Abstract
Magnetotactic bacteria are capable of forming nanosized, membrane-enclosed magnetosomes under iron-rich and oxygen-limited conditions. The complete genomic sequence of Magnetospirillum gryphiswaldense strain MSR-1 has been analyzed and found to contain five fur homologue genes whose protein products are predicted to be involved in iron homeostasis and the response to oxidative stress. Of these, only the MGMSRv2_3149 gene (irrB) was significantly downregulated under high-iron and low-oxygen conditions, during the transition of cell growth from the logarithmic to the stationary phase. The encoded protein, IrrB, containing the conserved HHH motif, was identified as an iron response regulator (Irr) protein belonging to the Fur superfamily. To investigate the function of IrrB, we constructed an irrB deletion mutant (ΔirrB). The levels of cell growth and magnetosome formation were lower in the ΔirrB strain than in the wild type (WT) under both high-iron and low-iron conditions. The ΔirrB strain also showed lower levels of iron uptake and H2O2 tolerance than the WT. Quantitative real-time reverse transcription-PCR analysis indicated that the irrB mutation reduced the expression of numerous genes involved in iron transport, iron storage, heme biosynthesis, and Fe-S cluster assembly. Transcription studies of the other fur homologue genes in the ΔirrB strain indicated complementary functions of the Fur proteins in MSR-1. IrrB appears to be directly responsible for iron metabolism and homeostasis and to be indirectly involved in magnetosome formation. We propose two IrrB-regulated networks (under high- and low-iron conditions) in MSR-1 cells that control the balance of iron and oxygen metabolism and account for the coexistence of five Fur homologues.
INTRODUCTION
Iron is an essential nutrient for most organisms. It is involved in crucial biological processes such as nitrogen fixation, oxygen transport, central metabolism, respiration, gene regulation, and DNA biosynthesis (1). Under aerobic conditions at a neutral pH, iron is metabolically unavailable because of its insoluble state (2, 3). In cells, ferrous iron (Fe2+) and ferric iron (Fe3+) function, respectively, as the electron donor and electron acceptor, maintaining a compatible redox potential for many biochemical reactions (3). Excess ferrous iron catalyzes the formation of reactive oxygen species (ROS) via the Fenton reaction, resulting in cell damage or death (4). Bacteria and other microorganisms have developed various systems to maintain iron homeostasis, the most studied of which is the ferric uptake regulator (Fur) system. In Escherichia coli and Pseudomonas aeruginosa, iron binds Fur so as to occupy its promoter and inhibit the expression of Fur-controlled genes (5, 6).
Magnetospirillum gryphiswaldense MSR-1, a Gram-negative alphaproteobacterium, is capable of synthesizing unique intracellular magnetic nanoparticles composed of Fe3O4, termed magnetosomes (7). The biosynthesis of magnetosomes, whose membrane-bound magnetite nanocrystals are aligned in chain-like structures within the cell, requires the cell to assimilate a large amount of iron from the environment (8) while avoiding the potential toxic effects of surplus intracellular iron. Our de novo sequencing of the MSR-1 genome revealed the presence of five Fur homologues differing from those of E. coli and other nonmagnetotactic bacteria (9). The five candidate Fur proteins are assignable to three different subfamilies of the Fur superfamily (10). Only one of the proteins (MGMSRv2_3137) is considered to be the real Fur protein; we have shown that Fur in M. gryphiswaldense directly regulates the expression of several key genes involved in iron transport and oxygen metabolism and also functions in magnetosome formation (11). Another of the five proteins (MGMSRv2_2136) belongs to the zinc uptake regulator (Zur) family (responsive to zinc). The other three proteins (MGMSRv2_1721 [IrrA], MGMSRv2_3149 [IrrB], and MGMSRv2_3660 [IrrC]) are most closely related to the iron response regulator (Irr) family (responsive to heme). The physiological functions of the five homologues and the relationships among the five Fur proteins remain poorly understood.
We recently generated magnetosome-forming and non-magnetosome-forming variants of MSR-1 cells by modulating the iron concentration in the medium and performed complete transcriptome analysis of the two variants. The expression of the MGMSRv2_3149 gene (irrB) was significantly downregulated under iron-rich and oxygen-poor (hypoxic) conditions. In the present study, to clarify the role of irrB in iron regulation and magnetite biomineralization in MSR-1, we successfully constructed its null strain and complemented strain. The mutant showed reduced magnetosome formation, a phenotype similar to that of the MGMSRv2_3137 gene (a fur-like gene) deletion mutant. In contrast to Fur, IrrB may indirectly regulate genes involved in heme biosynthesis, iron storage, or yet unknown pathways under high-iron conditions. We also performed quantitative real-time reverse transcription-PCR (RT-qPCR) in order to assess and compare the transcription levels of other fur-like genes in the mutant and wild-type (WT) cells. The findings indicate that IrrB in MSR-1 helps control both cell growth and magnetosome formation with varying iron concentrations. Coordination among the Fur proteins maintains the balance of iron and oxygen levels required for magnetosome formation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. M. gryphiswaldense strain MSR-1 was cultured in sodium lactate medium (SLM) at 30°C with shaking at 100 rpm (12). Sterile ferric citrate was added as an iron source after autoclaving. E. coli strains were cultured in Luria broth (LB) at 37°C. The antibiotics used were as follows: for E. coli, 100 μg/ml ampicillin (Amp), 50 μg/ml kanamycin (Km), and 20 μg/ml gentamicin (Gm); for M. gryphiswaldense, Km, Gm, tetracycline (Tc), and nalidixic acid (Nx), each at 5 μg/ml. The growth (expressed as the optical density at 565 nm [OD565]) and magnetic response (Cmag) of MSR-1 cells were measured as described previously (13).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| M. gryphiswaldense | ||
| MSR-1 | WT; Nxr | DSM 6361 |
| ΔirrB mutant | irrB-defective mutant; Nxr Gmr | This study |
| CF3149 | Complemented strain of the ΔirrB mutant; Nxr Gmr Tcr | This study |
| Δfur mutant | fur-defective mutant; Nxr Gmr | 11 |
| E. coli | ||
| DH5α | endA1 hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA (Nalr) recA1 Δ(lacZYA-argF)U169 deoR[ϕ80dlacZΔM15] | 41 |
| S17-1 | thi endA recA hsdR with RP4-2-Tc::Mu-Km::Tn7 integrated into the chromosome; Smr | 42 |
| Plasmids | ||
| pMD 18 T-simple | Cloning vector; Ampr | TaKaRa |
| pUCGm | pUC1918 carrying the aacC1 gene, Gmr | 43 |
| pUX19 | Suicide vector for M. gryphiswaldense MSR-1; Kmr | 44 |
| pUXF | pUX19 containing Gm cassette, irrB upstream and downstream regions; Kmr Gmr | This study |
| pRK415 | Broad-host-range cloning vector; Tcr | 45 |
| pRK415F | pRK415 containing irrB from M. gryphiswaldense MSR-1 | This study |
Construction of the irrB deletion mutant and its complemented strain.
For the construction of the irrB deletion mutant (ΔirrB), fragments 1,110 bp upstream and 1,114 bp downstream of irrB were amplified using primer sets F3149uF/F3149uR and F3149dF/F3149dR. The amplified upstream fragment digested by BamHI and SacI, the amplified downstream fragment digested by SacI and KpnI, and the gentamicin resistance cassette from pUCGm were ligated into the suicide vector pUX19 to form pUXF. By using E. coli S17-1 as the donor strain, pUXF was introduced into WT MSR-1 by biparental conjugation. Colonies were screened and were selected by Gmr and Nxr, and double crossover was confirmed by PCR (14, 15). To construct the complemented strain, irrB was amplified by primer set f3149-F/f3149-R and was then ligated into the HindIII and KpnI sites of pRK415. The resulting plasmid, pRKF, was introduced into the ΔirrB strain by biparental conjugation as described above. The Gmr Tcr Nxr colonies were confirmed by PCR, and the complemented strain was termed CF3149.
TEM.
Cells were cultured in SLM for 15 h, applied to copper grids, washed twice with double-distilled H2O, and observed directly by transmission electron microscopy (TEM) (model JEM-1230; JEOL; Tokyo, Japan). The numbers and diameters of magnetosomes were analyzed statistically using ImageJ (National Institutes of Health, Bethesda, MD, USA), a Java-based image-processing program (13).
Iron content.
The WT MSR-1, ΔirrB, and CF3149 strains were grown in SLM supplemented with 20, 40, or 60 μM ferric citrate at 30°C for 24 h and were harvested by centrifugation (8,000 × g, 5 min). The precipitate was digested by nitric acid (13, 14), and the total cellular iron content was measured by inductively coupled plasma–optical emission spectrometry (ICP-OES) (Optima 5300 DV system; Perkin-Elmer, Waltham, MA, USA).
H2O2 tolerance.
The three strains were cultured in SLM without ferric citrate, inoculated into 50 ml SLM containing 200, 300, 400, or 500 μM H2O2, and cultured for 24 h at 30°C with shaking. The OD565, reflecting cell density, was measured using a spectrophotometer. The experiments were performed in triplicate.
RNA extraction and RT-qPCR.
The WT and ΔirrB strains were grown in SLM without ferric citrate to an OD565 of ∼0.7. The culture was split; 30 μM 2,2-dipyridyl (DIPy) was added to one half and 60 μM ferric citrate to the other. Growth was continued for 2 h at 30°C, and the cells were harvested by centrifugation (12,000 × g, 1 min, 4°C). Total RNA was isolated using TRIzol reagent (Tiangen Biotech, Beijing, China) and was reverse transcribed into cDNA using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega, Madison, WI, USA) as described previously (13). mRNA transcripts were quantified by quantitative real-time RT-PCR (RT-qPCR), with the rpoC gene (which encodes RNA polymerase subunit β′) as an internal control. RT-qPCR was performed with a LightCycler 480 RT-PCR system and a LightCycler 480 SYBR green I Master kit (Roche, Mannheim, Germany) according to the manufacturer's instructions. Gene names and numbers, encoded proteins, and primer names and sequences are listed in Tables S1 and S2 in the supplemental material. The transcription levels of selected genes were determined by the 2−ΔΔCp method, where Cp is the crossing point, the point at which the fluorescense rises appreciably above the background fluorescence (13, 16).
RESULTS
Comparison of amino acid sequences of Fur proteins from various bacterial species.
We focused on irrB because transcriptome data analysis identified it as the gene with the most striking differential expression under high-iron versus low-iron conditions. The irrB gene in MSR-1 has 420 bp and encodes 139 amino acid residues. It is not located in the “magnetosome island” (MAI). In the MSR-1 genome, the neighbors of the IrrB protein are trimethylamine N-oxide (TMAO) reductase I (MGMSRv2_3148) and a conserved protein (MGMSRv2_3150) with unknown function. The bacterial localization prediction tool PSORTb (www.psort.org/psortb/) indicates that IrrB is located in the cytoplasm. Sequence alignment of Fur-like proteins using the MicroScope platform (www.genoscope.cns.fr/agc/microscope) showed 61.4%, 40.2%, 26.8%, and 15.0% sequence identity of IrrB with IrrA, IrrC, Fur, and Zur, respectively.
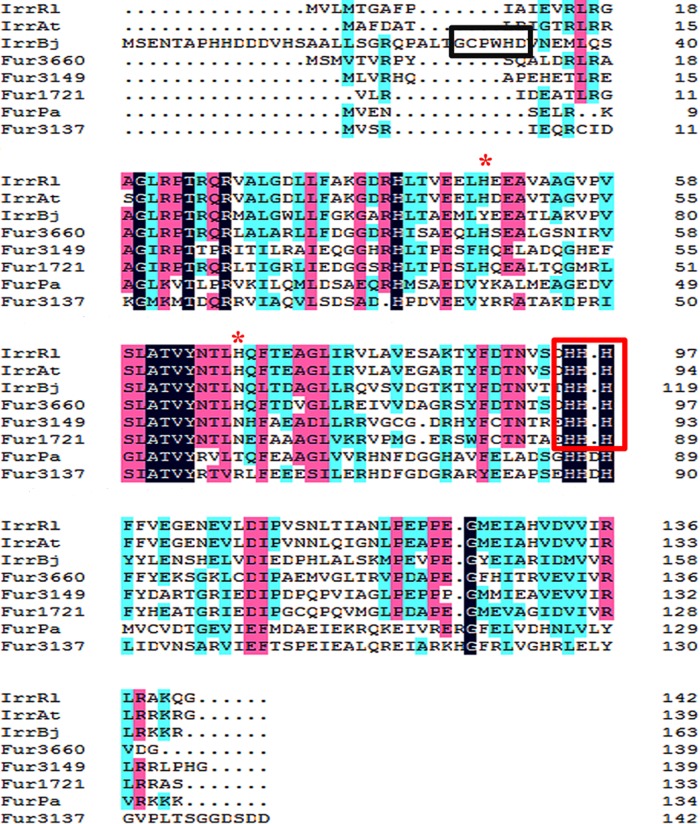
The amino acid sequences of Fur proteins from M. gryphiswaldense MSR-1 (including IrrA, IrrB, IrrC, and Fur) and P. aeruginosa (Fur-Pa) and Irr proteins from Bradyrhizobium japonicum (Irr-Bj), Rhizobium leguminosarum (Irr-Rl), and Agrobacterium tumefaciens (Irr-At) were aligned using the ClustalX software program (Conway Institute, University College Dublin, Dublin, Ireland). BLASTP analysis revealed high identities of the IrrB amino acid sequence with those of Irr-Rl and Irr-At (48%) and moderate identities with those of Irr-Bj (41%) and Fur-Pa (30%). Fur and Fur-Pa contain the characteristic Fe-binding motif His-His-Asp-His (Fig. 1), which is conserved in all Fur/Mur orthologues in alphaproteobacteria (17). IrrA, IrrB, and IrrC contain the histidine residue motif (HHH) (outlined in red in Fig. 1) that binds ferrous heme in Irr-Bj, Irr-At, and Irr-Rl, but not the heme regulatory motif (HRM) (amino acid residues GCPWHD; outlined in black in Fig. 1) that binds ferric heme in Irr-Rl (18). The HHH motif is conserved in most Irr proteins, indicating that IrrA, IrrB, and IrrC may belong to this group. Both Irr-Rl and Irr-At have a second heme-binding site that consists of H48 and H68 in R. leguminosarum and H45 and H65 in A. tumefaciens. H48 and H68 are involved in the oligomerization of Irr-Rl, whereas H45 and H65 are involved in the repression function of Irr-At (18, 19). IrrC retains the two histidine sites, but IrrA and IrrB have only H41 and H45, respectively, in the second heme-binding site (Fig. 1, red asterisks).
FIG 1.
Primary sequence alignment of Fur and Irr proteins by the ClustalW program. Four Fur homologues were from M. gryphiswaldense MSR-1: MGMSRv2_3137 (Fur; GenBank accession no. YP_008939008), MGMSRv2_1721 (IrrA; GenBank accession no. YP_008937596), MGMSRv2_3149 (IrrB; GenBank accession no. YP_008939020), and MGMSRv2_3660 (IrrC; GenBank accession no. YP_008939529). One related Fur sequence was from P. aeruginosa: Fur-Pa (GenBank accession no. NP_253452). Three related Irr sequences were from B. japonicum (Irr-Bj; accession no. YP_005605653), R. leguminosarum (Irr-Rl; accession no. WP_017962725), and A. tumefaciens (Irr-At; accession no. WP_003493135). Black, pink, or blue shading indicates that eight, six, or fewer than five proteins, respectively, share the same amino acids at a given site. The conserved histidine residue motif (HHH) is boxed in red. The heme regulatory motif is boxed in black. The second heme-binding site is indicated by red asterisks.
Reductions in the growth rate and the level of magnetosome formation in an irrB-deficient strain.
To examine the role of IrrB in cell growth and magnetosome formation, we constructed an irrB-deficient strain (ΔirrB) and a complemented strain with irrB (CF3149). The WT, ΔirrB, and CF3149 strains were cultured in a medium supplemented with 60 μM ferric citrate (high-iron condition) or 30 μM 2,2-dipyridyl (DIPy) (low-iron condition). Under low-iron conditions, the growth rate of the ΔirrB strain was roughly half that of the WT and CF3149 (Fig. 2A). After 36 h of incubation, the OD565 of the ΔirrB strain (0.685 ± 0.156) was roughly half those of the WT and CF3149. Under high-iron conditions, the growth rate of the ΔirrB strain was 1.46-fold lower than those of the WT and CF3149, and the highest OD565 value of the ΔirrB strain (0.934 ± 0.094) was somewhat lower than those of the WT (1.459 ± 0.019) and CF3149 (1.157 ± 0.017) (Fig. 2B). These findings indicate that irrB deletion results in a reduced growth rate, particularly under low-iron conditions. Iron may function as a signal to activate IrrB. The growth rate was inhibited by low iron concentrations and IrrB deficiency.
FIG 2.
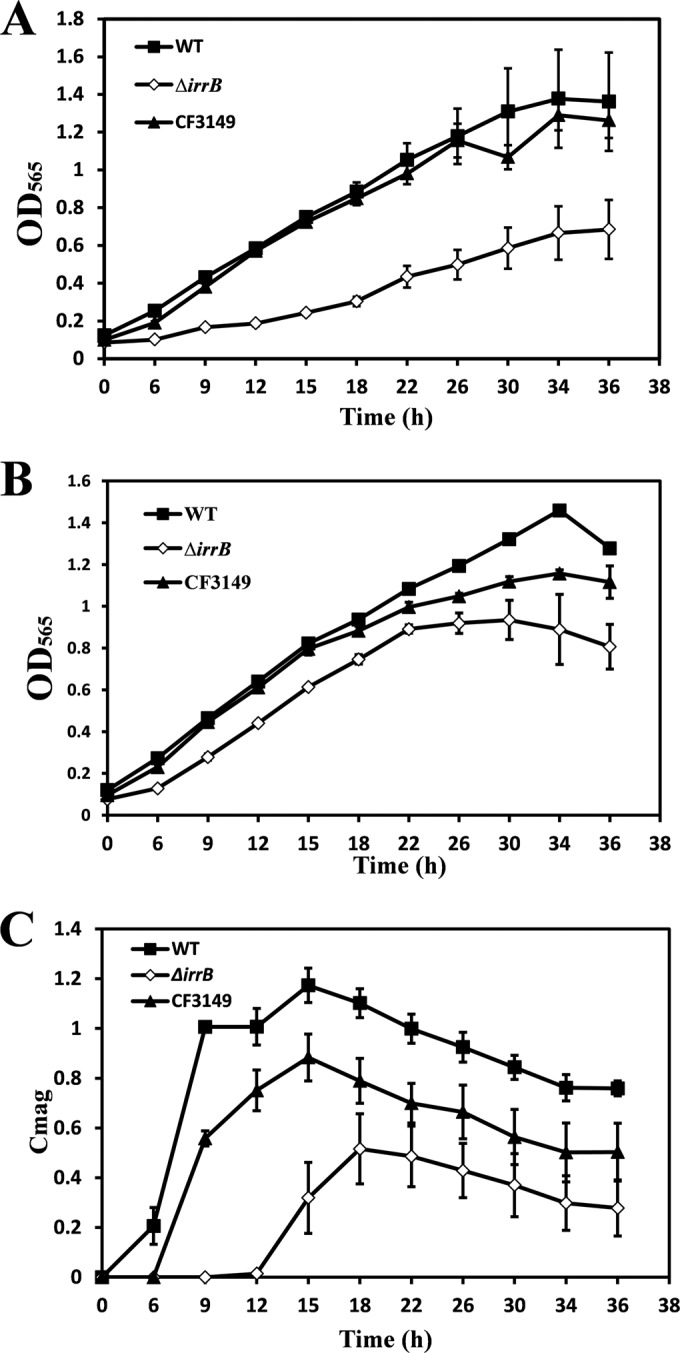
(A and B) Growth rates of the MSR-1 WT, ΔirrB (irrB-deficient mutant), and CF3149 strains in culture with 30 μM DIPy (low-iron conditions) (A) or 60 μM ferric citrate (high-iron conditions) (B). (C) Magnetic responses (Cmag) under high-iron conditions. The ΔirrB strain showed clear reductions in the levels of growth and magnetosome formation under low-iron conditions.
Under low-iron conditions, magnetic response (Cmag) values were zero for all three strains. Under high-iron conditions, the WT absorbed a large amount of iron, which was used for magnetosome synthesis; the maximal Cmag in these cells was ∼1.2. In contrast, the maximal Cmag of the ΔirrB strain was 0.516 ± 0.140 at 18 h, and this strain first produced magnetism at 12 h, much later than the WT and CF3149 (Fig. 2C). These findings indicate that irrB deletion inhibited magnetosome formation as well as the growth rate.
Magnetosome phenotype of the ΔirrB strain.
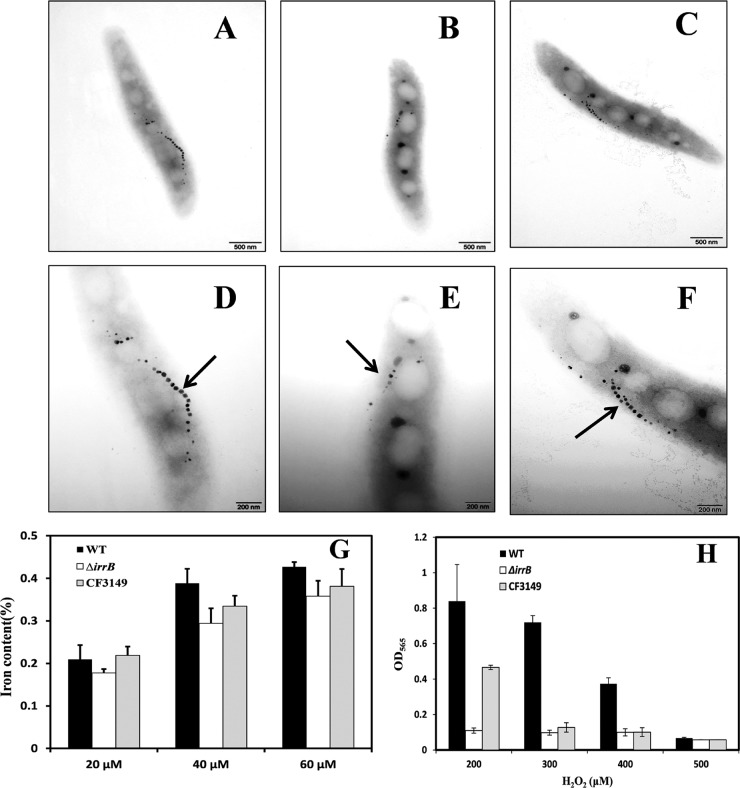
TEM observations showed that the mean number of magnetosomes (5.95 ± 2.27) was significantly (P < 0.05) lower, and their mean diameter (17.70 ± 5.97 nm) was significantly smaller, in the ΔirrB strain than in the WT and CF3149 (Fig. 3A to F; Table 2). The number and size of magnetosomes in the fur deletion mutant (Δfur) were also smaller than those in the WT and CF3149 (11). The diameter of magnetosomes of the Δfur strain (14.69 ± 5.39 nm) was smaller than that for the ΔirrB strain, but the Δfur strain had more magnetosomes (8.74 ± 4.34) (P < 0.05) (Table 2).
FIG 3.
TEM images, intracellular iron contents, and H2O2 tolerances of the three MSR-1 strains. (A to C) WT, ΔirrB, and CF3149 cells, respectively, viewed by conventional TEM. Bars, 500 nm. (D to F) WT, ΔirrB, and CF3149 cells, respectively, at a higher magnification. Bars, 200 nm. Arrows indicate magnetosomes. The level of magnetosome formation was notably reduced in the ΔirrB strain. (G) Cells were grown in SLM supplemented with 20, 40, or 60 μM ferric citrate, and the intracellular iron content was determined by ICP-OES. (H) Cells were grown in SLM supplemented with four different concentrations of H2O2, as shown, and the OD565 was measured by spectrophotometry. Growth in 200 μM H2O2 was normal for the WT, greatly reduced (13% of the WT value) for the ΔirrB strain, and partially restored for the complemented strain CF3149. In 300 μM H2O2, growth was close to normal for the WT (OD565, 0.7) but greatly reduced for the ΔirrB strain and CF3149. The growth of all three strains was completely inhibited in 500 μM H2O2. The experiments were performed in triplicate.
TABLE 2.
Diameters and numbers of magnetosomes in four MSR-1 strainsa
| Strain | Mean magnetosome diam (nm) | Mean no. of magnetosomes |
|---|---|---|
| WT | 24.69 ± 6.49 | 14.03 ± 2.12 |
| ΔirrB mutant | 17.70 ± 5.97 | 5.95 ± 2.27 |
| CF3149 | 24.60 ± 6.97 | 10.90 ± 3.31 |
| Δfur mutant | 14.69 ± 5.39 | 8.74 ± 4.34 |
Magnetite crystals were visualized by TEM in 45 to 60 cells per strain, and the sizes and numbers of magnetosomes were determined by using the ImageJ program. Both parameters differed significantly (P < 0.05) between the WT and the ΔirrB mutant.
These TEM observations were confirmed by culturing the three strains with 20, 40, and 60 μM ferric citrate and measuring the intracellular iron content by ICP-OES. With the three ferric citrate concentrations, the iron content in the ΔirrB strain was reduced by 15%, 24%, and 16%, respectively, from that in the WT, whereas the iron content in CF3149 was similar to that in the WT (Fig. 3G). The iron content in the Δfur mutant was reduced by ∼37% from that in the WT. The ratio of the iron content in the ΔirrB strain to that in the WT was slightly less. These findings indicate that intracellular iron content was reduced in the ΔirrB and Δfur strains. The large reduction in the Δfur strain may be due to disrupted transcription of the ferrous uptake system (feoAB) involved in magnetosome formation (11, 12, 14).
Hypersensitivity of the ΔirrB strain to oxidative stress.
Fur proteins are typically involved in the regulation of iron metabolism and responses to oxidative stress (6). To clarify the role of IrrB in oxygen metabolism, we evaluated the effects of four H2O2 concentrations on the growth of the WT, ΔirrB, and CF3149 strains. In a culture with 200 μM H2O2, WT growth was normal, whereas the growth of the ΔirrB strain was greatly reduced (OD565 value, ∼13% that of the WT) (Fig. 3H). The WT phenotype was partially complemented in CF3149. In a culture with 300 μM H2O2, the WT reached an OD565 of 0.7, whereas the growth of the ΔirrB strain and CF3149 was inhibited. In cultures with 500 μM H2O2, the growth of all three strains was completely inhibited. These findings indicate that irrB deletion results in hypersensitivity to oxidative stress, a phenotype similar to the Δfur phenotype. Sensitivity to H2O2 may result from the intensity of the Fenton reaction through the loss of fur genes. H2O2 promotes Irr degradation in the presence of iron, and Irr degradation in response to H2O2 is heme dependent (20). irrB deficiency may lead to a loss of cellular response to oxidative stress through the regulation of heme biosynthesis, which would account for the greatly reduced growth of the ΔirrB strain.
Transcription levels of irrA, irrC, and fur in the ΔirrB strain.
The irrB deletion did not eliminate magnetosome formation. To evaluate the effects of irrB deletion on other fur genes, we measured the transcription levels of irrA, irrC, and fur in the WT and ΔirrB strains under high- and low-iron conditions. The zur gene was excluded from this analysis because it showed low amino acid sequence identity to the other proteins, and its protein, Zur, had a different function (zinc homeostasis). The primers used for RT-qPCR are listed in Table S1 in the supplemental material.
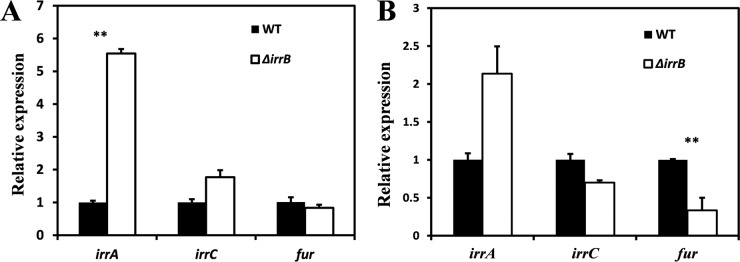
Under high-iron conditions, the transcription level of irrA in the ΔirrB strain was 5.5-fold higher (P < 0.05) than that in the WT (Fig. 4A). The transcription level of irrC was slightly higher, and that of fur was slightly lower, in the ΔirrB strain than in the WT (Fig. 4A). Under low-iron conditions, the transcription level of irrA was 2-fold higher in the ΔirrB strain than in the WT, whereas those of irrC and fur were lower in the ΔirrB strain (Fig. 4B). irrA is highly expressed in the ΔirrB strain and may be complementary to irrB deletion regardless of high- or low-iron conditions. Another possibility is that IrrB inhibits irrA expression. irrC showed increased expression only under high-iron conditions, and this expression was weaker than that of irrA. However, fur did not show any striking change in expression. A recent report indicates that Fur functions as a real Fur protein, like Fur-Pa, and differs from Irr proteins (21).
FIG 4.
Transcription levels of fur and two fur-like genes in the WT and ΔirrB strains under high-iron (A) and low-iron (B) conditions. Means were compared using Student's t test; differences with P values of <0.05 were considered significant. Under high-iron conditions, the irrA transcription level was 5.5-fold higher in the ΔirrB strain than in the WT (P < 0.05). Under low-iron conditions, the irrA level in the ΔirrB strain was 2-fold higher than that in the WT, but the irrC and fur levels were lower.
Regulation of IrrB.
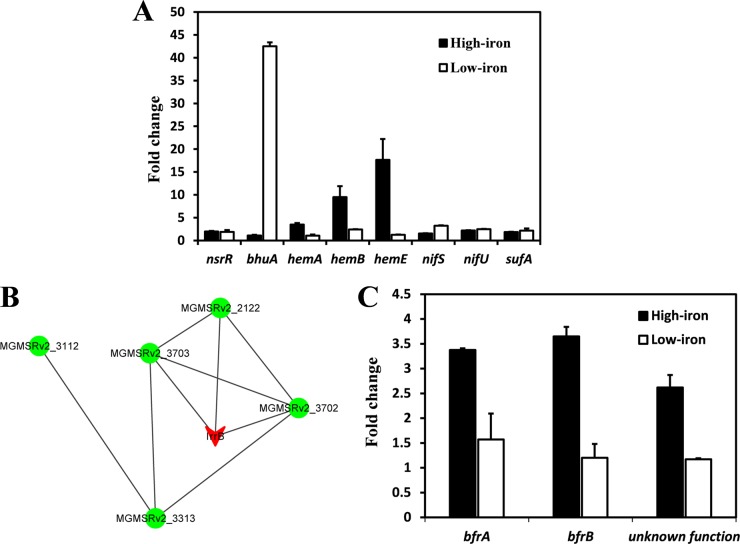
Iron response regulator (Irr) proteins bind conserved promoter sequences in the absence of iron and can activate or repress the expression of target genes (22). The conserved DNA in the promoter of target genes has 17 bp and is termed the iron control element (ICE) (23). Scanning of the entire MSR-1 genome did not reveal a gene promoter with an ICE box, suggesting that Irr proteins may recognize different ICE motifs in MSR-1. Comparison with Irr-regulated gene sequences in nonmagnetotactic bacteria (24, 25) revealed eight homologous genes in the MSR-1 genome. mRNAs transcribed from these genes were quantified by RT-qPCR. Under high-iron conditions, the ΔirrB strain showed 3.5-, 9.5-, and 17.6-fold-lower expression of three hem genes (related to heme biosynthesis), and slightly higher expression of the nifU gene (which encodes Fe-S cluster assembly scaffold protein NifU), than the WT (Fig. 5A, filled bars). Under low-iron conditions, the ΔirrB strain showed lower expression of genes involved in iron transport (bhuA), iron-sulfur assembly (sufA), and nitrogen fixation (nifS, encoding cysteine desulfurase) than the WT (Fig. 5A, open bars); in particular, the expression of bhuA (a putative TonB-dependent receptor gene) was ∼42-fold lower. These findings suggest that IrrB activates heme biosynthesis under high-iron conditions and iron transport under low-iron conditions. The expression levels of hemB and nifU were >2-fold lower in the ΔirrB strain than in the WT regardless of the presence or absence of iron, suggesting that these genes are not direct targets of IrrB and are more strongly induced in the absence of Irr proteins.
FIG 5.
Transcription patterns and predicted interactions among IrrB and related proteins. (A) Expression of eight genes in the WT relative to that in the ΔirrB strain (fold change) under high-iron (filled bars) and low-iron (open bars) conditions, as evaluated by RT-qPCR. (B) Interactions among IrrB and related proteins, predicted using the online tool STRING 9.1. The network nodes represent proteins encoded by the MGMSRv2_3149 (irrB; red), MGMSRv2_2122, MGMSRv2_3112, MGMSRv2_3313, MGMSRv2_3702, and MGMSRv2_3703 genes. Lines between nodes indicate predicted associations between the corresponding proteins. (C) Expression of bfrA, bfrB, and a gene with unknown function in the WT relative to that in the ΔirrB strain (fold change) under high-iron and low-iron conditions, as determined by RT-qPCR.
DISCUSSION
IrrB coordinates iron homeostasis and resistance to oxidative stress in MSR-1.
Irr proteins are the major transcriptional regulators of iron response reactions in rhizobia and accumulate in cells under low-iron conditions (25). In the MSR-1 genome, three proteins, encoded by the MGMSRv2_1721, MGMSRv2_3149, and MGMSRv2_3660 genes, were annotated as Irr homologues. To functionally characterize an Irr homologue, we constructed an irrB deletion mutant (ΔirrB) and its complemented strain CF3149.
Under low-iron conditions, the growth of the ΔirrB strain was significantly reduced and the bhuA gene was downregulated, findings similar to those for A. tumefaciens and Brucella abortus (26, 27). In MSR-1, bhuA is annotated as a putative TonB-dependent receptor that is located in the outer membranes and facilitates the acquisition of scarce resources in an energy-dependent manner (28). These findings suggest that IrrB is required for the induction of bhuA transcription in response to low-iron conditions and that irrB deletion disrupts iron uptake.
In nonmagnetotactic bacteria, under high-iron conditions, heme binds the HHH motif to promote the degradation of Irr and prevent the accumulation of a porphyrin precursor, whereas under low-iron conditions, the Irr protein activates the expression of genes involved in iron transport (23). The normal response to iron limitation presumably does not occur in the ΔirrB strain, resulting in a further reduction in the intracellular iron pool, similar to the decrease in total-iron content observed in B. japonicum (29), and striking growth inhibition. Thus, IrrB is responsible directly for iron homeostasis and indirectly for magnetosome formation.
Aside from the iron utilized for magnetosome formation (8) and redox reactions (30), the residual iron in the ΔirrB strain was sufficient to trigger the Fenton reaction under H2O2 toxicity stress. The increased H2O2 sensitivity observed in the ΔirrB strain may be related to disruption of heme biosynthesis. Lower expression of three hem genes showed that Fur activated heme biosynthesis. nifU and nifS are required for the formation of Fe-S clusters for the nitrogenase enzyme complex; NifU, in particular, provides Fe-S clusters to a variety of proteins (31). In the suf system, SufA carries Fe-S clusters and transfers them to target Fe-S apoproteins (32). Fe-S cluster assembly mediated by the suf system occurs under conditions of oxidative stress and iron limitation (33). Exposed Fe-S clusters are easily converted to unstable forms by oxygen species and disintegrate (34). Indirect regulation of IrrB by nifU, nifS, and sufA may therefore be involved in the sensing of the redox state and/or oxidative stress by cells.
Application of the online tool STRING 9.1 (http://string-db.org) predicted interactions between IrrB and five other proteins (Fig. 5B; see also Table S3 in the supplemental material). According to the predicted network scheme, IrrB has direct intrinsic interactions with bacterioferritin (MGMSRv2_3702 [BfrA], MGMSRv2_3703 [BfrB]) and a protein with unknown function (MGMSRv2_2122) and indirect interactions with a uroporphyrinogen decarboxylase (MGMSRv2_3313 [HemE]) related to heme biosynthesis (35) and a predicted DNA polymerase I (MGMSRv2_3112). Two bfr genes involved in iron storage and an unknown gene showed higher expression in the WT than in the ΔirrB strain in the presence of iron, but RT-qPCR did not reveal striking expression changes in the absence of iron (Fig. 5C). The two bfr genes encode Bfr subunits similar to those in E. coli and Magnetospirillum magnetotacticum MS-1, which contain heme-binding sites and display ferroxidase activity related to intracellular iron accumulation in bacterioferritin (36). The activation of bfr genes by IrrB under high-iron conditions facilitates the storage of surplus iron and helps maintain iron homeostasis.
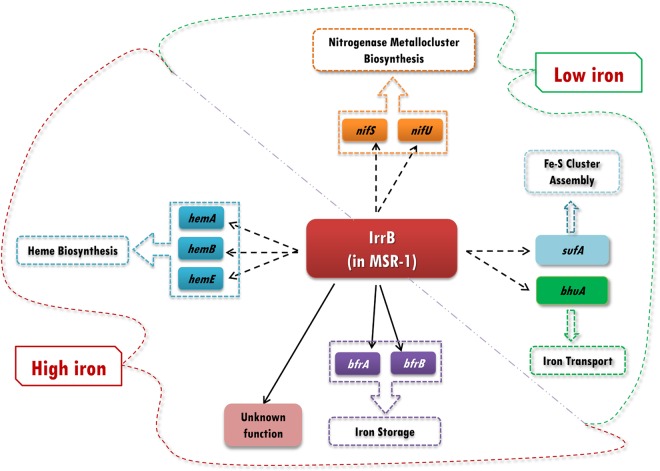
The iron level is closely associated with oxidative stress, because iron, as part of the Fenton reaction, potentiates oxygen toxicity (37). The IrrB-regulated networks proposed to function in MSR-1 cells under high-iron and low-iron conditions, based on results of the present and previous studies, are presented schematically in Fig. 6. Under high-iron conditions, IrrB indirectly regulates genes involved in iron storage (bfr), heme biosynthesis (hem), and an unknown pathway to increase gene expression, thereby reducing oxidative stress from surplus iron. In addition, ROS promote heme production for the degradation of IrrB, so as to reduce oxidative stress, through an unknown pathway. Under low-iron conditions, transcriptional activation by IrrB of the TonB-dependent receptor bhuA allows cells to take up scarce resources from the medium and avoid iron starvation. Upregulated expression of sufA (for Fe-S cluster assembly) may help cells maintain normal metabolism and avoid oxidative stress. nifS and nifU, two genes associated with the biosynthesis of nitrogenase metalloclusters, are also upregulated by IrrB.
FIG 6.
Proposed IrrB-regulated networks in MSR-1 cells. Under high-iron conditions, IrrB indirectly regulates genes for heme biosynthesis (hemA, hemB, hemC), iron storage (bfrA, bfrB), and an unknown pathway. Under low-iron conditions, IrrB regulates genes for iron transport (bhuA), Fe-S cluster assembly (sufA), and nitrogenase metallocluster biosynthesis (nifS, nifU).
We attempted to identify the gene-binding sequences of IrrB in MSR-1 on the basis of related information from nonmagnetotactic bacteria. Electrophoretic mobility shift assay (EMSA) results indicate that IrrB affects the expression of the genes shown in Fig. 6 in an indirect manner. It is possible that the regulon of IrrB differs from the ICE box in nonmagnetotactic bacteria and that the recognition sequence in the promoter is polytropic.
Proposed complementary roles of Fur homologues.
Members of the Fur protein family are abundant and widespread in Gram-negative bacteria; they include Fur and Fur-like proteins, such as Irr, Zur, Mur, and PerR (38). Nonmagnetotactic bacteria typically have two or three Fur homologues, e.g., Fur and Zur in E. coli and Fur, Zur, and PerR in Bacillus subtilis (6, 39). In striking contrast, the MSR-1 genome includes five coexisting Fur homologues. We considered possible explanations for this high number. Fur (encoded by the MGMSRv2_3137 gene) is a real Fur protein and regulates the expression of genes involved in iron and oxygen metabolism (11). It has a unique sulfur-rich center that contains Cys9, Met14, and Met16 and is oxygen sensitive (21). IrrB has Irr characteristics and may be stable under conditions of iron deficiency. Transcriptome analysis showed that irrB was notably downregulated in the stationary phase of cell growth. Recent studies showed that expression levels of the catalase gene katG and the thiol peroxidase gene tpx at 18 h were higher in the presence of 20 μM ferric citrate than in its absence and were strikingly higher in the log phase of magnetosome formation (13). Reduced expression of IrrB may lead to the synthesis of heme proteins such as catalases and peroxidases (which detoxify H2O2 and peroxides, respectively), and IrrB degradation may be related to oxidative stress (20). BLAST analysis showed a partial similarity of IrrC to the peroxide-responsive repressor (PerR) in Staphylococcus epidermidis, and IrrC has been implicated in the peroxide stress response (40). In the present study, irrA expression was upregulated under low-oxygen (0.5% dissolved oxygen) and iron-rich conditions. These findings, taken together, indicate that IrrA and IrrC play important roles in the regulation of oxygen balance.
The Fur and Fur-like proteins in MSR-1 presumably display a division of labor and cooperativity under various iron and oxygen concentrations, as suggested by RT-qPCR analysis of fur and fur-like gene transcription levels. Fur primarily regulates iron transport and catalase and superoxide dismutase activities during the oxidative stress response, whereas IrrB regulates heme biosynthesis, iron uptake, and iron storage. Ferrous iron (Fe2+), which is required for Fur activity, serves as a corepressor of Fur, whereas IrrB degradation may depend on iron. When one of the other Fur proteins is deleted, IrrA and IrrC may perform complementary functions in iron uptake and biomineralization. This proposed scenario would account for the coexistence of multiple Fur homologues in MSR-1.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (grant number 31270093), the National Undergraduate Innovation Program of China (grant number 201310019010), and the China Postdoctoral Science Foundation (grant number 2015M570175).
We are grateful to Zhongzhou Chen, Zengqin Deng, and Yuan Zhou (China Agricultural University) for technical help and to S. Anderson for English editing of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02585-15.
REFERENCES
- 1.Rudolph G, Hennecke H, Fischer HM. 2006. Beyond the Fur paradigm: iron-controlled gene expression in rhizobia. FEMS Microbiol Rev 30:631–648. doi: 10.1111/j.1574-6976.2006.00030.x. [DOI] [PubMed] [Google Scholar]
- 2.Guerinot ML. 1994. Microbial iron transport. Annu Rev Microbiol 48:743–772. doi: 10.1146/annurev.mi.48.100194.003523. [DOI] [PubMed] [Google Scholar]
- 3.Papanikolaou G, Pantopoulos K. 2005. Iron metabolism and toxicity. Toxicol Appl Pharmacol 202:199–211. doi: 10.1016/j.taap.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 4.da Silva Neto JF, Braz VS, Italiani VC, Marques MV. 2009. Fur controls iron homeostasis and oxidative stress defense in the oligotrophic alpha-proteobacterium Caulobacter crescentus. Nucleic Acids Res 37:4812–4825. doi: 10.1093/nar/gkp509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochsner UA, Vasil ML. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc Natl Acad Sci U S A 93:4409–4414. doi: 10.1073/pnas.93.9.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hantke K. 2001. Iron and metal regulation in bacteria. Curr Opin Microbiol 4:172–177. doi: 10.1016/S1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- 7.Jogler C, Schüler D. 2009. Genomics, genetics, and cell biology of magnetosome formation. Annu Rev Microbiol 63:501–521. doi: 10.1146/annurev.micro.62.081307.162908. [DOI] [PubMed] [Google Scholar]
- 8.Schüler D, Baeuerlein E. 1998. Dynamics of iron uptake and Fe3O4 biomineralization during aerobic and microaerobic growth of Magnetospirillum gryphiswaldense. J Bacteriol 180:159–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Wang Q, Zhang WJ, Wang YJ, Li L, Wen T, Zhang TW, Zhang Y, Xu J, Hu JY, Li SQ, Liu LZ, Liu JX, Jiang W, Tian JS, Li Y, Schüler D, Wang L, Li JL. 2014. Complete genome sequence of Magnetospirillum gryphiswaldense MSR-1. Genome Announc 2(2):e00171-14. doi: 10.1128/genomeA.00171-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uebe R, Voigt B, Schweder T, Albrecht D, Katzmann E, Lang C, Bottger L, Matzanke B, Schüler D. 2010. Deletion of a fur-like gene affects iron homeostasis and magnetosome formation in Magnetospirillum gryphiswaldense. J Bacteriol 192:4192–4204. doi: 10.1128/JB.00319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi L, Li J, Zhang WJ, Liu JN, Rong CB, Li Y, Wu LF. 2012. Fur in Magnetospirillum gryphiswaldense influences magnetosomes formation and directly regulates the genes involved in iron and oxygen metabolism. PLoS One 7:e29572. doi: 10.1371/journal.pone.0029572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rong CB, Huang YJ, Zhang WJ, Jiang W, Li Y, Li JL. 2008. Ferrous iron transport protein B gene (feoB1) plays an accessory role in magnetosome formation in Magnetospirillum gryphiswaldense strain MSR-1. Res Microbiol 159:530–536. doi: 10.1016/j.resmic.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q, Liu JX, Zhang WJ, Zhang TW, Yang J, Li Y. 2013. Expression patterns of key iron and oxygen metabolism genes during magnetosome formation in Magnetospirillum gryphiswaldense MSR-1. FEMS Microbiol Lett 347:163–172. doi: 10.1111/1574-6968.12234. [DOI] [PubMed] [Google Scholar]
- 14.Rong CB, Zhang C, Zhang YT, Qi L, Yang J, Guan GH, Li Y, Li JL. 2012. FeoB2 functions in magnetosome formation and oxidative stress protection in Magnetospirillum gryphiswaldense strain MSR-1. J Bacteriol 194:3972–3976. doi: 10.1128/JB.00382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding Y, Li JH, Liu JN, Yang J, Jiang W, Tian JS, Li Y, Pan YX, Li JL. 2010. Deletion of the ftsZ-like gene results in the production of superparamagnetic magnetite magnetosomes in Magnetospirillum gryphiswaldense. J Bacteriol 192:1097–1105. doi: 10.1128/JB.01292-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang WJ, Santini CL, Bernadac A, Ruan J, Zhang SD, Kato T, Li Y, Namba K, Wu LF. 2012. Complex spatial organization and flagellin composition of flagellar propeller from marine magnetotactic ovoid strain MO-1. J Mol Biol 416:558–570. doi: 10.1016/j.jmb.2011.12.065. [DOI] [PubMed] [Google Scholar]
- 17.Pohl E, Haller JC, Mijovilovich A, Meyer-Klaucke W, Garman E, Vasil ML. 2003. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol Microbiol 47:903–915. doi: 10.1046/j.1365-2958.2003.03337.x. [DOI] [PubMed] [Google Scholar]
- 18.Bhubhanil S, Ruangkiattikul N, Niamyim P, Chamsing J, Ngok-Ngam P, Sukchawalit R, Mongkolsuk S. 2012. Identification of amino acid residues important for the function of Agrobacterium tumefaciens Irr protein. FEMS Microbiol Lett 335:68–77. doi: 10.1111/j.1574-6968.2012.02638.x. [DOI] [PubMed] [Google Scholar]
- 19.White GF, Singleton C, Todd JD, Cheesman MR, Johnston AW, Le Brun NE. 2011. Heme binding to the second, lower-affinity site of the global iron regulator Irr from Rhizobium leguminosarum promotes oligomerization. FEBS J 278:2011–2021. doi: 10.1111/j.1742-4658.2011.08117.x. [DOI] [PubMed] [Google Scholar]
- 20.Yang J, Panek HR, O'Brian MR. 2006. Oxidative stress promotes degradation of the Irr protein to regulate haem biosynthesis in Bradyrhizobium japonicum. Mol Microbiol 60:209–218. doi: 10.1111/j.1365-2958.2006.05087.x. [DOI] [PubMed] [Google Scholar]
- 21.Deng ZQ, Wang Q, Liu Z, Zhang MF, Carolina A, Machado D, Chiu TP, Feng C, Zhang Q, Yu L, Qi L, Zheng JG, Wang X, Huo XM, Qi XX, Wu W, Rohs R, Li Y, Chen ZZ. 2015. Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator. Nat Commun 6:7642. doi: 10.1038/ncomms8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JW, Helmann JD. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- 23.Johnston AB, Todd J, Curson A, Lei S, Nikolaidou-Katsaridou N, Gelfand M, Rodionov D. 2007. Living without Fur: the subtlety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other α-proteobacteria. Biometals 20:501–511. doi: 10.1007/s10534-007-9085-8. [DOI] [PubMed] [Google Scholar]
- 24.Peuser V, Remes B, Klug G. 2012. Role of the Irr protein in the regulation of iron metabolism in Rhodobacter sphaeroides. PLoS One 7:e42231. doi: 10.1371/journal.pone.0042231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaggavarapu S, O'Brian MR. 2014. Differential control of Bradyrhizobium japonicum iron stimulon genes through variable affinity of the iron response regulator (Irr) for target gene promoters and selective loss of activator function. Mol Microbiol 92:609–624. doi: 10.1111/mmi.12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hibbing ME, Fuqua C. 2011. Antiparallel and interlinked control of cellular iron levels by the Irr and RirA regulators of Agrobacterium tumefaciens. J Bacteriol 193:3461–3472. doi: 10.1128/JB.00317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson ES, Paulley JT, Martinson DA, Gaines JM, Steele KH, Roop RM. 2011. The iron-responsive regulator Irr is required for wild-type expression of the gene encoding the heme transporter BhuA in Brucella abortus 2308. J Bacteriol 193:5359–5364. doi: 10.1128/JB.00372-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim BL. 2010. TonB-dependent receptors in nitrogen-fixing nodulating bacteria. Microbes Environ 25:67–74. doi: 10.1264/jsme2.ME10102. [DOI] [PubMed] [Google Scholar]
- 29.Yang JH, Sangwan I, Lindemann A, Hauser F, Hennecke H, Fischer H-M, O'Brian MR. 2006. Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol Microbiol 60:427–437. doi: 10.1111/j.1365-2958.2006.05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braun V. 2001. Iron uptake mechanisms and their regulation in pathogenic bacteria. Int J Med Microbiol 291:67–79. doi: 10.1078/1438-4221-00103. [DOI] [PubMed] [Google Scholar]
- 31.Poza-Carrión C, Jimenez-Vicente E, Navarro-Rodriguez M, Echavarri-Erasun C, Rubio LM. 2014. Kinetics of nif gene expression in a nitrogen-fixing bacterium. J Bacteriol 196:595–603. doi: 10.1128/JB.00942-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta V, Sendra M, Naik SG, Chahal HK, Huynh BH, Outten FW, Fontecave M, Ollagnier de Choudens S. 2009. Native Escherichia coli SufA, coexpressed with SufBCDSE, purifies as a [2Fe-2S] protein and acts as an Fe-S transporter to Fe-S target enzymes. J Am Chem Soc 131:6149–6153. doi: 10.1021/ja807551e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chahal HK, Dai YY, Saini A, Ayala-Castro C, Outten FW. 2009. The SufBCD Fe-S scaffold complex interacts with SufA for Fe-S cluster transfer. Biochemistry 48:10644–10653. doi: 10.1021/bi901518y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imlay JA. 2006. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol 59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- 35.Phillips JD, Whitby FG, Kushner JP, Hill CP. 2003. Structural basis for tetrapyrrole coordination by uroporphyrinogen decarboxylase. EMBO J 22:6225–6233. doi: 10.1093/emboj/cdg606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rui H, Rivera M, Im W. 2012. Protein dynamics and ion traffic in bacterioferritin. Biochemistry 51:9900–9910. doi: 10.1021/bi3013388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Touati D. 2000. Iron and oxidative stress in bacteria. Arch Biochem Biophys 373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 38.Rodionov DA, Gelfand MS, Todd JD, Curson ARJ, Johnston AWB. 2006. Computational reconstruction of iron- and manganese-responsive transcriptional networks in α-proteobacteria. PLoS Comput Biol 2:e163. doi: 10.1371/journal.pcbi.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann JD. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol 29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 40.Huang YJ, Zhang WJ, Jiang W, Rong CB, Li Y. 2007. Disruption of a fur-like gene inhibits magnetosome formation in Magnetospirillum gryphiswaldense MSR-1. Biochemistry (Mosc) 72:1247–1253. doi: 10.1134/S0006297907110119. [DOI] [PubMed] [Google Scholar]
- 41.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 42.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 43.Schweizer HD. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15:831–834. [PubMed] [Google Scholar]
- 44.Zhang Y, Pohlmann EL, Roberts GP. 2005. GlnD is essential for NifA activation, NtrB/NtrC-regulated gene expression, and posttranslational regulation of nitrogenase activity in the photosynthetic, nitrogen-fixing bacterium Rhodospirillum rubrum. J Bacteriol 187:1254–1265. doi: 10.1128/JB.187.4.1254-1265.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.