22). It is not clear why the abundances of known FeOB are reduced in these tundra iron mats. Environmental factors, such as the short growing season, 24 h of daylight, and intense niche competition, might all play roles in this, but further study is necessary to test which factor(s) may be involved. Alternatively, there may be competition with other as-yet-unrecognized FeOB.
One group of interest in this regard is the Comamonadaceae, which is consistently one of the most abundant lineages recovered from the TFS mats. On average, >50% of the total reads classified as Comamonadaceae from the TFS mats belonged to a single OTU, based on a 97% similarity score for the 16S rRNA gene. Oligotyping of this OTU revealed it was composed of seven different oligotypes. One of these oligotypes accounted for between 38 and 70% of the oligotypes from the five different TFS sites, while the remaining oligotypes had more variable distributions (Fig. 6). The functional role of this particular OTU remains unknown. The cultivated lineage most closely related to the abundant iron mat OTU is the iron-reducing bacterium Rhodoferax ferrireducens (23). However, the genus Rhodoferax is metabolically diverse, consisting of species that carry out anoxygenic photosynthesis and chemotrophy (24); furthermore, members of the family Comamonadaceae, which includes Leptothrix species, exhibit a wide range of physiologies (25). Thus, the metabolic function(s) of these prevalent Comamonadaceae OTUs is unclear, and given the diversity of different oligotypes, it is possible there are different strains with different metabolisms. It is certainly possible that they are FeRB; however, they might also represent a novel group of lithotrophic FeOB. If they are indeed a novel group of lithotrophic FeOB, it would be interesting to determine if they couple anoxygenic photosynthesis to iron oxidation or if they are microaerophilic Fe oxidizers capable of direct competition with the Gallionellaceae.
FIG 6.
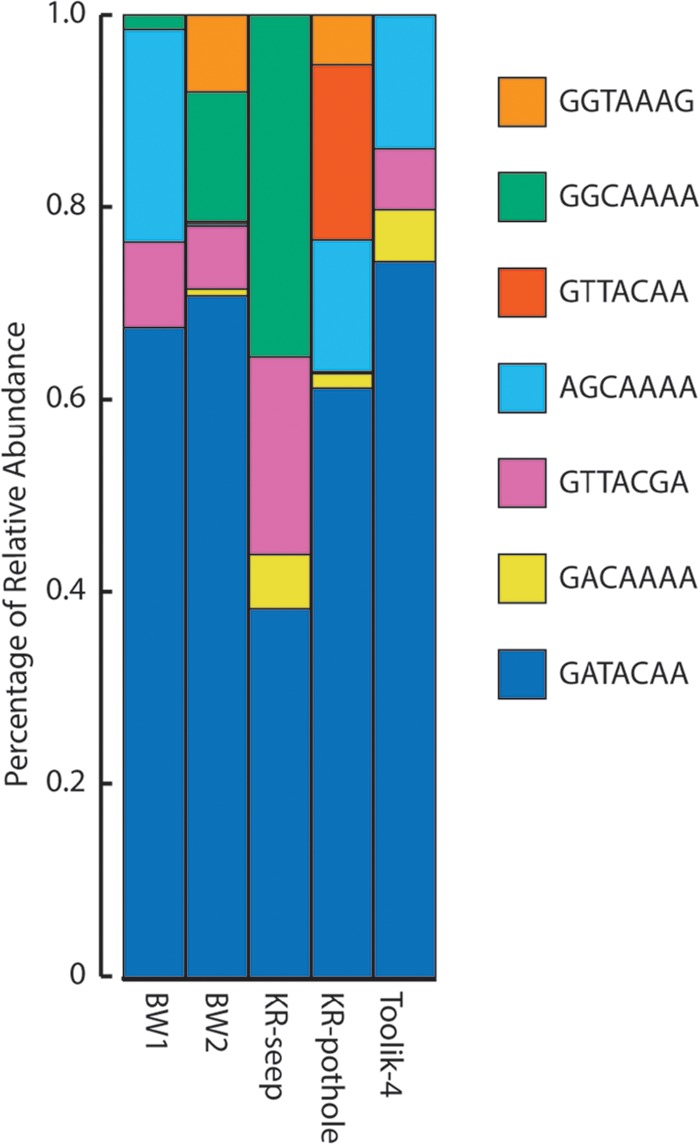
Oligotype analysis of the most abundant Comamonadaceae OTU found in the TFS samples, which were identified at the 97% similarity cutoff. The relative abundances of the seven different oligotypes that compose this OTU are shown.
Another interesting discovery about these tundra iron mats is the presence of active oxygenic photosynthetic microbes. Previous work has suggested that the presence of cyanobacteria or eukaryotic algae in chalybeate waters produce high O2 levels that strongly favor abiotic over biotic Fe oxidation (8). Molecular ecology studies of temperate iron mats generally report that cyanobacteria make up a small percentage of the population (26, 27). A recent study of an alpine spring (Fuschna Spring) in Switzerland found an interesting spatial relationship between a phototrophic iron mat that grew along the edge of the spring channel and an iron mat that grew in its center (28). Despite being separated by a centimeter or less, the populations in the phototrophic mat were quite unique from those in the iron mat community, indicating that there might be close physical proximity of the different communities without much intermixing of populations. In the tundra iron mats, in which both phototrophs and FeOB were observed, there was no discernible spatial separation. Oxygen measurements in some (but not all) mats (Fig. 3 and 4) detected evidence of oxygenic photosynthesis; however, the total O2 concentrations were typically <2-fold greater than the overlying water and never approached saturation. This suggests that the photosynthetic populations are limited in either light or some trace nutrient, for example, phosphorus; thus, O2 production is held in check to a degree that biotic iron oxidation can still compete with abiotic reactions. It is interesting to speculate that the phototrophs may derive some mutual benefit from the presence of FeOB by taking advantage of the large surface areas created by the physical matrices of the iron mat to position themselves in the water column where light is most available.
Methanotrophic and methylotrophic bacteria are other functional groups that have been recognized as being associated with freshwater iron mats (21, 29, 30). Relatives of these quite phylogenetically diverse groups of C-1 oxidizers can account for >10% of the OTUs in some habitats. It is not known whether there are any mutualistic associations between FeOB and methylotrophs or whether high-Fe(II) environments fed by anaerobic waters also tend to have high methane concentrations, leading to a coincidental overlap. In general, the TFS iron mats appeared to be deficient in methylotrophs, with four of the five iron mats having <2% of the total OTUs associated with methylotrophic groups. The one exception was the Kuparuk River seep, where the OTUs related to methylotrophs made up nearly 5% of the total sequences from mid-depth of the mat. This site also had the highest methane concentration (33 μM) that was measured in the overlaying waters, whereas at the other sites, the methane levels were <5 μM.
Iron cycling.
Microbial iron mats can be and often are sites of localized iron cycling, in which the iron oxides produced by FeOB are used by Fe-reducing bacteria (FeRB) as a terminal electron acceptor to carry out anaerobic respiration coupled to the mineralization of organic matter (15, 27, 31, 32). The results presented here confirm that these tundra iron mats harbor active populations of FeRB. Anaerobic incubation experiments showed that the addition of acetate to surficial iron mats resulted in detectable Fe reduction within 24 to 48 h, a response that is comparable to the most rapid response measured in other communities. Phylogenetic analysis identified several putative FeRB as being important members of these iron mat communities. For example, members of the Deltaproteobacteria were relatively abundant in all the mats and most prevalent in the depth profile at the Kuparuk River iron seep.
Relevance to biogeochemistry in tundra ecosystems.
The most extensive work on Fe reduction in tundra soils has been done by Lipson and colleagues (5, 33, 34), who investigated anaerobic processes in Arctic permafrost soils near Barrow, AK. These studies showed that anaerobic respiration coupled to Fe oxide reduction is a primary terminal electron-accepting process, accounting for 40 to 60% of the ecosystem respiration (5, 33, 34). Detailed mineralogical analysis by these authors revealed an abundance of poorly crystalline readily reducible iron oxides in these moist soils. A follow-on study that used metagenomics to investigate these Fe-reducing communities discovered that genes encoding decaheme cytochromes related to those found in Gallionella and Sideroxydans were present (35). Another study of a subarctic soil in Alaska (64°51′N, 163°39′W) identified a member of the Gallionellaceae as among the 10 most abundant OTUs in multiple soil cores in what was otherwise a soil community dominated by Acidobacteria, Alphaproteobacteria, and Actinobacteria (36). These findings suggest that lithotrophic iron oxidation is either happening directly in these soils, or there are Fe-oxidizing communities in close proximity. It is worth noting that the relative abundance of Acidobacteria in all the sampled TFS iron mats was low (<2%), and Actinobacteria were barely detected; yet, these phyla are generally abundant in tundra soils (36, 37). This indicates that soils that may be closely adjacent to iron mats harbor quite different communities that likely have different physiological requirements and functional capabilities. Nonetheless, the important point is that biogenic iron oxidation may be contributing readily reducible iron oxides to these soils and helping to stimulate anaerobic respiration coupled to Fe reduction, as has been shown in temperate systems (38).
In terms of contributing to the carbon cycle in the tundra, the overall contribution to primary production by Fe(II)-fueled chemolithoautotrophy is likely dwarfed by the associated tundra plant and algal communities. Nonetheless, chemosynthetic iron-oxidizing communities might play an important role in other aspects of biogeochemical cycling in the Arctic. As discussed above, rapid iron cycling might enhance the biomineralization of organic carbon and contribute to overall ecosystem respiration. This has important implications for the fate of the large amounts of organic carbon that are stored in Arctic soils (4). A major concern is that as the Arctic warms in the coming millennia, a significant fraction of this organic matter might be mineralized to methane under anaerobic conditions, thereby creating a positive feedback to greenhouse gas-driven global warming. Thermodynamic considerations show that iron reduction will outcompete methanogenesis as an anaerobic process (39), and in situ measurements bear this out (40–42). These relationships are further complicated by chemical and biological interactions of methanogens and Fe reducers (43). In any event, if iron oxidation and reduction are widespread in the Arctic tundra, the effects of iron cycling and its potential to suppress methanogenesis will need to be taken into account.
Finally, an indirect mechanism whereby microbial iron mats might be important in influencing the carbon cycle is through interaction with the phosphorus cycle. Phosphorus is an important limiting nutrient in tundra lakes and streams (44), and it is well known that iron oxides readily adsorb phosphates. Recent work has shown that the presence of biogenic iron oxides can be very effective at removing phosphorus from natural waters (7, 45, 46). Thus, microbial iron mats might serve as an important sink for the hydrologic transport of P and potentially cause limitation of primary P production. Of course, P that is bound to biogenic oxides may also be released upon iron reduction, adding another dimension of complexity to this biogeochemical process. In summary, there are a number of reasons that further investigation of the iron cycle in tundra soils is warranted.
Supplementary Material
ACKNOWLEDGMENTS
We thank Beth Orcutt for providing the methane analysis. D.E. thanks the TFS LTER (NSF/DEB/LTER-1026843) for financial support for travel and logistical support at the TFS.
This study was partially supported by NSF grant OCE-1155754.
All opinions, findings, conclusions, and recommendations expressed in this report are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02832-15.
REFERENCES
- 1.McGuire AD, Anderson LG, Christensen TR, Dallimore S, Guo L, Hayes DJ, Heimann M, Lorenson TD, Macdonald RW, Roulet N. 2009. Sensitivity of the carbon cycle in the Arctic to climate change. Ecol Monogr 79:523–555. doi: 10.1890/08-2025.1. [DOI] [Google Scholar]
- 2.Chapin FS III, Mcguire AD, Randerson J, Pielke R, Baldocchi D, Hobbie SE, Roulet N, Eugster W, Kasischke E, Rastteter EB, Zimov SA, Running SW. 2000. Arctic and boreal ecosystems of western North America as components of the climate system. Global Change Biol 6:211–223. doi: 10.1046/j.1365-2486.2000.06022.x. [DOI] [PubMed] [Google Scholar]
- 3.Hinzman LD, Bettez ND, Bolton WR, Chapin FS, Dyurgerov MB, Fastie CL, Griffith B, Hollister RD, Hope A, Huntington HP, Jensen AM, Jia GJ, Jorgenson T, Kane DL, Klein DR, Kofinas G, Lynch AH, Lloyd AH, McGuire AD, Nelson FE, Oechel WC, Osterkamp TE, Racine CH, Romanovsky VE, Stone RS, Stow DA, Sturm M, Tweedie CE, Vourlitis GL, Walker MD, Walker DA, Webber PJ, Welker JM, Winker KS, Yoshikawa K. 2005. Evidence and implications of recent climate change in northern Alaska and other Arctic regions. Clim Change 72:251–298. doi: 10.1007/s10584-005-5352-2. [DOI] [Google Scholar]
- 4.Tarnocai C, Canadell JG, Schuur EAG, Kuhry P, Mazhitova G, Zimov S. 2009. Soil organic carbon pools in the northern circumpolar permafrost region. Global Biogeochem Cycles 23:GB2023. doi: 10.1029/2008GB003327. [DOI] [Google Scholar]
- 5.Lipson DA, Jha M, Raab TK, Oechel WC. 2010. Reduction of iron (III) and humic substances plays a major role in anaerobic respiration in an Arctic peat soil. J Geophys Res 115:G00I06. doi: 10.1029/2009JG001147. [DOI] [Google Scholar]
- 6.Emerson D, Fleming EJ, McBeth JM. 2010. Iron-oxidizing bacteria: an environmental and genomic perspective. Annu Rev Microbiol 64:561–583. doi: 10.1146/annurev.micro.112408.134208. [DOI] [PubMed] [Google Scholar]
- 7.Rentz JA, Turner IP, Ullman JL. 2009. Removal of phosphorus from solution using biogenic iron oxides. Water Res 43:2029–2035. doi: 10.1016/j.watres.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 8.Emerson D, Weiss JV. 2004. Bacterial iron oxidation in circumneutral freshwater habitats: findings from the field and the laboratory. Geomicrobiol J 21:405–414. doi: 10.1080/01490450490485881. [DOI] [Google Scholar]
- 9.Stumm W, Lee GF. 1961. Oxygenation of ferrous iron. Ind Eng Chem 53:143–146. [Google Scholar]
- 10.Millero FJ, Sotolongo S, Izaguirre M. 1987. The oxidation kinetics of Fe(II) in seawater. Geochim Cosmochim Acta 51:793–801. doi: 10.1016/0016-7037(87)90093-7. [DOI] [Google Scholar]
- 11.Liang L, McNabb JA, Paulk JM, Gu B, McCarthy JF. 1993. Kinetics of iron (II) oxygenation at low partial pressure of oxygen in the presence of natural organic matter. Environ Sci Technol 27:1864–1870. doi: 10.1021/es00046a014. [DOI] [Google Scholar]
- 12.Jones BM, Kolden CA, Jandt R, Abatzoglou JT, Urban F, Arp DC. 2007. Fire behavior, weather, and burn severity of the 2007 Anaktuvuk River tundra fire, North Slope, Alaska. Arct Antarct Alp Res 41:309–316. [Google Scholar]
- 13.Maita TR, Lalli CM. 1984. A manual of chemical and biological methods for seawater analysis. Pergamon Press, New York, NY. [Google Scholar]
- 14.Stookey LL. 1970. Ferrozine—a new spectrophotometric reagent for iron. Anal Chem 42:779–781. doi: 10.1021/ac60289a016. [DOI] [Google Scholar]
- 15.Emerson D, Revsbech NP. 1994. Investigation of an iron-oxidizing microbial mat community located near Aarhus, Denmark: field studies. Appl Environ Microbiol 60:4022–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joye SB, Boetius A, Orcutt BN, Montoya JP, Schulz HN, Erickson MJ, Lugo SK. 2004. The anaerobic oxidation of methane and sulfate reduction in sediments from Gulf of Mexico cold seeps. Chem Geol 205:219–238. doi: 10.1016/j.chemgeo.2003.12.019. [DOI] [Google Scholar]
- 17.Scott JJ, Breier JA, Luther GW, Emerson D. 2015. Microbial iron mats at the Mid-Atlantic Ridge and evidence that Zetaproteobacteria may be restricted to iron-oxidizing marine systems. PLoS One 10:e0119284. doi: 10.1371/journal.pone.0119284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleming EJ, Cetinic I, Chan CS, King DW, Emerson D. 2014. Ecological succession among iron-oxidizing bacteria. ISME J 8:804–815. doi: 10.1038/ismej.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, Sogin ML. 2013. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4:1111–1119. doi: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleming EJ, Langdon AE, Martinez-Garcia M, Stepanauskas R, Poulton NJ, Masland EDP, Emerson D. 2011. What's new is old: resolving the identity of Leptothrix ochracea using single cell genomics, pyrosequencing and FISH. PLoS One 6:e17769. doi: 10.1371/journal.pone.0017769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quaiser A, Bodi X, Dufresne A, Naquin D, Francez A-J, Dheilly A, Coudouel S, Pedrot M, Vandenkoornhuyse P. 2014. Unraveling the stratification of an iron-oxidizing microbial mat by metatranscriptomics. PLoS One 9:e102561. doi: 10.1371/journal.pone.0102561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Sickinger M, Ciobota V, Herrmann M, Rasch H, Rösch P, Popp J, Küsel K. 2014. Revealing the microbial community structure of clogging materials in dewatering wells differing in physico-chemical parameters in an open-cast mining area. Water Res 63:222–233. doi: 10.1016/j.watres.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 23.Finneran KT, Johnsen CV, Lovley DR. 2003. Rhodoferax ferrireducens sp. nov., a psychrotolerant, facultatively anaerobic bacterium that oxidizes acetate with the reduction of Fe(III). Int J Syst Evol Microbiol 53:669–673. doi: 10.1099/ijs.0.02298-0. [DOI] [PubMed] [Google Scholar]
- 24.Kaden R, Sproer C, Beyer D, Krolla-Sidenstein P. 2014. Rhodoferax saidenbachensis sp. nov., a psychrotolerant, very slowly growing bacterium within the family Comamonadaceae, proposal of appropriate taxonomic position of Albidiferax ferrireducens strain T118T in the genus Rhodoferax and emended description of the genus Rhodoferax. Int J Syst Evol Microbiol 64:1186–1193. doi: 10.1099/ijs.0.054031-0. [DOI] [PubMed] [Google Scholar]
- 25.Willems A. 2014. The family Comamonadaceae, p 777–851. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes. 4th ed: alphaproteobacteria and betaproteobacteria Springer-Verlag, Berlin, Germany. [Google Scholar]
- 26.Bruun A-M, Finster K, Gunnlaugsson HP, Nørberg P, Friedrich MW. 2010. A comprehensive investigation on iron cycling in a freshwater seep including microscopy, cultivation and molecular community analysis. Geomicrobiol J 27:15–34. doi: 10.1080/01490450903232165. [DOI] [Google Scholar]
- 27.Roden EE, McBeth JM, Blothe M, Percak-Dennett EM, Fleming EJ, Holyoke RR, Luther G III, Emerson D, Schieber J. 2012. The microbial ferrous wheel in a neutral pH groundwater seep. Front Microbiol 3:172. doi: 10.3389/fmicb.2012.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hegler F, Losekann-Behrens T, Hanselmann K, Behrens S, Kappler A. 2012. Influence of seasonal and geochemical changes on the geomicrobiology of an iron carbonate mineral water spring. Appl Environ Microbiol 78:7185–7196. doi: 10.1128/AEM.01440-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fru EC, Piccinelli P, Fortin D. 2012. Insights into the global microbial community structure associated with iron oxyhydroxide minerals deposited in the aerobic biogeosphere. Geomicrobiol J 29:587–610. doi: 10.1080/01490451.2011.599474. [DOI] [Google Scholar]
- 30.Kato S, Chan C, Itoh T, Ohkuma M. 2013. Functional gene analysis of freshwater iron-rich flocs at circumneutral pH and isolation of a stalk-forming microaerophilic iron-oxidizing bacterium. Appl Environ Microbiol 79:5283–5290. doi: 10.1128/AEM.03840-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blöthe M, Roden EE. 2009. Microbial iron redox cycling in a circumneutral-pH groundwater seep. Appl Environ Microbiol 75:468–473. doi: 10.1128/AEM.01817-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato S, Kikuchi S, Kashiwabara T, Takahashi Y, Suzuki K, Itoh T, Ohkuma M, Yamagishi A. 2012. Prokaryotic abundance and community composition in a freshwater iron-rich microbial mat at circumneutral pH. Geomicrobiol J 29:896–905. doi: 10.1080/01490451.2011.635763. [DOI] [Google Scholar]
- 33.Lipson DA, Zona D, Raab TK, Bozzolo F, Mauritz M, Oechel WC. 2012. Water-table height and microtopography control biogeochemical cycling in an Arctic coastal tundra ecosystem. Biogeosciences 9:577–591. doi: 10.5194/bg-9-577-2012. [DOI] [Google Scholar]
- 34.Lipson DA, Raab TK, Goria D, Zlamal J. 2013. The contribution of Fe(III) and humic acid reduction to ecosystem respiration in drained thaw lake basins of the Arctic Coastal Plain. Global Biogeochem Cycles 27:399–409. doi: 10.1002/gbc.20038. [DOI] [Google Scholar]
- 35.Lipson DA, Haggerty JM, Srinivas A, Raab TK, Sathe S, Dinsdale EA. 2013. Metagenomic insights into anaerobic metabolism along an Arctic peat soil profile. PLoS One 8:e64659. doi: 10.1371/journal.pone.0064659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim HM, Jung JY, Yergeau E, Hwang CY, Hinzman L, Nam S, Hong SG, Kim O-S, Chun J, Lee YK. 2014. Bacterial community structure and soil properties of a subarctic tundra soil in Council, Alaska. FEMS Microbiol Ecol 89:465–475. doi: 10.1111/1574-6941.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jansson JK, Taş N. 2014. The microbial ecology of permafrost. Nat Rev Microbiol 12:414–425. doi: 10.1038/nrmicro3262. [DOI] [PubMed] [Google Scholar]
- 38.Weiss J, Emerson D, Megonigal J. 2004. Geochemical control of microbial Fe (III) reduction potential in wetlands: comparison of the rhizosphere to non-rhizosphere soil. FEMS Microbiol Ecol 48:89–100. doi: 10.1016/j.femsec.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 39.Megonigal JP, Hines ME, Visscher PT. 2004. Anaerobic metabolism: linkages to trace gases and aerobic processes, p 317–424. In Schlesinger WH. (ed), Biogeochemistry. Elsevier-Pergamon Press, Oxford, United Kingdom. [Google Scholar]
- 40.Jäckel U, Schnell S. 2000. Suppression of methane emission from rice paddies by ferric iron fertilization. Soil Biol Biochem 32:1811–1814. [Google Scholar]
- 41.Lueders T, Friedrich MW. 2002. Effects of amendment with ferrihydrite and gypsum on the structure and activity of methanogenic populations in rice field soil. Appl Environ Microbiol 68:2484–2494. doi: 10.1128/AEM.68.5.2484-2494.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Bodegom P, Stams F, Mollema L, Boeke S, Leffelaar P. 2001. Methane oxidation and the competition for oxygen in the rice rhizosphere. Appl Environ Microbiol 67:3586–3597. doi: 10.1128/AEM.67.8.3586-3597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bond DR, Lovley DR. 2002. Reduction of Fe(III) oxide by methanogens in the presence and absence of extracellular quinones. Environ Microbiol 4:115–124. doi: 10.1046/j.1462-2920.2002.00279.x. [DOI] [PubMed] [Google Scholar]
- 44.Slavik K, Peterson BJ, Deegan LA, Bowden WB, Hershey AE, Hobbie JE. 2004. Long-term response of the Kuparuk River ecosystem to phosphorus fertilization. Ecology 83:939–954. [Google Scholar]
- 45.Baken S, Salaets P, Desmet N, Seuntjens P, Vanlierde E, Smolders E. 2015. Oxidation of iron causes removal of phosphorus and arsenic from stream water in groundwater-fed lowland catchments. Environ Sci Technol.150206152358002. [DOI] [PubMed] [Google Scholar]
- 46.Baken S, Verbeeck M, Verheyen D, Diels J, Smolders E. 2015. Phosphorus losses from agricultural land to natural waters are reduced by immobilization in iron-rich sediments of drainage ditches. Water Res 71:160–170. doi: 10.1016/j.watres.2015.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.