Abstract
(R)-Specific enoyl-coenzyme A (enoyl-CoA) hydratases (PhaJs) are capable of supplying monomers from fatty acid β-oxidation to polyhydroxyalkanoate (PHA) biosynthesis. PhaJ1Pp from Pseudomonas putida showed broader substrate specificity than did PhaJ1Pa from Pseudomonas aeruginosa, despite sharing 67% amino acid sequence identity. In this study, the substrate specificity characteristics of two Pseudomonas PhaJ1 enzymes were investigated by site-directed mutagenesis, chimeragenesis, X-ray crystallographic analysis, and homology modeling. In PhaJ1Pp, the replacement of valine with isoleucine at position 72 resulted in an increased preference for enoyl-coenzyme A (CoA) elements with shorter chain lengths. Conversely, at the same position in PhaJ1Pa, the replacement of isoleucine with valine resulted in an increased preference for enoyl-CoAs with longer chain lengths. These changes suggest a narrowing and broadening in the substrate specificity range of the PhaJ1Pp and PhaJ1Pa mutants, respectively. However, the substrate specificity remains broader in PhaJ1Pp than in PhaJ1Pa. Additionally, three chimeric PhaJ1 enzymes, composed from PhaJ1Pp and PhaJ1Pa, all showed significant hydratase activity, and their substrate preferences were within the range exhibited by the parental PhaJ1 enzymes. The crystal structure of PhaJ1Pa was determined at a resolution of 1.7 Å, and subsequent homology modeling of PhaJ1Pp revealed that in the acyl-chain binding pocket, the amino acid at position 72 was the only difference between the two structures. These results indicate that the chain-length specificity of PhaJ1 is determined mainly by the bulkiness of the amino acid residue at position 72, but that other factors, such as structural fluctuations, also affect specificity.
INTRODUCTION
Polyhydroxyalkanoates (PHAs), which are synthesized by some bacterial taxa, act as energy and carbon stores for use during starvation and are desirable for use as bio-based polymer materials (1–3). PHA biosynthesis requires a number of specific enzymes, such as PHA synthases and monomer-supplying enzymes. PHA synthase polymerizes the (R)-3-hydroxyalkanoate moiety of (R)-3-hydroxyacyl coenzyme A (3HA-CoA) into PHA, whereas monomer-supplying enzymes synthesize (R)-3HA-CoA from various metabolic intermediates. To control the material properties of PHAs, it is necessary to regulate the PHA monomer composition, which depends strongly on the enzymatic properties of PHA synthases and monomer-supplying enzymes.
(R)-Specific enoyl-CoA hydratase [PhaJ, (R)-hydratase] is a monomer-supplying enzyme that was first found to be involved in the biosynthesis of PHA from fatty acids in Aeromonas caviae (4, 5). Trans-2-enoyl coenzyme A (enoyl-CoA), an intermediate in fatty acid β-oxidation, undergoes stereospecific hydration by the function of PhaJ, resulting in the formation of (R)-3HA-CoAs. Currently, four types of phaJ genes (phaJ1Pa to phaJ4Pa) have been identified in the genome of Pseudomonas aeruginosa DSM1707; their translational products were characterized in terms of substrate specificity using heterologous expression systems (6, 7). Of all the phaJ gene products, PhaJ1Pa shows relatively narrow substrate specificity toward enoyl-CoAs, preferring acyl chain lengths from 4 to 6, whereas PhaJ2Pa-PhaJ4Pa shows broad substrate specificity toward enoyl-CoAs, preferring acyl chain lengths from 6 to 12. Other Pseudomonas strains were found to have phaJ genes (8–10). In the case of P. putida KT2440, this strain has three possible phaJ genes, and one such gene product, PhaJ4Pp, has been suggested to supply monomers from β-oxidation to PHA biosynthesis in P. putida cells cultured on fatty acids (9, 10). In addition, phaJ1Pp and phaJ4Pp from P. putida were expressed heterologously in recombinant Escherichia coli, and the properties of their translational products were evaluated. Interestingly, in vivo evaluation revealed that despite sharing 67% amino acid sequence identity between PhaJ1Pp and PhaJ1Pa, PhaJ1Pp showed significantly broader substrate specificity than PhaJ1Pa (10).
To date, several (R)-hydratases have been crystallographically characterized (11–15). Of the bacterial PhaJs, PhaJAc from A. caviae was crystallized and its properties have been extensively investigated (11, 16). Based on structural analyses, the amino acid residues that define the substrate specificity of PhaJAc were identified, and several mutant enzymes with altered substrate specificities were created by using site-directed mutagenesis (16). Additionally, chimeragenesis, a useful technique for revealing target enzyme properties and for creating enzymes with altered properties, has been adopted for analyses of PHA synthases to create mutants with higher activities and altered substrate specificities (17, 18).
To identify the factors that determine the substrate specificity of PhaJ1Pp and PhaJ1Pa, site-directed mutagenesis and chimeragenesis of PhaJ1 were performed. Furthermore, crystallographic analyses of PhaJ1 were attempted and the structure of PhaJ1Pa was determined. However, because of difficulties in the large-scale production and purification of recombinant PhaJ1Pp, its structure was constructed by homology modeling. Structural information from these models was used to elucidate the contribution of an amino acid residue at the bottom of the substrate-binding pocket to the substrate specificity of PhaJ1 enzymes.
MATERIALS AND METHODS
Site-directed mutagenesis.
The site-directed mutagenesis of PhaJ1Pp was performed by PCR. The following primers were used for the V72A mutation: F01 (5′-GCAGGTCGACTCTAGAAATAATTTTG-3′) and F02 (5′-AGTGCGGCAGCGGCCTGCACCC-3′) for the forward direction and R01 (5′-CGGTACCCGGGGATCCCACTTC-3′) and R02 (5′-GTGCAGGCCGCTGCCGCACTGATC-3′) for the reverse direction. Primers used for the V72L mutation were F01 and F03 (5′-AGTGCGGCACTGGCCTGCACCC-3′) for the forward direction and R01 and R03 (5′-GTGCAGGCCAGTGCCGCACTGATC-3′) for the reverse direction. The underlined regions correspond to mutation sites. First, two sets of PCR were performed using pUCJ1Pp as a template: one using primer pairs F01/R02 and F02/R01 to generate the DNA fragments containing the V72A mutation (VA1 and VA2), and the second using primer pairs F01/R03 and F03/R01 to generate the DNA fragments containing the V72L mutation (VL1 and VL2). Each amplified DNA fragment was purified and subjected to a second round of PCR, which was performed using primers F01/R01 with the VA1 and VA2 templates for the V72A mutant and the VL1 and VL2 templates for the V72L mutant. After PCR amplification, the DNA fragments were purified and cloned into XbaI/BamHI sites of the pUC19 vector, producing the plasmids pUCJ1PpV72A and pUCJ1PpV72L. The resulting plasmids were sequenced to confirm open reading frames with the introduced mutations.
To produce the V72I and reverse I72V substitutions in PhaJ1Pp and PhaJ1Pa, respectively, synthetic genes with nucleotide changes at the corresponding sites were constructed. In phaJ1Pp, the GTG codon was replaced by ATC and vice versa in phaJ1Pa.
Construction of chimeric phaJ1 genes.
To construct the expression plasmids pUCJ1ch67 and pUCJ1ch172, the N-terminal and C-terminal truncated fragments of phaJ1Pa and phaJ1Pp were amplified by PCR (18). For N-terminal fragments, PCR was performed using pUCJ1 (7), which harbors phaJ1Pa from P. aeruginosa, as a template with the following oligonucleotide primers: for the forward direction, 5′-CAGGTCGACTCTAGAAATAATTTTGTTTAA-3′ (J1ch-F); for the reverse direction of phaJ1ch67, 5′-GCTGGTGTACTCGGCCTTTTGCCCG-3′ (J1ch-R67), or phaJ1ch172, 5′-GCGTTCCTTGAACTGGGTGGTGGCC-3′ (J1ch-R172). The underlined sequence in J1ch-F indicates the XbaI site. The reverse primers were phosphorylated at the 5′ end. For the C-terminal fragments, PCR was performed using pUCJ1Pp (10), which harbors phaJ1Pp, as a template with the following oligonucleotide primers: for the forward direction of phaJ1ch67, 5′-TCCGTTGAAGAACGCGACATCCAGC-3′ (J1chF-67), or phaJ1ch372, 5′-ATTGCCCATGGCATGTTCAGCGGCG-3′ (J1chF-172); reverse direction, 5′-ATTCGAGCTCGGTACCCGGGGATCC-3′ (J1ch-R). The underlined sequence in J1ch-R indicates the BamHI site, and the forward primers were phosphorylated at the 5′ end. Two pairs of truncated fragments were ligated to generate chimeric gene fragments. Chimeric genes were amplified by PCR using the ligated fragments as a template and the J1ch-F/J1ch-R primer pair. The PCR products were digested with XbaI and BamHI, and the resultant fragments were inserted into similarly digested pUC19, yielding the plasmids pUCJ1ch67 and pUCJ1ch172. For phaJ1ch372, the entire region of the plasmid pUCJ1 was amplified using the inverse PCR method (19) and with the following primers: forward, 5′-TATCGCCGCCGAACTTTGTGGCGAGCTGAAGTGGGATCCCCGGGTACCGAGCTCGAATTC-3′; reverse, 5′-CCAGCTCGACGGTCTGCTGCTTGCGCGGCGCCAGGATCTCGGCTTCACCCGCGACCACCAGCTCGTCGTTCTGGTTG-3′. Except for six nucleotides at the 3′ end of the reverse primer, the two primers have DNA sequences that are nearly identical to that of the phaJ1Pp gene. The PCR products were allowed to self-ligate, yielding the expression plasmid pUCJ1ch372.
In vitro hydratase assays.
In vitro hydratase assays were performed using the soluble fraction of recombinant E. coli DH5α or JM109 harboring phaJ1 genes. The recombinant strains were transformed with pUC-based expression plasmids harboring phaJ1 genes and were cultured in LB media (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter NaCl) at 37°C. Isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) was added at the beginning of growth for the DH5α strains and after 5 h of cultivation for the JM109 strains. After 24 h, cells were harvested by centrifugation and resuspended in 50 mM Tris-HCl buffer (pH 8.0). The cells were lysed by sonication, and the supernatants (after centrifugation) were used for hydratase activity measurements.
Hydratase activity assays were performed in 50 mM Tris-HCl (pH 8.0) containing 25 μM crotonyl-CoA or octenoyl-CoA at 30°C (5, 7). Crotonyl-CoA was purchased from Sigma-Aldrich, MO. As described previously, octenoyl-CoA was synthesized from trans-2-octenoic acid (Tokyo Kasei, Tokyo, Japan) and CoA (Oriental Yeast, Osaka, Japan) (20) and purified using Sep-Pak C18 columns (Waters, MA) (21). To initiate the hydration reaction, a 5-μl aliquot of protein solution was added to quartz cells (1.0-cm path length) containing the enoyl-CoA solution. The reaction was monitored using a UV-visible spectrophotometer at 263 nm, which describes the absorbance maximum of the enoyl-thioester bond. The ε263 of the enoyl-thioester bond is 6.7 × 103 M−1 cm−1. One unit of hydratase activity was defined as the amount of enzyme required to catalyze a decrease of 1 μmol of substrate in 1 min.
Protein concentrations were determined using a Quant-iT protein assay kit (Invitrogen, CA) according to the manufacturer's instructions.
SDS-PAGE was performed according to standard procedures using 13% gels. The gels were stained with Coomassie brilliant blue.
In vivo hydratase assays using PHA-producing recombinant E. coli.
To evaluate the substrate specificity of the products of phaJ genes in vivo, recombinant E. coli LS5218 [fadR601, atoC2 (Con)] (22–24) cells harboring the plasmid pPPAC (25, 26), which carries the phaC1Ps gene encoding a broad-substrate-specific PHA synthase from Pseudomonas sp. strain 61-3, were transformed with pUC19-based expression plasmids harboring the phaJ genes (described above). The recombinant E. coli LS5218 strains were inoculated into 100 ml of M9 media (17.1 g/liter Na2HPO4 · 12H2O, 3 g/liter KH2PO4, 0.5 g/liter NH4Cl, 0.5 g/liter NaCl, 2 ml/liter 1 M MgSO4, and 0.1 ml/liter 1 M CaCl2) containing sodium dodecanoate (0.25%, wt/vol), ampicillin (100 mg/liter), kanamycin (50 mg/liter), and isopropyl-β-d-thiogalactopyranoside (1 mM; IPTG) with Brij-35 (0.4%, wt/vol). The cells were cultivated for 96 h at 37°C in 500-ml flasks on a reciprocal shaker (130 strokes/min). The content and composition of accumulated PHAs in dry cells were evaluated after methanolysis by gas chromatography (GC) as previously described (27).
Structural determination of PhaJ1Pa.
Recombinant PhaJ1Pa was overexpressed in E. coli BL21(DE3) harboring pETJ1 (7), which carries phaJ1Pa, and was purified according to previously described procedures (11). The protein was concentrated to 10 mg/ml in 20 mM Tris-HCl (pH 7.5). Enzyme crystals were obtained by sitting-drop vapor diffusion using mother liquor containing 15 to 20% (wt/vol) polyethylene glycol 3350, 20% (vol/vol) glycerol, and 0.1 M Bis-Tris, pH 6.0 to 6.5. The crystals belonged to the orthorhombic space group P212121 with cell dimensions a = 63.5 Å, b = 65.7 Å, c = 77.5 Å, and it contained one dimeric molecule per asymmetric unit. The Matthews coefficient (28) and solvent content were calculated to be 2.41 Å3/Da and 49%, respectively. Native data were collected up to a resolution of 1.7 Å from a single crystal using synchrotron radiation at BL26B1, SPring-8, Harima, Japan. Prior to data collection, the crystal was flash-cooled in a nitrogen gas stream at 90K. The data were integrated and scaled in HKL2000 (29).
Initial phases were obtained by molecular replacement in PHENIX (30) using coordinates of PhaJAc (PDB code 1IQ6) (11) as a search model. The initial model was rebuilt and refined in PHENIX (30). Models of Ser2-Gln5 and Gly156 were manually built in COOT (31). The current model (two chains of Ser2-Gly156, eight glycerol molecules, and 233 water molecules) converged to a crystallographic R factor of 0.173 and free R (32) of 0.219 (calculated from 5.7% of total reflections). Met1 was not visible in the electron density map, probably due to posttranslational truncation (5). The structure was validated using PROCHECK (33) and MOLPROBITY (34).
Homology modeling.
The structural model of PhaJ1Pp was constructed by homology modeling in the HOMER server (36) using the model of PhaJ1Pa. The main-chain conformation of the model was essentially identical to that of PhaJ1Pa and was not subject to energy minimization. Because PhaJ1Pp and PhaJ1Pa share ∼67% amino acid sequence homology, it is expected that the overall conformation of PhaJ1Pp is highly similar to that of PhaJ1Pa with a predicted root-mean-square deviation (RMSD) of approximately 0.74 Å for the core main-chain atoms (37).
Protein structure accession number.
Coordinates and structure factors for PhaJ1 from P. aeruginosa were deposited in the Protein Data Bank (35) under accession number 5CPG.
RESULTS
Site-directed mutagenesis at residue 72 in PhaJ1Pp and PhaJ1Pa.
To assess the contribution of residue 72 to the substrate specificities of PhaJ1Pp and PhaJ1Pa, Leu, Ile, and Ala substitution mutants and a Val substitution mutant were created for PhaJ1Pp and PhaJ1Pa, respectively. Their substrate specificities were evaluated by both in vitro and in vivo analyses.
In the in vitro analyses, the E. coli JM109 strain, which harbors lacIq, was used as the host for mutant gene expression, as the strain previously used in heterologous hydratase assays (6), E. coli DH5α (lacIq negative), did not grow in LB media after transformation with the phaJ1Pp V72A gene. Substrate specificities of the enzymes were measured using enoyl-CoAs with C4 and C8 acyl chain lengths. Table 1 shows the results of in vitro hydratase activity assays of parental phaJ1 genes and their mutants. The soluble extracts containing parental PhaJ1Pp showed hydratase activities of 149 and 93 U/mg toward C4 and C8 substrates, respectively, resulting in a high C8/C4 activity ratio of 0.62. The soluble extracts containing the V72L or V72I PhaJ1Pp mutants showed significant hydratase activity, whereas those containing the V72A mutant showed weak hydratase activity. The weak hydratase activity of the PhaJ1Pp V72A mutant may be explained by low levels of soluble protein, which were barely detectable by SDS-PAGE analysis (data not shown). The hydratase activity of the V72L PhaJ1Pp extracts toward C4 was 173 U/mg and is similar to that of the parental extracts. However, its activity toward C8 was only 0.20 U/mg, which is 465-fold lower than that of the parental extracts. Subsequently, its C8/C4 activity ratio is only 0.0012. The hydratase activities of the V72I PhaJ1Pp extracts toward C4 and C8 (50.7 and 2.2 U/mg, respectively) were lower than those of the parental extracts, and its C8/C4 ratio (0.043) was one magnitude lower than that of the parental extracts. The hydratase activities of the V72A PhaJ1Pp extracts toward C4 and C8 did not show a marked difference and resulted in a higher C8/C4 activity ratio of 0.69. The hydratase activity of the I72V PhaJ1Pa extracts toward C4 (143 U/mg) was similar to that of the PhaJ1Pp extracts, while toward C8 its hydratase activity was 8.5-fold lower than that of the PhaJ1Pp extracts, resulting in significant differences in the C8/C4 activity ratios.
TABLE 1.
Hydratase activity assay of E. coli JM109 strains harboring parental or Val72/Ile72 mutant genes of phaJ1a
| Plasmid (relevant marker) | Sp act (U/mg) with: |
C8/C4 activity ratio | |
|---|---|---|---|
| Crotonyl-CoA (C4) | Octenoyl-CoA (C8) | ||
| pUC19 (none) | Not detected | Not detected | Not determined |
| pUCJ1Pp (phaJ1Pp) | 149 ± 42 | 93 ± 12 | 0.62 |
| pUCJ1PpV72A (phaJ1Pp V72A) | 0.29 ± 0.05 | 0.20 ± 0.07 | 0.69 |
| pUCJ1PpV72I (phaJ1Pp V72I) | 50.7 ± 2.6 | 2.2 ± 0.3 | 0.043 |
| pUCJ1PpV72L (phaJ1Pp V72L) | 173 ± 30 | 0.20 ± 0.04 | 0.0012 |
| pUCJ1I72V (phaJ1Pa I72V) | 143 ± 9.0 | 11 ± 2.1 | 0.077 |
All values are averages and standard deviations from triplicate tests.
The in vivo substrate specificity of the PhaJ1 enzymes was evaluated by analyzing the monomeric composition of PHAs accumulated in the recombinant E. coli LS5218 strain that coexpressed phaJ1 and phaC1Ps (encoding a broad-substrate-specific PHA synthase functioning as a C4-C12 monomer-polymerizing enzyme) (6, 7). The results are shown in Table 2. The levels of PHAs in the strains containing V72I or V72L PhaJ1Pp were 31% (wt/wt) and 30% (wt/wt) of PHAs, respectively. These levels are 1.6-fold higher than the levels found in the parental PhaJ1Pp-containing strain. PHAs accumulated in the strain containing these mutants were composed of 67 to 69 mol% of the 3HHx fraction, whereas those in the strain harboring parental phaJ1Pp were composed of 36 and 37 mol% of the 3HHx and 3HO fractions, respectively. The strain containing the PhaJ1Pp V72A mutant accumulated 7% (wt/wt) of PHAs with monomeric compositions similar to that of the control pUC19 strain, suggesting that PhaJ1Pp V72A does not function as a monomer-supplying enzyme in vivo. The level of PHAs in the strain containing the I72V PhaJ1Pa mutant was 32% (wt/wt) of PHAs, which was composed of 60 mol% of 3HHx fraction. The accumulation level of PHAs in this strain was similar to that in the parental PhaJ1Pa-containing strain; however, the monomeric composition of PHAs accumulated in this strain was different from those in the strains containing parental PhaJ1Pp or PhaJ1Pa.
TABLE 2.
PHA accumulation in recombinant E. coli LS5218 harboring parental or Val72/Ile72 mutant genes of phaJ1a
| Plasmids (relevant marker[s]) | Dry cell wt (g/liter) | PHA content (%, wt/wt) | PHA compositionb (mol%) |
||||
|---|---|---|---|---|---|---|---|
| 3HB (C4) | 3HHx (C6) | 3HO (C8) | 3HD (C10) | 3HDD (C12) | |||
| pPPAc, pUC19c (phaC1Ps) | 0.39 ± 0.09 | 4 ± 2 | 1 | 21 | 49 | 19 | 10 |
| pPPAC, pUCJ1Ppc (phaC1Ps, phaJ1Pp) | 0.54 ± 0.08 | 19 ± 2 | 9 | 36 | 37 | 11 | 7 |
| pPPAC, pUCJ1PpV72A (phaC1Ps, phaJ1Pp V72A) | 0.66 ± 0.09 | 7 ± 1 | 5 | 21 | 42 | 19 | 13 |
| pPPAC, pUCJ1PpV72I (phaC1Ps, phaJ1Pp V72I) | 0.58 ± 0.04 | 31 ± 3 | 12 | 67 | 14 | 4 | 3 |
| pPPAC, pUCJ1PpV72L (phaC1Ps, phaJ1Pp V72L) | 0.74 ± 0.02 | 30 ± 2 | 12 | 69 | 10 | 5 | 4 |
| pPPAC, pUCJ1 (phaC1Ps, phaJ1Pa) | 0.73 ± 0.02 | 32 ± 2 | 13 | 74 | 8 | 3 | 2 |
| pPPAC, pUCJ1Pa I72V (phaC1Ps, phaJ1Pa I72V) | 0.58 ± 0.16 | 32 ± 2 | 13 | 60 | 20 | 4 | 3 |
Cells harboring PHA synthase 1 gene (phaC1Ps) from Pseudomonas sp. strain 61-3 were cultivated for 96 h at 37°C in M9 medium containing sodium dodecanoate (0.25%, wt/vol) as the sole carbon source. The surfactant Brij-35 (0.4%, wt/vol) was added to each medium to solubilize dodecanoate. All values are averages and standard deviations from triplicate tests.
Abbreviations: 3HB, 3-hydroxybutyrate; 3HHx, 3-hydroxyhexanoate; 3HO, 3-hydroxyoctanoate; 3HD, 3-hydroxydecanoate; 3HDD, 3-hydroxydodecanoate.
Data were retrieved from reference 10.
Substrate specificity of chimeric enzymes.
Because PhaJ1Pp and PhaJ1Pa share 67% amino acid sequence identity, we expected that chimeras of the two enzymes would be active. Based on the PhaJAc structure, three types of chimeric PhaJ1 enzymes were designed (Fig. 1), and their substrate specificities were assessed in vitro and in vivo.
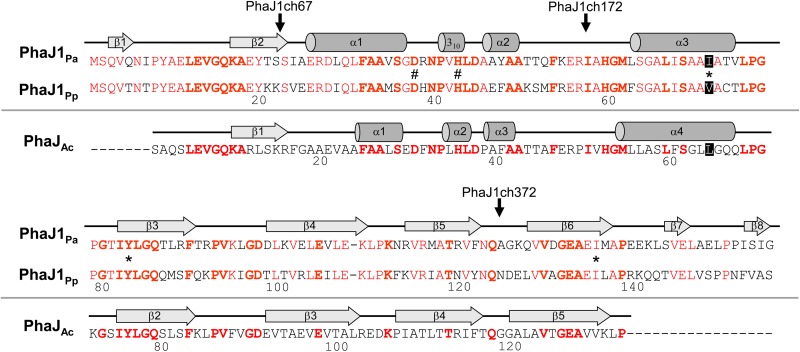
FIG 1.
Amino acid sequence alignment of PhaJ1Pa from P. aeruginosa DSM1707 and PhaJ1Pp from P. putida KT2440, with PhaJAc from A. caviae. Secondary structures of PhaJ1Pa and PhaJ1Ac are indicated along with each of their sequences. Helices and strands are shown by cylinders and arrows, respectively. Conserved residues in PhaJ1Pa and PhaJ1Pp are shown in red, whereas those in all three enzymes are indicated in boldface. Active-site residues Asp38 and His43 are indicated by the number sign, and residues Ile/Val72, Tyr83, and Ile136, which constitute the acyl-chain-binding pocket in PhaJ1, are indicated by asterisks. Vertical arrows indicate the junction points of the chimeric PhaJ1 enzymes (P. aeruginosa PhaJ1 is the N terminus of the chimera, and P. putida PhaJ1 is the C terminus).
Table 3 presents the results of hydratase activity assays of the recombinant E. coli DH5α strains transformed with the phaJ1 genes. Soluble fractions from these strains showed significant hydratase activity, while no activity was observed in the pUC19-transformed strain (negative control). Although every strain harboring phaJ1 genes showed higher hydratase activity toward C4 substrates than C8 substrates, there was a wide range in the C8/C4 activity ratio. The highest C8/C4 activity ratio was 0.68, observed in the phaJ1Pp-harboring strain, whereas the lowest was 0.0012, observed in the phaJ1Pa-harboring strain. The strains containing chimeric phaJ1 genes showed ratios between those of the strains harboring phaJ1Pp and phaJ1Pa. The strains harboring phaJ1ch67 and phaJ1ch172 exhibited C8/C4 activity ratios of 0.40 and 0.33, respectively, whereas the strain harboring phaJ1ch372 showed a C8/C4 activity ratio of 0.042, which is an order of magnitude lower than those of the other chimeras.
TABLE 3.
Hydratase activity assay of E. coli DH5α strains harboring parental or chimeric phaJ1 genesa
| Plasmidb (relevant marker) | Sp act (U/mg) with: |
C8/C4 activity ratio | |
|---|---|---|---|
| Crotonyl-CoA (C4) | Octenoyl-CoA (C8) | ||
| pUC19 (none) | 0.01 ± 0.01 | 0.02 ± 0.00 | NDc |
| pUCJ1Pp (phaJ1Pp) | 120 ± 17 | 81 ± 18.6 | 0.68 |
| pUCJ1ch67 (phaJ1ch67) | 12 ± 0.5 | 4.8 ± 0.27 | 0.40 |
| pUCJ1ch172 (phaJ1ch172) | 2.7 ± 0.13 | 0.88 ± 0.13 | 0.33 |
| pUCJ1ch372 (phaJ1ch372) | 15 ± 3.6 | 0.63 ± 0.11 | 0.042 |
| pUCJ1 (phaJ1Pa) | 490 ± 35 | 0.57 ± 0.03 | 0.0012 |
All values are averages and standard deviations from triplicate tests.
phaJ1ch67, chimeric gene of phaJ1Pa (nucleotide location 1 to 66) and phaJ1Pp (nucleotide location 67 to 471); phaJ1ch172, chimeric gene of phaJ1Pa (nucleotide location 1 to 171) and phaJ1Pp (nucleotide location 172 to 471); phaJ1ch372, chimeric gene of phaJ1Pa (nucleotide location 1 to 371) and phaJ1Pp (nucleotide location 372 to 471).
ND, not determined.
Table 4 shows PHA accumulation and composition from sodium dodecanoate in recombinant E. coli LS5218 strains that coexpressed phaJ1 and phaC1Ps genes. The strains containing PhaJ1Pp or PhaJ1Pa accumulated 19% (wt/wt) and 32% (wt/wt) PHA in dry cells, respectively, whereas the strains containing chimeric PhaJ1 accumulated 18 to 30% (wt/wt) PHA in dry cells. This indicates that the chimeric PhaJ1 enzymes demonstrate monomer-supplying activities in E. coli cells. The monomeric compositions of accumulated PHAs differed according to the chimeric phaJ1 gene expressed. Strains containing chimeric PhaJ1ch67 or PhaJ1ch172 accumulated PHAs composed predominantly of 3HHx and 3HO fractions (50 and 30 to 32 mol%, respectively), with slight differences in the 3HD and 3HDD fraction compositions. The compositions of the PHAs accumulated in these strains were similar to those in strains containing PhaJ1Pp and PhaJ1Pa, indicating that in vivo, these chimeric PhaJ1 enzymes show substrate specificities between those displayed by the parental PhaJ1 enzymes. The strain containing chimeric PhaJ1ch372 accumulated PHAs composed of a high 3HHx fraction (75 mol%), resembling the strain containing PhaJ1Pa in terms of PHA accumulation level. Contrary to in vitro observations, these results indicate that the substrate specificity of chimeric PhaJ1ch372 in vivo is similar to that of PhaJ1Pa.
TABLE 4.
PHA accumulation in recombinant E. coli LS5218 harboring chimeric phaJ1 genesa
| Plasmidsb (relevant markers) | Dry cell wt (g/liters) | PHA content (%, wt/wt) | PHA compositionc (mol%) |
||||
|---|---|---|---|---|---|---|---|
| 3HB (C4) | 3HHx (C6) | 3HO (C8) | 3HD (C10) | 3HDD (C12) | |||
| pPPAC, pUCJ1ch67 (phaC1Ps, phaJ1ch67) | 0.54 ± 0.11 | 18 ± 4 | 14 | 50 | 30 | 4 | 2 |
| pPPAC, pUCJ1ch172 (phaC1Ps, phaJ1ch172) | 0.51 ± 0.09 | 22 ± 3 | 14 | 50 | 32 | 3 | 1 |
| pPPAC, pUCJ1ch372 (phaC1Ps, phaJ1ch372) | 0.80 ± 0.05 | 30 ± 3 | 14 | 75 | 8 | 2 | 1 |
Cells harboring PHA synthase 1 gene (phaC1Ps) from Pseudomonas sp. strain 61-3 were cultivated for 96 h at 37°C in M9 medium containing sodium dodecanoate (0.25%, wt/vol) as the sole carbon source. The surfactant Brij-35 (0.4%, wt/vol) was added to each medium to solubilize dodecanoate. All values are averages and standard deviations from triplicate tests.
phaJ1ch67, phaJ1ch172, and phaJ1ch372 were chimeric (R)-hydratase genes (see footnote b to Table 3).
Abbreviations: 3HB, 3-hydroxybutyrate; 3HHx, 3-hydroxyhexanoate; 3HO, 3-hydroxyoctanoate; 3HD, 3-hydroxydecanoate; 3HDD, 3-hydroxydodecanoate.
Crystallographic analysis of PhaJ1Pa. (i) Overall structure.
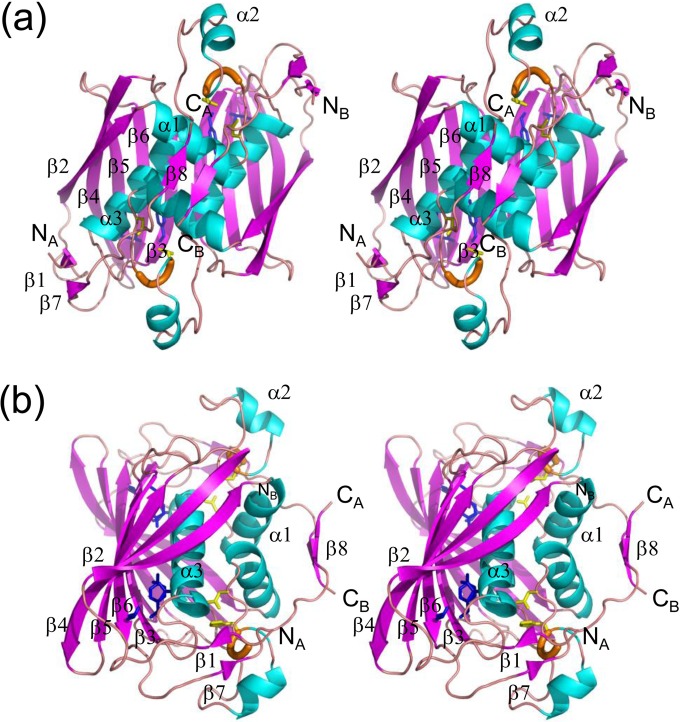
The crystal structure of PhaJ1Pa was determined at a resolution of 1.7 Å by molecular replacement using coordinates of PhaJAc (PDB entry 1IQ6) (11) as a search model (Fig. 2). Statistics of data collection and structural refinement are summarized in Table 5. PhaJ1Pa forms a dimer in which the monomers are related by 2-fold symmetry. Each monomer consists of all 155 amino acid residues from Ser2 to Gly156. The monomer structure represents a hotdog fold containing a five-stranded antiparallel β-sheet “bun” (β2-β6) and an α-helix “sausage” (α3), with an (R)-hydratase-specific overhang segment containing two α-helices, α1 and α2, between strand β2 and helix α3. The overall fold is similar to that of PhaJAc, with RMSD of 1.0 to 1.3 Å. In addition, compared to PhaJAc, PhaJ1Pa has N- and C-terminal extensions 6 and 17 residues in length, respectively. The C-terminal extension exhibits an extended conformation, forming an arched bridge over the central helix α3 and reaching over to helix α1. The N-terminal extension exhibits an extended conformation and contains strand β1, which forms a short antiparallel β-sheet with strand β7 in the C-terminal extension. In the dimeric structure, the five-stranded β-sheets of the two monomers associate via hydrogen bonding interactions at strands β3 in a side-by-side manner to form a ten-stranded β-sheet. An additional short antiparallel β-sheet is formed by two strands of β8 in the C-terminal extensions of both monomers. Between these major and minor β-sheets, helices α1 and α3 from both monomers are held together mainly by hydrophobic interactions. Thus, the dimeric molecule represents a special four-layered hotdog structure, that is, the bun, sausage, sauce (helices α1 and α2), and topping (β8) layers (Fig. 1b), and is unique among the hotdog fold enzymes that have been structurally characterized thus far.
FIG 2.
Stereo diagrams of the crystal structure of the PhaJ1Pa dimer. α-Helices, β-strands, and 310-helices are indicated by helices (blue), arrows (magenta), and thick tubes (orange), respectively. The catalytic dyad residues Asp38 and His43 are represented by stick models (yellow). Residues located at the bottom of the acyl-chain-binding pocket (Ile72, Tyr83, and Ile136) also are represented by stick models (blue). N and C termini of each chain are labeled. Secondary structure annotations are shown only for chain A. (a) A front view of the structure along the 2-fold axis of symmetry. Two subunits adopt an almost identical conformation, with an RMSD of 0.38 Å for Cα atoms. (b) A side view of the structure, generated by 90° rotation relative to the image shown in panel a, highlighting the four-layered structure.
TABLE 5.
Data collection and refinement statistics
| Parameter | Value(s)a |
|---|---|
| Data collection statistics | |
| Wavelength (Å) | 1.0000 |
| Resolution range (Å) | 39.3–1.69 (1.75–1.69) |
| Space group | P212121 |
| Unit cell dimensions (a, b, c) (Å) | 63.5, 65.7, 77.5 |
| Mosaicity | 0.53–0.58 |
| Total no. of reflections | 503,743 |
| Unique no. of reflections | 35,564 (3,133) |
| Multiplicity | 14.0 |
| Completeness (%) | 96.9 (86.8) |
| Mean I/σ(I) | 37.3 (1.9) |
| Wilson B factor (Å2) | 13.1 |
| Rmerge | 0.073 |
| Rmeas | 0.075 |
| Structure refinement statistics | |
| Rwork | 0.17 (0.24) |
| Rfree | 0.22 (0.30) |
| Number of nonhydrogen atoms | 2,656 |
| Protein | 2,375 |
| Glycerol | 48 |
| Water | 233 |
| RMSD from ideal values | |
| Bond length (Å)/bond angles (°) | 0.013/1.46 |
| Ramachandran plot | |
| Favored/allowed/outliers (%) | 99.7/0.3/0 |
| Clash score | 3.86 |
| Average B factor (Å2) | 17.80 |
| Protein | 16.20 |
| Glycerol | 36.60 |
| Water | 29.70 |
Statistics for the highest-resolution shell are shown in parentheses.
(ii) Active site and acyl-chain-binding pocket.
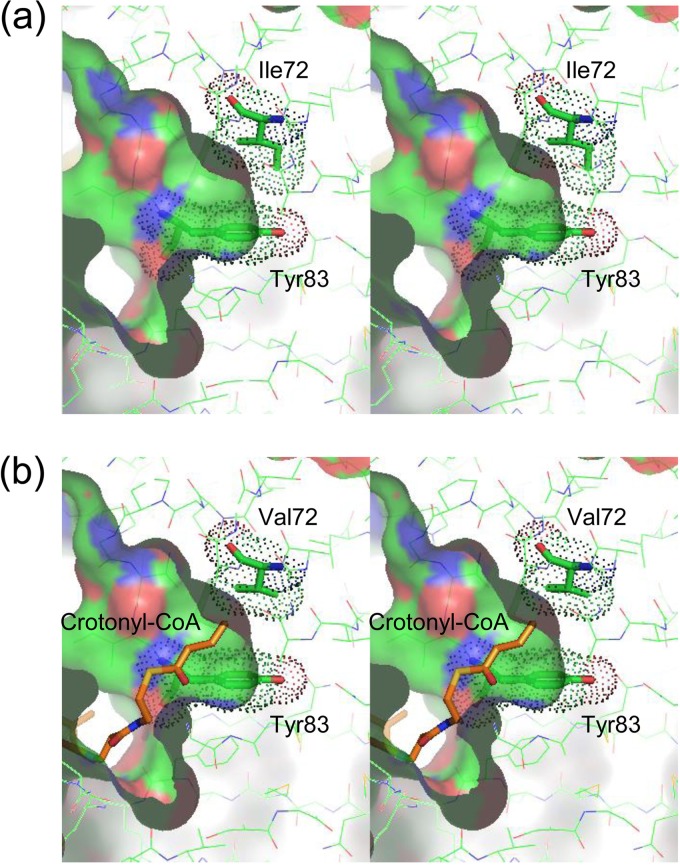
The current structural model represents a substrate-free form of the enzyme. The location of the active site and acyl-chain-binding pocket can be inferred on the basis of the sequence and structure similarities with PhaJAc, for which the active site has been identified (11). Two putative active sites related by the molecular 2-fold symmetry are formed at the interface of the two monomers. Each active site is located deep within the putative substrate-binding tunnel, which is contributed by both of the monomers. The end of the substrate-binding tunnel is a predominantly hydrophobic pocket which should serve as an acyl-chain-binding region. The active site is formed by Asp38 and His43, which come from one subunit, whereas the acyl-chain-binding pocket is formed by side chains of hydrophobic residues, such as Ile72, Thr81, Tyr83, and Ile136, as well as main-chain atoms of residues, such as Ser69, from the other subunit. Of these residues, Tyr83 adopts a side-chain conformation with dihedral angles for the Cα-Cβ bond (χ1) and for the Cβ-Cγ bond (χ2) of 208.8° to 211.7° and 5.8° to 6.8°, respectively, which is deviated from the observed range in the χ1-χ2 plot (Fig. 3). This energetically unfavorable conformation is stabilized by a hydrogen bond with the side chain of Gln86 and a van der Waals contact with the side chain of Phe90′ (the prime sign indicates that the residue is from the neighboring subunit). The molecular surface of the acyl-chain-binding pocket (Fig. 4a), calculated with a probe radius of 1.7 Å, which is approximately equal to the van der Waals radius of a methyl group, is similar in size and shape to PhaJAc, indicating that the acyl-chain-binding pocket is sufficiently deep to accommodate acyl chains with a length of up to six carbon atoms. This is consistent with biochemical results for the substrate specificity of PhaJ1Pa (10). The hydrophobic residues contributing the acyl-chain-binding pocket play another important role in forming the hydrophobic core of the protein structure with several other hydrophobic residues formed between helix α3 and the major β-sheet.
FIG 3.
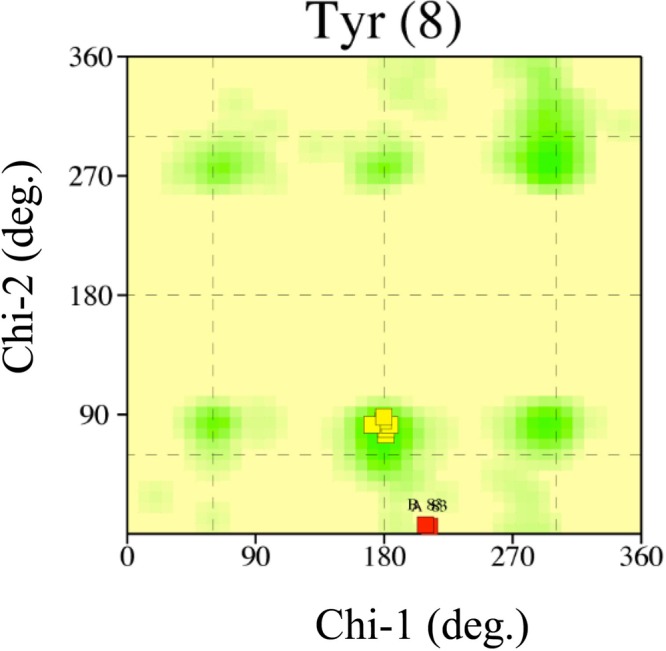
χ1-χ2 plot for eight tyrosine residues in PhaJ1Pa generated by PROCHECK (33). Commonly observed side-chain conformations for tyrosine residues are shown as green regions. Tyr83 residues in chains A and B of PhaJ1Pa are indicated by red boxes. The other six residues are indicated by yellow boxes.
FIG 4.
Stereo diagrams of the molecular surfaces of the acyl-chain binding pockets of PhaJ1Pa (a) and PhaJ1Pp (b). Molecular surfaces, calculated with a probe radius of 1.7 Å, are represented by a solid surface using a cutaway view. Residues are represented by thin-stick models with carbon, oxygen, nitrogen, and sulfur atoms in green, red, blue, and yellow, respectively. Residues Ile72 and Tyr83 are represented by thick-stick models. van der Waals surfaces of these residues are shown as dots. (b) Crotonyl-CoA, represented by an orange stick model, is manually docked in the substrate-binding pocket of PhaJ1Pp by using the binding mode of (R)-3-hydroxydecanoyl-CoA bound to fungal hydratase 2 (PDB entry 1PN4) (12) as a reference model.
Structural model of PhaJ1Pp by homology modeling.
The structural model of PhaJ1Pp was constructed by homology modeling using coordinates of PhaJ1Pa as a template. Because PhaJ1Pp and PhaJ1Pa have the same polypeptide length and share high sequence homology (∼67% identity), the structure of PhaJ1Pa is a good template for the homology modeling of PhaJ1Pp. Accordingly, the model was reliable with few clashes around the acyl-chain-binding region as calculated by MolProbity (34).
The residues contributing to the acyl-chain-binding pocket of PhaJ1Pp are essentially identical to those of PhaJ1Pa. The only difference is the residue at position 72; this residue is valine in PhaJ1Pp, whereas it is isoleucine in PhaJ1Pa. In spite of this difference, the molecular surface calculation with a probe radius of 1.7 Å showed that the acyl-chain-binding pockets of PhaJ1Pp and PhaJ1Pa have almost identical shape and depth (Fig. 4). The manual docking of an acyl-CoA substrate to the acyl-chain-binding pocket based on the binding mode of (3R)-hydroxydecanoyl-CoA with fungal enoyl-CoA hydratase 2 (PDB entry 1PN4) (12) also indicated that the enzyme could not bind enoyl-CoA with acyl chains longer than six carbon atoms (Fig. 4b). These observations suggest that the substrate specificity of PhaJ1 is not determined simply by the bulkiness of residue 72 (see Discussion). Residues forming the hydrophobic core are almost identical to those of PhaJ1Pa with a few exceptions. Residues 88 and 117 are methionine and isoleucine, respectively, in PhaJ1Pp, whereas they are leucine and methionine, respectively, in PhaJ1Pa.
DISCUSSION
In our previous study, in vivo substrate specificity assessment suggested that PhaJ1Pp has broader substrate specificity than PhaJ1Pa (10). In this study, in vitro substrate specificity assessment showed remarkable differences in the substrate specificities of PhaJ1Pp and PhaJ1Pa (Table 3). Based on sequence comparison of these enzymes around the acyl-chain-binding pocket and with aid from the structure of PhaJAc (11), we targeted residue 72 of PhaJ1Pa and PhaJ1Pp, located at the end of the acyl-chain-binding tunnel, to investigate the substrate specificity determinants of PhaJ1.
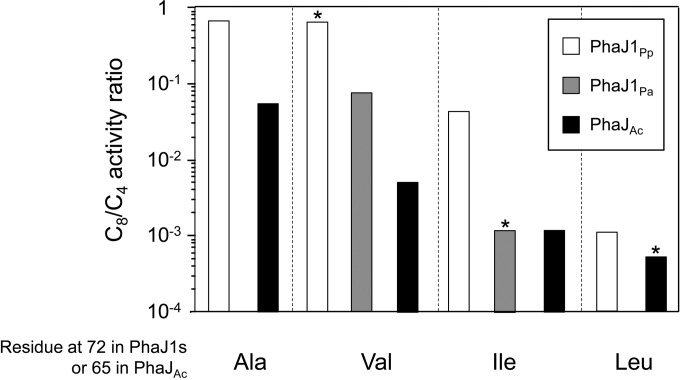
Figure 5 compares the C8/C4 ratios of the wild types and mutants of PhaJ1Pp and PhaJ1Pa, as well as those of PhaJAc (data are from reference 16), showing the dependence of the C8/C4 ratio on the amino acid at residue 72 (at residue 65 for PhaJAc). It may be reasonable to assume that the acyl-chain-length preference of the enzyme is related to the size and shape of the acyl-chain-binding pocket. Accordingly, these data indicate that an enzyme with a bulkier amino acid at residue 72 has a smaller acyl-chain-binding pocket; thus, it prefers the shorter-chain-length enoyl-CoA.
FIG 5.
Comparison of C8/C4 activity ratios. Activity ratios are represented as white bars for PhaJ1Pp and its mutants, gray bars for PhaJ1Pa and its mutant, and black bars for PhaJAc and its mutants. Wild-type enzymes are indicated by an asterisk. Data for PhaJAc and its mutants were retrieved from reference 16.
However, the crystal structure of PhaJ1Pa (as well as the structural model of PhaJ1Pp) poses an apparently contradictory situation. The Cδ atom of Ile72 did not contribute to the surface of the acyl-chain-binding pocket in PhaJ1Pa, as revealed in the calculation of the molecular surface of the pocket with a probe radius of 1.7 Å. In fact, the calculated molecular surface of the acyl-chain-binding pocket of the I72V mutant is almost identical to that of the wild type in size and shape (data not shown). This indicates that the I72V mutant should have a substrate specificity spectrum similar to that of the wild-type enzyme, which, however, is not consistent with the biochemical data. The same holds for PhaJ1Pp.
This apparent discrepancy may be explained by a potential conformational change of Tyr83 upon binding of substrates or due to the thermal fluctuation. Tyr83, situated at the opening of the acyl-chain-binding pocket, adopts a side-chain conformation with χ1 and χ2 angles deviating from the commonly observed range in the χ1-χ2 plot (Fig. 3). This suggests that the side chain of Tyr83 tends to change its conformation upon the binding of substrates, which results in expanding the acyl-chain-binding pocket. The difference in the bulkiness at residue 72 may influence the expanded volumes of the pocket and affect the range of substrate that can be accommodated.
On the other hand, comparison of the C8/C4 values between enzymes with the same amino acid (valine or isoleucine) at residue 72, derived from PhaJ1Pa and PhaJ1Pp, indicates that the acyl-chain-length preference is not determined only by the bulkiness of that residue. It should be noted that PhaJ1Pp and PhaJ1Pa share almost identical residues that form the hydrophobic core with residue 72, as indicated by the homology modeling of PhaJ1Pp. Therefore, the V72I mutant of PhaJ1Pp, for example, is essentially identical to wild-type PhaJ1Pa in terms of the structure of the acyl-chain-binding pocket and the adjacent hydrophobic core. However, this is inconsistent with the biochemical data showing that the mutant represents a C8/C4 value an order of magnitude larger than that of wild-type PhaJ1Pa (Fig. 5).
This peculiarity might be accounted for by the intrinsic conformational fluctuation, which may differ in magnitude between PhaJ1Pa and PhaJ1Pp. Larger conformational fluctuation of PhaJ1Pp may effectively expand the acyl-chain-binding pocket expected from the static structure and enable longer enoyl-CoA acyl chains to be accommodated in the pocket.
The results of the C8/C4 ratio for the chimeric proteins can be explained by this fluctuation hypothesis rather than by the single-site difference. If the substrate specificity can be determined only by the bulkiness of the amino acid residue at position 72, the specificities of the chimeric proteins would be discontinuous at the chimeric junction point before or after residue 72. However, the activity measurement for these chimeric proteins showed a gradual change in substrate specificity, correlated with the composition of the two parental proteins (Table 3). This indicates that the composition of the parental proteins, which bear different degrees of thermal fluctuation, affects the degree of conformational fluctuation in the resulting chimeric protein.
In conclusion, the chain-length specificity of PhaJ1 from Pseudomonas is determined mainly by the bulkiness of the amino acid residue at position 72; however, other factors, such as structural fluctuation specific for each protein, also may affect the specificity. These insights would be helpful to design PhaJs with altered substrate specificity as metabolic tools for the synthesis of tailor-made PHA copolymers.
ACKNOWLEDGMENTS
We thank Surekha Kanagarajan for technical support in the purification and crystallization of PhaJ1Pa. We thank the staff at BL26B1, SPring-8, for help with data collection.
This work was supported by KAKENHI (to T.T. and T.H.) and JST, CREST (to T.T.).
REFERENCES
- 1.Madison LL, Huisman GW. 1999. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev 63:21–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lenz RW, Marchessault RH. 2005. Bacterial polyesters: biosynthesis, biodegradable plastics and biotechnology. Biomacromolecules 6:1–8. doi: 10.1021/bm049700c. [DOI] [PubMed] [Google Scholar]
- 3.Sudesh K, Abe H, Doi Y. 2000. Synthesis, structure and properties of polyhydroxyalkanoate: biological polyesters. Prog Polym Sci 25:1503–1555. doi: 10.1016/S0079-6700(00)00035-6. [DOI] [Google Scholar]
- 4.Fukui T, Doi Y. 1997. Cloning and analysis of the poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) biosynthesis genes of Aeromonas caviae. J Bacteriol 179:4821–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukui T, Shiomi N, Doi Y. 1998. Expression and characterization of (R)-specific enoyl coenzyme A hydratase involved in polyhydroxyalkanoate biosynthesis by Aeromonas caviae. J Bacteriol 180:667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsuge T, Fukui T, Matsusaki H, Taguchi S, Kobayashi G, Ishizaki A, Doi Y. 2000. Molecular cloning of two (R)-specific enoyl-CoA hydratase genes from Pseudomonas aeruginosa and their use for polyhydroxyalkanoate synthesis. FEMS Microbiol Lett 184:193–198. doi: 10.1111/j.1574-6968.2000.tb09013.x. [DOI] [PubMed] [Google Scholar]
- 7.Tsuge T, Taguchi K, Taguchi S, Doi Y. 2003. Molecular characterization and properties of (R)-specific enoyl-CoA hydratases from Pseudomonas aeruginosa: metabolic tools for synthesis of polyhydroxyalkanoates via fatty acid β-oxidation. Int J Biol Macromol 31:195–205. doi: 10.1016/S0141-8130(02)00082-X. [DOI] [PubMed] [Google Scholar]
- 8.Fiedler S, Steinbüchel A, Rehm BHA. 2002. The role of the fatty acid β-oxidation multienzyme complex from Pseudomonas oleovorans in polyhydroxyalkanoate biosynthesis: molecular characterization of the fadBA operon from P. oleovorans and of the enoyl-CoA hydratase genes phaJ from P. oleovorans and Pseudomonas putida. Arch Microbiol 178:149–160. doi: 10.1007/s00203-002-0444-0. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, Nomura CT. 2010. Monitoring differences in gene expression levels and polyhydroxyalkanoate (PHA) production in Pseudomonas putida KT2440 grown on different carbon sources. J Biosci Bioeng 110:653–659. doi: 10.1016/j.jbiosc.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Sato S, Kanazawa H, Tsuge T. 2011. Expression and characterization of (R)-specific enoyl coenzyme A hydratases making a channeling route to polyhydroxyalkanoate biosynthesis in Pseudomonas putida. Appl Microbiol Biotechnol 90:951–959. doi: 10.1007/s00253-011-3150-5. [DOI] [PubMed] [Google Scholar]
- 11.Hisano T, Tsuge T, Fukui T, Iwata T, Miki K, Doi Y. 2003. Crystal structure of the (R)-specific enoyl-CoA hydratase from Aeromonas caviae involved in polyhydroxyalkanoate biosynthesis. J Biol Chem 278:617–624. [DOI] [PubMed] [Google Scholar]
- 12.Koski MK, Haapalainen AM, Hiltunen JK, Glumoff T. 2004. A two-domain structure of one subunit explains unique features of eukaryotic hydratase 2. J Biol Chem 279:24666–24672. doi: 10.1074/jbc.M400293200. [DOI] [PubMed] [Google Scholar]
- 13.Koski MK, Haapalainen AM, Hiltunen JK, Glumoff T. 2005. Crystal structure of 2-enoyl-CoA hydratase 2 from human peroxisomal multifunctional enzyme type 2. J Mol Biol 345:1157–1169. doi: 10.1016/j.jmb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Johansson P, Castell A, Unge T, Jones TA, Bäckbro K. 2006. Structure and function of Rv0130, a conserved hypothetical protein from Mycobacterium tuberculosis. Protein Sci 15:2300–2309. doi: 10.1110/ps.062309306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Zhang K, Zhu J, Song W, Zhao L, Zhang X. 2013. Structure reveals regulatory mechanisms of a MaoC-like hydratase from Phytophthora capsici involved in biosynthesis of polyhydroxyalkanoates (PHAs). PLoS One 8:e80024. doi: 10.1371/journal.pone.0080024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsuge T, Hisano T, Taguchi S, Doi Y. 2003. Alteration of chain length substrate specificity of Aeromonas caviae R-enantiomer-specific enoyl-coenzyme A hydratase through site-directed mutagenesis. Appl Environ Microbiol 69:4830–4836. doi: 10.1128/AEM.69.8.4830-4836.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niamsiri N, Delamarre SC, Kim YR, Batt CA. 2004. Engineering of chimeric class II polyhydroxyalkanoate synthases. Appl Environ Microbiol 70:6789–6799. doi: 10.1128/AEM.70.11.6789-6799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumoto K, Takase K, Yamamoto Y, Doi Y, Taguchi S. 2009. Chimeric enzyme composed of polyhydroxyalkanoate (PHA) synthases from Ralstonia eutropha and Aeromonas caviae enhances production of PHAs in recombinant Escherichia coli. Biomacromolecules 10:682–685. doi: 10.1021/bm801386j. [DOI] [PubMed] [Google Scholar]
- 19.Imai Y, Matsushima Y, Sugimura T, Terada M. 1991. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res 19:2785. doi: 10.1093/nar/19.10.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fong JC, Schulz H. 1981. Short-chain and long-chain enoyl-CoA hydratase from pig heart muscle. Methods Enzymol 71:390–398. [DOI] [PubMed] [Google Scholar]
- 21.Valentin HE, Steinbüchel A. 1994. Application of enzymatically synthesized short-chain-length hydroxyl fatty acid coenzyme A thioesters for assay of polyhydroxyalkanoic acid synthases. Appl Microbiol Biotechnol 40:699–709. doi: 10.1007/BF00173332. [DOI] [Google Scholar]
- 22.Jenkins LS, Nunn WD. 1987. Genetic and molecular characterization of the genes involved in short-chain fatty acid degradation in Escherichia coli: the ato system. J Bacteriol 169:42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenkins LS, Nunn WD. 1987. Regulation of the ato operon by the atoC gene in Escherichia coli. J Bacteriol 169:2096–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhie HG, Dennis D. 1995. Role of fadR and atoC(Con) mutations in poly(3-hydroxybutyrate-co-3-hydroxyvalerate) synthesis in recombinant pha+ Escherichia coli. Appl Environ Microbiol 61:2487–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsusaki H, Manji S, Taguchi K, Kato M, Fukui T, Doi Y. 1998. Cloning and molecular analysis of the poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyalkanoate) biosynthesis genes in Pseudomonas sp. strain 61-3. J Bacteriol 180:6459–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taguchi K, Aoyagi Y, Matsusaki H, Fukui T, Doi Y. 1999. Coexpression of 3-ketoacyl-ACP reductase and polyhydroxyalkanoate synthase genes induces PHA production in Escherichia coli HB101 strain. FEMS Microbiol Lett 176:183–190. doi: 10.1111/j.1574-6968.1999.tb13660.x. [DOI] [PubMed] [Google Scholar]
- 27.Kato M, Bao HJ, Kang CK, Fukui T, Doi Y. 1996. Production of a novel copolymer of 3-hydroxybutyric acids and medium-chain-length 3-hydroxyalkanoic acids by Pseudomonas sp. 61-3 from sugars. Appl Microbiol Biotechnol 45:363–370. doi: 10.1007/s002530050697. [DOI] [Google Scholar]
- 28.Matthews BW. 1968. Solvent content of protein crystals. J Mol Biol 33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brünger AT. 1997. Free R value: Cross-validation in crystallography. Methods Enzymol 277:366–396. doi: 10.1016/S0076-6879(97)77021-6. [DOI] [PubMed] [Google Scholar]
- 33.Laskowski RA, MacArthur MW, Moss DS, Thorton JM. 1993. PROCHECK–a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 34.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GG, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The Protein Data Bank. Nucleic Acids Res 28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tosatto SCE. 2005. The Victor/FRST function for model quality estimation. J Comput Biol 12:1316–1327. doi: 10.1089/cmb.2005.12.1316. [DOI] [PubMed] [Google Scholar]
- 37.Chothia C, Lesk AM. 1986. The relation between the divergence of sequence and structure in proteins. EMBO J 5:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]